Abstract
The expression of SREBP-1 (sterol-regulatory-element-binding protein-1) isoforms differs between tissues and cultured cell lines in that SREBP-1a is the major isoform in established cell lines, whereas SREBP-1c predominates in liver and most other human tissues. SREBP-1c is transcriptionally less active than SREBP-1a, but is a main mediator of hepatic insulin action and is selectively up-regulated by LXR (liver X receptor) agonists. LXR-mediated transactivation is co-activated by PGC-1α (peroxisome-proliferator-activated receptor-γ co-activator-1α), which displays deficient expression in skeletal-muscle-derived cell lines. In the present paper, we show that PGC-1α expression is also deficient in HepG2 cells and in a human brown adipocyte cell line (PAZ6). In transient transfection studies, PGC-1α selectively amplified the LXR-mediated transcription from the human SREBP-1c promoter in HepG2 and PAZ6 cells via two LXR-response elements with extensive similarity to the respective murine sequence. Mutational analysis showed that the human LXR-response element-1 (hLXRE-1) was essential for co-activation of LXR-mediated SREBP-1c gene transcription by PGC-1α. Ectopic overexpression of PGC-1α in HepG2 cells enhanced basal SREBP-1c and, to a lesser extent, -1a mRNA expression, but only SREBP-1c expression was augmented further in an LXR/RXR (retinoic X receptor)-dependent fashion, thereby inducing mRNA abundance levels of SREBP-1c target genes, fatty acid synthase and acetyl-CoA carboxylase. These results indicate that PGC-1α contributes to the regulation of SREBP-1 gene expression, and can restore the SREBP-1 isoform expression pattern of HepG2 cells to that of human liver.
Keywords: co-activator, gene expression, liver X receptor (LXR), peroxisome-proliferator-activated receptor-γ co-activator-1α (PGC-1α), sterol-regulatory-element-binding protein (SREBP)
Abbreviations: AAV, adeno-associated virus; ACC, acetyl-CoA carboxylase; CMV, cytomegalovirus; FAS, fatty acid synthase; FOXO1, forkhead transcription factor 1; 22(R)HC, 22(R)-hydroxycholesterol; Luc, luciferase; LXR, liver X receptor; LXRE, liver X receptor response element; hLXRE, human LXRE; PGC-1α, peroxisome-proliferator-activated receptor-γ co-activator-1α; 9cRA, 9-cis retinoic acid; RT, reverse transcriptase; RXR, retinoic X receptor; SREBP, sterol-regulatory-element-binding protein; TK, thymidine kinase
INTRODUCTION
The three SREBPs (sterol-regulatory-element-binding proteins), 1a, 1c and 2, belong to the basic helix–loop–helix leucine zipper family of transcription factors and control transcriptional programmes that regulate the biosynthesis of lipids [1]. They are synthesized as membrane proteins of the endoplasmic reticulum, and require a two-step proteolytic cleavage for their nuclear translocation and transactivation function [1,2]. SREBP-1a and SREBP-1c isoforms are transcribed from two distinct transcriptional start sites of a single gene, thereby producing alternative forms of exon 1, designated 1a and 1c [3,4], whereas SREBP-2 is encoded by a separate gene [4,5]. Although the three SREBP family members display overlapping specificities for their target genes, the genes related to the cholesterol-synthesis pathway are activated more strongly by SREBP-2, while the genes related to the fatty-acid-synthesis pathway are mainly activated by SREBP-1 [6–8]. Due to its shorter acidic transcription-activation domain, SREBP-1c is a weaker transcriptional activator than SREBP-1a [9], but is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes [10–13].
The relative amounts of SREBP-1 transcripts containing exons 1a or 1c vary significantly among different animal and human tissues, and cultured cells [14]. In most mammalian tissues, including liver, muscle, brain and adrenal gland as well as white and brown adipose tissues, SREBP-1c is much more abundant than SREBP-1a. Among human and mouse tissues, the highest molar ratio of SREBP-1c/SREBP-1a has been reported for liver. Only in spleen tissue is SREBP-1a the predominant isoform. In contrast, in all cultured cell lines examined, including HepG2, NIH-3T3 and HEK-293 cells, SREBP-1a transcripts are more abundant than SREBP-1c transcripts. The predominance of the SREBP-1a isoform in cultured cells has been explained by the increased demand of these rapidly growing cells for cholesterol and fatty acid biosynthesis during cell proliferation [9]. Recent cell culture and animal studies indicate that SREBP-1c, but not SREBP-1a, gene expression is regulated by the nuclear hormone receptor LXR (liver X receptor) [15–18]. A potent LXRE (LXR-response element) complex consisting of two LXR-binding motifs has been identified in the mouse SREBP-1c promoter [15]. The functional significance of LXR-mediated SREBP-1c transactivation has been demonstrated in mice lacking both LXRα and LXRβ that were characterized by markedly reduced levels of SREBP-1c and its lipogenic target enzymes [18,19]. We have reported previously that PGC-1α (peroxisome-proliferator-activated receptor-γ co-activator-1α) acts as a transcriptional co-activator of LXRs [20]. PGC-1α gene expression has been shown to be much lower in skeletal muscle cell lines than in the respective tissues [21]. We therefore reasoned that reduced transcriptional activity of LXRs due to diminished PGC-1α expression may contribute to the low SREBP-1c mRNA abundance and the high SREBP-1a/SREBP-1c ratio observed in cultured cells, and asked whether or not PGC-1α overexpression can, at least in part, rescue SREBP-1c gene expression in these cells.
MATERIALS AND METHODS
Human tissue samples
Tissue samples were obtained from musculus rectus abdominis, liver and intraperitoneal adipose tissue of 35–55-year-old healthy male subjects who underwent an elective surgical procedure for the repair of hernias. Tissue biopsies were taken at the beginning of the surgical procedure, divided into aliquots and frozen at −70 °C. Study subjects provided informed consent, and the study was approved by the local ethics committee.
Plasmid constructs
The SREBP-1c–Luc plasmid in which the region from −422 to −186 of the human SREBP-1 gene drives the promoterless firefly luciferase (Luc) gene was generated using 5′-CAGGTACCGAGTTCTGGTGTGTTGGGCCA-3′ (−422 to −402) and 5′-ACTGCTAGCCGCGCTGCCGCCTCGCTAG-3′ (−205 to −186) as forward and reverse primers respectively to amplify a 236 bp DNA fragment that was cloned into the TK (thymidine kinase)–Luc vector described previously [20]. For construction of the SREBP-1a–Luc plasmid, a 4532 bp fragment was amplified using 5′-CAGGTACCTCCTGTCCACCCCATTCCTCCC-3′ (−4443 to −4422) and 5′-ACTACTGCTAGCCAGTCGCCGCCGCCGCTCC-3′ (+71 to +89) as forward and reverse primers respectively. The resulting fragment was ligated into the pGL3-Promoter Vector (Promega, Madison, WI, U.S.A.). KpnI and NheI restriction endonuclease sites introduced into the primer sequences are indicated in bold. Numbers in parentheses refer to primer positions relative to the transcriptional start site (GenBank® accession number NT_010718).
The hLXRE-1mut (where hLXRE is human LXRE and mut is mutant) construct was generated by site-directed mutagenesis of the SREBP-1c–Luc plasmid using the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, U.S.A.) and 5′-GAGGGCCAGAGTCCGCCAGATTCCCCGGCAGA-3′ and 5′-TCTGCCGGGGAATCTGGCGGACTCTGGCCCTC-3′ as forward and reverse mutagenic primers respectively. For the construction of the hLXRE-2mut plasmid 5′-GGCGGAAGTCCGCTAGATTCCCCAACCCCATT-3′ and 5′-AATGGGGTTGGGGAATCTAGCGGACTTCCGCC-3′ were used as forward and reverse mutagenic primers respectively. Mutated nucleotide positions are in bold. All constructs described were verified by dye-terminator cycle-sequencing using the ABI Prism™ 310 Genetic Analyser (Applied Biosystems, Foster City, CA, U.S.A.). The PGC-1α expression plasmid was described previously [22].
An AAV (adeno-associated virus) expression vector containing the human cDNA for PGC-1α (pAAV-PGC) was generated by RT (reverse transcriptase)-PCR amplification of the entire PGC-1α coding region (GenBank® accession number AF106698). A 2400 bp cDNA fragment was amplified from human kidney total RNA using 5′-TTCAGGATCCGGATGGCGTGGGACATGTGC-3′ (+119 to +135) and 5′-TGCTCGAGGTTACCTGCGCAAGCTTCTGTGAGCTTC-3′ (+2501 to +2518) as forward and reverse primers respectively, and ligated into the pAAV-MCS vector (Invitrogen, Carlsbad, CA, U.S.A.). BamHI and XhoI restriction enzymes sites introduced into the primer sequences are in bold. Numbers in parentheses refer to primer positions relative to the transcriptional start site.
Cell-culture, transfections and AAV infections
HEK-293 cells derived from human embryonic kidney and human HepG2 hepatoma cells were grown as recommended by the supplier (A.T.C.C., Manassas, VA, U.S.A.). PAZ6 cells derived from human brown adipose tissue were grown and differentiated as described previously [23,24]. Drugs at concentrations of 10 μM for 22(R)HC [22(R)-hydroxycholesterol] and 9cRA (9-cis retinoic acid) were added immediately before transfection.
HepG2 cells and differentiated PAZ6 cells cultured in 24-well dishes were transfected using the LIPOFECTAMINE 2000 reagent (Invitrogen). Unless otherwise indicated, we used 1 μg of reporter plasmids, 0.5 μg of expression plasmids and 1 ng of pRL-CMV (cytomegalovirus) plasmid (Promega) as transfection control. For transfection of the SREBP-1a–Luc reporter constructs, 20 ng of pRL-TK plasmid was used as an internal control. Cells were collected 24 h after transfection and firefly and Renilla luciferase activities were measured with a luminometer (Anthos Labtec Instruments, Salzburg, Austria) using the Dual-Luciferase Reporter Assay System (Promega). Histograms are representative of two experiments, each performed in quadruplicate, with results given as means±S.D. Analysis of variance and Tukey's post-hoc test were used to compare results of transactivation assays.
Recombinant PGC-1α virions (rAAV-PGC-1α) were generated using the AAV helper-free system (Invitrogen). Briefly, the cis plasmid pAAV-PGC (containing the AAV inverted terminal repeats), the trans plasmid pAAV-RC (containing the AAV Rep and Cap genes), and the pHelper plasmid (containing an essential region from the adenovirus genome) were co-transfected into HEK-293 cells at a proportion of 1:1:2 by calcium phosphate precipitation (Promega). Cells were harvested 96 h post-transfection, recombinant virions were purified using the BD Adeno-X™ Virus Purification Kit (Clontech, Palo Alto, CA, U.S.A.) and virus titres were estimated by real-time quantitative PCR. A 185 bp DNA fragment of the CMV promoter contained within the AAV expression vector was amplified using 5′-CTGACCGCCCAACGACCC-3′ and 5′-GCCATTTACCGTCATTGA-3′ as forward and reverse primers respectively, and the SYBR Green PCR Master Mix (Applied Biosystems). For the generation of a standard curve, real-time PCR analyses with serial dilutions of the pAAV-MCS plasmid were performed.
HepG2 cells (1×106) were infected with approx. 1×108 infectious units of rAAV-PGC-1α virions. PGC-1α mRNA expression was approx. 3-fold higher in virion-infected HepG2 cells. Control infections with LacZ-virions at a similar MOI (multiplicity of infection) revealed that 30% of cells expressed the transgene. After infection, cells were incubated with drugs as indicated, harvested 24 h post-infection and used for isolation of total RNA.
RNA isolation, RNase protection assays and quantitative real-time RT-PCR
Total RNA was isolated from the respective human tissue samples and cell lines using the Qiagen RNeasy Mini kit (Qiagen, Hilden, Germany). All RNA samples were digested with DNase I to eliminate any contaminating DNA. The integrity of RNA samples was ascertained by their electrophoretic mobility pattern in formaldehyde gels. RNA concentrations were determined by absorbance measurements at 260 nm using a DU 640 Spectrophotometer (Beckman, Palo Alto, CA, U.S.A.). SREBP-1 RNase protection assays have been described in detail in [25]. Briefly, 32P-labelled antisense and 3H-labelled sense RNA were transcribed from a cloned fragment encompassing part of exon 1a and exon 2 of the human SREBP-1 gene with the Riboprobe Combination System-SP6/T7 (Promega) and used for absolute quantification of SREBP-1a and SREBP-1c gene expression levels in muscle tissue. For real-time PCR experiments, a standard curve was generated by serial dilutions of this muscle cDNA.
Equal amounts of total RNA (0.5 μg/assay) were reverse-transcribed using 200 units of Moloney murine leukaemia virus reverse transcriptase (Gibco BRL Life Technologies, Grand Island, New York, NY, U.S.A.), 10 mM Tris/HCl, pH 8.3, 50 mM KCl, 5 mM MgCl2, 2.5 μM random hexamers, 1 mM dNTP and 20 units of RNasin® (Promega) in a volume of 20 μl. cDNAs were amplified and quantified using the SYBR Green PCR Master Mix, according to the manufacturer's instructions. Real-time PCR reaction mixtures contained 1 μl of cDNA, 1 μl of 10 μM forward primer, 1 μl of 10 μM reverse primer, 2.5 μl of PCR 10× SYBR Green PCR Master Mix and 19.5 μl of water. Primers used for quantification, amplicon sizes and annealing temperatures are listed in Table 1. Melting curves were analysed to ensure that fluorescence signals solely reflected specific amplicons. Reactions were amplified in quadruplicate with the iCycler iQ Multicolour Real-Time PCR Detector (Bio-Rad, Hercules, CA, U.S.A.) for 40 cycles in optical 96-well reaction plates (Bio-Rad) with denaturation at 94 °C for 30 s, annealing for 30 s, and extension at 72 °C for 30 s. CT, the PCR cycle at which an increase in reporter fluorescence above the baseline signal is detected, was used for quantification. SREBP-1a and SREBP-1c mRNAs in unknown samples were quantified using the slopes generated from the CTs in samples with known content of the respective mRNAs. Results for FAS (fatty acid synthase), ACC (acetyl-CoA carboxylase) and PGC-1α mRNA abundance were normalized for the expression of ARP (acidic ribosomal protein p0) and are given in arbitrary units. The fold induction value for quadruplicate wells was averaged, and results are presented as means±S.D.
Table 1. Primer sequences, annealing temperatures and amplicon sizes of quantitative real-time PCR assays.
| Gene | GenBank® accession number | Forward primer | Reverse primer | Annealing temperature (°C) | Amplicon Size (bp) |
|---|---|---|---|---|---|
| PGC-1α | NM_013261 | 5′-GAGCCGAGCTGAACAAGCAC-3′ | 5′-AGACACATTGAACAATGAATAGGATG-3′ | 57 | 238 |
| SREBP-1c | NM_004176 | 5′-TCAGCGAGGCGGCTTTGGAGCAG-3′ | 5′-CATGTCTTCGATGTCGGTCAG-3′ | 55 | 80 |
| SREBP-1a | NM_004176 | 5′-GGAGGGGTAGGGCCAACGGCCT-3′ | 5′-CATGTCTTCGAAAGTGCAATCC-3′ | 55 | 80 |
| FAS | NM_004104 | 5′-CGGTACGCGACGGCTGCCTG-3′ | 5′-GCTGCTCCACGAACTCAAACACCG-3′ | 60 | 231 |
| ACC | NM_000664 | 5′-TGATGTCAATCTCCCCGCAGC-3′ | 5′-TTGCTTCTTCTCTGTTTTCTCCCC-3′ | 60 | 353 |
| ARP | NM_001002 | 5′-GGCACCATTGAAATCCTGAGTGAT-3′ | 5′-TTGCGGACACCCTCCAGGAAGC-3′ | 60 | 215 |
RESULTS AND DISCUSSION
To confirm previous findings on SREBP-1 isoform expression in human tissues and cell lines, we quantified SREBP-1a and SREBP-1c mRNA levels in human liver, adipose tissue and skeletal muscle, as well as in HepG2 and PAZ6 cells, using quantitative real-time RT-PCR. Consistent with previous results [25], SREBP-1c mRNA levels exceeded those of SREBP-1a in all tissues samples studied [14]. SREBP-1c/SREBP-1a ratios were 9.8±3.8, 5.9±2.9 and 4.6±2.3 in liver, white adipose tissue and skeletal muscle respectively. In HepG2 cells, expression levels of SREBP-1c and SREBP-1a isoforms were >200-fold and >20-fold lower than in liver tissue samples and the SREBP-1c/SREBP-1a ratio was 0.72±0.2. In PAZ6 cells, expression levels of SREBP-1c and SREBP-1a isoforms were several orders of magnitude lower compared with adipose tissue samples and the SREBP-1c/SREBP-1a ratio was 0.67±0.2.
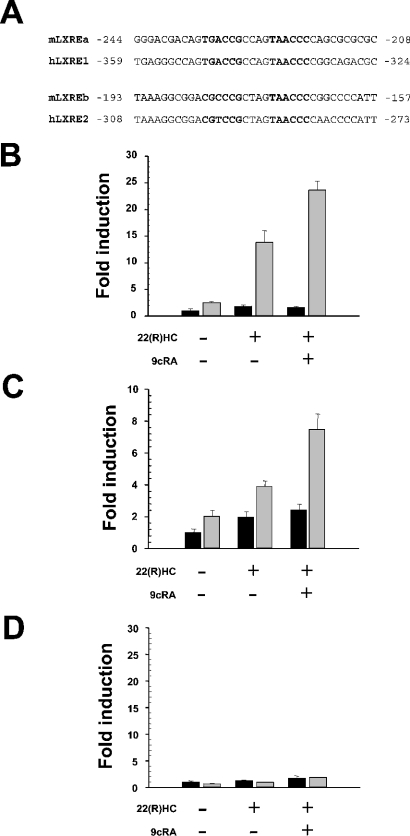
Computational analyses of the human SREBP-1a and SREBP-1c promoters revealed two putative LXREs at −283 to −298 (hLXRE2) and −333 to −348 (hLXRE1) in the SREBP-1c upstream regulatory region (Figure 1A) showing extensive similarity to mouse LXREs described previously [15]. In contrast, no putative LXR-binding sites were identified in the promoter region of SRBEP-1a. Since PGC-1α acts a transcriptional co-activator of LXR [20], we measured PGC-1α mRNA abundance in human liver and HepG2 cells, and observed an approx. 4-fold lower expression level in the hepatoma line (1.0±0.2 compared with 0.26±0.07, arbitrary units). In PAZ6 cells, PGC-1α was almost not detectable (results not shown). To assess directly the role of PGC-1α and LXR in the transcriptional regulation of SREBP-1a and SREBP-1c isoforms in humans, we transiently transfected HepG2 and PAZ6 cells with the reporter constructs SREBP-1a–Luc, harbouring an approx. 4.5 kb fragment comprising the promoter region of the human SREBP-1a gene, and SREBP-1c–Luc, containing the two LXREs of the human SREBP-1c promoter region. In both cell lines, basal activity of SREBP-1c–Luc was stimulated slightly by the LXR-specific ligand 22(R)HC or a combination of the LXR/RXR ligands 22(R)HC and 9cRA. Co-transfections with a PGC-1α expression plasmid resulted in increased basal transcription activity, and additional increases in activity of SREBP-1c–Luc were observed upon stimulation of cells with 22(R)HC alone or in combination with 9cRA. No increase in basal luciferase activity of the SREBP-1a–Luc construct was detected with 22(R)HC or 22(R)HC/9cRA. Co-transfection of the PGC-1α expression plasmid did not enhance basal or LXR-mediated luciferase activity of SREBP-1a–Luc (Figures 1B–1D).
Figure 1. PGC-1α potentiates LXR-mediated transcription of the SREBP-1c gene.
(A) Sequence comparison of two previously characterized LXREs in the mouse SREBP-1c promoter with the respective region of the human SREBP-1c gene identifies LXREs with extensive similarities. (B) Differentiated PAZ6 cells were transiently transfected with the SREBP-1c–Luc reporter plasmid encompassing two putative LXREs (black bars) or co-transfected with a human PGC-1α expression construct (grey bars). SREBP-1c-driven firefly luciferase activity levels were standardized to pRL-CMV driven Renilla luciferase levels used as transfection controls. Cells were incubated with 22(R)HC, or 22(R)HC and 9cRA, 10 μM each. (C) HepG2 cells were used in transient transfection experiments as described above. (D) Differentiated PAZ6 cells incubated with 22(R)HC or with 22(R)HC and 9cRA were transfected with the SREBP-1a–Luc reporter plasmid encompassing approx. 4.5 kb of the human SREBP-1a promoter or co-transfected with PGC-1α. Fold induction refers to basal SREBP-1c–Luc or SREBP-1a–Luc reporter gene expression levels in the absence of PGC-1α expression plasmid and drugs.
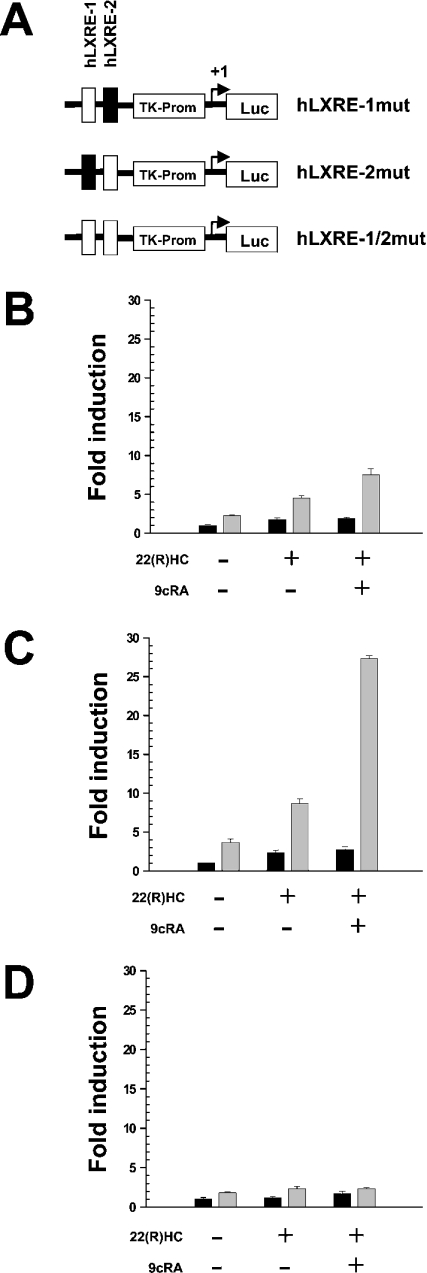
Mutational analyses were performed to delineate further the functional effects of the two LXREs, hLXRE-1 and hLXRE-2. Using site-directed mutagenesis, three different reporter constructs were generated that harboured mutations in hLXRE-1 (hLXRE-1mut), hLXRE-2 (hLXRE-2mut) or both (hLXRE-1/2mut). Reporter plasmids were co-transfected into PAZ6 cells together with the PGC-1α expression construct. Mutation of hLXRE-1 resulted in a profound loss of PGC-1α-mediated transcriptional activity of the reporter construct in the presence of 22(R)HC or a combination of 22(R)HC and 9cRA. Mutation of hLXRE-2 only slightly reduced LXR-mediated transcriptional responses in co-transfection studies with PGC-1α arguing for a minor role of hLXRE-2 in LXR-mediated transcriptional regulation of SREBP-1c. Mutation of both LXREs completely abolished PGC-1α-mediated transcriptional responses in the presence and the absence of LXR/RXR ligands (Figures 2A–2D).
Figure 2. The hLXRE-1 binding site is essential for PGC-1α-mediated transcriptional co-activation of the human SREBP-1c gene.
(A) Schematic representation of the hLXRE-1mut, hLXRE-2mut and hLXRE-1/2mut constructs. Mutated binding sites are indicated as white boxes. (B) Differentiated PAZ6 cells were transfected with hLXRE-1mut or co-transfected with PGC-1α (grey bars) and incubated with 22(R)HC, or 22(R)HC and 9cRA, 10 μM each. Differentiated PAZ6 cells were transfected with hLXRE-2mut (C) or hLXRE-1/2mut (D) or co-transfected with PGC-1α (grey bars) in the presence or absence of 22(R)HC, or 22(R)HC and 9cRA. Fold induction refers to basal (black bars) hLXRE-1mut, hLXRE-2mut and hLXRE-1/2mut expression levels respectively, in the absence of PGC-1α expression plasmid and drugs.
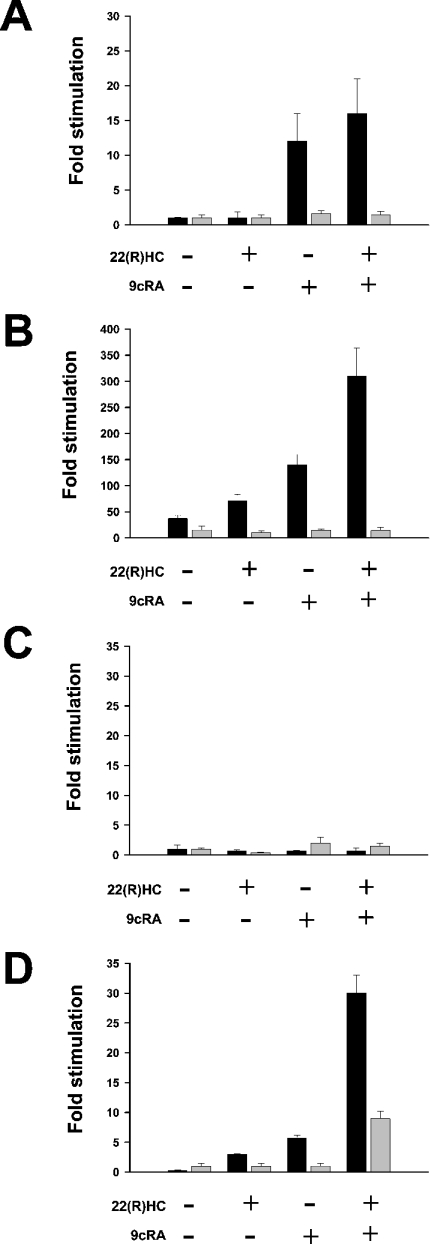
We used an AAV vector system to augment PGC-1α gene expression in HepG2 cells. mRNA expression levels of SREBP-1a and SREBP-1c in control or virion-infected cultures were determined by real-time RT-PCR in both the presence and the absence of LXR/RXR ligands. Stimulation of control cells with 9cRA or a combination of 22(R)HC and 9cRA increased SREBP-1c gene expression without any effects on SREPB-1a mRNA abundance (Figure 3A). Strikingly, in comparison with control HepG2 cells, basal expression levels of SREBP-1a and SREBP-1c in infected cells were increased approx. 6- and 35-fold respectively. SREBP-1c gene expression in these cells was stimulated further by the addition of 22(R)HC or 9cRA and a more than additive effect of both ligands was noted (Figure 3B). Conversely, these drugs had no stimulatory effect on SREBP-1a mRNA abundance levels. Comparison of SREBP-1a and SREBP-1c expression levels in human liver and HepG2 cells revealed that ectopic expression of PGC-1α and stimulation with LXR/RXR ligands augmented the abundance of SREBP-1c mRNA in HepG2 cells up to levels nearly equivalent to those observed in human liver tissue (Table 2). Expression levels of the two SREBP-1c target genes, FAS and ACC, were determined in HepG2 cells with or without ectopic expression of PGC-1α. No or minimal induction of FAS or ACC gene expression with LXR/RXR ligands was noted in control cells, but virus-induced overexpression of PGC-1α enhanced FAS and ACC gene expression in an LXR/RXR-dependent fashion (Figures 3C and 3D).
Figure 3. Ectopic expression of PGC-1α in HepG2 cells potentiates the transcription of SREBP-1c and SREBP-1c target genes.
(A) Real-time RT-PCR analyses of SREBP-1a (grey bars)/SREBP-1c (black bars) gene expression in control HepG2 cells stimulated with 22(R)HC, 9cRA, or a combination of 22(R)HC and 9cRA. Results are means±SD for experiments performed in quadruplicate normalized for the expression of ARP (acidic ribosomal protein p0). Fold induction refers to basal SREBP-1a/SREBP-1c expression in the absence of drugs. (B) HepG2 cells were infected with recombinant PGC-1α virions at a MOI (multiplicity of infection) of >100 and stimulated with drugs as indicated. SREBP-1a (grey bars)/SREBP-1c (black bars) gene expression was quantified using real-time RT-PCR. (C) Analyses of gene expression of FAS (black bars) and ACC (grey bars) in HepG2 cells in the absence or presence of 22(R)HC, 9cRA, or a combination of 22(R)HC and 9cRA. Fold induction refers to basal FAS or ACC expression in the absence of drugs. (D) HepG2 cells were infected with recombinant PGC-1α virions, and FAS (black bars) and ACC (grey bars) mRNA abundance was determined as described above.
Table 2. mRNA expression levels of SREBP-1a and SREBP-1c in human liver and HepG2 cells.
Results are means±S.D. of experiments performed in four human liver samples and HepG2 cell preparations. HepG2 cells and PGC-1α virion-infected HepG2 cells were cultured in the absence (basal) or presence (stimulated) of 22(R)HC and 9cRA, 10 μmol each.
| SREBP-1a (amol/μg RNA) | SREBP-1c (amol/μg RNA) | SREBP-1c/SREBP-1a ratio | |
|---|---|---|---|
| Liver | 0.39±0.18 | 3.81±0.44 | 9.77±3.84 |
| HepG2 | 0.018±0.003 | 0.013±0.004 | 0.72±0.21 |
| PGC-1α virion-infected HepG2 cells, basal | 0.11±0.07 | 0.27±0.08 | 2.45±1.03 |
| PGC-1α virion-infected HepG2 cells, stimulated | 0.10±0.03 | 2.29±0.43 | 22.9±8.29 |
SREBPs are regulated at the post-transcriptional and transcriptional level. At the post-transcriptional level, the sterol content of the endoplasmic reticulum determines the rate of SREBP cleavage and nuclear translocation [2,26]. Transcriptional regulation includes a feed-forward loop that is mediated by SREs in the 5′-regulatory regions of each gene [27,28]. Furthermore, SREBP-1c transcription is selectively up-regulated by LXRs [17,18] which are co-activated by PGC-1α [20].
Our ectopic overexpression studies of PGC-1α in HepG2 cells, deficient in both PGC-1α and SREBP-1 gene expression, demonstrated that PGC-1α increased SREBP-1c mRNA abundance, which was augmented further by LXR/RXR ligands. This result is consistent with transient transfections showing amplification of LXR-mediated transactivation of the SREBP-1c promoter by PGC-1α. In contrast with transient transfection studies using a SREPB-1a reporter construct, ectopic overexpression of PGC-1α also increased SREBP-1a abundance levels, which may have resulted from a feed-forward mechanism mediated by SREBP-1c via sterol-regulatory elements in the SREBP-1a promoter. Other potential mechanisms for SREBP-1a induction include an increased need for membrane constituents stemming from PGC-1α-induced mitochondrial biogenesis.
HepG2 cells have been used extensively for studies on lipoprotein biosynthesis and secretion. Compared with primary hepatocytes or hepatic perfusates, these cells display much lower rates of triacylglycerol secretion [29]. HepG2 cells secrete triacylglycerol-rich LDL (low-density lipoprotein) as the major apoB-containing particle, while primary hepatocyte cultures, hepatic perfusates and whole liver secrete less-buoyant VLDL (very-low-density lipoprotein) as main apoB particles [30]. Moreover, exogenous fatty acids modulate the rate of apoB secretion in HepG2 cells [31], but have little effect in primary hepatocytes and liver [32–34]. Our experiments show that LXR-mediated induction of SREBP-1c alone was not sufficient to enhance the mRNA abundance of the SREBP-1c targets FAS and ACC in HepG2 cells. Ectopic expression of PGC-1α together with LXR/RXR activation resulted in substantially increased SREBP-1c expression and in enhanced FAS and ACC mRNA abundance levels, suggesting that SREBP-1c expression was rate-limiting. Thus some discrepancies between HepG2 cells and primary liver cells may reflect differences in the SREBP-1c expression level and its effect on endogenous triacylglycerol synthesis.
PGC-1α has been implicated in insulin-regulated hepatic gluconeogenesis by co-activating hepatocyte nuclear factor 4α and FOXO1 (forkhead transcription factor 1) [35,36]. While insulin does not appear to have a direct effect on hepatic PGC-1α expression, it induces Akt-mediated phosphorylation of FOXO1. As a result, FOXO1 is sequestered in the cytoplasm and the FOXO1–PGC-1α interaction is disrupted. SREBP-1c has also been shown to mediate hepatic effects of insulin [12,13]. Its mRNA and protein expression is selectively decreased in livers of fasted mice and increased after refeeding the animals a high-carbohydrate diet. Changes of SREBP-1c mRNA expression, observed in livers and hepatocyte cultures of rats with streptozotocin-induced diabetes, are correlated with changes in FAS and ACC mRNA levels [12]. The induction of FAS mRNA synthesis is abolished upon overexpression of a dominant-negative SREBP-1 protein, and overexpression of a transcriptionally active form of SREBP-1c in the liver of diabetic mice induces glucokinase and lipogenic enzyme expression and represses the expression of phosphoenolpyruvate carboxykinase, thereby increasing hepatic glycogen and triacylglycerol content [13,37]. Thus PGC-1α appears not only to contribute to insulin-regulated hepatic gluconeogenesis, but also enhances insulin effects on hepatic lipogenesis by co-activation of LXR-mediated SREBP-1c expression.
Acknowledgments
This study was supported by grants from the Oesterreichische Nationalbank (Project No. 10678), the Medizinische Forschungsgesellschaft Salzburg and the Verein für medizinische Fortbildung Salzburg.
References
- 1.Horton J. D., Goldstein J. L., Brown M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown M. S., Goldstein J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 3.Yokoyama C., Wang X., Briggs M. R., Admon A., Wu J., Hua X., Goldstein J. L., Brown M. S. SREBP-1, a basic-helix–loop–helix–leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 4.Hua X., Wu J., Goldstein J. L., Brown M. S., Hobbs H. H. Structure of the human gene encoding sterol regulatory element binding protein-1 (SREBF1) and localization of SREBF1 and SREBF2 to chromosomes 17p11.2 and 22q13. Genomics. 1995;25:667–673. doi: 10.1016/0888-7543(95)80009-b. [DOI] [PubMed] [Google Scholar]
- 5.Hua X., Yokoyama C., Wu J., Briggs M. R., Brown M. S., Goldstein J. L., Wang X. SREBP-2, a second basic-helix–loop–helix–leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horton J. D., Shimomura I., Brown M. S., Hammer R. E., Goldstein J. L., Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Invest. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimano H., Horton J. D., Hammer R. E., Shimomura I., Brown M. S., Goldstein J. L. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J. Clin. Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horton J. D., Shah N. A., Warrington J. A., Anderson N. N., Park S. W., Brown M. S., Goldstein J. L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimano H., Horton J. D., Shimomura I., Hammer R. E., Brown M. S., Goldstein J. L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foretz M., Pacot C., Dugail I., Lemarchand P., Guichard C., Le Liepvre X., Berthelier-Lubrano C., Spiegelman B., Kim J. B., Ferré P., Foufelle F. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol. Cell. Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guillet-Deniau I., Mieulet V., Le Lay S., Achouri Y., Carre D., Girard J., Foufelle F., Ferré P. Sterol regulatory element binding protein-1c expression and action in rat muscles: insulin-like effects on the control of glycolytic and lipogenic enzymes and UCP3 gene expression. Diabetes. 2002;51:1722–1728. doi: 10.2337/diabetes.51.6.1722. [DOI] [PubMed] [Google Scholar]
- 12.Shimomura I., Bashmakov Y., Ikemoto S., Horton J. D., Brown M. S., Goldstein J. L. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foretz M., Guichard C., Ferré P., Foufelle F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 1997;99:838–845. doi: 10.1172/JCI119247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshikawa T., Shimano H., Amemiya-Kudo M., Yahagi N., Hasty A. H., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., et al. Identification of liver X receptor–retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell. Biol. 2001;21:2991–3000. doi: 10.1128/MCB.21.9.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshikawa T., Shimano H., Yahagi N., Ide T., Amemiya-Kudo M., Matsuzaka T., Nakakuki M., Tomita S., Okazaki H., Tamura Y., et al. Polyunsaturated fatty acids suppress sterol regulatory element-binding protein 1c promoter activity by inhibition of liver X receptor (LXR) binding to LXR response elements. J. Biol. Chem. 2002;277:1705–1711. doi: 10.1074/jbc.M105711200. [DOI] [PubMed] [Google Scholar]
- 17.DeBose-Boyd R. A., Ou J., Goldstein J. L., Brown M. S. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRα and LXRβ. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., Mangelsdorf D. J. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 20.Oberkofler H., Schraml E., Krempler F., Patsch W. Potentiation of liver X receptor transcriptional activity by peroxisome-proliferator-activated receptor γ co-activator 1α. Biochem. J. 2003;371:89–96. doi: 10.1042/BJ20021665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michael L. F., Wu Z., Cheatham R. B., Puigserver P., Adelmant G., Lehman J. J., Kelly D. P., Spiegelman B. M. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oberkofler H., Esterbauer H., Linnemayr V., Strosberg A. D., Krempler F., Patsch W. Peroxisome proliferator-activated receptor (PPAR) γ coactivator-1 recruitment regulates PPAR subtype specificity. J. Biol. Chem. 2002;277:16750–16757. doi: 10.1074/jbc.M200475200. [DOI] [PubMed] [Google Scholar]
- 23.Zilberfarb V., Pietri-Rouxel F., Jockers R., Krief S., Delouis C., Issad T., Strosberg A. D. Human immortalized brown adipocytes express functional β3-adrenoceptor coupled to lipolysis. J. Cell Sci. 1997;110:801–807. doi: 10.1242/jcs.110.7.801. [DOI] [PubMed] [Google Scholar]
- 24.Esterbauer H., Schneitler C., Oberkofler H., Ebenbichler C., Paulweber B., Sandhofer F., Ladurner G., Hell E., Strosberg A. D., Patsch J. R., et al. A common polymorphism in the promoter of UCP2 is associated with decreased risk of obesity in middle-aged humans. Nat. Genet. 2001;28:178–183. doi: 10.1038/88911. [DOI] [PubMed] [Google Scholar]
- 25.Oberkofler H., Fukushima N., Esterbauer H., Krempler F., Patsch W. Sterol regulatory element binding proteins: relationship of adipose tissue gene expression with obesity in humans. Biochim. Biophys. Acta. 2002;1575:75–81. doi: 10.1016/s0167-4781(02)00279-8. [DOI] [PubMed] [Google Scholar]
- 26.Shimomura I., Bashmakov Y., Shimano H., Horton J. D., Goldstein J. L., Brown M. S. Cholesterol feeding reduces nuclear forms of sterol regulatory element binding proteins in hamster liver. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12354–12359. doi: 10.1073/pnas.94.23.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato R., Inoue J., Kawabe Y., Kodama T., Takano T., Maeda M. Sterol-dependent transcriptional regulation of sterol regulatory element-binding protein-2. J. Biol. Chem. 1996;271:26461–26464. doi: 10.1074/jbc.271.43.26461. [DOI] [PubMed] [Google Scholar]
- 28.Amemiya-Kudo M., Shimano H., Yoshikawa T., Yahagi N., Hasty A. H., Okazaki H., Tamura Y., Shionoiri F., Iizuka Y., Ohashi K., et al. Promoter analysis of the mouse sterol regulatory element-binding protein-1c gene. J. Biol. Chem. 2000;275:31078–31085. doi: 10.1074/jbc.M005353200. [DOI] [PubMed] [Google Scholar]
- 29.Dixon J. L., Ginsberg H. N. Regulation of hepatic secretion of apolipoprotein B-containing lipoproteins: information obtained from cultured liver cells. J. Lipid Res. 1993;34:167–179. [PubMed] [Google Scholar]
- 30.Wang S. R., Pessah M., Infante J., Catala D., Salvat C., Infante R. Lipid and lipoprotein metabolism in Hep G2 cells. Biochim. Biophys. Acta. 1988;961:351–363. doi: 10.1016/0005-2760(88)90082-3. [DOI] [PubMed] [Google Scholar]
- 31.Dixon J. L., Furukawa S., Ginsberg H. N. Oleate stimulates secretion of apolipoprotein B-containing lipoproteins from Hep G2 cells by inhibiting early intracellular degradation of apolipoprotein B. J. Biol. Chem. 1991;266:5080–5086. [PubMed] [Google Scholar]
- 32.Patsch W., Tamai T., Schonfeld G. Effect of fatty acids on lipid and apoprotein secretion and association in hepatocyte cultures. J. Clin. Invest. 1983;72:371–378. doi: 10.1172/JCI110977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis R. A., Boogaerts J. R. Intrahepatic assembly of very low density lipoproteins: effect of fatty acids on triacylglycerol and apolipoprotein synthesis. J. Biol. Chem. 1982;257:10908–10913. [PubMed] [Google Scholar]
- 34.Salam W. H., Wilcox H. G., Heimberg M. Effects of oleic acid on the biosynthesis of lipoprotein apoproteins and distribution into the very-low-density lipoprotein by the isolated perfused rat liver. Biochem. J. 1988;251:809–816. doi: 10.1042/bj2510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature (London) 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 36.Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., Spiegelman B. M. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature (London) 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 37.Becard D., Hainault I., Azzout-Marniche D., Bertry-Coussot L., Ferré P., Foufelle F. Adenovirus-mediated overexpression of sterol regulatory element binding protein-1c mimics insulin effects on hepatic gene expression and glucose homeostasis in diabetic mice. Diabetes. 2001;50:2425–2430. doi: 10.2337/diabetes.50.11.2425. [DOI] [PubMed] [Google Scholar]