Summary
Protein biogenesis within the endoplasmic reticulum (ER) is crucial for organismal function. Errors during protein folding necessitate the removal of faulty products. ER-associated protein degradation and ER-phagy target misfolded proteins for proteasomal and lysosomal degradation. The mechanisms initiating ER-phagy in response to ER proteostasis defects are not well understood. By studying mouse primary cells and patient samples as a model of ER storage disorders (ERSDs), we show that accumulation of faulty products within the ER triggers a response involving SESTRIN2, a nutrient sensor controlling mTORC1 signaling. SESTRIN2 induction by XBP1 inhibits mTORC1’s phosphorylation of TFEB/TFE3, allowing these transcription factors to enter the nucleus and upregulate the ER-phagy receptor FAM134B along with lysosomal genes. This response promotes ER-phagy of misfolded proteins via FAM134B-Calnexin complex. Pharmacological induction of FAM134B improves clearance of misfolded proteins in ERSDs. Our study identifies the interplay between nutrient signaling and ER quality control, suggesting therapeutic strategies for ERSDs.
Keywords: endoplasmic reticulum, autophagy, TFEB, ER-phagy, mTORC1, quality control, collagen, alpha(1)-antitrypsin Z (alpha(1)-ATZ), ER storage disorders, FAM134B
Graphical abstract

Highlights
-
•
ER-phagy is enhanced in cellular models of ER storage disorders (ERSDs)
-
•
ER-phagy induction is dependent on FAM134B transcriptional activation via TFEB/TFE3
-
•
The ER stress sensor XBP1 promotes TFEB/TFE3 nuclear translocation via SESTRIN2
-
•
Pharmacological activation of FAM134B represents a therapeutic treatment for ERSDs
ER-phagy is activated in response to misfolded protein accumulation in the ER. De Leonibus, Maddaluno, et al. characterize an ER-phagy response that is dependent on SESTRIN2 and FAM134B transcriptional induction, which can be pharmacologically targeted to enhance misfolded protein degradation in ER storage disorders.
Introduction
The endoplasmic reticulum (ER) coordinates the synthesis, folding, and maturation of a significant portion of the human proteome. Folding-defective proteins or disruption of ER functions can lead to the accumulation of unfolded proteins, triggering ER stress. In humans, several diseases are characterized by the accumulation of misfolded proteins within the ER lumen.1,2
The ER-associated protein degradation (ERAD) pathway recognizes misfolded proteins and facilitates their cytosolic translocation, ultimately leading to degradation in proteasomes.3 There are, however, numerous misfolded polypeptides that do not qualify for the ERAD pathway, likely due to structural constraints or their inherent propensity to form large aggregates. Consequently, these “ERAD-resistant” clients are degraded through alternative mechanisms involving lysosomes, collectively known as ER-to-lysosome-associated degradation (ERLAD) pathways.4
Among the ERLAD pathways, macro-ER-phagy is involved in the removal of aberrant proteins, preserving the integrity of the ER.5 ER-phagy is initiated with the formation of ER vesicles that are captured by autophagosomes, facilitating their delivery to lysosomes for efficient degradation.5 Alternatively, ER-derived vesicles can fuse with or invaginate into lysosomes through processes referred to as LC3-dependent vesicular transport and micro-ER-phagy, respectively.4,6 Several disease-associated ER misfolded proteins, such as mutant alpha(1)-antitrypsin Z (alpha(1)-ATZ) protein,7 type I procollagen,8 and proinsulin,9,10 are targets of ERLAD/ER-phagy pathways (reviewed in Reggiori and Molinari11).
The ER-phagy receptors FAM134A,12 FAM134B,13 FAM134C,12 CCPG1,14 RTN3,15 ATL3,16 SEC62,17 and TEX26418,19 are key ER-phagy initiators.20 They are ER membrane proteins that bind to the autophagy LC3/GABARAP family of proteins, facilitating the delivery of ER fragments to the autophagy-lysosome pathway. Some of these receptors also participate in the recognition of misfolded cargo. For instance, the ER-phagy receptor FAM134B forms a complex with the ER chaperone Calnexin (CANX), which, through its luminal domain, binds to glycosylated misfolded cargoes like ATZ and collagens.7,8 The CCPG1 receptor interacts directly with its cargoes, such as the prolyl 3-hydroxylase family member 4 (P3H4), through luminal cargo-interacting regions.21 The ER protein PGRMC1 captures misfolded prohormones (e.g., proinsulin) for RTN3-dependent ER-phagy.22
During starvation-induced ER-phagy, ER-phagy receptors are activated by transcriptional and post-translational mechanisms.20 For example, FAM134B isoform 2 is typically expressed at low levels but is transcriptionally induced during starvation by the transcription factors TFEB, TFE3, and C/EBPβ.23,24 Post-translation modifications, such as phosphorylation,25,26 ubiquitination,27,28 and acetylation,29 favor receptor oligomerization and interaction with the autophagy machinery. Whether similar activation mechanisms also operate to promote ER-phagy in response to ER storage of misfolded molecules is unknown. In this work, we hypothesize the existence of an “ER storage response” that is activated upon misfolded protein accumulation within the ER lumen that stimulates ER-phagy. Characterizing this response paves the way for therapeutic approaches to target diseases characterized by the accumulation of misfolded proteins in the ER lumen, collectively called ER storage disorders (ERSDs).30
Results
ER storage activates ER-phagy
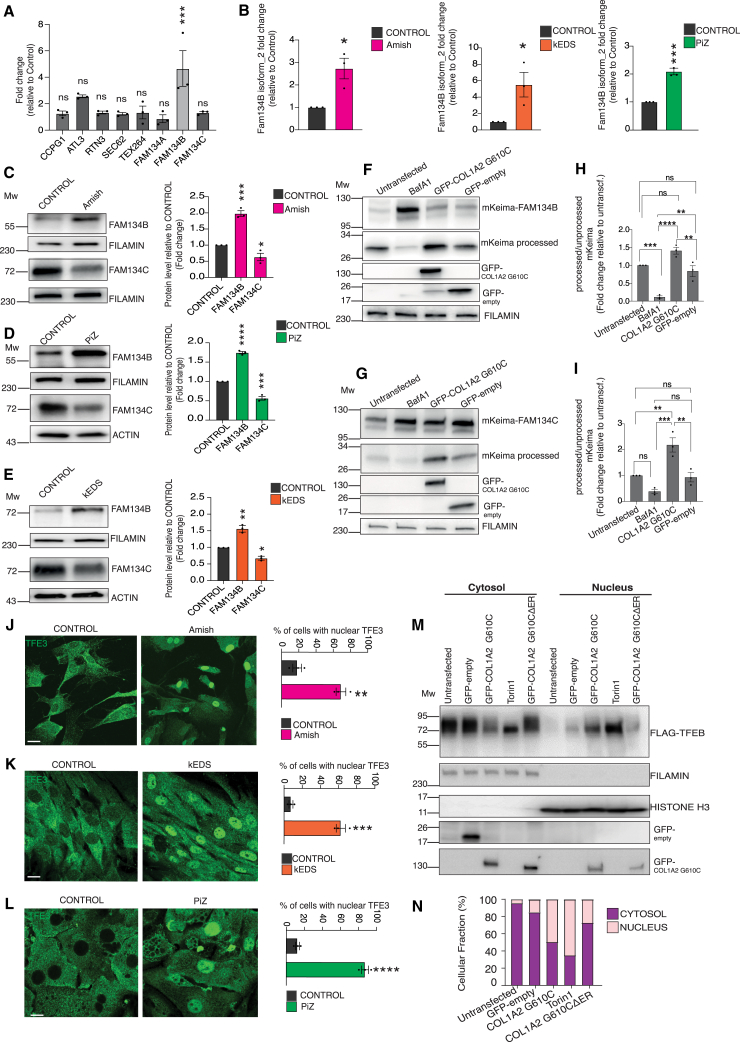
To investigate ER-phagy in response to the accumulation of misfolded cargoes within the ER, we study various cellular models of ERSDs. These models are characterized by the build-up of misfolded proteins within the ER lumen, leading to its enlargement. The first model is represented by human fibroblasts from individuals affected by kyphoscoliotic Ehlers-Danlos syndrome (kEDS). These cells are characterized by ER enlargement (Figure S1A) caused by defective type III and VI collagen folding within the ER due to point mutations in FKBP prolyl isomerase 14 (FKBP14).31 The second model is primary osteoblasts derived from a mouse model with a dominant form of osteogenesis imperfecta (OI), known as the Amish mouse.32 These osteoblasts carry a heterozygous G610C substitution in the α2 chain of collagen I (Col1a2+/G610C), resulting in the accumulation of misfolded collagen I within the ER.33 The third model is primary hepatocytes obtained from a mouse model of α1-antitrypsin deficiency (PiZ mouse).34 These hepatocytes express a E342K mutant version of human SERPINA1 (ATZ) that accumulates in the ER lumen.35
To assess ER-phagy activity, both control and disease cells were transduced with a fluorescence ER-phagy tandem reporter (ssRFP-GFP-KDEL).18 The formation of red-only puncta, which represents the ER delivered to lysosomes, was quantified as a measure of ER-phagy (Figure 1A). We observed a significant increase in ER-phagy in Amish osteoblasts, PiZ hepatocytes, and kEDS fibroblasts compared with their respective controls (Figures 1B–1G). An induction of ER-phagy also occurred after transfection of ATZ, COL1A2 G610C, or COL2A1 harboring a Arg789-to-Cys substitution (COL2A1 R789C), mutated in spondyloepiphyseal dysplasia36 (Figures S1B–S1D). To ascertain that the induction of ER-phagy was not a response to cell stress, we generated a COL1A2 G610C mutant lacking the N-terminal signal peptide (COL1A2 G610CΔER) fused with GFP (Figure 1H). Although COL1A2 G610C co-localized with the ER marker CANX, the COL1A2 G610CΔER mutant exhibited a cytosolic localization pattern (Figure 1I). ER-phagy, quantified by monitoring the delivery of the ER membrane protein CLIMP63 to the lysosome, was induced upon the expression of the COL1A2 G610C but not the COL1A2 G610CΔER mutant (Figures 1J and 1K). Similarly, although COL2A1 R789C expression induced ER-phagy, COL2A1 R789CΔER did not (Figure S1E). To corroborate these findings, we evaluated ER-phagy activity in rat chondrosarcoma (RCS) cells stably expressing the mKeima reporter37 fused with the ER membrane protein RAMP4. Monitoring the appearance of mKeima proteolytically cleaved from RAMP4 reflects the lysosomal processing of RAMP4-decorated ER fragments. The overexpression of COL1A2 G610C led to a substantial increase in processed mKeima levels, confirming ER-phagy induction (Figure S1F).
Figure 1.
ER storage activates ER-phagy
(A) Schematic representation of the ssRFP-GFP-KDEL ER-phagy reporter composed of an N-terminal ER signal sequence, a tandem monomeric RFP and GFP, and the ER retention sequence KDEL. Delivery of ssRFP-GFP-KDEL to lysosomes quenches the GFP signal, while RFP is relatively stable. Quantification of the red-only puncta provides a measure of ER-phagy induction.
(B) Representative fluorescence microscopy of primary osteoblasts derived from wild-type (control) and Amish mice expressing the ssRFP-GFP-KDEL plasmid. Scale bars, 10 μm.
(C) Bar graph showing the ratio of red-only puncta/total cells in (B). Mean ± standard error of mean (SEM) of N = 3 biological replicates. n = 33 cells were counted. Student’s unpaired t test: ∗∗∗p < 0.0005.
(D) Representative fluorescence microscopy of primary hepatocytes, isolated from control and PiZ mice, transfected with ssRFP-GFP-KDEL. Scale bars, 10 μm.
(E) Bar graph showing the ratio of red-only puncta/total cells in (D). Mean ± SEM of N = 3 biological replicates. n = 17 cells were counted. Student’s unpaired t test: ∗∗p < 0.005.
(F) Representative fluorescence microscopy of primary human fibroblasts, isolated from control subjects and kEDS patients, transfected with ssRFP-GFP-KDEL. Scale bars, 10 μm.
(G) Bar graph showing the ratio of red-only puncta/total cells in (F). Mean ± SEM of N = 3 biological replicates of n = 21 cells. Student’s unpaired t test: ∗p < 0.05.
(H) GFP-COL1A2 G610C accumulates as a misfolded protein in the ER. In the absence of the N-terminal signal peptide the GFP-COL1A2 G610CΔER accumulates as a misfolded protein in the cytosol.
(I) Immunofluorescence analysis of GFP-COL1A2 G610C and GFP-COL1A2 G610CΔER by confocal microscopy. COL1A2 G610C (GFP, green) shows co-localization with the ER-marker Calnexin (red). Conversely, COL1A2 G610CΔER shows a cytosolic localization. Scale bars, 5 μm.
(J) Immunofluorescence staining of CLIMP63 (green) and lysosomes (LAMP1, red) in U2OS cells transfected with GFP-COL1A2 G610C or GFP-COL1A2 G610CΔER, or GFP-empty plasmid as a transfection control. Starvation with Hank's Balanced Salt Solution (HBSS) was used as a positive control for ER-phagy induction. Cells were treated with BafA1 (100 nM, 4 h). Scale bars, 5 μm. Insets show magnification of CLIMP63 localization in lysosomes.
(K) Bar graph showing data quantification of (J). Mean ± SEM of N = 3 independent experiments. n = 18 cells were counted. One-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test: ∗p < 0.05; ∗∗p < 0.005; ns ≥ 0.05.
We also monitored mitophagy and cytosolic (bulk) autophagy by using fluorescent tandem (RFP-GFP-COX8 and RFP-GFP-LDHB, respectively) and mKeima reporters (mKeima-COX8 and mKeima-LDHB, respectively). Although mitophagy was not activated, an induction, albeit limited, of cytosolic autophagy was present upon COL1A2 G610C overexpression (Figures S2A–S2D).
These results suggest that the accumulation of misfolded proteins within the ER lumen primarily triggers the activation of ER-phagy.
ER storage activates ER-phagy via TFE3/TFEB-mediated transcriptional induction of FAM134B
To elucidate the mechanisms underlying ER-phagy activation in ER storage diseases (ERSDs), we examined the expression levels of ER-phagy receptors. We observed an increase in the mRNA levels of FAM134B (isoform 2) upon overexpression of mutant collagens. In contrast, other members of the FAM134 family (FAM134A and FAM134C) and other ER-phagy receptors (CCPG1, ATL3, SEC62, TEX264, and RTN3) were largely unaffected (Figures 2A and S2E). Overexpression of COL1A2 G610CΔER did not upregulate FAM134B, suggesting that FAM134B induction is a specific response to ER storage (Figure S2F). Amish primary osteoblasts, PiZ primary hepatocytes, and kEDS human fibroblasts exhibited elevated FAM134B at both transcriptional and protein levels compared with their respective control cells (Figures 2B–2E). Conversely, FAM134C protein levels were lower in Amish primary osteoblasts, PiZ primary hepatocytes, and kEDS human fibroblasts compared with their respective controls (Figures 2C–2E). FAM134C is not transcriptionally induced by misfolded proteins (Figures 2A and S2E), but it physically interacts with FAM134B during ER-phagy27 and undergoes rapid degradation during starvation-induced ER-phagy.25 To monitor the degradation rate of FAM134B and FAM134C, we generated cell lines expressing FAM134B and FAM134C fused to the mKeima reporter. The overexpression of the COL1A2 G610C mutant significantly increased the delivery of FAM134B and FAM134C proteins to lysosomes, as demonstrated by the levels of fragmented mKeima (Figures 2F–2I). These observations provide biochemical evidence of FAM134B and FAM134C-mediated ER-phagy activation in ERSD cells and suggest that higher levels of FAM134B in ERDS cells are due to FAM134B transcriptional induction.
Figure 2.
ER storage induces TFE3/TFEB nuclear translocation and transcriptional induction of FAM134B
(A) Quantitative real-time PCR analysis of ER-phagy receptors in U2OS cells expressing COL1A2 G610C. Values were normalized to HPRT gene expression and shown as fold change relative to untransfected cells (control). Mean ± standard error of mean (SEM) of N = 3 biological replicates. One-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test: ∗∗∗p < 0.0005; ns ≥ 0.05.
(B) Quantitative real-time PCR analysis of FAM134B isoform 2 in Amish, osteoblasts, kEDS fibroblasts, and PiZ hepatocytes. Values are normalized to S16 (osteoblasts and hepatocytes) and HPRT (fibroblasts) and shown as fold change relative to their corresponding controls. Mean ± SEM of N = 3 biological replicates. Student’s unpaired t test: ∗p < 0.05; ∗∗∗p < 0.0005.
(C–E) Western blot analysis (left) of FAM134B and FAM134C proteins in control and Amish, osteoblasts (C), PiZ hepatocytes (D), and kEDS fibroblasts (E), and their corresponding controls. FILAMIN and actin were used as a loading control. Bar graphs showing mean ± SEM of N = 3 biological replicates. One-way ANOVA with Dunnett’s multiple comparisons test: ∗p < 0.05; ∗∗p < 0.005, ∗∗∗p < 0.0005; ∗∗∗∗p < 0.0001.
(F–I) Western blot analysis and quantification of cells expressing mKeima-FAM134B (F) and mKeima-FAM134C (G), transfected with GFP-COL1A2 G610C or GFP-empty plasmid (control) and treated with BafA1 (50 nM, 12 h) where indicated. FILAMIN was used as a loading control. Bar graphs showing quantification of mKeima/mKeima-FAM134B (H) and FAM134C (I) ratio relative to untransfected cells. Mean ± SEM of N = 3 biological replicates. One-way ANOVA with Tukey’s multiple comparisons test: ∗∗p < 0.005; ∗∗∗p < 0.0005; ∗∗∗∗p < 0.0001; ns ≥ 0.05.
(J–L) Immunofluorescence analysis (left panels) of TFE3 (green) subcellular localization in control and Amish, primary osteoblasts (J), human kEDS fibroblasts (K), and PiZ hepatocytes (L) and their corresponding controls. Scale bars, 20 μm. Bar graphs showing quantification of the percentage of cells with nuclear TFE3 (right). Mean ± SEM of N = 3 independent experiments. n = 245 control and Amish osteoblasts; n = 312 control and kEDS fibroblasts; n = 421 control and PiZ hepatocytes. Student’s unpaired t test: ∗∗p < 0.005; ∗∗∗p < 0.0005; ∗∗∗∗p < 0.0001.
(M) Nuclear and cytosolic levels of FLAG-TFEB in U2OS cells transfected with GFP-COL1A2 G610C and GFP-COL1A2 G610CΔER. GFP-empty plasmid was used as control. Torin1 was used as a positive control. FILAMIN and Histone H3 were used as a loading control.
(N) Quantification the data in (M). TFEB localization was expressed as % of cytosolic and nuclear fractions relative to total. Mean of N = 4 independent experiments.
The transcriptional induction of FAM134B can be mediated by TFEB/TFE3 factors through their direct binding to the CLEAR consensus sequence in the FAM134B promoter.23 Normally localized in the cytoplasm, TFEB and TFE3 translocate to the nucleus upon dephosphorylation.38 The nuclear accumulation of endogenous TFE3 was significantly enhanced in Amish primary osteoblasts, kEDS human fibroblasts, and PiZ primary hepatocytes compared with control cells (Figures 2J–2L). Transient overexpression of mutant collagens and ATZ, but not wild-type (WT) or ΔER collagens, also promoted the nuclear translocation of TFEB (Figures 2M, 2N, and S2G–S2K). These findings collectively suggest that ER storage of misfolded proteins triggers the activation of TFEB and TFE3.
To demonstrate the regulatory role of TFEB/TFE3 in FAM134B transcriptional induction, we expressed COL1A2 G610C, COL2A1 R789C, and ATZ in U2OS osteoblasts, RCS chondrocytes, and mouse embryonic fibroblasts (MEFs), respectively, where TFEB and TFE3 had been deleted using CRISPR-Cas9 technology (TFEB/TFE3 DKO cells). Mutant protein overexpression failed to transcriptionally induce FAM134B in TFEB/TFE3 DKO cells, confirming that TFEB/TFE3 are the major regulators of FAM134B transcriptional induction in response to protein misfolding in the ER (Figures 3A–3C). RNA sequencing (RNA-seq) analysis (GSE239527) of WT and TFEB/TFE3 DKO RCS cells with or without COL2A1 R789C expression demonstrated that several genes were induced upon ER storage in a TFEB- and TFE3-dependent manner, including endosomal, lysosomal, and autophagy genes (Figures 3D, 3E, and S3A; Tables S1, S2, and S3).
Figure 3.
ER storage activates ER-phagy via TFEB/TFE3-mediated transcriptional induction of FAM134B
(A–C) Quantitative real-time PCR analysis showing transcriptional levels of FAM134B isoform 2 in control and TFEB/TFE3 DKO cell lines expressing the indicated proteins. Values were normalized to HPRT and shown as fold change relative to untransfected cells (control). Mean ± standard error of mean (SEM) of N = 3 biological replicates. One-way analysis of variance (ANOVA) with Šídák’s multiple comparisons test: ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005; ns ≥ 0.05.
(D) Venn diagram showing specific and common genes identified by transcriptomic analysis performed in WT and TFEB/3 DKO RCS cells with or without the expression of the COL2A1 R789C mutant protein.
(E) Heatmap on 24 lysosomal genes induced by COL2A1 R789C overexpression in a TFE3- and TFEB-dependent manner.
(F) Immunofluorescence analysis of FLAG-COL2A1 R789C (blue inset) in WT RCS, FAM134B KO RCS, and TFEB/3 DKO RCS overexpressing ER-phagy tandem reporter (ssRFP-GFP-KDEL). HBSS treatment was used as a positive control. Bottom insets show magnification of the boxed areas. Scale bars, 5 μm.
(G) Quantification of the data in (F). The bar graph represents mean ± standard error of mean (SEM) of N = 3 independent experiments. n = 21 cells were counted for each condition. One-way ANOVA with Šídák’s multiple comparisons test: ∗∗∗∗p < 0.0001; ns ≥ 0.05.
(H) Western blot analysis in WT, FAM134B KO, and TFEB/3 DKO RCS cells expressing mKeima-RAMP4, transfected with GFP-COL1A2 G610C or GFP-empty plasmid (control). Actin was used as a loading control.
(I) Immunofluorescence staining of Calnexin (green) and lysosomes (LAMP1, red) in U2OS cells transfected with GFP-COL1A2 G610C or GFP-COL1A2 G610CΔER, or GFP-empty plasmid (control). Cells were treated with BafA1 (100 nM, 4 h). Scale bars, 5 μm. Insets show magnification of Calnexin localization in lysosomes. Bar graph (bottom) showing data quantification of Calnexin fluorescence intensity in lysosomes. Mean ± SEM of N = 3 independent experiments. n = 21 cells were counted. One-way ANOVA with Dunnett’s multiple comparisons test: ∗∗p < 0.005; ns ≥ 0.05.
(J and K) Western blot analysis showing immunoprecipitation (IP) experiment of endogenous FAM134B in U2OS cells overexpressing GFP-COL1A2 G610C (J) or GFP-TFEB (K). BafA1 (200 nM, 4 h) was supplied where indicated. Actin was used as a loading control.
We then investigated whether TFEB/TFE3 and FAM134B are involved in the activation of ER-phagy in ERSD cells. Overexpression of the COL2A1 R789C protein significantly increased ER-phagy in cells expressing the ER-phagy reporters (ssRFP-GFP-KDEL or mKeima-RAMP4). However, these effects were not observed in FAM134BKO or TFEB/TFE3 DKO RCS cells (Figures 3F–3H). These findings suggest a crucial role of the TFEB/TFE3-FAM134B axis in the activation of ER-phagy in ERSD cells.
Furthermore, the lysosomal delivery of mCHERRY-COL2A1 R789C and ATZ was blunted in MEFs lacking TFEB/TFE3, FAM134B, and the autophagy gene ATG7. Notably, deletion of the early autophagy protein ATG9 completely inhibited collagen delivery to lysosomes and partially affected the delivery of ATZ (Figures S3B–S3E). These data indicate that the activation of FAM134B via TFEB/TFE3 participates in both macro-autophagy-dependent and independent (LC3-dependent vesicular transport) pathways of protein delivery to lysosomes for degradation.
FAM134B forms a complex with CANX to deliver misfolded cargo to lysosomes.7,8 We observed that cells accumulating COL1A2 G610C or overexpressing TFEB have enhanced (FAM134B-CANX) complex formation (Figures 3J and 3K), leading to increased delivery of endogenous CANX to lysosomes (Figure 3I). These data suggest that the induction of FAM134B by TFEB promotes the formation of the FAM134B-CANX complex, which is involved in the disposal of misfolded cargoes from the ER.
To study mutant collagen delivery to lysosomes, we generated U2OS cells expressing COL1A2 G610C fused with GFP-RFP (tandem collagen assay). At steady state, the ssGFP-RFP-COL1A2 G610C showed ER-localization with the appearance of red-only puncta that colocalize with the lysosomal marker LAMP1, indicating ER-phagy of mutant collagen (Figures S3F and S3G). FAM134B interference was more effective than the silencing of other ER-phagy receptors in reducing collagen delivery to lysosomes. Similarly, Fam134b silencing was more efficient than the silencing of other ER-phagy receptors in limiting ER-phagy activation in response to COL1A2 G610C overexpression (Figures S4A–S4E).
Collectively, these data demonstrate that the activation of the TFEB/TFE3-FAM134B axis promotes protein clearance from the ER via ER-phagy.
The ER storage response activates the TFEB-FAM134B axis via Sestrin2 induction
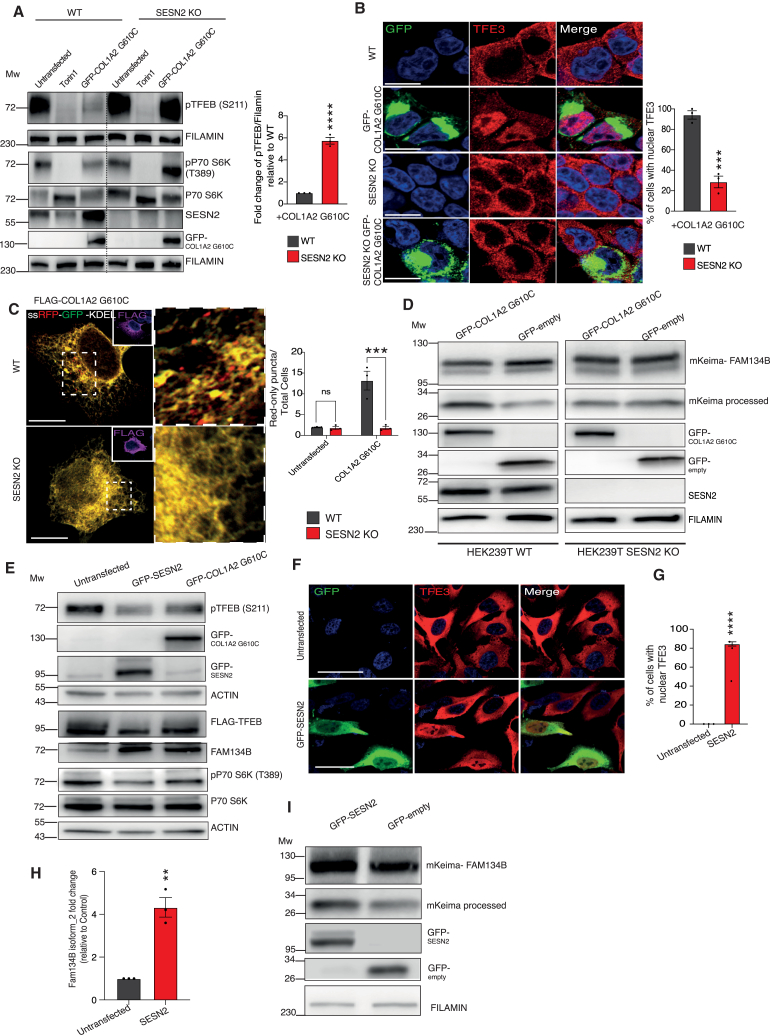
To investigate the mechanisms of TFEB/TFE3 activation in cells affected by ER storage, we compared two transcriptome analyses from RCS cells expressing the COL2A1 R789C protein (Table S1, GSE239527) and HeLa cells expressing COL1A2 G610C (Table S4, GSE239525). We identified a set of 73 differentially expressed genes in common between the two datasets (Table S5; Figure 4A). We silenced 40 out of the 46 commonly upregulated genes and evaluated GFP-TFEB nuclear translocation by high-content fluorescent microscopy after transfection with COL1A2 G610C. The most significant impact on GFP-TFEB nuclear translocation was observed upon the silencing of SESN2, SRSF11, USP36, CHAC1, and CEP95 (Figures 4B and S5A), suggesting that their induction contributes to TFEB nuclear translocation upon ER storage. Notably, upregulation of SESN2, but not of SRSF11, USP36, CHAC1, or CEP95, was also detectable in Amish primary osteoblasts, PiZ primary hepatocytes, and kEDS primary human fibroblasts compared with control cells, suggesting that SESN2 upregulation represents a general response to ER storage (Figures 4C, S5B, and S5C).
Figure 4.
ER storage promotes TFEB nuclear translocation via the upregulation of the leucine sensor Sestrin2
(A) Venn diagram showing that 73 genes were commonly regulated in RCS and HeLa cells upon COL2A1 R789C and COL1A2 G610C expression, respectively.
(B) Heatmap of the genes whose silencing significantly reverted TFEB nuclear localization in HeLa GFP-TFEB cells expressing COL1A2 G610C. A low score of nuclear TFEB localization is represented in violet, a high score in pink. Values represent the mean of N = 8 biological replicates.
(C)Quantitative real-time PCR analysis of Sestrin2 (SESN2) in Amish, PiZ, and kEDS primary cells. Values were normalized to CYC (Amish and PiZ) and HPRT (kEDS and PiZ) gene expression and shown as fold change relative to their corresponding controls. Mean ± standard error of mean (SEM) of N = 3 biological replicates in Amish, N = 4 biological replicates in PiZ, and N = 3 biological replicates in kEDS. Student’s unpaired t test: ∗p < 0.05; ∗∗p < 0.005.
(D–F) Western blot analysis of (D) phospho-TFEB (S211) and phosho-P70S6K (T389) in HeLa cells and (E) phospho-ULK1 (S757) in U2OS cells expressing COL1A2 G610C. Torin 1 was used as a positive control. Actin and FILAMIN were used as loading controls. In (F), the bar graphs show quantification of the indicated proteins as fold change relative to untransfected cells. Mean ± SEM of N = 3 independent experiments. One-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test: ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005; ∗∗∗∗p < 0.0001.
(G) Immunofluorescence staining of mTORC1 (green) and lysosomes (LAMP1, red) in U2OS cells expressing GFP-COL1A2 G610C. Inset, GFP-COL1A2 G610C immunostaining. Small right panels are magnifications of the boxed areas. Scale bars, 10 μm. The bar graph shows co-localization of mTORC1 with LAMP1 (expressed as Manders’ coefficient). Mean ± SEM of N = 3 independent experiments. n = 20 control and GFP-COL1A2 G610C transfected cells. Student’s unpaired t test: ∗∗p < 0.005.
(H) Subcellular localization of endogenous TFE3 (red) in WT and KICSTOR KO HEK293T cells transfected with GFP-COL1A2 G610C (green) or treated with Torin 1. Scale bars, 20 μm. The bar graph (bottom) shows quantification of cells with nuclear TFE3. Mean ± SEM of N = 3 independent experiments. n = 40 cells. One-way ANOVA with Šídák’s multiple comparisons test: ∗∗∗∗p < 0.0001; ns ≥ 0.05.
(I) Representative immunofluorescence of FLAG-TFEB (red) subcellular localization in U2OS cells expressing GFP-COL1A2 G610C cells (green) without or with leucine (1.2 mM, 1 h). The dotted circle indicates nuclei. Scale bars, 7 μm. The bar graph shows quantification of % cells with nuclear TFEB relative to control. Mean ± SEM of N = 3 independent experiments. n = 40 cells for each treatment. One-way ANOVA with Dunnett's multiple comparisons test: ∗∗∗p < 0.0005; ns ≥ 0.05.
(J) Immunofluorescence analysis of TFE3 (green) subcellular localization in PiZ hepatocytes untreated (control) or treated with leucine (4 mM, 3 h). The dotted circle indicates nuclei. Scale bars, 20 μm. Bar graph showing quantification of the percentage of cells with nuclear TFE3 (right). Mean ± SEM of N = 3 independent experiments. n = 292 control and PiZ hepatocytes. Student’s unpaired t test: ∗∗∗∗p < 0.0001.
(K) Representative fluorescence microscopy analysis of PiZ hepatocytes transfected with the ssRFP-GFP-KDEL plasmid, left untreated (control) or treated with leucine (4 mM, 3 h). Scale bars, 10 μm. Bar graph shows the ratio of red-only puncta/total cells. Mean ± SEM of N = 3 biological replicates. n = 19 control and PiZ hepatocytes. Student’s unpaired t test: ∗∗∗p < 0.0005.
SESN2 is a leucine-binding protein and plays a crucial role in the regulation of mTORC1 via the GATOR complexes.39,40 When SESN2 binds to GATOR2, GATOR1 is active and acts as a GTPase-activating protein (GAP) toward the RagA/B GTPases. Guanosine diphosphate (GDP)-loaded RagA and RagB GTPases prevent mTORC1 from being recruited to the lysosomal membrane and hinder its kinase activity. When leucine is present, it binds to SESN2 leading to its dissociation from GATOR2, which can now bind and inhibit GATOR1.41 Thus, leucine inhibition of SESN2 binding to GATOR2 ultimately leads to the activation of mTORC1 on the lysosome.
The following experiments demonstrated the role of SESN2 in TFEB-FAM134B activation in response to ER storage. First, we observed a strong reduction in the phosphorylation levels of TFEB on Serine 211, which is phosphorylated by mTORC1 on lysosome upon COL1A2 G610C expression compared with non-transfected cells. Notably, the phosphorylation of the other mTORC1 substrates, P70S6K on Threonine 389, and of ULK1 on Serine 757, were only partially influenced by COL1A2 G610C expression (Figures 4D–4F; see also Figure 5A), suggesting that ER storage modulate primarily mTORC1 phosphorylation of TFEB/TFE3. Second, we observed that mTORC1 lysosomal localization was significantly reduced in cells expressing COL1A2 G610C compared with control (Figure 4G). Third, the inhibition of GATOR1 activity by depletion of the SZT2 subunit of the KICSTOR complex (which is required for GATOR1 association with lysosomal membranes40) prevented TFEB nuclear translocation upon COL1A2 G610C expression (Figure 4H); fourth, supplementing the media with leucine reverted TFEB/TFE3 nuclear accumulation and ER-phagy activation in COL1A2 G610C expressing cells and, more importantly, in ERSD primary cells (Figures 4I–4K and S5D–S5F). Fifth, downregulation of SESN2 by small interfering RNA (siRNA) blunted TFEB dephosphorylation and nuclear translocation in response to COL1A2 G610C overexpression (Figures S5G–S5I).
Figure 5.
ER storage response activates ER-phagy via Sestrin2 induction
(A) Western blot analysis of phosho-TFEB (S211) and phosho-P70S6K (T389) in WT and SESN2 KO HEK283T cells transfected with GFP-COL1A2 G610C or treated with Torin 1. FILAMIN was used as a loading control. Bar graph shows quantification of phospho-TFEB levels. Mean ± standard error of mean (SEM) of N = 3 biological replicates. Student’s unpaired t test: ∗∗∗∗p < 0.0001.
(B) Representative images of immunofluorescence staining of TFE3 (red) subcellular localization in WT and SESN2 KO HEK283T cells expressing or not GFP-COL1A2 G610C. Scale bars, 8 μm. Bar graph shows quantification of % cells with nuclear TFE3. Mean ± SEM of N = 3 biological replicates. n = 70 WT and SESN2 KO cells. Student’s unpaired t test: ∗∗∗p < 0.0005.
(C) Representative images of WT and SESN2 KO cells expressing FLAG-COL1A2 G610C and the ssRFP-GFP-KDEL plasmid. Scale bars, 8 μm. Right: bar graph showing the ratio of red-only puncta/total cells. Mean ± SEM of N = 3 biological replicates. n = 45 WT and SESN2 KO cells. Two-way analysis of variance (ANOVA) with Šídák’s multiple comparisons test: ∗∗∗p < 0.0005; ns ≥ 0.05.
(D) Western blot analysis of WT and SESN2 KO HEK283T cells overexpressing mKeima-FAM134B transfected with GFP-COL1A2 G610C or GFP-empty plasmid as a control. FILAMIN was used as a loading control.
(E) Western blot analysis of phosho-TFEB (S211) and phosho-P70S6K (T389) in U2OS cells with GFP-COL1A2 G610C and GFP-SESN2 overexpression. Actin was used as a loading control.
(F) Immunofluorescence staining of TFE3 (red) subcellular localization in U2OS cells overexpressing GFP-SESN2. Scale bars, 8 μm.
(G) Bar graph shows % of cells with nuclear TFE3 with GFP-SESN2 overexpression relative to untransfected cells. Mean ± SEM of N = 3 biological replicates. n = 45 cells. Student’s unpaired t test: ∗∗∗∗p < 0.0001.
(H) Quantitative real-time PCR analysis of FAM134B isoform 2 in U2OS cells overexpressing GFP-SESN2 relative to untransfected cells (control). Mean ± standard error of mean (SEM) of N = 3 biological replicates. Values were normalized to HPRT gene expression and are shown as fold change relative to control. Student’s unpaired t test: ∗∗p < 0.005.
(I) Western blot analysis in HEK283T cells overexpressing mKeima-FAM134B transfected with GFP-SESN2 or GFP-empty plasmid. FILAMIN was used as a loading control.
Next, we analyzed ER-phagy responses in cell lines lacking SESN2 (SESN2 KO). COL1A2 G610C overexpressing SESN2 KO cells showed persistent cytosolic localization of TFEB/TFE3, mTORC1 association with lysosomes, and increased phosphorylation levels of TFEB on Serine 211 (Figures 5A, 5B, and S5J). FAM134B transcription was significantly reduced in SESN2 KO cells compared with control upon overexpression of COL1A2 G610C (Figure S5K), and a marked decrease in ER-phagy was observed in SESN2 KO cells expressing COL1A2 G610C compared with their respective controls (Figures 5C and 5D). Conversely, the solely SESN2 overexpression was sufficient to induce TFEB dephosphorylation and nuclear translocation, Fam134B transcriptional induction, and ER-phagy activation (Figures 5E–5I). Collectively, these data demonstrate that SESN2 induction in response to ER storage is both necessary and sufficient to activate ER-phagy.
Activation of XBP1 promotes Sestrin 2 activation and TFEB nuclear translocation
Sestrin proteins are upregulated during stress although the mechanisms of this regulation appear to be context and cell specific.42,43,44 The three main signaling branches that characterize the unfolded protein response (UPR) are IRE1α-XBP1, PERK-eIF2-ATF4, and ATF6.45 We found that the accumulation of misfolded proteins in the ER lumen elicited solely a mild activation of the IRE1-XBP1 branch of the UPR (Figure 6A). We pharmacologically inhibited IRE1α-XBP1, PERK-eIF2-ATF4, and ATF6 and found that treatment with an IRE1 ribonuclease inhibitor (4μ8C), which blocks the RNase domain of IRE1,46 reversed TFEB nuclear translocation in response to COL1A2 G610C expression (Figures 6B and 6C). Milder effects were observed using the integrated stress response inhibitor (ISRIB), which blocks the PERK-mediated activation of ATF4,47 or a competitive inhibitor of sterol regulatory element-binding protein (SREBP) site 1 protease (PF-429242), which blocks ATF6 activation48 (Figures 6B and 6C). The primary target of IRE1 is the transcription factor XBP1. Therefore, we investigated its role in SESN2-mediated activation of TFEB and TFE3. Upon silencing XBP1, we observed a significant reversal of TFEB nuclear translocation, which we analyzed using high-content fluorescent microscopy, in response to COL1A2 G610C expression (Figure 6D). Milder, but significant, effects were observed also silencing ATF4 and ATF6. These data suggest that XBP1 activation is a major contributor of TFEB activation in response to ER storage of misfolded proteins. We then investigated whether XBP1 mediates the transcriptional induction of SESN2. Consistent with this hypothesis, XBP1 silencing inhibited the transcriptional induction of SESN2, as well as its protein accumulation in response to COL1A2 G610C overexpression (Figures 6E–6H). These data indicate a cascade of events where the progressive accumulation of misfolded proteins in the ER triggers XBP1-mediated activation of SESN2 that promotes the activation of the TFEB/TFE3-FAM134B axis (Figure 6I).
Figure 6.
XBP1 activation triggers SESN2 induction and TFEB nuclear translocation in response to ER storage
(A) Quantitative real-time PCR analysis of the expression of indicated gene in U2OS cells transfected with COL1A2 G610C. Values were normalized to HPRT and are shown as fold change relative to untransfected cells (control). Mean ± standard error of mean (SEM) of N = 3 biological replicates. One-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test: ∗∗p < 0.005; ns ≥ 0.05.
(B) Cytosolic and nuclear levels of FLAG-TFEB in U2OS cells transfected or not with GFP-COL1A2 G610C and treated with the indicated drugs or DMSO as control. Torin1 was used as a positive control for TFEB nuclear translocation. FILAMIN and histone H3 served as loading controls.
(C) Quantification of TFEB localization in (B) was expressed as % of cytosolic and nuclear fractions relative to the total. Mean of N = 4 independent experiments.
(D) Quantification of TFEB nuclear levels by high-content fluorescent microscopy in cells transfected with COL1A2 G610C and treated with scramble, XBP1, ATF6, and ATF4 siRNAs. Mean ± SEM of N = 6 biological replicates. One-way ANOVA with Dunnett’s multiple comparisons test: ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005.
(E and F) Quantitative real-time PCR analysis of the spliced form of XBP1 (SP XBP1) (E) and SESN2 (F) in HeLa cells expressing COL1A2 G610C and treated with scramble or XBP1 siRNA at different time points. Mean ± standard error of mean (SEM) of N = 5 biological replicates. Values were normalized to HPRT gene expression and are shown as fold change relative to untransfected cells (control). Two-way ANOVA with Tukey’s multiple comparisons test: ∗p < 0.05; ∗∗p < 0.005; ∗∗∗p < 0.0005.
(G) Western blot analysis of SESN2 levels in cells transfected with GFP-COL1A2 G610C and with scramble or XBP1 SiRNAs. Tubulin was used as a loading control.
(H) Bar graph showing quantification of SESN2 levels relative to loading control. Mean ± standard error of mean (SEM) of N = 3 biological replicates. Student’s unpaired t test: ∗p < 0.05.
(I) Proposed model of ER-phagy activation in ERSDs. When misfolded proteins accumulation induces IRE1-XBP1 mediated transcriptional induction of SESN2 (I). By limiting mTORC1 activity on lysosome, SESN2 promotes TFEB/TFE3 activation (II). As a consequence, there is a transcriptional induction of FAM134B, as well as lysosomal and autophagy genes (III), promoting ER-phagy-mediated clearance of misfolded proteins (IV).
(J) Western blot analysis of soluble and insoluble ATZ polymers in PiZ hepatocytes in which FAM134B was silenced. FILAMIN and histone H3 were used as a loading control.
(K) Bar graph showing quantification of insoluble ATZ polymers. Mean ± standard error of mean (SEM) of N = 3 biological replicates. Student’s unpaired t test: ∗p < 0.05.
(L) Western blot analysis of soluble and insoluble ATZ polymers in PiZ hepatocytes in which HA-FAM134B was overexpressed. FILAMIN and histone H3 were used as a loading control.
(M) Bar graph showing quantification of insoluble ATZ polymers. Mean ± standard error of mean (SEM) of N = 3 biological replicates. Student’s unpaired t test: ∗∗p < 0.005.
Pharmacological activation of FAM134B as a therapy for ER-storage disorders
The above data suggested that upregulation of FAM134B could hold promise as a therapeutic approach for ERSDs. Consistent with this, we found that overexpression of FAM134B significantly reduced, whereas its silencing augmented, the accumulation of ATZ polymeric insoluble aggregates in PiZ primary hepatocytes (Figures 6J–6M). We employed Gene2drug, a computational bioinformatic methodology for drug repositioning,49 to identify compounds capable of transcriptionally inducing FAM134B. Among the 70 compounds identified by Gene2drug (Table S6), we tested 48 for their ability to induce ER-phagy in RFP-GFP KDEL expressing A549 cells by high content screening microscopy (Figures S6A and S6B). Twenty compounds were selected and further tested in U20S and RCS cells expressing the RFP-GFP KDEL reporter. Fluphenazine (FPZ) and tetrandrine (TET) exhibited the most potent induction of ER-phagy in both cell lines, analyzing the RFP-GFP KDEL and the mKeima RAMP4 reporters (Figures 7A, 7B, S6C, and S6D). FPZ and TET induced TFEB dephosphorylation at S211 and S142, facilitating its nuclear localization. FPZ and TET induced FAM134B expression in a TFEB- and TFE3-dependent manner, as it was significantly inhibited in TFEB/TFE3 DKO U2OS cells (Figures 7C–7F and S6E). FPZ and TET did not activate ER-stress responses (UPR activation) or Sestrin2, indicating that they do not perturb ER homeostasis (Figure S6F). We found that FPZ and TET interfere with the ability of mTORC1 to associate with lysosomes in response to nutrients, hence explaining their potent effect on TFEB/TFE3 phosphorylation, that is occurring on lysosomal membrane50 (Figure S6G). Notably, FPZ and TET did not impair the ability of mTORC1 to phosphorylate other substrates, such as ULK1 and P70S6K (Figures S6H and S6I). Importantly, FPZ and TET showed modest and no activation of cytosolic and mitochondria autophagy (mKeima-LDHB and mKeima-COX8), respectively, suggesting that they primarily regulate ER-phagy (Figures S6J and S6K).
Figure 7.
Pharmacological induction of FAM134B promotes ER-cargo clearance via ER-phagy
(A) Representative fluorescence microscopy analysis of U2OS cells stably expressing the ssRFP-GFP-KDEL reporter and treated with fluphenazine (FPZ) or tetrandrine (TET) (10 μM, 12 h). Torin1 (500 nM for 12 h) and DMSO were used as positive control and vehicle, respectively. Scale bars, 10 μm. Bar graph showing the ratio of red-only puncta/total cells. Mean ± standard error of mean (SEM) of N = 3 independent experiments. n = 21 cells were counted. One-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test: ∗∗p < 0.005; ∗∗∗p < 0.0005; ∗∗∗∗p < 0.0001.
(B) Western blot analysis in RCS cells expressing mKeima-RAMP4, treated with FPZ, TET (10 μM, 12 h), or BafA1 (100 nM, 4 h). Torin1 (500 nM for 12 h) and DMSO were used as positive control and vehicle, respectively. FILAMIN was used as a loading control.
(C) TFE3 (green) subcellular localization in U2OS cells treated with FPZ and TET (10 μM, 12 h). Scale bars, 30 μm. Quantitative analysis of nuclear TFE3 (expressed as %) Torin 1 (500 nM for 12 h) was used as a positive control. Mean ± SEM of N = 3 independent experiments. n = 85 cells were counted. One-way ANOVA with Dunnett’s multiple comparisons test: ∗∗∗∗p < 0.0001.
(D and E) Western blot analysis of phospho-TFEB (S211 and S142) and total TFEB in U2OS cells treated with FPZ (E) or TET (F) (10 μM) for the indicated time points. Torin1 (500 nM for 12 h) and DMSO were used as positive control and vehicle, respectively. Actin was used as a loading control.
(F)Quantitative real-time PCR analysis of FAM134B isoform 2 in WT and TFEB/3 DKO RCS cells treated with Torin1 (500 nM for 12 h), and FPZ and TET (10 μM, 12 h). Mean ± standard error of mean (SEM) of N = 4 independent experiments. Values were normalized to cyclophilin (CYC) gene expression and expressed as fold change relative to DMSO cells. One-way ANOVA with Šídák’s multiple comparisons test: ∗p < 0.05; ∗∗∗∗p < 0.0001.
(G) Fluorescence microscopy analysis in U2OS cell with indicated genotype expressing tandem ssRFP-GFP-COL1A2 G610C and treated with FPZ or TET (10 μM, 12 h). Scale bars, 5 μm. Insets show magnification of boxed area. Bar graph showing the ratio of red-only puncta/total cells. n = 26 cells were counted. Two-way ANOVA with Tukey’s multiple comparisons test: ∗p < 0.05; ∗∗p < 0.005; ∗∗∗∗p < 0.0001.
(H) Western blot analysis of GFP-COL1A2 G610C levels in WT, shFAM134B, TFEB/3 DKO, and ATG7 KO U2OS cells treated with the indicated drugs. Actin was used as a loading control.
(I) Western blot analysis of insoluble ATZ polymers of PiZ primary hepatocytes treated with the indicated compounds. FILAMIN and histone H3 were used as a loading control. Quantitative analysis of indicated proteins levels (on right). Mean ± SEM of N = 3 independent experiments. One-way ANOVA with Šídák’s multiple comparisons test: ∗∗∗∗p < 0.0001; ns ≥ 0.05.
Subsequently, we tested the ability of FPZ and TET to promote the clearance of misfolded cargo from the ER. Control, TFEB/TFE3 DKO, and shFAM134B U2OS cells that stably express the tandem collagen I reporter (ssRFP-GFP-COL1A2 G610C) were treated for 12 h with vehicle, FPZ, or TET. We found that FPZ and TET significantly enhanced COL1A2 G610C delivery to lysosomes compared with vehicle in control, but not in TFEB/TFE3 DKO, shFAM134B, or ATG7KO U2OS cells (Figures 7G and S6L). Consistent with this, TET and FPZ administration reduced GFP-COL1A2 G610C protein levels in control but not in FAM134B-, TFEB/TFE3-, and ATG7-depleted cells (Figure 7H). Furthermore, TET, but not FPZ, significantly reduced the levels of insoluble ATZ from PIZ hepatocytes (Figure 7I). Collectively, these data indicate that the transcriptional induction of FAM134B could represent a therapeutic approach to promote ER clearance in ERSDs.
Discussion
In this study, we reported an increase in ER-phagy across various cellular models of ERSDs, which are characterized by the build-up of distinct misfolded proteins within the ER lumen. This suggests that the induction of ER-phagy represents a response to protein misfolding, potentially operating as a complementary or alternative system alongside the ER-associated degradation (ERAD) pathway.
Although the regulation of ER-phagy has been explored predominantly in the context of the starvation response,13,23,25,27 understanding the mechanisms initiating ER-phagy in response to misfolded protein accumulation remains limited. Our results indicate that the transcriptional activation of FAM134B plays a role in driving ER-phagy in response to protein misfolding. Other post-translational activation mechanisms, such as phosphorylation,26 ubiquitination,27 and UFMylation,51 which have been implicated in ER-phagy activation, have not been investigated in our study. As regards general autophagy regulation, it is likely that transcriptional and post-translational regulatory mechanisms coexist and cooperate to achieve appropriate cellular ER-phagy responses.
The upregulation of FAM134B could facilitate the degradation of misfolded proteins by interacting with CANX. The FAM134B-CANX complex complex seems to identify misfolded clients ineligible for ERAD, including alpha(1)-ATZ protein and collagens.7,8,52 Most likely, CANX’s involvement is subordinate to FAM134B induction due to the typically low expression levels of FAM134B. Consistently, the transcriptional induction of FAM134B promotes the formation of the FAM134B-CANX complex, essential for efficient ER-phagy-mediated protein clearance.27,28
Our study reveals that FAM134B transcriptional induction is primarily mediated by TFEB/TFE3 activation. This implies that TFEB/TFEB triggers both the initiation of ER-phagy and subsequent steps such as the lysosomal degradation of ER cargo. An induction of the ER-phagy receptor FAM134B can lead to the formation of ER vesicles, which are either sequestered by double-membrane autophagosomes prior to lysosomal digestion8 or directly fuse with the endo/lysosome compartment.7 Given the lysosome’s role as the ultimate destination for cargo removal, both pathways benefit from TFEB activation.
Prior research demonstrated that pharmacological activation of UPR with a prolonged tunicamycin administration triggers TFEB/TFE3 dephosphorylation and nuclear translocation via PERK-mediated calcineurin phosphatase activation, but independently of mTORC1.53 Furthermore, pharmacologically inducing UPR also promotes the activation of other ER-phagy receptors, such as CCPG1.14 However, in our model systems, the accumulation of mutant proteins has limited effect on UPR, activating mildly XBP1, that promotes the transcriptional activation of FAM134B, but not of other ER-phagy receptors. These data suggest that we have identified an early response to altered ER proteostasis, which can eventually be followed by a more potent and generalized ER-phagy induction mediated by multiple UPR pathways and ER-phagy receptors.
Recent studies have demonstrated that the ER is the primary substrate for autophagy during periods of starvation.54,55 A significant discovery from our investigation is that protein misfolding within the ER triggers ER-phagy through a signaling cascade that partially overlaps with the response to nutrient deprivation. However, although starvation inhibits the mTORC1-induced phosphorylation of all its substrates, ER stress predominantly impairs mTORC1’s ability to phosphorylate TFEB/TFE3. Recent studies have identified a non-canonical mechanism by which mTORC1 phosphorylates TFEB/TFE3, occurring on the lysosome and mediated by amino acid signaling rather than growth factors.50 Our findings suggest that a finely regulated induction of SESN2 in response to ER stress primarily inhibits non-canonical mTORC1 signaling at the lysosome. The phosphorylation of ULK1 observed in cells experiencing ER stress, or those treated with FPZ and TET, could explain a limited activation of bulk autophagy. These findings may elucidate the role of mTORC1 in selective autophagy and provide insights into the therapeutic benefits of nutrient starvation in managing ERSDs.
The identification of FAM134B transcriptional activators led us to the discovery of potential ER-phagy inducers, such as TET and FPZ, among FDA-approved drugs. FPZ, a phenothiazine compound similar to carbamazepine, has been tested in a C. elegans model of A1AD.56 Our data provide a mechanistic explanation for the beneficial effects of this class of compounds in ERSDs. Similarly, TET, a calcium channel blocker known for its autophagy-inducing properties,57 emerged as an ER-phagy inducer capable of clearing misfolded cargo from the ER. These findings offer future prospects for drug repurposing in treating ERSDs diseases.
We uncovered a signaling mechanism governing ER-phagy initiation in response to protein misfolding, involving the interplay between SESN2-dependent nutrient sensing and ER quality control with the autophagy machinery. Importantly, our work underscores the potential therapeutic value of enhancing the FAM134B-dependent ER-phagy pathway in managing ERSDs that are characterized by aberrant protein folding within the ER.
Limitations of the study
Although our study demonstrates the potential role of SESN2 activation in promoting protein clearance in response to ER storage, several limitations warrant further investigation. First, additional research is needed to understand the selectivity of ER-phagy in response to ER storage via SESN2. It is possible that additional factors or post-translational modifications of FAM134B and its partners promote the recruitment of the autophagy machinery at specific ER subdomains. Recent work has shown that the assembly of the vATPase at the ER exit sites favors ER-phagy of ATZ by promoting the recruitment of autophagy factors.58 Notably, genes encoding subunits of the vATPase are among the most responsive targets of TFEB/TFE3,59 suggesting that TFEB/TFE3 activation might promote ER-phagy also through the vATPase.
Second, future studies utilizing animal models of diseases characterized by protein misfolding will be necessary to provide valuable insights into the therapeutic potential of targeting this pathway in various tissues of the body.
Finally, our observations raise questions regarding the degradation mechanism of misfolded proteins cleared via ER-phagy in a FAM134B-independent manner. Further investigation of alternative pathways involved in the clearance of misfolded proteins is necessary to fully understand cellular quality control mechanisms.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Polyclonal LAMP1 antibody | Abcam | Cat ab24170; RRID: AB_775978_775978 |
| Rabbit Polyclonal CLIMP63 antibody | Proteintech | Cat 16686-1-AP; RRID: AB_2276275 |
| Mouse Monoclonal antibody HA.11 | BioLegend | Cat 901501; RRID: AB_2565006 |
| Rat Monoclonal antibody LAMP1 Antibody 1D4B | Santa Cruz Biotechnology | Cat sc-19992; RRID: AB_2134495 |
| Rabbit Polyclonal REEP5 antibody | Proteintech | Cat14643-1-AP;AB_2178440 |
| Rabbit Polyclonal CALNEXIN antibody | Enzo Life Sciences | Cat ADI-SPA-860-D; RRID: AB_2038898 |
| Mouse Monoclonal antibody Flag-M2 | Sigma-Aldrich | Cat F1804; RRID: AB_262044 |
| Mouse TFEB Monoclonal Antibody | MyBioSource | Cat MBS120432; RRID: AB_2271743 |
| Rabbit Polyclonal TFE3 antibody | Sigma-Aldrich | Cat HPA023881; RRID: AB_1857931 |
| Rabbit Polyclonal TFE3 antibody | Cell Signaling | Cat #14779; RRID: AB_2687582 |
| Rabbit Monoclonal mTOR (7C10) Antibody | Cell Signaling | Cat #2983; RRID: AB_2105622 |
| Rabbit Monoclonal phospho-p70 S6 Kinase (Thr389) (108D2) Antibody | Cell Signaling | Cat #9234S; RRID: AB_2269803 |
| Rabbit Polyclonal p70 S6 Kinase Antibody | Cell Signaling | Cat #9202S; RRID:AB_331676 |
| Rabbit Monoclonal phospho-ULK1 (Ser757) (D7O6U) Antibody |
Cell Signaling | Cat #14202; RRID: AB_2665508 |
| Rabbit Monoclonal ULK1 (D9D7) Antibody |
Cell Signaling | Cat #6439; RRID:AB_11178933 |
| Mouse Monoclonal monomeric Keima-Red Antibody | MBL Life Science | Cat M126-3M; RRID: AB_10210643 |
| Rabbit Polyclonal FAM134B Antibody | Sigma-Aldrich | Cat HPA012077; RRID: AB_2668621 |
| Rabbit Polyclonal FAM134C Antibody | Sigma-Aldrich | Cat HPA016492; RRID: AB_1853027 |
| Mouse Monoclonal anti-beta-Actin Antibody (AC-15) | Novus Biologicals | Cat NB600-501; RRID: AB_10077656 |
| Rabbit Polyclonal FILAMIN A Antibody | Cell Signaling | Cat #4762; RRID: AB_2106408 |
| Mouse Monoclonal Vinculin A Antibody | Sigma-Aldrich | Cat V9264; RRID: AB_10603627 |
| Rabbit Polyclonal SESTRIN2 Antibody | Proteintech | Cat 10795-1-AP; RRID: AB_2185489 |
| Rabbit Polyclonal phosho-TFEB S142 Antibody | EMD Millipore | Cat ABE1971; RRID: AB_2928101 |
| Rabbit Monoclonal Phospho-TFEB (Ser211) (E9S8N) antibody | Cell Signaling | Cat #37681; RRID: AB_2799117 |
| Rabbit Polyclonal Histone-H3 Antibody | EMD Millipore | Cat 07-690; RRID:AB_417398 |
| Rabbit Polyclonal GFP Antibody | Novus Biologicals | Cat NB600-308; RRID: AB_10003058 |
| Mouse Monoclonal mCHERRY(1C51) Antibody | Novus Biologicals | Cat NBP1-96752; RRID: AB_11034849 |
| Rabbit Polyclonal Alpha-1-antitrypsin Antibody | Dako | Cat A0012; RRID: AB_2335672 |
| Mouse monoclonal alpha-1-antitrypsin 2C1 antibody | Hycult | Cat HM2289 |
| Bacterial and virus strains | ||
| Lentiviral vector pCW57-CMV-ssRFP-GFP-KDEL | Addgene | #128257 |
| Retroviral vector PCMVneo-mKeima-RAMP4 | Di Lorenzo et al.25 | https://doi.org/10.1126/sciadv.abo1215 |
| pLJC5-Tmem192-3xHA | Addgene | #102930 |
| pHAGE mKeima-LDHB | Gift from Stefano Santaguida. IEO, Milan, Italy | This study |
| pHAGE mKeima-COX8 | Addgene | #131626 |
| Chemicals, peptides, and recombinant proteins | ||
| Bafilomycin A1 | Sigma-Aldrich | B1793 |
| Torin 1 | Cell Signaling | #14379 |
| L-Leucine | Sigma-Aldrich | L8000 |
| Fluphenazine dihydrochloride | MCE (MedChemExpress) | HY-A0081 |
| Tetrandrine | MCE (MedChemExpress) | HY-13764 |
| ISRIB | Sigma-Aldrich | SML0843 |
| 4u8C | Sigma-Aldrich | SML0949 |
| PF-429242 | Sigma-Aldrich | SML0667 |
| Puromycin | Sigma-Aldrich | P9620-10ML |
| Doxycycline | Sigma-Aldrich | D9891-5G |
| Neomyin (G418) | Sigma-Aldrich | A1720 |
| Blasticidine | Sigma-Aldrich | 15205-100MG |
| Critical commercial assays | ||
| In-Fusion00E2 HD Cloning Kit | Takara Bio | #638920 |
| BP Clonase Reaction Kit; LR Clonase Reaction Kit | Invitrogen | #11789020; #11791020 |
| Lipofectamine00E4 3000 Transfection Reagent | Thermo Fisher Scientific | Cat#3000015 |
| Lipofectamine00E4 and Plus Reagent | Thermo Fisher Scientific | Cat#15338030 |
| Lipofectamine00E4 RNAiMax Reagent | Thermo Fisher Scientific | Cat#13778030 |
| BCA Protein Assay | Thermo Fisher Scientific | Cat#23225 |
| Deposited data | ||
| Transcriptome profile of RCS WT and TFEB/3 DKO cells transfected with mutant COL2A1 R789C compared to control | Tables S1, S2, and S3 | GEO: GSE239527 |
| Transcriptome profile of HeLa COL1A2 G610C transfected cells compared to control cells | Table S4 | GEO: GSE239525 |
| Genes differentially expressed genes commonly regulated in the two datasets (Superseries GSE239528 includes the two datasets) | Table S5 | GEO: GSE239528 |
| FDA-approved drugs identified as FAM134B transcriptional inducers | Table S6 | Gene2drug.tigem.it |
| Uncropped western blot data | Mendeley Data, V1 | [https://doi.org/10.17632/gg7cnt2fyd.1] |
| Experimental models: Cell lines | ||
| RCS: Swarm chondrosarcoma chondrocyte line | Cinque et al.23 | TIGEM, Pozzuoli, Italy |
| RCS FAM134B KO and TFEB/TFE3 DKO | Cinque et al.23 | TIGEM, Pozzuoli, Italy |
| U2OS shFAM134B and ATG7 KO cell lines | Khaminets et al.13 | N/A |
| Wild-type U2OS TRex cells | Stephen Blacklow from Brigham and Women’s Hospital and Harvard Medical School,USA | N/A |
| Wild-type and TFEB/TFE3 DKO MEFs | Martina et al.53 | N/A |
| HeLa stably expressing GFP-TFEB | Settembre et al.38 | N/A |
| KICSTOR KO HEK293T | gift from David Sabatini. IOCB Prague, Czech Republic | N/A |
| Wild-type and SESN2 KO HEK293T | gift from Roberto Zoncu. Department of Molecular and Cellular Biology, University of California at Berkeley, USA | N/A |
| Experimental models: Organisms/strains | ||
| Primary osteoblasts extracted from wild type and Amish mice | From Antonella Forlino. Department of Molecular Medicine, University of Pavia, Italy | This study |
| Punch biopsies of the skin of FKBP14-kEDS patients | From Cecilia Giunta. Biobank of the Division of Metabolism at the Children’s Hospital, Zurich, Switzerland. | This study |
| Primary hepatocytes isolated from the liver of 6-week-old PiZ mice | From Pasquale Piccolo. TIGEM, Pozzuoli, Italy | This study |
| Oligonucleotides | ||
| siGENOME SMARTpool SiRNAs, Dharmacon Thermo Scientific, SiRNA 4Duplexes, LP_151890, G-CUSTOM-866712). | Dharmacon Thermo Scientific | This study |
| siRNA ON-TARGET plus human ATF4 | Dharmacon Thermo Scientific | L-005125-00-0005 |
| siRNA ON-TARGET plus human ATF6 | Dharmacon Thermo Scientific | L-009917-00-0005 |
| siRNA ON-TARGET plus human XBP1 | Dharmacon Thermo Scientific | L-009552-00-0005 |
| siRNA ON-TARGET plus human SESTRIN 2 | Dharmacon Thermo Scientific | L-019134-02-0005 |
| Primers for mutagenesis: COL1A2 G610CΔER – Fw 5′GGAATTCGTAGACATGGTGAGCAAGGGCGAGG 3’. | Eurofins genomics | This study |
| Primers for mutagenesis: COL2A1 R789CΔER – 5′ GTGAACCGTCAGATCCATGGCCATCATCAAGG 3′; | Eurofins genomics | This study |
| Primers for generation of U2OS TFEB/3 DKO: sgRNA sequence Fw – 5’ CCCAGAAGCGAGAGCTCAC 3’. | Eurofins genomics | This study |
| ON-TARGETplus Human ATL3 (25923) siRNA - SMARTpool, 5 nmol. ON-TARGETplus Human SEC62 (7095) siRNA - SMARTpool, 5 nmol. ON-TARGETplus Human TEX264 (51368) siRNA - SMARTpool, 5 nmol. ON-TARGETplus Human RETREG2 (79137) siRNA - SMARTpool, 5 nmol. ON-TARGETplus Human RETREG3 (162427) siRNA - SMARTpool, 5 nmol. ON-TARGETplus Human CCPG1 (9236) siRNA - SMARTpool, 5 nmol. ON-TARGETplus Human RTN3 (10313) siRNA - SMARTpool, 5 nmol. |
Dharmacon-Horizon | L-010656-01-0005; L-010218-01-0005; L-010662-01-0005; L-018422-02-0005; L-01845.L-013998; L-013998-00-0005; L-020088-01-0005. |
| Recombinant DNA | ||
| Retroviral vector iTAP MSCV-N-FLAG-HA mKEIMA-FAM134B-PURO | This paper | N/A |
| Retroviral vector iTAP MSCV-N-FLAG-HA mKEIMA-FAM134C-PURO | This paper | N/A |
| Lentiviral vector pcW57-ssRFP-GFP- COL1A2 G610C | This paper | N/A |
| pHAGE ssRFP-GFP- LDHB | This paper | N/A |
| pcDNA3.1 3XFLAG COL1A2 G610C | This paper | N/A |
| Software and algorithms | ||
| Fiji-ImageJ | Java | https://imagej.nih.gov/ij/ |
| Zen Blue software | ZEISS microscopy sotwere | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| Harmony software (PerkinElmer) | Harmony high-content imaging and analysis software | https://www.revvity.com/it-en/category/cellular-imaging-software |
| ChemiDoc-lt imaging system (Uvitec sotware) | Uvitec Alliance | https://www.uvitec.co.uk/alliance-q9-advanced/ |
| LightCycler 480 (Roche) software | Roche diagnostics | https://diagnostics.roche.com/global/en/products/instruments/lightcycler-480-ins-445.html |
| BD FACSAria sotware | DB Bioscience | https://www.bdbiosciences.com/en-ca/products/instruments/flow-cytometers/research-cell-sorters/bd-facsaria-iii |
| QuantSeq 3′ mRNA sequencing data processing and analysis | NEGEDIA (Next Generation Diagnostic srl) | PMID: 25260700 |
| Data visualization | Biorender | https://app.biorender.com/ |
| Statistics | GraphPad PRISM software | https://www.graphpad.com/features |
| Other | ||
| FLAG-TFEB and GFP-TFEB plasmids | Settembre et al.38 TIGEM, Pozzuoli, Italy | N/A |
| FAM134B-HA expression plasmid | Khaminets et al.13 | N/A |
| Wild-type and mutant COL1A2 G610C (GFP and mApple-COL1A2 G610C) plasmids | Addgene | #119826; #119827 |
| Wild-type and mutant COL2A1 R789C | Forrester et al.8 | N/A |
| AAT-HA and ATZ-HA plasmids | Gift from Maurizio Molinari. Faculty of Biomedical Sciences, Institute for Research in Biomedicine, Bellinzona, Switzerland | N/A |
| GFP-SESN2 plasmid | Addgene | #100519 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Carmine Settembre (Settembre@tigem.it).
Materials availability
Primary osteoblasts extracted from wild type and Amish mice are generated by Professor Antonella Forlino laboratory from Department of Molecular Medicine, University of Pavia, Italy. Skin fibroblasts from biopsy of kEDS patients deposited in the Biobank of the Division of Metabolism at the Children’s Hospital Zurich, Switzerland are a gift from Professor Cecilia Giunta laboratory. Primary hepatocytes are generated from Pasquale Piccolo laboratory from Telethon Institute of Genetics and Medicine (TIGEM), Pozzuoli, Italy.
Wild-type and SESN2 KO HEK293T cells are a gift from Professor Roberto Zoncu from the Department of Molecular and Cellular Biology, University of California at Berkeley, CA USA.
The plasmid pHAGE mKeima-LDHB is a gift from Professor Stefano Santaguida from Istituto Europeo di Oncologia (IEO), Milan, Italy. AAT-HA and ATZ-HA plasmids are a gift from Professor Maurizio Molinari from Faculty of Biomedical Sciences, Institute for Research in Biomedicine, Bellinzona, Switzerland.
Cloning methods for other plasmid constructs based on the available plasmids are described in the method details section. All materials generated in this study are available from the lead contact with a completed Material Transfer Agreement.
Data and code availability
RNA-seq data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under the series accession numbers GSE239527; GSE239525; GSE239528 (GEO: GSE239527; GSE239525; GSE239528) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All data reported in this paper and any additional information required to reanalyze the data are available from the lead contact upon request.
Experimental model and subject participant details
Cultured cell sample preparation
Primary osteoblasts extracted from wild type and Amish mice were isolated from 1-day-old pup calvariae using 200 U/mL collagenase type II (GIBCO) at 37°C for 15–20 min. A detailed protocol can be found in Bianchi et al., 2015.60 Mouse primary osteoblasts were cultured in αMEM supplemented with 10%FCS and 50 μg/mL sodium ascorbate.
As part of the diagnostic workup of kEDS, punch biopsies of the skin of FKBP14-kEDS patients were obtained in the presence of their signed informed consent and with the approval of Swiss Ethics (KEK-ZH-Nr. 2019-00811) for the establishment of fibroblast cultures. The culture of fibroblasts was done according to routine procedures as published previously31. The biological material was stored in the Biobank of the Division of Metabolism at the Children’s Hospital Zurich. Skin fibroblasts were cultured in DMEM.
Primary hepatocytes were isolated from the liver of 6-week-old PiZ mice by a modified protocol based on Pronase/collagenase digestion. In brief, mouse livers were perfused through the inferior vena cava with an EGTA solution followed by enzymatic digestion with Pronase (P5147 Sigma–Aldrich) and then collagenase type D (11088866001 Roche Applied Science). Next, livers were harvested, and liver cells were disassociated by digestion with Pronase/collagenase solution and filtered through a 70 μm cell strainer to remove undigested tissues and debris. The resulting cell suspension was centrifuged at 50 × g for 3 min at 4 °C. The cell pellet was washed three times with Williams' medium E (Gibco) and centrifuged at 50 × g for 3 min at 4 °C to obtain hepatocytes.
Method details
Cell Culture
RCS, MEF, HeLa, and HEK293T cell lines were cultured in DMEM (Euroclone) supplemented with 10% fetal bovine serum (FBS from Euroclone) and 1% penicillin/streptomycin at 37°C in 5% CO2. U2OS cells were purchased from ATCC and cultured in McCoy's medium (Euroclone) with 10% FBS and 1% penicillin/streptomycin at 37°C in 5% CO2. Complete starvation was performed in Hank's Balanced Salt Solution (HBSS) (Euroclone, ECB4006L) and amino acid starvation was performed in amino acid–free RPMI (US Biological, R9010-01) supplemented with 10% dialyzed FBS (Invitrogen, Thermo Fisher Scientific).
The RCS cell line was a Swarm chondrosarcoma chondrocyte line. RCS FAM134B KO and TFEB/TFE3 DKO cells were previously described in Cinque et al.23. U2OS shFAM134B and ATG7 KO cell lines were previously described13. Wild-type U2OS TRex cells were provided by Stephen Blacklow (Brigham and Women’s Hospital and Harvard Medical School). Wild-type and TFEB/TFE3 DKO MEFs were described in Martina et al.53. HeLa cells stably expressing GFP-TFEB were previously described61. KICSTOR KO HEK293T cells were a gift from David Sabatini. Wild-type and SESN2 KO HEK293T cells were a gift from Roberto Zoncu.
Transfections, siRNAs, and Stable Cell Lines
Cells were transfected with Lipofectamine LTX and Plus reagent (Invitrogen) and Lipofectamine 3000 following a reverse transfection protocol according to the manufacturer's instructions. For SiRNA experiments, siGENOME SMARTpool siRNAs (Dharmacon Thermo Scientific) were transfected to a final concentration of 100 nM and cells harvested 48/72 h after transfection. A SiRNA library of differentially expressed genes identified by transcriptomic analysis was designed and purchased from Dharmacon (siGENOME SMARTpool SiRNAs, Dharmacon Thermo Scientific, SiRNA 4Duplexes, LP_151890, G-CUSTOM-866712).
Stable cell lines were generated using retroviral or lentiviral viruses, as previously described. For lentivirus, 3.3 μg of plasmid with pPAX2 packaging plasmid (2.7 μg) and pMD2.G envelope plasmid (1 μg) were co-transfected in HEK293T cells in a six-well plate. The cells were incubated with a lentiviral suspension containing polybrene (8 μg/ml; Merck, #TR-1003-G) for 24 hours. Confluent-infected cell lines were then selected with the relevant antibiotics.
Plasmids
Lentiviral vector pCW57-CMV-ssRFP-GFP-KDEL was used for transient transfection in primary cells and for generating stable clones in different cell lines (S7 A-E)18 Retroviral vector PCMVneo-mKeima was used as previously described in Di Lorenzo et al.25.
TMEM192-HA was a gift from David Sabatini; FLAG-TFEB and GFP-TFEB plasmids were previously described in Settembre et al.38. The FAM134B-HA expression plasmid was previously described13. Mutant COL1A2 G610C (GFP and mApple-COL1A2 G610C) and COL2A1 R789C and their corresponding wild-type plasmids were purchased from Addgene (Plasmid #119826; #119827)8. GFP-SESN2 was purchased from Addgene (Plasmid #100519). The AAT-HA was a gift from Maurizio Molinari and ATZ-HA was previously described4. The mKeima-LDHB plasmid was a gift from Stefano Santaguida. mKeima-COX8 was purchased from Addgene (Plasmid #131626).
The plasmids mKeima-FAM134B and mKeima-FAM134C were cloned into the pDONR223 vector (Invitrogen) using the BP Clonase Reaction Kit (Invitrogen, #11789020) and further re-combined, through the LR Clonase Reaction Kit (Invitrogen, #11791020), into the GATEWAY destination vectors iTAP MSCV-N-FLAG-HA IRES-PURO.
The mutant COL1A2 G610C and COL2A1 R789C plasmids lacking the N-terminal signal peptide (COL1A2 G610CΔER and COL2A1 R789CΔER) were generated by site-directed mutagenesis using the Agilent QuikChange XL Site-Directed mutagenesis kit using the mCHERRY-PC2 and eGFP-PC1 backbone. Primer sequences were designed with PrimerX online software and were the following:
R789C Fw: 5′ GTGAACCGTCAGATCCATGGCCATCATCAAGG 3′;
R789C Rev: 3′ CCTTGATGATGGCCATGGATCTGACGGTTCAC 5′;
G610CFw: 5′ GGAATTCGTAGACATGGTGAGCAAGGGCGAGG 3′;
G610C Rev: 5′ CCTCGCCCTTGCTCACCATGTCTACGAATTCC 3′.
Flag-COL1A2 G610C was subcloned in pcDNA3.1 3XFLAG plasmid (Sigma, E4401) by restriction digestion and PCR amplification using primer sequences:
Flag-COL1A2 G610C Fw: 5’ GCTTGGTACCGAGCTCGATGCTCAGCTTTGTG 3’;
Flag- COL1A2 G610C Rev (1): 3’ GTCATCCTTGTAATCCATTTGGCATGTTGCTAGG 5’;
Flag- COL1A2 G610C Rev (2): 3’ TATCCTTGTACTTCTTGTCATCGTCATCCTTG 5’.
FLAG-COL2A1 R789C was cloned in the pcDNA3.1 3XFLAG plasmid by PCR amplification using primer sequences:
COL2A1 R789C Fw: 5’ CGGTCTGCCTGGGCAATGTGGTGAGAGAGGATTC 3’;
COL2A1 R789C Rev: 3’ GAATCCTCTCT CACCACATTGCCCAGGCAGACCG 5’.
The tandem ssRFP-GFP-COL1A2 G610C plasmid was cloned in eGFP-proa(I)G610C and in the lentiviral pcW57-ssRFP-GFP-KDEL plasmid by PCR amplification to add the collagen signal peptide (SP) and RFP/GFP sequence using primer sequences:
SS Fw: 5’ ACATGCCAAGATATCATGGCCTCCTCCGAGG 3’;
SS Rev: 3’ CTTGCTCACGATATCGGCGCCGGTGGAGTG 5’;
RFP-GFP-COL1A2 Fw: 5’ CACTCCACCGGCGCCGATATCGTGAGCAAG 3’;
RFP-GFP-COL1A2 Rev: 3’ GTCAGAATAGATATCCTTGTACAGCTCGTC 5’.
The tandem ssRFP-GFP-LDHB plasmid was cloned in lentiviral pHAGE plasmid by PCR amplification using primer sequences:
RFP-GFP Fw: 5’ CATAGAAGACACCGGCGGCCATGGCCTCCTCCGAGGACG 3’;
RFP-GFP Rev: 3’ TAAGAGTTGCCATCTTGTACAGCTCGTCCAT 5’;
LDHB Fw: 5’ CTGTACAAGATGGCAACTCTTAAGGAAAAACTCA 3’;
LDHB Rev: 3’ GGGGGGGGGCGGAATTTCACAGGTCTTTTAGGTCCTTCTG 5’.
Generation of CRISPR clones
The U2OS TFEB/3 DKO cell line was generated as follows. Gene disruption was performed using clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology. 1 × 106 U2OS cells were transfected with 5 μg of all-in-one vector containing the sgRNA of interest:
TFEB sgRNA sequence: CCCAGAAGCGAGAGCTCAC;
TFE3 sgRNA sequence: GCGCGTTGGGTTCTCCAGAT.
The all-in-one vector contains the U6 promoter, a recombinant form of Cas9 protein under the control of the CMV promoter, and eGFP or mCHERRY reporter gene under the control of the SV40 promoter (Sigma-Aldrich). Forty-eight hours after transfection, putative clones were FACS-sorted for the eGFP fluorescence using the BD FACSAria. Sorted cells were kept in culture until confluence and then subjected to PCR analysis followed by Sanger sequencing to identify mutations. Selected clones were validated by Western blotting analysis of the protein of interest.
Chemicals
Bafilomycin A1 (BafA1; Sigma-Aldrich) was used at a final concentration of 100-200 nM for 4-6 hours or 50 nM for 12 hours. Torin 1 (Cell Signaling #14379) was used at 1 μM for 2 hours unless otherwise indicated. Torin1 500 nM for 12 hours was used as control for mKeima blots and drug treatments (FPZ and TET). L-Leucine (Sigma-Aldrich L8000, ID PubChem: 24896488) supplementation was performed at 1.2 mM for 1 hour or 4 mM for 3 hours in amino acid–free RPMI (US Biological, R9010-01).
Drugs were identified by the Gene2drug tool as FAM134B inducers and all compounds were purchased from MCE (MedChemExpress). FPZ and TET were used at 10 μM for 8h and 12h. A dose and time dependency of FPZ and TET was checked by using different concentrations (1-5-10 μM) and time points (2-4-6-8h) (S7 F-H).
Immunofluorescence
Cells were seeded on coverslips at least 24 hours before treatment and fixed for 10 min in 4% PFA in PBS and permeabilized for 30 min in blocking buffer (0.05% (w/v) saponin, 0.5% (w/v) BSA, 50 mM NH4Cl and 0.02% NaN3 in PBS, pH 7.4). Cells were incubated in a humid chamber for 1 hour at room temperature with primary antibodies: Lamp1 (Abcam ab24170 1:200); CLIMP63 (Proteintech 16686-1-AP 1:200); HA (BioLegend 901501 1:500); Lamp1 (Santa Cruz Biotechnology sc-19992 1:500); Reep5 (Proteintech, 14643-1-AP, 1:200), CANX (Enzo Life Sciences ADI-SPA-860-D, 1:200); Flag M2 (Sigma, F1804 1:200). After washing three times in PBS, they were incubated for 1 hour at room temperature with the secondary antibody (Alexa Fluor–labeled goat anti-rabbit A11011/A11008 or goat anti-mouse A11001, A11004; Life Technologies, Thermo Fisher Scientific), washed three times in PBS, incubated for 20 min with 1 μg/ml Hoechst 33342, and finally mounted in Mowiol (Sigma-Aldrich) or Vectashield (Vector Laboratories) supplemented with 4′,6-diamidino-2-phenylindole (DAPI).
TFEB, TFE3 and mTOR lysosomal localization immunofluorescence
RCS chondrocytes were fixed for 15 min in 4% PFA in PBS and permeabilized for 30 min in 0.02% Triton X-100 in PBS. Cells were incubated in a humid chamber for 1 hour in blocking buffer (0.1% Triton X-100, 10% goat serum in PBS) and then with primary antibodies (TFEB MyBioSource MBS120432 1:50; TFE3 Sigma-Aldrich HPA023881 1:200) diluted in 0.1% Triton X-100 and 5% goat serum in PBS overnight at 4°C. Alexa Fluor-conjugated secondary antibodies (1:400) were incubated for 1 hour at room temperature in 0.1% Triton X-100 and 1% goat serum in PBS. Nuclei were stained with DAPI 1:1,000 in PBS for 20 min at room temperature. Cells were washed with PBS, once in Milli-Q water and mounted with Mowiol.
For detection of TFE3 in HeLa and U2OS cell lines, fixed cells were permeabilized with 0.1% Triton-X100 in PBS for 5 min, and then incubated with blocking buffer (3% BSA plus 0.02 % saponin in PBS) for 45 min. Subsequently, coverslips were incubated with TFE3 (Cell Signaling, #14779 1:100) in blocking buffer for 12 hours at 4 °C, then rinsed three times with PBS and incubated with Alexa-Fluor conjugated secondary antibodies (Invitrogen) in blocking buffer for 1 hour at room temperature in the dark, and then washed four times with PBS. Slides were mounted on glass coverslips using Mowiol and imaged on a Leica SPE confocal microscope.
For mTOR lysosomal localization: cells were grown on Lab--Tek chamber slides and the day of the experiment they were rinsed with PBS and incubated in amino acid--free RPMI supplemented with 10% dialyzed FBS for 1 hour, and then left untreated or stimulated with a 3X amino acid (3x-AA) mixture for 30 min. Then, slides were rinsed with PBS once and fixed for 15 min with 4% PFA in PBS at room temperature (RT). After fixation, slides were rinsed twice with PBS and cells were permeabilized with 0.1% Triton in PBS 5 min. After rinsing three times with PBS, the slides were incubated with blocking buffer (3% BSA; 0.02% Saponin in PBS) for 1 hour at RT. Cells are incubated with primary antibody (mTOR, Cell signalling 7C10 and LAMP1 1:200) in blocking buffer for 2 hours at room temperature, rinsed four times with PBS, and incubated with secondary antibodies (diluted 1:200) for 1 hour at room temperature in the dark, then washed four times with PBS. Slides were mounted on glass coverslips using Vectashield (Vector Laboratories) and imaged on a confocal microscope.
Tandem reporter analysis
Cell lines were infected or, where indicated, transiently transfected with: I) lentivirus pCW57-CMV-ssRFP-GFP-KDEL to generate ER-phagy reporter-inducible cell lines and gene expression was induced with doxycycline (4 μg/ml; 24 hours). II) pHAGE RFP-GFP-LDHB to generate bulk-autophagy cell line. III) pHAGE RFP-GFP-COX8 to generate mitophagy cell line. Cells were collected in PBS and the fluorescence was analyzed with BD FACSAria III or fixed in 4% PFA for 15 min and the number of red-only puncta was quantified after image acquisition using ImageJ plugins.
Confocal and super resolution microscopy
Scanning laser confocal experiments were acquired using a Zeiss LSM 800 or Leica TCS SP5 confocal microscope equipped with a 63× 1.4 numerical aperture oil objective. Airyscan microscopy was performed using a Zeiss LSM 880 confocal microscope, equipped with Plan-Apochromat 63×/1.4 numerical aperture oil objective and pixel size of 8.7 nm. Images were subjected to post-acquisition Airyscan processing. Image acquisition and processing were performed with Zen Blue software and co-localization analysis and image presentation was performed using ImageJ FIJI software or Photoshop (Adobe). All quantifications were performed used ImageJ plugins.
Transmission electron microscopy
For routine EM analysis, the cells were fixed with 1% glutaraldehyde (GA) prepared in 0.2 M HEPES buffer (pH 7.4) for 30 min at room temperature (RT). All specimens were then post-fixed as described in Polishchuk and Polishchuk (2019). After dehydration, the specimens were embedded in epoxy resin and polymerized at 60 °C for 72 hours. Thin 60-nm sections were cut on a Leica EM UC7 microtome. EM images were acquired from thin sections using a FEI Tecnai-12 electron microscope equipped with a VELETTA CCD digital camera (FEI, Eindhoven, The Netherlands) for digital image acquisition.
High content screening by OPERA
The silencing protocol was developed by the TIGEM High Content Facility. The protocols use a liquid handler (Hamilton STARlet) to dispense SiRNA, Optimem, and Lipofectamine RNAimax reagent (ThermoFIscher). Then, the cells were seeded and silenced for 48 hours and transfected with mApple-COL1A2 G610C for the last 24 hours. At the end of silencing, cells were fixed and images were acquired with the automated confocal Opera Phenix® High-Content Screening System (PerkinElmer). The acquired images were then analyzed with the Harmony software (PerkinElmer) to define cytoplasmic and nuclear intensity.
Western blotting
Cells were washed twice with PBS and then scraped in RIPA lysis buffer (20 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate) supplemented with PhosSTOP and EDTA-free protease inhibitor tablets (1× final concentration) (Roche, Indianapolis, IN, USA). Cell lysates were incubated on ice for 20 min; then, the soluble fraction was isolated by centrifugation at 18,000 g for 20 min at 4 °C. Total protein concentration in cellular extracts was measured using the colorimetric BCA protein assay kit (Pierce Chemical Co, Boston, MA, USA). Protein extracts, separated by SDS–PAGE and transferred onto PVDF, were probed with primary antibodies overnight against phospho-P70S6K (Cell Signaling Technology 9234S 1:1000); P70S6K (Cell Signaling Technology 9202S 1:1000); phospho-ULK1 (Cell Signaling Technology 14202S 1:1,000); monomeric Keima-Red (MBL Life Science M126-3M 1:1000); FAM134B (Sigma-Aldrich HPA012077 1:1,000); FAM134C (Sigma-Aldrich HPA016492 1:1000); B-actin (Novus Biologicals NB600-501 1:5000); FILAMIN (Cell Signaling Technology 4762 1:1000); Vinculin (Sigma-Aldrich V9264 1:3000) SESTRIN-2 (Proteintech 10795-1-AP 1:1000); human-specific TFEB antibody (Cell Signaling Technology BL12896_15 1:1000); phospho-TFEB S142 (ABE1971 EMD Millipore 1:10000); Phospho-TFEB S211 (E9S8N, Cell Signaling 1:1000); FLAG M2 (Sigma-Aldrich F1804, 1:1000); Histone H3 (EMD Millipore 07-690 1:5000); GFP (Novus Biologicals NB600-308 1:1000) and mCherrry (Novus Biologicals NBP1-96752 1:1000). Proteins of interest were detected with HRP-conjugated goat anti-mouse or anti-rabbit IgG antibody (1: 2,000, Vector Laboratories) and visualized with the ECL Star Enhanced Chemiluminescent Substrate (Euroclone) according to the manufacturer's protocol. The Western blotting images were acquired using the ChemiDoc-lt imaging system (UVP).
Nuclear-cytosolic fractionation and hepatocyte soluble/insoluble fractions
For nuclear-cytosolic fractionation, cells were seeded at 50% confluence and were harvested after 24/72 hours. Subcellular fractionation was carried out as follows: briefly, cells were lysed in 0.5 Triton X-100 lysis buffer (50mM Tris-HCl, 0.5% Triton, 137.5 mM NaCl, 10% glycerol, 5 mM EDTA supplemented with fresh protease and phosphatase inhibitors). After 15 min the lysate was centrifuged and the supernatant was collected as the cytosolic fraction while the pellet (nuclear fraction) was washed twice and lysed in 0.5 Triton X-100 buffer 0.5% SDS and sonicated.
Preparation of soluble and insoluble fractions from livers was performed according to previous studies .62 Briefly, cell lysates were prepared in 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2 mM KCl, 2 mM MgCl2, 0.5% Triton X-100, 0.5% sodium deoxycholate, and 2 mM N-ethyl-maleimide supplemented with protease inhibitors and mechanically homogenized on ice. Insoluble material was recovered by centrifugation at 16,000 g for 20 min. Pellets were washed once and solubilized in 50 mM Tris-HCl (pH 6.8), 5% SDS, and 10% glycerol with 1 min of sonication and then 10 min of boiling. Immunoblotting of soluble fractions was performed using antibodies against human Alpha-1-antitrypsin (Dako A0012, 1:5000) and FILAMIN as control. Immunoblotting of insoluble fractions was performed using antibodies against polymeric forms of human alpha-1-antitrypsin, (mAb 2C1 Hycult HM2289, 1:1000) and Histone H3 as control.
FAM134B Immunoprecipitation
U2OS cells were transiently transfected with plasmids encoding GFP-COL1A2 G610C and GFP-TFEB. After 24 hours, the cells were detached with trypsin–EDTA and centrifuged. The cell pellets were washed three times with ice-cold PBS and then resuspended in lysis buffer (50 mM Tris-HCl pH8.0, 150 mM NaCl, 1% NP-40). The lysates were incubated on ice for 20 min with gentle swirling and centrifuged at 13,000 rpm for 20 min. The supernatants were collected and subjected to protein quantification using BCA protein assay kit (Pierce Chemical). 1 mg of each lysate was incubated overnight at 4°C with the FAM134B antibody (Sigma-Aldrich HPA012077), previously conjugated to Protein A Sepharose beads (Thermo Fisher Scientific, 9424). Beads were then washed three times in 0.1% (v/v) NP-40. Proteins were eluted by boiling in Laemmli buffer and loaded on SDS-PAGE for Western blot analysis.
qPCR analysis
Cells were harvested for RNA extraction using a RNeasy Mini Kit (Cat No./ID: 74106 (250), Qiagen) according to the manufacturer's protocol. 1 μg of total RNA was used for reverse transcription using QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions. qPCR was performed in triplicate using LightCycler 480 SYBER Green I Master (Roche) and analyzed by LightCycler 480 (Roche). The Ct values were normalized to cyclophilin or HPRT gene expression, and the expression of each gene was represented as 2(−ddCt) relative to control.
Primers used were as follows:
Rat
FAM134A: Fw 5′-CAGAACAGCAGGGTCCCATA-3′;
FAM134A: Rev 5′-TCCACTTTAGACCCTGGCTG-3′
FAM134B_2: Fw 5′- ACAGGAGGCAGTCACTTTGG -3′;
FAM134B_2: Rev 5′- TGCTTGCCACAACTCAGACA -3′;
FAM134C: Fw 5′-CCCAGTCTTGTCCCCTGAAT-3′;
FAM134C: Rev 5′-TTGCCTGTAGTACCACCCTG-3′
SEC62: Fw 5′-TCTGGCCAGCAGAAATGAGA-3′;
SEC62: Rev 5′-CAGTCAGGTTTGGCAGGAAC-3′
ATL3: Fw 5′-ACCCCTGCAGTTCTGTTCAC-3′;
ATL3: Rev 5′-CCCAGCTCAAGATACTGCCC-3′
RTN3: Fw 5′-TCTCACACACTACAGCAGCA-3′;
RTN3: Rev 5′- TGAGCGATGTTCACTCCTGT-3′
CCPG1: Fw 5′-TCTTGTGGCTGGACTGTCAT-3′;
CCPG1: Rev 5′-TTTGCACTGCTTTCTCCACC-3′
TEX264: Fw 5′-GTGCCAGAGGTGAAGGAGAC-3′;
TEX264: Rev 5′-TTGCTTGCCCCAGGAGAAAA-3′
Human
FAM134A: Fw 5′- GAATCCAGCTCAGTTCTGCG -3′;
FAM134A: Rv 5′- TGCCTTCATGCTGTAGTCCA-3′
FAM134B_2: Fw 5′-TTCATTCAAGGGAGGCAGGC-3′;
FAM134B _2: Rv 5′-CACCTGCTAACCACGGCTAA-3′
FAM134C: Fw 5′- AGGGAATTGGCCATCACAGA -3′;
FAM134C: Rv 5′- AAGTCTGGAAGGTCTCTGGC -3′
TEX264: Fw 5′- CGCTCCATCGCTGTCTACTA -3′;
TEX264: Rev 5′- AATGGTGGTGTAGGGGAAGG -3′;
ATL3: Fw 5′- CAAGAGGAGCAGATGATGCCA- 3’;
ATL3: Rev 5’- TCATCCATTGCCAGACGACC -3’;
RTN3: Fw 5’- GCCATGGTGCACATCAACAG -3’;
RTN3: Rev 5’- CTCGGGCGATGCCAACATA -3’;
CCPG1: Fw 5’- TTCTGTGACCCCCACTGACA-3’;
CCPG1: Rev 5’- TTGGCTGCTTTCTCCTTGCT-3’;
SESN2: Fw 5’- AGACATGCTGTGCTTTGTGG-3’;
SESN2: Rev 5’- TGTGCATGGCGATGGTATTG-3’
SRSF11: Fw 5’- GGGGCTCCTACTCTTGATCC -3’;
SRSF11: Rev 5’- GGGACTGTGCTTCTCGTACT-3’;
USP36: Fw 5’- CCAGCCACCTCTCCCTTTTA-3’;
USP36: Rev 5’- ACAGCACAGTCGTCTCAGAA-3’;
CHAC1: Fw 5’- AAGATGCTCCTGACCAACCA -3’;
CHAC1: Rev 5’- CCACAGAGCTGCATGAAGTC -3’;
CEP95: Fw 5’- CCCAGAGGCCAAGAAAGAGA -3’;
CEP95: Rev: 5’- GATGCCATCACTGTGCTGAG -3’;
HES1: Fw 5’- TGAAAGTCTGAGCCAGCTGA-3’;
HES1: Rev 5’-GTCACCTCGTTCATGCACTC-3’
PRPF38B: Fw 5’- GGAGATCTCTGAGTCCACGG-3’;
PRPF38B: Rev 5’-GGGATCTTCGCCGTTCTTTC-3’.
ATF6: Fw 5’- AATTCTCAGCTGATGGCTGT-3’;
ATF6: Rev 5’-TGGAGGATCCTGGTGTCCAT-3’;
ATF4: Fw 5’- GTTCTCCAGCGACAAGGCTA-3’;
ATF4: Rev 5’-ATCCTCCTTGCTGTTGTTGG-3’;
XBP1 Total: Fw 5’-TGGCCGGGTCTGCTGAGTCCG-3’;
UNSPLICED XBP1: Fw 5’-CAGCACTCAGACTACGTGCA-3’;
SPLICED XBP1: Fw 5’-CTGAGTCCGAATCAGGTGCAG-3’;
XBP1: Rev 5’-ATCCATGGGGAGATGTTCTGG-3’.
Mouse
FAM134B -2: Fw 5′-CATAATAGTCCACTCCTCGGCTTC-3′
FAM134B -2: Rev 5′- CTCAGTCTGGCTCTTTCATCTG-3′
SESN2: Fw 5′- CCTTCTCCACACCCAGACAT -3′;
SESN2: Rev 5′- GTGCATGGCGATGGTGTTAT -3′
SRSF11: Fw 5’- ACCTTTGCCAGTCTCCTCTC -3’
SRSF11: Rev: 5’- ATCTGGGTAAGTGGGTTGGG -3’
USP36: Fw 5’- CGCAGGACCTAATTCAGCAC -3’;
USP36: Rev 5’- CCCCAAGCAGTCACTAGGAA -3’;
CHAC1: Fw 5’- CCGGATCCTATGCACAGACA - 3’;
CHAC1: Rev 5’- CCAAGCCCTGTACAGAGACA -3’;
CEP95: Fw 5’ – AGATGGTCGCCTGAAGTCAA -3’;
CEP95: Rev 5’ – ACAAGAGCAGTGAGGAGGTC -3’.
QuantSeq 3′ mRNA sequencing library preparation
Wild type and TFEB/TFE3-CRISPR-KO RCS chondrocytes (three biological replicates/condition) were reverse transfected with the mCHERRY-COL2A1 R789C plasmid and mCHERRY-empty plasmid as control. Twenty-four hours after transfection cells were FACS-sorted for the mCHERRY fluorescence using the BD FACSAria. Total RNA was extracted from N = 3, for each condition. Total RNA from sorted cells was extracted according to the manufacturer's instructions (RNeasy Mini Kit, Cat No./ID: 74106 (250), Qiagen); RNA extracted from both cell lines was used as control. The extracted RNA was quantified and mixed at 50 ng/μl. Total RNA (100 ng) from each sample was prepared. Wild-type HeLa were transfected with mApple-COL1A2 G610C and FACS-sorted for the mCHERRY fluorescence using the BD FACSAria after 24h. Total RNA was extracted from N = 5 for each condition. Cells were harvested for RNA extraction using RNAeasy Mini Kit (Qiagen) according to the manufacturer's instuctions. RNA extracted from untreated cells was used as a control. RNA extracted was quantified and mixed at 50 ng/ul. Total RNA was quantified using the Qubit 4.0 fluorimetric Assay (Thermo Fisher Scientific). Libraries were prepared from 125 ng of total RNA using the NEGEDIA Digital mRNA-seq research grade sequencing service (Next Generation Diagnostic srl) (https://doi.org/10.1038/s41598-017-14892-x) which included library preparation, quality assessment and sequencing on a NovaSeq 6000 sequencing system using a single-end, 100 cycle strategy (Illumina Inc.).
QuantSeq 3′ mRNA sequencing data processing and analysis
Illumina NovaSeq 6000 base call (BCL) files were converted in fastq file through bcl2fastq; Trimming and cleaning with bbduk; Alignment was performed with STAR 2.6.0a. The expression levels of genes were determined with HTseq-counts 0.9.1. The reference genomes were Hg38 and rn6. The raw data were analyzed by Next Generation Diagnostic srl proprietary NEGEDIA Digital mRNA-seq pipeline (v2.0) which involves a cleaning step by quality filtering and trimming, alignment to the reference genome and counting by gene (PMID: 25260700). The raw expression data were normalized, analyzed, and visualized by Rosalind HyperScale architecture OnRamp BioInformatics, Inc.
Data availability and accession codes
The transcriptome data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE239527 and GSE239525. The Series GSE239527 has been named: Transcriptome profile of RCS WT and TFEB/3 DKO cells transfected with mutant COL2A1 R789C compared to control. The Series GSE239525 has been named: Transcriptome profile of HeLa COL1A2 G610C transfected cells compared to control cells. The Superseries GSE239528 includes the two datasets. Source data and quantifications given in the main text have associated raw data. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Quantification and statistical analysis
Functional analysis of transcriptomics data
The threshold for the statistical significance of gene expression was FDR < 0.05. Gene Ontology enrichment analysis (GOEA) was performed on induced and inhibited genes, separately, using the DAVID Bioinformatic tool restricting the output to Biological Process (BP), Cellular Compartments (CC) terms. The threshold for statistical significance of GOEA was FDR < 0.1 and Enrichment Score ≥ 1.5.
Statistics
Statistics were performed in GraphPad PRISM software. A two-tailed, paired and unpaired Student's t-test was performed when comparing the same cell population with two different treatments or cells with different genotypes, respectively. One-way ANOVA with Dunnett's post hoc test was performed when comparing more than two groups relative to a single factor (treatment) and Šídák's multiple comparisons test for multiple comparisons. Two-way ANOVA with Tukey’s post hoc test was performed when comparing more than two groups relative to two independent factors. A P-value of 0.05 or less was considered statistically significant.
Acknowledgments
We thank G. Diez Roux, A. Ballabio, C. Wilson, and R. Zoncu for critical reading of the manuscript. We thank R. Zoncu (University of Berkeley, USA); Paolo Grumati (TIGEM, Italy), D. Sabatini (IOCB Prague, Czech Republic), and R. Puertollano (NIH, USA); and M. Molinari (Institute for Research in Biomedicine, Switzerland) for reagents.
We thank the high content screening, advanced microscopy and imaging, and the bioinformatic core facilities at TIGEM.
C.S. acknowledges the Italian Telethon funding agency (GSA21F005), the Alpha-1 Foundation (1030868), the European Research Council (ERC-101045285-AUTOSELECT), and the Italian Ministry of Health (PNRR-MAD-2022-12376672) and PRIN 20223C8C5B to A.F. and C.S.
Author contributions
C.D.L. and M.M. performed most of the experiments. S.G. generated data in Figure 7. R.F. and P.P. generated PIZ hepatocytes. R.B. and A.F. generated Amish osteoblasts. P.J.L., M.R., and C.G. generated kEDS patient’s fibroblasts. R.D.C. and D.D.B. performed bioinformatic analysis. S.M., D.L.M., M.M., and A.P. performed high content screening microscopy. E.P. performed TEM analysis in kEDS fibroblasts. L.C. performed FPZ and TET signaling experiments. M.I. generated mKeima-LDHB and RFP-GFP-LDHB cells. C.S. and C.D.L. designed the study. C.S., C.D.L., and M.M. prepared figures and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: August 1, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2024.07.004.
Supplemental information
(A) Analysis restricting the output to cellular component term (“CC”).
(B) Analysis restricting the output to biological pathway term (“BP”).
The list is restricted to transcription factor terms (TFT).
References
- 1.Schwarz D.S., Blower M.D. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016;73:79–94. doi: 10.1007/s00018-015-2052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oakes S.A., Papa F.R. The Role of Endoplasmic Reticulum Stress in Human Pathology. Annu. Rev. Pathol. 2015;10:173–194. doi: 10.1146/annurev-pathol-012513-104649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu X., Rapoport T.A. Mechanistic insights into ER-associated protein degradation. Curr. Opin. Cell Biol. 2018;53:22–28. doi: 10.1016/j.ceb.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fregno I., Molinari M. Proteasomal and lysosomal clearance of faulty secretory proteins: ER-associated degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathways. Crit. Rev. Biochem. Mol. Biol. 2019;54:153–163. doi: 10.1080/10409238.2019.1610351. [DOI] [PubMed] [Google Scholar]
- 5.Ferro-Novick S., Reggiori F., Brodsky J.L. ER-Phagy, ER Homeostasis, and ER Quality Control: Implications for Disease. Trends Biochem. Sci. 2021;46:630–639. doi: 10.1016/j.tibs.2020.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Leonibus C., Cinque L., Settembre C. Emerging lysosomal pathways for quality control at the endoplasmic reticulum. FEBS Lett. 2019;593:2319–2329. doi: 10.1002/1873-3468.13571. [DOI] [PubMed] [Google Scholar]
- 7.Fregno I., Fasana E., Bergmann T.J., Raimondi A., Loi M., Soldà T., Galli C., D’Antuono R., Morone D., Danieli A., et al. ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J. 2018;37 doi: 10.15252/embj.201899259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forrester A., De Leonibus C., Grumati P., Fasana E., Piemontese M., Staiano L., Fregno I., Raimondi A., Marazza A., Bruno G., et al. A selective ER-phagy exerts procollagen quality control via a Calnexin-FAM134B complex. EMBO J. 2019;38 doi: 10.15252/embj.201899847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parashar S., Chidambaram R., Chen S., Liem C.R., Griffis E., Lambert G.G., Shaner N.C., Wortham M., Hay J.C., Ferro-Novick S. Endoplasmic reticulum tubules limit the size of misfolded protein condensates. eLife. 2021;10 doi: 10.7554/eLife.71642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham C.N., Williams J.M., Knupp J., Arunagiri A., Arvan P., Tsai B. Cells Deploy a Two-Pronged Strategy to Rectify Misfolded Proinsulin Aggregates. Mol. Cell. 2019;75:442–456.e4. doi: 10.1016/j.molcel.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reggiori F., Molinari M. ER-phagy: mechanisms, regulation, and diseases connected to the lysosomal clearance of the endoplasmic reticulum. Physiol. Rev. 2022;102:1393–1448. doi: 10.1152/physrev.00038.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reggio A., Buonomo V., Berkane R., Bhaskara R.M., Tellechea M., Peluso I., Polishchuk E., Di Lorenzo G., Cirillo C., Esposito M., et al. Role of FAM134 paralogues in endoplasmic reticulum remodeling, ER-phagy, and Collagen quality control. EMBO Rep. 2021;22 doi: 10.15252/embr.202052289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khaminets A., Heinrich T., Mari M., Grumati P., Huebner A.K., Akutsu M., Liebmann L., Stolz A., Nietzsche S., Koch N., et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 14.Smith M.D., Harley M.E., Kemp A.J., Wills J., Lee M., Arends M., von Kriegsheim A., Behrends C., Wilkinson S. CCPG1 Is a Non-canonical Autophagy Cargo Receptor Essential for ER-Phagy and Pancreatic ER Proteostasis. Dev. Cell. 2018;44:217–232.e11. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grumati P., Morozzi G., Hölper S., Mari M., Harwardt M.I., Yan R., Müller S., Reggiori F., Heilemann M., Dikic I. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife. 2017;6 doi: 10.7554/eLife.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Q., Xiao Y., Chai P., Zheng P., Teng J., Chen J. ATL3 Is a Tubular ER-Phagy Receptor for GABARAP-Mediated Selective Autophagy. Curr. Biol. 2019;29:846–855.e6. doi: 10.1016/j.cub.2019.01.041. [DOI] [PubMed] [Google Scholar]
- 17.Fumagalli F., Noack J., Bergmann T.J., Cebollero E., Pisoni G.B., Fasana E., Fregno I., Galli C., Loi M., Soldà T., et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat. Cell Biol. 2016;18:1173–1184. doi: 10.1038/ncb3423. [DOI] [PubMed] [Google Scholar]
- 18.Chino H., Hatta T., Natsume T., Mizushima N. Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol. Cell. 2019;74:909–921.e6. doi: 10.1016/j.molcel.2019.03.033. [DOI] [PubMed] [Google Scholar]
- 19.An H., Ordureau A., Paulo J.A., Shoemaker C.J., Denic V., Harper J.W. TEX264 Is an Endoplasmic Reticulum-Resident ATG8-Interacting Protein Critical for ER Remodeling during Nutrient Stress. Mol. Cell. 2019;74:891–908.e10. doi: 10.1016/j.molcel.2019.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iavarone F., Di Lorenzo G., Settembre C. Regulatory events controlling ER-phagy. Curr. Opin. Cell Biol. 2022;76 doi: 10.1016/j.ceb.2022.102084. [DOI] [PubMed] [Google Scholar]
- 21.Ishii S., Chino H., Ode K.L., Kurikawa Y., Ueda H.R., Matsuura A., Mizushima N., Itakura E. CCPG1 recognizes endoplasmic reticulum luminal proteins for selective ER-phagy. Mol. Biol. Cell. 2023;34 doi: 10.1091/mbc.E22-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y.-J., Knupp J., Arunagiri A., Haataja L., Arvan P., Tsai B. PGRMC1 acts as a size-selective cargo receptor to drive ER-phagic clearance of mutant prohormones. Nat. Commun. 2021;12 doi: 10.1038/s41467-021-26225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cinque L., De Leonibus C., Iavazzo M., Krahmer N., Intartaglia D., Salierno F.G., De Cegli R., Di Malta C., Svelto M., Lanzara C., et al. MiT/TFE factors control ER-phagy via transcriptional regulation of FAM134B. EMBO J. 2020;39 doi: 10.15252/embj.2020105696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohno S., Shiozaki Y., Keenan A.L., Miyazaki-Anzai S., Miyazaki M. An N-terminal-truncated isoform of FAM134B (FAM134B-2) regulates starvation-induced hepatic selective ER-phagy. Life Sci. Alliance. 2019;2 doi: 10.26508/lsa.201900340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Lorenzo G., Iavarone F., Maddaluno M., Plata-Gómez A.B., Aureli S., Quezada Meza C.P., Cinque L., Palma A., Reggio A., Cirillo C., et al. Phosphorylation of FAM134C by CK2 controls starvation-induced ER-phagy. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang X., Wang X., Ding X., Du M., Li B., Weng X., Zhang J., Li L., Tian R., Zhu Q., et al. FAM134B oligomerization drives endoplasmic reticulum membrane scission for ER-phagy. EMBO J. 2020;39 doi: 10.15252/embj.2019102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.González A., Covarrubias-Pinto A., Bhaskara R.M., Glogger M., Kuncha S.K., Xavier A., Seemann E., Misra M., Hoffmann M.E., Bräuning B., et al. Ubiquitination regulates ER-phagy and remodelling of endoplasmic reticulum. Nature. 2023;618:394–401. doi: 10.1038/s41586-023-06089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foronda H., Fu Y., Covarrubias-Pinto A., Bocker H.T., González A., Seemann E., Franzka P., Bock A., Bhaskara R.M., Liebmann L., et al. Heteromeric clusters of ubiquitinated ER-shaping proteins drive ER-phagy. Nature. 2023;618:402–410. doi: 10.1038/s41586-023-06090-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X., Jiang X., Li B., Zheng J., Guo J., Gao L., Du M., Weng X., Li L., Chen S., et al. A regulatory circuit comprising the CBP and SIRT7 regulates FAM134B-mediated ER-phagy. J. Cell Biol. 2023;222 doi: 10.1083/jcb.202201068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rutishauser J., Spiess M. Endoplasmic reticulum storage diseases. Swiss Med. Wkly. 2002;132:211–222. doi: 10.4414/smw.2002.09861. [DOI] [PubMed] [Google Scholar]
- 31.Baumann M., Giunta C., Krabichler B., Rüschendorf F., Zoppi N., Colombi M., Bittner R.E., Quijano-Roy S., Muntoni F., Cirak S., et al. Mutations in FKBP14 Cause a Variant of Ehlers-Danlos Syndrome with Progressive Kyphoscoliosis, Myopathy, and Hearing Loss. Am. J. Hum. Genet. 2012;90:201–216. doi: 10.1016/j.ajhg.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daley E., Streeten E.A., Sorkin J.D., Kuznetsova N., Shapses S.A., Carleton S.M., Shuldiner A.R., Marini J.C., Phillips C.L., Goldstein S.A., et al. Variable bone fragility associated with an Amish COL1A2 variant and a knock-in mouse model. J. Bone Miner. Res. 2010;25:247–261. doi: 10.1359/jbmr.090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garibaldi N., Contento B.M., Babini G., Morini J., Siciliani S., Biggiogera M., Raspanti M., Marini J.C., Rossi A., Forlino A., et al. Targeting cellular stress in vitro improves osteoblast homeostasis, matrix collagen content and mineralization in two murine models of osteogenesis imperfecta. Matrix Biol. 2021;98:1–20. doi: 10.1016/j.matbio.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlson J.A., Rogers B.B., Sifers R.N., Finegold M.J., Clift S.M., DeMayo F.J., Bullock D.W., Woo S.L.C. Accumulation of PiZ α1-antitrypsin causes liver damage in transgenic mice. J. Clin. Invest. 1989;83:1183–1190. doi: 10.1172/JCI113999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pastore N., Blomenkamp K., Annunziata F., Piccolo P., Mithbaokar P., Maria Sepe R., Vetrini F., Palmer D., Ng P., Polishchuk E., et al. Gene transfer of master autophagy regulator TFEB results in clearance of toxic protein and correction of hepatic disease in alpha-1-anti-trypsin deficiency. EMBO Mol. Med. 2013;5:397–412. doi: 10.1002/emmm.201202046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan D., Taylor T.K., Cole W.G. Characterization of an arginine 789 to cysteine substitution in alpha 1 (II) collagen chains of a patient with spondyloepiphyseal dysplasia. J. Biol. Chem. 1993;268:15238–15245. [PubMed] [Google Scholar]
- 37.Sun N., Malide D., Liu J., Rovira I.I., Combs C.A., Finkel T. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat. Protoc. 2017;12:1576–1587. doi: 10.1038/nprot.2017.060. [DOI] [PubMed] [Google Scholar]
- 38.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfson R.L., Chantranupong L., Saxton R.A., Shen K., Scaria S.M., Cantor J.R., Sabatini D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfson R.L., Chantranupong L., Wyant G.A., Gu X., Orozco J.M., Shen K., Condon K.J., Petri S., Kedir J., Scaria S.M., et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543:438–442. doi: 10.1038/nature21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goul C., Peruzzo R., Zoncu R. The molecular basis of nutrient sensing and signalling by mTORC1 in metabolism regulation and disease. Nat. Rev. Mol. Cell Biol. 2023;24:857–875. doi: 10.1038/s41580-023-00641-8. [DOI] [PubMed] [Google Scholar]
- 42.Ding B., Parmigiani A., Divakaruni A.S., Archer K., Murphy A.N., Budanov A.V. Sestrin2 is induced by glucose starvation via the unfolded protein response and protects cells from non-canonical necroptotic cell death. Sci. Rep. 2016;6 doi: 10.1038/srep22538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brüning A., Rahmeh M., Friese K. Nelfinavir and bortezomib inhibit mTOR activity via ATF4-mediated sestrin-2 regulation. Mol. Oncol. 2013;7:1012–1018. doi: 10.1016/j.molonc.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saveljeva S., Cleary P., Mnich K., Ayo A., Pakos-Zebrucka K., Patterson J.B., Logue S.E., Samali A. Endoplasmic reticulum stress-mediated induction of SESTRIN 2 potentiates cell survival. Oncotarget. 2016;7:12254–12266. doi: 10.18632/oncotarget.7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 46.Cross B.C.S., Bond P.J., Sadowski P.G., Jha B.K., Zak J., Goodman J.M., Silverman R.H., Neubert T.A., Baxendale I.R., Ron D., et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. USA. 2012;109:E869–E878. doi: 10.1073/pnas.1115623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sidrauski C., Acosta-Alvear D., Khoutorsky A., Vedantham P., Hearn B.R., Li H., Gamache K., Gallagher C.M., Ang K.K.-H., Wilson C., et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife. 2013;2 doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lebeau P., Byun J.H., Yousof T., Austin R.C. Pharmacologic inhibition of S1P attenuates ATF6 expression, causes ER stress and contributes to apoptotic cell death. Toxicol. Appl. Pharmacol. 2018;349:1–7. doi: 10.1016/j.taap.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 49.Napolitano F., Carrella D., Mandriani B., Pisonero-Vaquero S., Sirci F., Medina D.L., Brunetti-Pierri N., di Bernardo D. gene2drug: a computational tool for pathway-based rational drug repositioning. Bioinformatics. 2018;34:1498–1505. doi: 10.1093/bioinformatics/btx800. [DOI] [PubMed] [Google Scholar]
- 50.Napolitano G., Di Malta C., Esposito A., de Araujo M.E.G., Pece S., Bertalot G., Matarese M., Benedetti V., Zampelli A., Stasyk T., et al. A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome. Nature. 2020;585:597–602. doi: 10.1038/s41586-020-2444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang J.R., Lingeman E., Luong T., Ahmed S., Muhar M., Nguyen T., Olzmann J.A., Corn J.E. A Genome-wide ER-phagy Screen Highlights Key Roles of Mitochondrial Metabolism and ER-Resident UFMylation. Cell. 2020;180:1160–1177.e20. doi: 10.1016/j.cell.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fregno I., Fasana E., Soldà T., Galli C., Molinari M. N-glycan processing selects ERAD-resistant misfolded proteins for ER-to-lysosome-associated degradation. EMBO J. 2021;40 doi: 10.15252/embj.2020107240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martina J.A., Diab H.I., Brady O.A., Puertollano R. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 2016;35:479–495. doi: 10.15252/embj.201593428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hickey K.L., Swarup S., Smith I.R., Paoli J.C., Miguel Whelan E., Paulo J.A., Harper J.W. Proteome census upon nutrient stress reveals Golgiphagy membrane receptors. Nature. 2023;623:167–174. doi: 10.1038/s41586-023-06657-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahashi S., Saito C., Koyama-Honda I., Mizushima N. Quantitative 3D correlative light and electron microscopy of organelle association during autophagy. Cell Struct. Funct. 2022;47:89–99. doi: 10.1247/csf.22071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J., Pak S.C., O’Reilly L.P., Benson J.A., Wang Y., Hidvegi T., Hale P., Dippold C., Ewing M., Silverman G.A., et al. Fluphenazine reduces proteotoxicity in C. elegans and mammalian models of alpha-1-antitrypsin deficiency. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wong V.K.W., Zeng W., Chen J., Yao X.J., Leung E.L.H., Wang Q.Q., Chiu P., Ko B.C.B., Law B.Y.K. Tetrandrine, an Activator of Autophagy, Induces Autophagic Cell Death via PKC-α Inhibition and mTOR-Dependent Mechanisms. Front. Pharmacol. 2017;8 doi: 10.3389/fphar.2017.00351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun Y., Wang X., Yang X., Wang L., Ding J., Wang C.C., Zhang H., Wang X. V-ATPase recruitment to ER exit sites switches COPII-mediated transport to lysosomal degradation. Dev. Cell. 2023;58:2761–2775.e5. doi: 10.1016/j.devcel.2023.10.007. [DOI] [PubMed] [Google Scholar]
- 59.Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 60.Bianchi L., Gagliardi A., Maruelli S., Besio R., Landi C., Gioia R., Kozloff K.M., Khoury B.M., Coucke P.J., Symoens S., Marini J.C., Rossi A., Bini L., Forlino A. Altered cytoskeletal organization characterized lethal but not surviving Brtl+/- mice: insight on phenotypic variability in osteogenesis imperfecta. Human molecular genetics. 2015;24:6118–6133. doi: 10.1093/hmg/ddv328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt B.Z., Perlmutter D.H. Grp78, Grp94, and Grp170 interact with alpha1-antitrypsin mutants that are retained in the endoplasmic reticulum. American journal of physiology. Gastrointestinal and liver physiology. 2005;289:G444–G455. doi: 10.1152/ajpgi.00237.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Analysis restricting the output to cellular component term (“CC”).
(B) Analysis restricting the output to biological pathway term (“BP”).
The list is restricted to transcription factor terms (TFT).
Data Availability Statement
RNA-seq data generated in this study have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) under the series accession numbers GSE239527; GSE239525; GSE239528 (GEO: GSE239527; GSE239525; GSE239528) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. All data reported in this paper and any additional information required to reanalyze the data are available from the lead contact upon request.
The transcriptome data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession codes GSE239527 and GSE239525. The Series GSE239527 has been named: Transcriptome profile of RCS WT and TFEB/3 DKO cells transfected with mutant COL2A1 R789C compared to control. The Series GSE239525 has been named: Transcriptome profile of HeLa COL1A2 G610C transfected cells compared to control cells. The Superseries GSE239528 includes the two datasets. Source data and quantifications given in the main text have associated raw data. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.