Abstract
Doublecortin (DCX) is a 40 kDa microtubule-associated protein required for normal neural migration and cortical layering during development. Mutations in the human DCX gene cause a disruption of cortical neuronal migration. Defects in cdk5 (cyclin-dependent kinase 5) also cause defects in neural migration and cortical layering. DCX is a substrate for cdk5 in vitro and in vivo and the major site of in vitro phosphorylation is Ser-297. We used a highly developed MS strategy to identify the cdk5 phosphorylation sites and determine the major and minor sites. Several phosphopeptides were identified from a tryptic digest of 32P-labelled, cdk5-phosphorylated DCX using a combination of off-line HPLC and matrix-assisted laser-desorption ionization-MS with alkaline phosphatase treatment. Tandem MS/MS enabled the identification of seven phosphorylation sites for cdk5. Monitoring of 32P label indicated that there was one major site, Ser-28, at the N-terminus, and a major site, Ser-339, in the serine/proline-rich domain at the C-terminus. Five other sites, Ser-287, Thr-289, Ser-297, Thr-326 and Ser-332, were also found in the tail. Site-directed mutagenesis largely supported these findings. Single mutation of Ser-28 reduced but did not abolish phosphorylation. Double, rather than single, mutation for Ser-332 and Ser-339 was required to reduce overall phosphorylation, suggesting an interaction between these sites. Truncations of the tail produced a significant reduction in cdk5 phosphorylation of DCX. These results do not support Ser-297 as the major cdk5 phosphorylation site in DCX, but indicate that DCX is subject to complex multisite phosphorylation. This illustrates the importance of a well-developed MS strategy to identify phosphorylation sites.
Keywords: cyclin-dependent kinase 5 (cdk5), doublecortin (DCX), lissencephaly, MS, mutagenesis, phosphorylation
Abbreviations: cdk5, cyclin-dependent kinase 5; DCX, doublecortin; ESI, electrospray ionization; GSH, reduced glutathione; GST, glutathione S-transferase; JNK, c-Jun N-terminal kinase; MALDI, matrix-assisted laser-desorption ionization; MAP, microtubule-associated protein; NUDEL, nudE-like; TFA, trifluoroacetic acid; wt, wild-type
INTRODUCTION
Doublecortin (DCX) is an X-linked, 40 kDa MAP (microtubule-associated protein) [1–3], which is essential for migration of neurons during the development of the cerebral cortex [4]. Mutations in the human DCX gene cause lissencephaly in males and the milder (X-linked) subcortical laminar heterotopia (or double-cortex syndrome) in females [5,6]. Lissencephaly is a severe cortical malformation disorder with massive disorganization of neurons. The affected cortex has a disordered four-layered structure with neurons that either fail to begin or complete migration. Females develop double cortex due to X-inactivation leading to two populations of migrating neurons. One population expresses the normal allele and migrates correctly, whereas the other expresses the mutant allele and terminates migration to form the heterotopic band of neurons. Both lissencephaly and subcortical laminar heterotopia are associated with severe cognitive impairment and epilepsy [4].
Disruption of one allele of the LIS1 gene also causes lissencephaly. LIS1 is a 45 kDa ubiquitously expressed protein with seven WD40 repeats that mediate protein–protein interactions. LIS1 associates with NUDEL (nudE-like), a brain-enriched homologue of the Aspergillus nidulans nuclear disruption factor nudE found at centrosomes and neuronal growth cones, and which interacts with cytoplasmic dynein. NUDEL is a substrate of cdk5 (cyclin-dependent kinase 5), a proline-directed serine/threonine kinase that is abundant in brain and is critical for neuronal migration [7,8]. Phosphorylation increases its association with the 14-3-3 epsilon protein, which protects it from dephosphorylation and sustains the effects of cdk5 phosphorylation [9].
The discovery that mutations in LIS1 and DCX cause almost indistinguishable phenotypes in humans raises the question of their molecular connection. Both proteins are involved in microtubule regulation. Two tandem repeats of DCX form a functional microtubule-binding domain [10,11]. LIS1 purifies with polymerized microtubules from brain, binds tubulin and inhibits microtubule catastrophe events [12]. It also binds DCX and the two proteins enhance tubulin polymerization in an additive manner [13]. This was the first identified interaction between two of the prominent pathways that regulate neuronal migration.
Mutations in other human or mouse genes have also been found to cause defects in cortical neuronal migration. These include Reelin, Disabled, cdk5 and others. Reelin is a protein secreted into the extracellular matrix by the earliest neurons in brain. It often appears to act as a stop signal for neuronal migration [14]. Reelin mutations in humans cause lissencephaly and cerebellar malformations [14], suggesting strong links with the other lissencephaly genes. Lis1 has recently been shown to be a downstream target of Reelin signalling [15]. Mutations in the Disabled (Dab1) gene in the mouse show phenotypes that are indistinguishable from the Reelin mutation [16]. Dab1 is a cytoplasmic adaptor protein in the Reelin-signalling pathway and is also a substrate for cdk5 [17–19].
A potential common link between all these neuronal migration defects is cdk5. Mutations to the cdk5 gene result in disruption of neuronal migration [20]. It is an attractive candidate for regulating DCX or LIS1 phosphorylation and function and for the proteins of the Reelin/Dab1 pathway. Cdk5 binds with and is activated by a neuronal-specific activator, p35, which also plays a number of roles in both neuronal development and degeneration. Targeted disruption of the p35 gene also causes serious neuronal migration defects in mice [21]. Cdk5 phosphorylates a number of MAPs and many proteins involved in neuronal migration. It was proposed that cdk5 might phosphorylate LIS1 or DCX in addition to NUDEL [7] and Dab1 [22].
DCX was initially found to be a phosphoprotein [1,2], since treatment of DCX with alkaline phosphatase caused a downward mobility shift of an SDS/PAGE band detected by a DCX antibody [1]. Sequence analysis has suggested that DCX has a potential Abl tyrosine kinase site near the N-terminus and several mitogen-activated protein kinase sites near the C-terminus within a region rich in Ser-Pro and Thr-Pro motifs [6]. Protein kinase C and casein kinase II have also been suggested as possible kinases involved in DCX phosphorylation [5]. However, there has been no evidence that DCX is a substrate for any of these kinases. Owing to the similarity in disease phenotypes, cdk5 has been often linked indirectly to DCX [22–25]. Three recent reports have revealed the kinases that phosphorylate DCX. MAP/microtubule affinity regulator kinase and protein kinase A phosphorylate DCX at Ser-47 [26] and JNK (c-Jun N-terminal kinase) phosphorylates DCX on Thr-321, Thr-331 and Ser-334 (which are in the equivalent positions to Thr-326, Thr-336 and Ser-339 in the mouse sequence used in the present study) in the C-terminus [27]. DCX was also shown to be a substrate for cdk5 [28]. Ser-297 was identified as the sole major site by MS analysis of a tryptic digest from an in vitro phosphorylation. No other site was observed using this simple screening method and it was therefore claimed that it indicated Ser-297 as the major site. Utilizing a phospho-specific antibody and cdk5−/− as well as dcx−/− mice, Ser-297 was also found to be phosphorylated on this site in vivo.
In contrast with the discovery of Ser-297 as the major cdk5 site in DCX, the discovery of a phosphorylation site in a protein does not usually reveal whether it is the sole or major site. Detection of phosphorylation by MS remains a challenging process because of the low ionization efficiency of some phosphopeptides [29]. Non-phosphopeptides present in tryptic digests may suppress the ionization of the phosphopeptides because of a higher gas-phase proton affinity [30]. Separation of the phosphopeptides from interfering non-phosphopeptides improves phosphopeptide detection [31]. Off-line HPLC separation was recently shown to be more effective for the enrichment of phosphopeptides [32], when compared with the more widely used immobilized metal-affinity chromatography [31]. The use of 32P labelling, where possible, reduces the number of collected fractions that need to be analysed and also provides information on the relative amounts of incorporated phosphate. Radioactive fractions can be analysed by Edman degradation [33] or MS [34]. In the present study, we incorporated these methodologies into a well-developed strategy aimed at determining whether Ser-297 is the sole or major in vitro site for cdk5.
In this study, we confirm the phosphorylation of Ser-297 in DCX by cdk5, but demonstrate that it is not the major site and that cdk5 phosphorylates six additional sites. The phosphorylation sites were identified using a combination of off-line HPLC, radioactive detection and hybrid QqTOF-MS/MS (quadrupole time-of-flight tandem mass spectrometry) after in vitro phosphorylation of DCX with cdk5 using [γ-32P]ATP. Six sites were found in the serine/proline-rich domain near the C-terminus and a single site was near the N-terminus. The N-terminal Ser-28 and one of the C-terminal sites, Ser-339, were particularly good substrates, as observed from incorporation of 32P. The effect of site-directed mutagenesis at Ser-28 was supportive of the MS data, but for Ser-339 the result indicated that an interaction with an adjacent phosphorylation site(s) was also important. The results significantly expand on those found previously [28] and define novel phosphorylation sites in DCX. The results illustrate that direct examination of a tryptic digest [28] is inappropriate for finding the major phosphorylation sites, and we illustrate an appropriately developed strategy for the identification of phosphorylation sites in DCX.
METHODS
Chemicals and DNA constructs
All chemicals were of analytical grade or higher. MilliQ water was used in all experiments (MilliQ UF PLUS; Millipore, Billerica, MA, U.S.A.). Tris was from Amresco (Solon, OH, U.S.A.). Hydrochloric acid, magnesium sulphate and formic acid were from BDH Laboratory Supplies (U.K.). EGTA, Tween 80, ATP, ammonium bicarbonate, TFA (trifluoroacetic acid) and activated charcoal were from Sigma (St. Louis, MO, U.S.A.). [γ-32 or 33P]ATP was from PerkinElmer Life Sciences (Boston, MA, U.S.A.) and trypsin (Gold-MS grade, porcine) was from Promega (Madison, WI, U.S.A.). Restriction enzymes and calf intestinal alkaline phosphatase were from New England Biolabs (Beverly, MA, U.S.A.). The acetonitrile was from Burdick and Jackson (Muskegon, MI, U.S.A.). GST (glutathione S-transferase)-cdk5 plasmid was from J. Wang (Hong Kong University, Hong Kong) and the GST-p25 plasmid was from L.-H. Tsai (Harvard, U.S.A.) as we have described previously [35].
Isolation and cloning of murine DCX
A murine DCX clone was isolated from a foetal mouse (embryonic day 19) cDNA library by OriGene (OriGene Technologies, Rockville, MD, U.S.A.) by screening with DCX-specific primers F408 (5′-AGCCAGAGAGAACAAGGAC-3′) and F409 (5′-GGACCACAAGCAATGAAC-3′). The isolated DCX clone (CDA001 in a pCMV6-XL3 vector) was digested with SacI and KpnI to produce a 3 kb fragment of the DCX coding region with approx. 2 kb of 3′ untranslated region. This fragment was subcloned into the SacI and KpnI sites of a pGEM-3Z vector (Promega). The translated amino acid sequence of this clone has up to two amino acid differences and is one amino acid longer (GenBank® accession number AY560329) than other published sequences, probably representing polymorphisms. The full-length coding region of DCX (approx. 1.1 kb) was amplified from the pGEM-3Z-DCX clone using primers F439 (5′-AAAAGTTGCCGCGGTTCCAC-3′) and F440 (5′-GGCTTGAATTCAGCCTATCATC-3′), which introduce SacII and EcoRI sites respectively. The PCR contained 5–10 ng of template DNA, 1 unit of Taq DNA polymerase (Roche, Basel, Switzerland), enzyme buffer supplied by the manufacturer, 0.2 mM of each dNTP, 0.4 μM of each primer and amplified on a Hybaid PCR Express thermocycler (Thermo Hybaid, Middlesex, U.K.) using the following parameters: initial denaturation at 93 °C for 3 min; 30 cycles at 93 °C for 30 s, 55 °C for 30 s and 72 °C for 45 s and a final extension at 72 °C for 5 min. The PCR product was subcloned into a pGEM-T-Easy (Promega) vector, which takes advantage of the adenosine overhangs on the PCR products introduced during amplification by Taq DNA polymerase.
Generation of GST-fusion constructs with wt (wild-type) and mutant DCX
DCX, full-length and C-terminal truncations were cloned into a GST vector to express a GST-fusion protein. The full-length DCX and the DCX1–300 truncation were subcloned from the pGEMT-DCX plasmid by digestion with SacII and EcoRI (full-length) or SacII and BpmI (DCX1–300), the overhangs were modified to blunt ends with the Klenow fragment of DNA polymerase I (Promega), according to the manufacturer's instructions. These fragments were cloned into a pGEX-6P-3 vector (Amersham Biosciences, Uppsala, Sweden) that had been linearized with the blunt-end cutter SmaI. The DCX1–274 and DCX1–236 fragments were amplified from the pGEMT-DCX plasmid using the following primers: DCX1–274 with F439 and F436 (5′-TGTGGGTGTAGAGATAGGAG-3′) primers and DCX1–236 with F439 and F441 (5′-ATGGAGACAGTCGACCTGCT-3′) primers. The PCR conditions were as those described above, except that a high-fidelity polymerase, Pfu DNA polymerase (Stratagene, La Jolla, CA, U.S.A.), was used. PCR products were digested with SacII and RcaI for DCX1–274, and SacII and SalI for DCX1–236. The overhangs were modified to blunt ends by incubation with Klenow fragment and cloned into a pGEX-6P-3 vector linearized with SmaI. Plasmids containing GST-fusion constructs were transformed into BL21(DE3) competent cells, containing a thioredoxin expression plasmid (pT-Trx) [36].
Key cdk5 phosphorylation sites were mutated to alanine in the pGEX-DCX(wt) vector using Stratagene's QuikChange™ Mutagenesis kit (Stratagene) according to the manufacturer's instructions. The complete coding region for GST–DCX was sequenced in each of the mutant plasmids (Westmead DNA, Westmead, NSW, Australia). This ensured that the target site was mutated and that there were no undesirable mutations at other locations in the sequence introduced by the PCR-based mutagenesis.
Purification of recombinant GST-fusion proteins
The bacterial cells were induced to express GST-fusion proteins by incubation with 0.2 mM isopropyl β-D-thiogalactoside for 2–4 h at 30 °C. Recombinant protein was purified on GSH (reduced glutathione)–Sepharose beads (Amersham Biosciences). For cdk5, bacterial pellets from BL21/Trx cells expressing GST-cdk5 and GST-p25 were combined before purification of the fusion proteins on GSH–Sepharose beads. Bacterial cells were lysed with 100 μg·ml−1 lysozyme in the presence of proteinase inhibitors [20 μg·ml−1 leupeptin, 1 mM PMSF and Roche EDTA-free Complete proteinase inhibitor tablet (1 tablet/litre of culture)] followed by three rounds of freeze and thaw, and sonication using a Branson Sonifier 250 (Branson Ultrasonics, Danbury, CT, U.S.A.). Triton X-100 at a final concentration 1% (v/v) was added to the lysate at 4 °C for 30 min, followed by centrifugation at 20000 g for 30 min. GSH–Sepharose beads (500 μl bead volume/litre of culture) were added to the supernatant, incubated with mixing at 4 °C for 1 h and then the beads were washed three times in an excess volume of PBS. Beads with recombinant protein were stored at −20 °C in 30% (v/v) glycerol. Recombinant fusion proteins were eluted with several washes of GSH buffer (20 mM GSH/100 mM Tris/HCl, pH 8/120 mM NaCl). GST-cdk5/p25 was dialysed and concentrated using Microcon YM10 spin columns (Millipore) against the phosphorylation buffer (30 mM Tris/HCl, pH 7.4/1 mM magnesium sulphate/1 mM EGTA, pH 7.4).
Phosphorylation of GST–DCX
GST–DCX(wt) and GST–DCX(mutant) proteins were phosphorylated at 37 °C with GST-cdk5/p25 in 15–80 μl of reaction volume for 5–90 min (depending on individual experiments) using the following conditions: substrate protein (0.1– 4 μg), 1:100 to 1:250 dilution of the prepared GST-cdk5/p25 [each fresh batch was tested for optimal activity against GST–DCX(wt) protein] and phosphorylation buffer (as described above), 0.05% (v/v) Tween 80/40 μM ATP/0.063–0.125 μCi·μl−1 [γ-32 or 33P]ATP. Reactions were stopped by the addition of 0.5 vol. of 3× SDS sample buffer [0.2 M Tris/HCl, pH 6.8/28% glycerol/6% (w/v) SDS/0.07% (w/v) Bromophenol Blue/12% (v/v) 2-mercaptoethanol] and the samples were heated in a boiling water bath for 5 min. Samples were analysed by SDS/PAGE and autoradiography.
Phosphorylation of wt GST–DCX with GST-cdk5/p25 for MS
GST–DCX (4 μg) was phosphorylated for 10 min by GST-cdk5/p25 in an 80 μl volume as described above. Four samples of the same amount of GST–DCX were phosphorylated for 90 min in the absence of [γ-32P]ATP with increased concentrations of magnesium sulphate and ATP (20 mM and 800 μM respectively). Acrylamide was added to a final concentration of 2% (w/v) to alkylate reduced cysteine residues and the samples were incubated at room temperature (20–22 °C) for 30 min and then the proteins were resolved by SDS/PAGE. Gels were stained with zinc [37].
Relative phosphorylation of wt and mutant GST–DCX
Approx. 1 μg of wt or mutant GST–DCX fusion protein was phosphorylated by GST-cdk5/p25 in the presence of radiolabelled ATP for 5 min in a 15 μl reaction volume. Samples were separated by SDS/PAGE (10% gel) and stained with Coomassie Brilliant Blue. Protein loading was assessed using the GS-800 Calibrated Densitometer and using QuantityOne v4.2.2 software (Bio-Rad Laboratories, Hercules, CA, U.S.A.). Dried gels were exposed to a phosphorimaging screen and the incorporated radioactivity was quantified on a STORM 860 PhosphorImager using ImageQuant v5.2 software (Molecular Dynamics; Amersham Biosciences). Incorporation of 32P was corrected for protein loading and an additional correction was made for the truncations for equimolar representation of the data.
Tryptic digestion and HPLC
Gel bands containing DCX were cut out after zinc staining and destained in 150 μl of 1×Tris/glycine/SDS buffer (Bio-Rad Laboratories). Each band was washed four times in 1 ml of water for 10 min before incubation with 0.3 μg of trypsin in 30 μl of 50 mM ammonium bicarbonate (pH 8.0) for 16 h at 37 °C. The digestion solution from each sample was removed and combined. A second extraction of tryptic peptides was performed using 30 μl of 5% (v/v) formic acid in acetonitrile. The two-step extraction collected approx. 85% of the phosphopeptides in the gel piece, as determined by measuring the Cerenkov radiation. The combined solution was concentrated to <5 μl in a rotary vacuum concentrator (ALPHA-IR RVC, Christ, Germany) and then made up to 50 μl with 0.1% (v/v) TFA aqueous solution before injection on to a HPLC (SMART system; Amersham Biosciences, Little Chalfont, Bucks., U.K.). The peptides were separated by reversed-phase chromatography using a 2.1 mm×100 mm column [μRPC C2/C18, 3 μm, 120 Å (1 Å=0.1 nm); Amersham Biosciences] at 100 μl·min−1. The gradient was from 100% phase A (0.1% TFA aqueous solution) to 40% phase B (0.1% TFA in acetonitrile) in 25 min and then to 100% phase B in 10 min. Fractions (50 μl) were collected every 30 s. Cerenkov radiation emitted from each fraction was measured using a Minaxi Tricarb 4430 scintillation counter (PerkinElmer Packard Instruments, Boston, MA, U.S.A.). Fractions were then dried to <5 μl.
Alkaline phosphatase treatment and MALDI (matrix-assisted laser-desorption ionization)-MS
To 500 nl of each sample, 2 μl of a solution containing 50 mM ammonium bicarbonate, 1 mM magnesium sulphate and 0.25 unit·μl−1 of alkaline phosphatase was added. The sample was incubated for 3 h at 37 °C. Analysis of the samples before and after alkaline phosphatase treatment was performed on a Voyager DE-Pro MALDI-MS (Applied Biosystems, Boston, MA, U.S.A.). On a MALDI plate, 500 nl of each sample or 2.5 μl of alkaline-phosphatase-treated sample was spotted, followed by 500 nl of matrix solution [0.3% TFA, 60% (v/v) acetonitrile aqueous solution containing 10 mg·ml−1 α-cyano-4-hydroxycinnamate]. Samples were analysed in linear or reflectron mode using external calibration.
Tandem MS/MS
Peptide fragmentation was performed using a QSTAR XL QqTOF MS (Applied Boisystems/MDS Sciex, Concord, ON, Canada) with either the NanoSpray™ or oMALDI2™ ion sources. Samples analysed using the NanoSpray™ were made up to 0.1% formic acid, 50% acetonitrile aqueous solution and were sprayed off-line using a 1 μm inside diameter EconoTip™ (New Objective, Woburn, MA, U.S.A.). The ion spray voltage was 1000–1200 V. Samples analysed using oMALDI2™ were made up to 2% formic acid solution and concentrated using a microcolumn manufactured from a GELoader™ (Eppendorf, Hamburg, Germany) pipette tip [38]. The microcolumn was packed with the hydrophobic resins POROS R2, POROS R3 (Applied Biosystems) or with activated charcoal [39], depending on the hydrophilicity of the peptides. After loading the sample in 10 μl, the microcolumn was washed with 10 μl of 2% formic acid solution and the bound material was eluted on to a MALDI plate in matrix solution.
Statistical analysis
A two-tailed t test that assumed unequal variance was performed on the phosphorylation by cdk5 of GST–DCX mutants and wt. Values greater than 2 S.D. from the mean were omitted from the analysis. P<0.05 was considered significant.
RESULTS
Phosphorylation of DCX
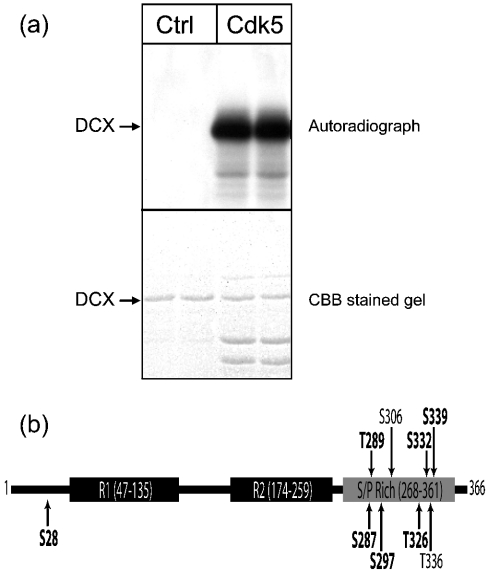
Recombinant DCX (murine sequence) was phosphorylated by incubation with cdk5 in the presence of [γ-32P]ATP. Autoradiography of the dried gel confirmed the incorporation of phosphate (Figure 1a). Phospho-amino acid analysis by thin layer electrophoresis [40] revealed that cdk5 phosphorylation is predominantly on serine residues (93%) relative to threonine (7%) or tyrosine (0%, results not shown). Cdk5 is a proline-directed serine/threonine protein kinase [41]; therefore examining the DCX protein sequence for Ser-Pro or Thr-Pro motifs indicated the presence of nine potential cdk5 phosphorylation target residues (Figure 1b), one at the N-terminus preceding the tandem DCX domains and eight in the serine/proline-rich region at the C-terminus. The requirement of nearby basic residues was not strictly applied in arriving at this number. However, all the sites have at least one basic residue within five residues of the serine/threonine residue.
Figure 1. DCX is a substrate for phosphorylation by cdk5 on multiple sites.
(a) In vitro phosphorylation of DCX. GST–DCX was phosphorylated for 10 min at 37 °C in the presence of GST-cdk5/p25. The upper panel shows an autoradiograph, whereas the lower panel shows the Coomassie Brilliant Blue (CBB)-stained gel from the same experiment. DCX was incubated without any protein kinase (lanes 1 and 2) or with cdk5 (lanes 3 and 4). Results are shown in duplicate from one of six experiments. (b) Schematic representation of mouse DCX, indicating the location of R1 and R2 domains and the serine/proline-rich region. Arrows indicate the location of potential cdk5 phosphorylation sites and those identified in the present study are shown in bold text.
MS analysis of cdk5 phosphorylation sites on wt DCX
Initially, a tryptic digest of DCX phosphorylated with cdk5 was directly examined by MALDI-MS, using both POROS R2 and graphite microcolumns packed in GELoader™ tips for de-salting [38] (results not shown). No phosphopeptides of significant intensity matching the DCX sequence were identified. Next, immobilized metal-affinity chromatography [31] was used before microcolumn cleanup. Three possible phosphopeptides were found in the MALDI-MS spectra using this method and an extra phosphopeptide was implicated after alkaline phosphatase treatment (results not shown). However, it was clear from the spectra that many non-phosphopeptides derived from DCX were still present in the sample. The presence of these peptides added ambiguity to the results of the alkaline phosphatase treatment, made selection of ions for tandem MS/MS difficult and was suspected of being responsible for suppression of ionization of the observed phosphopeptides and others still hidden.
To overcome these problems, a combination of approaches was used to identify the phosphorylation sites. DCX was phosphorylated to high stoichiometry with unlabelled ATP, but the samples were ‘spiked’ with low stoichiometry radiolabelled DCX to trace the peptides containing the maximum 32P incorporation. Thus DCX was first phosphorylated by cdk5 in vitro using [γ-32P]ATP for 10 min. Another aliquot of DCX was then phosphorylated with a higher concentration of Mg2+ and unlabelled ATP for 90 min to maximize the incorporation of phosphate. The phosphorylated DCX from both the reactions was pooled after SDS/PAGE and trypsin digestion. This spiked sample approach provided key information on the level of 32P incorporation during the short reaction time, but also supplied a relatively high stoichiometry of incorporated phosphate for the MS analyses. The DCX phosphorylation sites that are most important under physiological conditions are probably the same sites that incorporate most of the 32P in an in vitro reaction with the conditions of limited Mg and ATP in a short reaction time. In contrast, long phosphorylation times under optimal in vitro conditions, probably produce sites not necessarily of in vivo relevance.
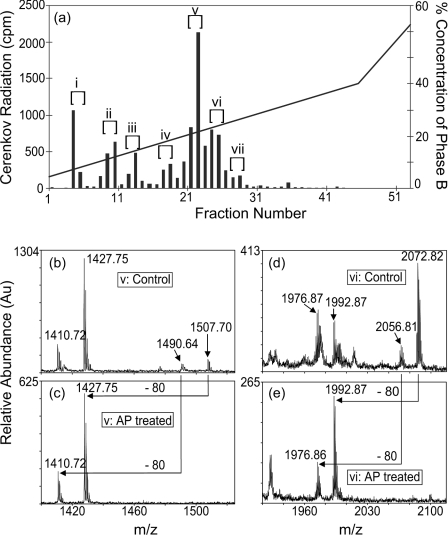
To separate the phosphopeptides derived from phospho-DCX, the trypsin-digested sample was applied to reversed-phase HPLC and each eluted fraction was counted for 32P (Figure 2a). Significant amounts of radiation were detected across the elution profile, indicating multiple phosphorylation sites in DCX. Fourteen fractions containing high counts were combined into seven pools, i–vii (Figure 2a), which probably contain a single phosphopeptide each. Each pooled sample was concentrated in a rotary vacuum concentrator. Samples prepared for MALDI-MS/MS were further concentrated by applying them to reversed-phase microcolumns packed in GELoader™ tips. Although the fractions from the first reversed-phase separation are purified, the extra step allows the concentration of the sample to <500 nl, resulting in an improved signal-to-noise ratio. Each of the pooled samples was examined by MALDI-MS before and after alkaline phosphatase treatment. When dephosphorylation was observed (a decrease in mass of 80 units on the m/z scale), the peptide was chosen for further analysis by tandem MS/MS. Figures 2(b), 2(c) and 2(d), 2(e) show two examples of MALDI-MS spectra from samples v and vi before and after alkaline phosphatase treatment respectively. Samples i–iv and vii are not shown. This approach revealed five distinct phosphopeptides from DCX. However, the sequence of each peptide (except ii) contained more than one serine or threonine and hence it was not possible to assign specifically the phospho-acceptor site in most of the cases. Therefore each pooled fraction was further analysed by tandem MS/MS using either nanoESI (electrospray ionization) or MALDI, depending on which ionization method gave the highest intensity or least interference from other peptides or impurities. Tandem MS/MS unequivocally identified the phospho-acceptor site in each case.
Figure 2. Distribution of the cdk5 phosphorylation sites in DCX.
(a) DCX was phosphorylated for 10 min in the presence of [γ-32P]ATP and for 90 min in the presence of excess non-radioactive ATP and the samples were mixed. After SDS/PAGE, DCX was digested with trypsin and the fragments were separated by HPLC. A plot of Cerenkov radiation versus fraction number for the reversed-phase HPLC separation is shown. The percentage concentration of the organic phase (phase B) is also shown on the right axis. The fractions that showed a specific increase in phosphorylation were pooled into seven (i–vii) samples. (b–e) Part of the MALDI-MS spectra of samples v and vi before (Control) and after their treatment with alkaline phosphatase to dephosphorylate the peptides. Sample v before (b) and after (c) dephosphorylation. An 80 Da mass shift from m/z 1507.70 to 1427.75 and m/z 1490.64 to 1410.64 indicates the loss of a phosphate group. The dephosphorylated peptides match the mass of DCX331–344 and the N-terminal pyroglutamic acid modified DCX331–344 respectively. A region of the MALDI-MS spectra from sample vi is shown before (d) and after (e) alkaline phosphatase treatment. The 80 Da mass shift from m/z 2072.82 to 1992.87 and m/z 2056.81 to 1976.86 indicates loss of a phosphate group. The dephosphorylated peptides match oxidized and non-oxidized DCX23–39 respectively.
Sample i
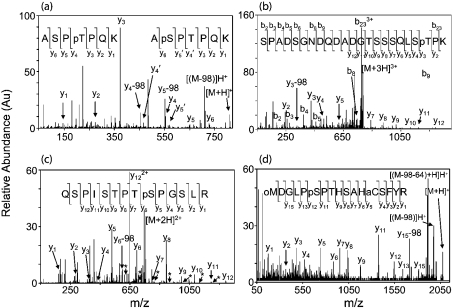
MALDI-MS analysis of sample i showed no peaks that could match any theoretical DCX phosphopeptide. However, after alkaline phosphatase treatment, a new peak appeared at m/z 728.45 (results not shown). This peak matched the mass of the pseudo-molecular ion ([M+H]+) of ASPTPQK in DCX (theoretical m/z 728.39). The sample was further analysed by ESI–MS/MS. The singly charged ion at m/z 808.35, corresponding to the phosphorylated form (+80 Da) of the above sequence (theoretical m/z 808.36), but not present in the MALDI-MS spectrum was observed in the ESI–MS spectrum. This ion was selected for fragmentation to produce the product ion spectrum shown in Figure 3(a), which has peaks corresponding to two separate, and complete, series of y ions. The two series correspond to both possible phosphopeptides of DCX286–292, ApSPTPQK and ASPpTPQK, where pS or pT represent phospho-serine or phospho-threonine respectively. These phosphopeptides share an overlapping ion series for y1 to y3 and also share y6, but differ in the mass of their y4 and y5 ions. In Figure 3(a), the transition that shows the phospho-Thr-289 (181.0 Da) is from y3 to y4, whereas the transition that shows phospho-Ser-287 (167.0 Da) is from y5′ to y6. This result shows that both Ser-287 and Thr-289 are phosphorylated by cdk5. No doubly phosphorylated peptide was observed; however, the result does not reveal whether this peptide may be doubly phosphorylated. Ion intensities in mass spectra do not reflect the amounts of molecules present in the original sample, due to differences in the efficiency of ionization and the transition from solution phase (ESI) or solid-phase (MALDI) to gas-phase ions. Therefore it is not possible to determine which phosphorylated species was more abundant [29].
Figure 3. Direct sequencing of the phosphorylation sites using tandem MS/MS.
(a) ESI–MS/MS spectrum of the fragmentation of the singly charged peptide at m/z 808.35 from sample i (Figure 2a). Two separate, and complete, series of y ions describe the phosphopeptides DCX286–292, where either Ser-287 or Thr-289 is phosphorylated. Ions marked with a prime are exclusive to the sequence containing phospho-Ser-287. The rest of the y ions are overlapping. Ions demonstrating neutral loss of phosphoric acid (−98 Da) are also shown (indicated by [(M-98)H]+). (b) ESI–MS/MS spectrum of the fragmentation of the triply charged m/z 782.32 from sample iii (Figure 2a). The spectrum matches the sequence for DCX306–328, where Thr-326 is phosphorylated and Asn-317 has been deamidated. (c) ESI–MS/MS spectrum of the fragmentation of the doubly charged m/z 754.41 from sample v (Figure 2a). The sequence derived from this spectrum matches DCX331–344, where Ser-339 is phosphorylated. There is also a second very low abundance y ion series in the spectrum (marked with *). This series best describes DCX331–334, where Ser-332 is phosphorylated. (d) MALDI-MS/MS spectrum of the singly charged parent ion at m/z 2072.86 from sample vi (Figure 2a). The spectrum reveals the sequence of DCX23–39, where Met-23 is oxidized (oM), Asn-24 has been deamidated, Cys-35 has been modified by acrylamide (aC) and Ser-28 is phosphorylated. The neutral loss of phosphoric acid and the abundant further neutral loss of methane sulphenic acid (CH3SOH, −64 Da) are also shown.
Sample ii
MALDI-MS analysis of sample ii showed peaks at m/z 724.29 and 740.28 that could be dephosphorylated to m/z 644.32 and 660.30 respectively. The latter corresponded to DCX297–302 (SPGPMR) and its oxidized form respectively (theoretical m/z 644.32 and 660.31). This sequence contains a single residue that can be phosphorylated, Ser-297. This site has recently been identified as a cdk5 phosphorylation site by Tanaka et al. [28]. This sample was analysed by ESI–MS/MS. Selecting the parent ion at m/z 724.34, the phosphopeptide was confirmed as having the sequence pSPGPMR (results not shown).
Sample iii
MALDI-MS analysis of sample iii showed a peak at m/z 2344.91 that could be dephosphorylated to m/z 2265.94. This peak is approx. 1 unit larger than the phosphorylated peptide DCX306–328 (SPADSGNDQDANGTSSSQLSTPK, theoretical m/z 2263.96). When an asparagine residue is followed by a glycine, the asparagine is highly susceptible to deamidation to aspartic acid or iso-aspartic acid under alkaline conditions [42]. The phosphopeptide was probably deamidated during the weakly alkaline conditions of the tryptic digestion, resulting in an overall increase in mass of 1 Da. The phosphopeptide was fragmented using ESI–MS/MS. The triply charged parent ion at m/z 782.32 was selected to produce the product ion spectrum shown in Figure 3(b). This shows a series of y ions describing half of the C-terminus and a series of b ions describing nearly half of the N-terminus of the phosphopeptide DCX306–328 (SPADSGNDQDADGTSSSQLSpTPK). The transition from y11 to y12 matched an aspartic acid residue. This shows that Asn-317 had been deamidated. Alternatively, the transition may also be to an iso-aspartic acid residue, since they are not distinguishable by MS. The y ions from y2 to y3 describe the loss of phospho-Thr-326. There was no evidence that any of the other serine or threonine residues were phosphorylated, including the N-terminal serine, which matches the motif for cdk5 substrates. Therefore Thr-326 is phosphorylated by cdk5.
Sample iv
MALDI-MS of sample iv revealed a peak at m/z 1635.80, which could be dephosphorylated to m/z 1556.86 (results not shown). The latter peak matches DCX331–345 (QSPISTPTSPGSLRK, theoretical m/z 1555.84), containing a single trypsin mis-cleavage. Using MALDI-MS/MS, fragmentation of the parent ion at m/z 1635.79 produced a product ion spectrum, which was consistent with singly phosphorylated DCX331–345 (results not shown). However, the site of phosphorylation was ambiguous due to the low abundance of the spectrum.
Sample v
The MALDI-MS spectrum of sample v, before and after alkaline phosphatase treatment, is shown in Figures 2(b) and 2(c). There are two peaks at m/z 1507.70 and 1490.64, which could be dephosphorylated to m/z 1427.75 and 1410.72 respectively. The peak at m/z 1427.75 matched the peptide DCX331–344 (QSPISTPTSPGSLR, theoretical m/z 1427.75), whereas that at m/z 1410.72 could not be directly matched to DCX (but is probably the same peptide with an N-terminal glutamine modified to pyroglutamic acid, see below). Sample v was analysed by ESI–MS/MS. The doubly charged parent ion at m/z 754.41, corresponding to the peak detected at m/z 1507.70, was fragmented to produce the product ion spectrum as shown in Figure 3(c). The spectrum shows a complete y ion series for the predicted phosphopeptide DCX331–344. The transition from y5 to y6 shows the loss of phospho-Ser-339. Therefore Ser-339 is phosphorylated by cdk5. However, there is also a second low intensity, but nonetheless high signal-to-noise (ranging from 5 to 20) y ion series in the spectrum (marked with asterisks). This very low abundance series best describes DCX331–334, where Ser-332 is phosphorylated. Since many of the possible assignments have overlapping theoretical ion series, it is not possible to rule out that phospho-Ser-335, Thr-336 and Ser-338 may also be present at very low abundance.
The phosphopeptide at m/z 1490.64 (and its dephosphorylated counterpart at m/z 1410.72) does not match the mass of any peptide predicted from the DCX sequence. However, a simple explanation is that the peptide QSPISTPTSPGSLR may have had its N-terminal glutamine modified to pyroglutamic acid, accounting for the mass shift of −17 Da (http://www.abrf.org/index.cfm/dm.home?AvgMass=all). This common modification occurs under the same alkaline conditions as deamidation [42].
Sample vi
The MALDI-MS spectrum of sample vi before and after alkaline phosphatase treatment is shown in Figures 2(d) and 2(e). Two peaks at m/z 2072.82 and 2056.81 could be dephosphorylated to m/z 1992.87 and 1976.86 respectively. These latter peaks exactly match oxidized DCX23–39 (MNGLPSPTHSAHCFYR) and its non-oxidized form respectively, where the Asn-24 has been deamidated (+1 Da, similar to sample iii above) and Cys-35 has been modified by acrylamide (+71 Da, this modification was performed before SDS/PAGE, see the Methods section): MDGL-PpSPTHSAHaCFYR=1904.85 ([M+H]+)+71.04+0.98+79.97=2056.84 and MoxDGLPpSPTHSAHaCFYR + 2056.84+16.00=2072.84. Using MALDI-MS/MS, the larger, oxidized parent ion at m/z 2072.82 was selected for fragmentation to produce the product ion spectrum in Figure 3(d). A nearly complete y ion series was detected to identify unambiguously the peptide as oxidized DCX23–39, where Ser-28 is phosphorylated and Met-23 is oxidized. The transition from y11 to y12 shows the loss of phospho-Ser-28. A feature of this spectrum is a strong peak showing the neutral loss of phosphoric acid (−98 Da). A more abundant peak showing a further neutral loss of methane sulphenic acid (CH3SOH, −64 Da) is a signature mass shift for an oxidized methionine [43,44]. The peak matching non-oxidized DCX23–39 (m/z 2056.81) was not examined further.
Sample vii
MALDI-MS analysis of sample vii showed a peak at m/z 2054.94, which could be dephosphorylated to m/z 1974.99 (results not shown). The latter peak did not match any predicted DCX tryptic peptide. However, MALDI-MS/MS analysis revealed that the phosphopeptide matched the sequence DCX23–39, where both Met-23 is oxidized (+16 Da) and Asn-24 has been converted into a succinimide intermediate (−17 Da), resulting in an overall loss in molecular mass of −1 Da (results not shown). The formation of succinimide is a result of deamidation of asparagine and this intermediate is stable at low pH [45] (see also http://www.abrf.org/index.cfm/dm.home?AvgMass=all). It was probably able to persist due to the acidic sample storage conditions (0.1% formic acid aqueous solution) used in the present study.
A relatively small amount of Cerenkov radiation was observed in fraction 35 (Figure 2a). This fraction was analysed using the same treatment as for samples i–vii. A phosphopeptide was detected at m/z 1895.98. This ion was sequenced by MALDI-MS/MS (results not shown). The sequence was unambiguously determined to be SDLEVLFQGPLGpSPNSR. Although this phosphopeptide is part of the GST–DCX sequence, it does not include any specific DCX sequence, but is derived from the multiple cloning site of the pGEX-6P-3 vector in which DCX was cloned to produce the GST–DCX fusion protein.
The tandem MS/MS peptide sequencing, combined with the Cerenkov radiation measurements, allowed the determination of the DCX phosphorylation sites that were most readily phosphorylated by cdk5. The identified phosphorylation sites and the amount of Cerenkov radiation associated with those sites are shown in Table 1. Cerenkov radiations from samples iv and v were summed, since they are associated with the same phosphorylation site. Similarly, counts for samples vi and vii were also summed. The counts from fraction 23, which eluted between samples v and vi, were equally divided between the two phosphorylation sites (arbitrarily). The counts indicated that Ser-339 (43%) and Ser-28 (24%) are the major phosphorylation sites for cdk5 in vitro. Since the duration of the phosphorylation with radiolabelled ATP was short, these sites may be important for the in vivo phosphorylation of DCX. It is important to note that in the present study, we used GST-tagged DCX and cdk5/p25 and that these tags may have introduced non-native steric interactions.
Table 1. Incorporation of 32P into specific in vitro phosphorylation sites in DCX by cdk5.
| Cerenkov radiation (c.p.m.) | % c.p.m. | Summed fractions | Phosphorylation site | Sequence (phospho-site in bold) | HPLC fraction (Figure 2a) |
|---|---|---|---|---|---|
| 4360 | 43 | 17–23(1/2) | Ser-339, Ser-332 (minor) | …KSKQSPISTPTSPGSL… | iv, v |
| 2399 | 24 | 23(1/2)–28 | Ser-28 | …NGLPSPTHS… | vi, vii |
| 1289 | 13 | 4–5 | Ser-287, Thr-289 | …GPKASPTPQKT… | i |
| 1269 | 13 | 8–10 | Ser-297 | …TSAKSPGPM… | ii |
| 783 | 8 | 12–14 | Thr-326 | …SQLSTPKSK… | iii |
In vitro phosphorylation of mutant DCX by cdk5
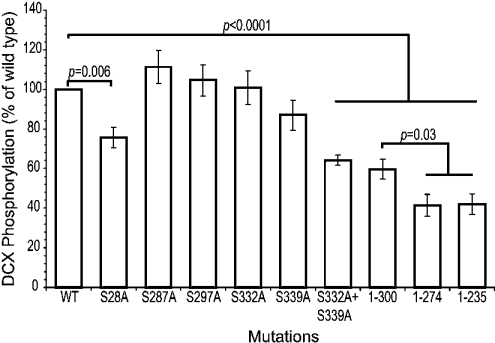
The significance of key in vitro cdk5 target residues in DCX was further assessed by mutating these sites to alanine by site-directed mutagenesis. Mutation of any of the putative cdk5 target threonines (Thr-289, Thr-326 or Thr-336) to alanine did not show significant changes in the level of phosphorylation (results not shown). This is consistent with the low ratio of phosphorylated threonine to serine in DCX (Table 1 and phosphoamino acid analysis). Key serine residues shown to be significantly phosphorylated using MS were also mutated and the proteins were phosphorylated in vitro for 5 min with cdk5 (Figure 4). The single mutations S287A (Ser-287-Ala), Ser-297-Ala or Ser-332-Ala did not significantly reduce phosphorylation. However, the single mutations of Ser-28-Ala or the double mutation of Ser-332-Ala plus Ser-339-Ala showed significant reduction in phosphorylation to 75% (P=0.006) and 64% (P<0.0001) of wt phosphorylation respectively (Figure 4). These results are highly consistent with the results in Table 1, except that a greater loss in phosphorylation of Ser-339-Ala was expected.
Figure 4. Comparative phosphorylation of wt and mutant DCX by cdk5.
GST-fusion protein (approx. 1 μg) of wt DCX, serine to alanine mutants and DCX truncations was phosphorylated by recombinant GST-cdk5/p25 in the presence of [γ-32 or 33P]ATP for 5 min at 37 °C and then analysed by SDS/PAGE and autoradiography. Phosphorylation was quantified by a STORM PhosphorImager. Results are represented as a percentage of wt DCX for the corrected activity (see the Methods section). Means±S.E.M. for six or seven replicated reactions per protein are presented.
Truncated forms of DCX were also assessed, since C-terminal truncations show a severe phenotype in patients (e.g. R303X and Q235X, where X is the introduction of a stop codon) [6]. All the truncated proteins showed significantly reduced phosphorylation by cdk5 (Figure 4), consistent with the loss of a number of putative cdk5 target sites. The DCX1–300 truncation loses three of the identified sites, whereas both the DCX1–274 and DCX1–236 truncations lose six identified sites. Comparison of the DCX1–300 with the Ser-332-Ala plus Ser-339-Ala double mutant showed no significant difference in the level of phosphorylation (Figure 4). This indicates that the loss of Ser-332 and Ser-339 is sufficient to account for the reduction in this truncation. Comparison between the truncations showed that there was a further significant reduction in phosphorylation when DCX was truncated further to either DCX1–274 or DCX1–236 (P=0.03), indicating that Ser-287, Thr-289 or Ser-297 contributed to phosphorylation in vitro. The results support the MS sequencing data and show that when all cdk5 phosphorylation sites in the C-terminal tail are removed, Ser-28 at the N-terminus is still accessible.
DISCUSSION
We used a well-developed MS strategy that was capable of detecting and identifying multiple phosphorylation sites for cdk5 in DCX. First, the strategy was tailored to overcome common problems associated with detection of phosphopeptides by MS, i.e. signal suppression and low efficiency of ionization. The phosphopeptides were separated into fractions so that the number and amount of interfering non-phosphorylated peptides were decreased. This reduces the suppression of ionization of phosphopeptides and increases their intensity. Also, phosphopeptides can be selected for MS/MS without unintentional selection of another peptide with similar m/z. The samples were highly concentrated first by simply reducing the volume of the fractions and secondly by using microcolumns before MS. This enables high-quality MS/MS spectra to be obtained, with an increased signal-to-noise ratio for y or b ions, reducing the ambiguity of assignment of phosphorylation sites. Secondly, non-radio-labelled-phosphorylated DCX was run simultaneously with radiolabelled DCX both to increase the amount of phosphopeptide available for MS and to reveal the relative abundance of each phosphopeptide by direct counting of radiation. Using this strategy, we have confirmed that DCX is a substrate for cdk5 in vitro and showed that DCX is phosphorylated on seven sites (of nine predicted sites) by cdk5 in vitro (Figure 1b, sites in bold), rather than on one as reported previously. Six of the sites are in the serine/proline-rich C-terminal tail region, whereas one site is near the N-terminus. Five of the seven sites are phospho-Ser, which correlates with phospho-amino acid analysis data.
The relative abundance of an ionized peptide in a mass spectrum is not a quantitative measure of the relative amount of that peptide in a sample, nor is the absence of an ion definitive evidence for the absence of that peptide in a sample. This is due to the effects of suppression and differing efficiencies of ionization and detection. To address this limitation, we measured the 32P incorporation into each fraction from the HPLC and sequenced the phosphopeptides in that fraction. Since only one unique phosphorylation site was found in most fractions, the relative level of phosphorylation of each site could be estimated.
Mutational analysis in an in vitro assay is sufficient to show whether a residue is a potential target for a particular kinase, but on the face value, not its importance. Tanaka et al. [28] showed that more than one residue was phosphorylated when these sites were individually mutated to alanine, co-transfected with cdk5/p35 into 293T cells and cell lysates examined for a shift in DCX mobility on gel electrophoresis. However, 293T cells have been shown to express endogenous JNK under stress conditions [46] and JNK is known to target some of the same residues as cdk5 [27]. JNK expression or the potential expression of other mitogen-activated protein kinases that are known to be proline-directed serine/threonine protein kinases and target DCX (T. C. Tan and P. J. Robinson, unpublished work), would give a similar result in this type of experiment. This group also mutated DCX, so that there was a single residue remaining as the target in an in vitro cdk5 phosphorylation assay. However, as the authors point out, mutation of a large number of residues in a single protein, potentially changes the conformational structure of that protein and therefore the efficiency of phosphorylation by a kinase; hence it cannot be the sole measure of the relative importance of a particular site. By measuring the change in signal intensity from 32P incorporation as a function of individual or double site-directed mutations (Figure 4) and comparing these with measured Cerenkov radiation from tryptic peptides of wt DCX (Table 1), we have established the relative importance of each of the potential cdk5 phosphorylation sites in vitro.
We provide evidence to suggest that Ser-339 at the C-terminus is the major in vitro phosphorylation site. The peptide containing this site had the highest level of Cerenkov radiation, and tandem MS/MS sequencing clearly identified a phospho-Ser-339 residue, whereas there was very low signal-to-noise ratio on other potential phospho-residues on that peptide (namely phospho-Ser-332) even though the sample was phosphorylated to maximum levels (90 min incubation in the presence of high ATP and Mg). Whereas the mutation of Ser-339-Ala alone did not show a significant reduction in phosphorylation, mutation of both Ser-332 and Ser-339 to alanine significantly lowered the level of phosphorylation by cdk5 compared with wt DCX. The apparent redundancy of Ser-339 strongly suggests a hierarchy of phosphorylation sites, where the next site in the hierarchy can be highly phosphorylated if a more favourable site is phosphorylated first, or in this case, mutated. The result may also be explained by conformational changes in DCX as a result of the mutation that may increase access of other cdk5 target residues, including Ser-28, resulting in a compensatory increase in phosphorylation of these other sites. Other sites found by MS were phosphorylated to a lesser extent (as determined by 32P incorporation) and, in the mutational analysis, did not significantly alter the level of DCX phosphorylation when substituted with alanine. Our results do not exclude an in vivo role for these sites.
It has been shown that phosphorylation of DCX with cdk5 or microtubule affinity regulator kinase/protein kinase A reduces its ability to polymerize microtubules in vitro, decreases its affinity for microtubules both in vitro and in vivo and affects neuronal migration [26,28]. Phosphorylation with JNK increases the velocity and reduces pause time for migration in primary cerebellar neurons [27]. JNK is reported to phosphorylate DCX in the tail region on Thr-321, Thr-331 and Ser-334 (which are in equivalent positions to Thr-326, Thr-336 and Ser-339 in the mouse sequence used in the present study) [27]. Phosphorylation of these sites was also demonstrated in cultured neurons, supporting the physiological relevance of these sites. Ser-339 was also a major site for cdk5 in this study. Therefore Ser-339 may be a target for at least two protein kinases, cdk5 and JNK. Of the two other nearby sites phosphorylated by JNK, Thr-326 is also a cdk5 substrate, whereas Thr-336 was not. This suggests that the integration of different signal-transduction pathways may occur in the regulation of DCX function. Phosphorylation of Ser-339 by JNK had clear effects on neurite outgrowth and velocity and pause time of migrating neurons [27]. Results from our study raise the possibility that cdk5 may produce similar functional effects via this site.
The identification of the Ser-28 site at the N-terminus as the second most prominent site of 32P incorporation was highly correlative between the MS data, the level of Cerenkov radiation from DCX phosphopeptides and mutational analysis. In a recent work by Tanaka et al. [28], mutation of eight out of nine putative cdk5 sites, while leaving Ser-28 as the sole target for cdk5, resulted in levels of phosphorylation of the mutant protein that were slightly reduced. This drop was not quantified, but matches our observations. It remains unknown whether this site may be phosphorylated in vivo. The in vitro phosphorylation sites identified by this study may also have implications for the regulation of DCX function in microtubule polymerization and stability. Co-assembly of DCX with taxol-stabilized microtubules is increased by 1–268 fragments of DCX compared with wt or 47–260 fragments [11]. In addition, the 1–171 fragments containing the R1 domain as well as the first 46 amino acids also showed evidence of weak co-assembly with microtubules [11]. This suggests that the 1–46 region preceding the microtubule-binding domains R1 and R2, where the putative phospho-Ser-28 resides, may regulate binding to microtubules.
An MAP with similar spatial and temporal expression compared with DCX is present in mammalian brain [47,48]. This MAP has similarity with DCX and Ca2+/calmodulin-dependent protein kinase and is known as doublecortin-like kinase. DCX-like kinase associates with microtubules in cultured neuronal cells at the growth cone and increases microtubule polymerization [48,49]. The DCX-like region has >80% amino acid sequence similarity [47], which includes a very high degree of sequence identity at sites shown to be major cdk5 target sites in DCX (results not shown). Of the seven sites identified in this study, five are conserved (Ser-28, Ser-297, Thr-326, Ser-332 and Ser-339) and two are not (Ser-287 and Thr-289). This raises the possibility that DCX-like kinase may also be a target for cdk5 phosphorylation.
The process of neuronal migration involves a number of steps. Post-mitotic neurons must leave the ventricular zone, migrate along radial fibres, pass previous waves of migrated neurons then find their position in the cortex [4]. The present study shows that cdk5 may act as the controller for a number of processes in neuronal migration. It has been established that cdk5 is involved in positioning of neurons with its involvement in the Reelin-signalling pathway. Cdk5- or p35-deficient mice has a reeler-like phenotype [20,21] and cdk5 has recently been shown to phosphorylate Dab1 in vivo, a down-stream target of Reelin signalling [18]. Cdk5/p39 phosphorylates tau, another MAP, during brain development, reducing its affinity for microtubules [50]. Cdk5 also phosphorylates NUDEL [7], which interacts with Lis1. A direct relationship between DCX and cdk5 is of major significance in understanding the neuronal migration and development of the brain. A significant reduction in the level of phosphorylation was evident when we phosphorylated DCX truncated at residue 300. The truncation mutation R303X, where a stop codon replaces the arginine codon at amino acid position 303, is found in a number of unrelated patients and results in a severe phenotype both in males (agyria) and females (thick heterotopic band and parchygyria), resulting in severe mental retardation and epilepsy [51,52]. The R303X mutation would result in the loss of the putative cdk5 phosphorylation sites Ser-332 and Ser-339.
Our results highlight the importance of a well-designed strategy for the identification of phosphorylation sites by MS. In particular, it reinforces the concept that failure to detect a phosphopeptide in a tryptic digest is an insufficient criterion for concluding that the remaining site is the major one. Additional results, such as comparison with 32P counts, are essential. We have shown that DCX is multiply phosphorylated by cdk5 in vitro and postulate that phosphorylation of these sites in vivo controls neuronal locomotion and potentially other processes. Phosphorylation of DCX affects its interaction with microtubules in vitro and displaces it from microtubules in cultured neurons [26]. Further work is required to determine if the novel sites identified in the present study affect microtubule polymerization and stability, as well as to establish their effect on neuronal migration.
Acknowledgments
We thank Professor P. Rowe for helpful discussion and comments on the manuscript and Dr M. Larsen for technical advice and helpful discussions. P.L.J. and P.J.R. are supported by the National Health and Medical Research Council of Australia.
References
- 1.Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M. C., Friocourt G., McDonnell N., Reiner O., Kahn A., et al. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- 2.Gleeson J. G., Lin P. T., Flanagan L. A., Walsh C. A. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 3.Horesh D., Sapir T., Francis F., Wolf S. G., Caspi M., Elbaum M., Chelly J., Reiner O. Doublecortin, a stabilizer of microtubules. Hum. Mol. Genet. 1999;8:1599–1610. doi: 10.1093/hmg/8.9.1599. [DOI] [PubMed] [Google Scholar]
- 4.Marin O., Rubenstein J. L. Cell migration in the forebrain. Annu. Rev. Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 5.des Portes V., Pinard J. M., Billuart P., Vinet M. C., Koulakoff A., Carrie A., Gelot A., Dupuis E., Motte J., Berwald-Netter Y., et al. A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell (Cambridge, Mass.) 1998;92:51–61. doi: 10.1016/s0092-8674(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 6.Gleeson J. G., Allen K. M., Fox J. W., Lamperti E. D., Berkovic S., Scheffer I., Cooper E. C., Dobyns W. B., Minnerath S. R., Ross M. E., et al. Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell (Cambridge, Mass.) 1998;92:63–72. doi: 10.1016/s0092-8674(00)80899-5. [DOI] [PubMed] [Google Scholar]
- 7.Niethammer M., Smith D. S., Ayala R., Peng J., Ko J., Lee M. S., Morabito M., Tsai L. H. NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 8.Smith D. S., Tsai L. H. Cdk5 behind the wheel: a role in trafficking and transport? Trends Cell Biol. 2002;12:28–36. doi: 10.1016/s0962-8924(01)02181-x. [DOI] [PubMed] [Google Scholar]
- 9.Toyo-Oka K., Shionoya A., Gambello M. J., Cardoso C., Leventer R., Ward H. L., Ayala R., Tsai L. H., Dobyns W., Ledbetter D., et al. 14-3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller–Dieker syndrome. Nat. Genet. 2003;34:274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 10.Sapir T., Horesh D., Caspi M., Atlas R., Burgess H. A., Wolf S. G., Francis F., Chelly J., Elbaum M., Pietrokovski S., et al. Doublecortin mutations cluster in evolutionarily conserved functional domains. Hum. Mol. Genet. 2000;9:703–712. doi: 10.1093/hmg/9.5.703. [DOI] [PubMed] [Google Scholar]
- 11.Taylor K. R., Holzer A. K., Bazan J. F., Walsh C. A., Gleeson J. G. Patient mutations in doublecortin define a repeated tubulin-binding domain. J. Biol. Chem. 2000;275:34442–34450. doi: 10.1074/jbc.M007078200. [DOI] [PubMed] [Google Scholar]
- 12.Sapir T., Elbaum M., Reiner O. Reduction of microtubule catastrophe events by LIS1, platelet-activating factor acetylhydrolase subunit. EMBO J. 1997;16:6977–6984. doi: 10.1093/emboj/16.23.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caspi M., Atlas R., Kantor A., Sapir T., Reiner O. Interaction between LIS1 and doublecortin, two lissencephaly gene products. Hum. Mol. Genet. 2000;9:2205–2213. doi: 10.1093/oxfordjournals.hmg.a018911. [DOI] [PubMed] [Google Scholar]
- 14.Rice D. S., Curran T. Role of the reelin signaling pathway in central nervous system development. Annu. Rev. Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- 15.Assadi A. H., Zhang G., Beffert U., McNeil R. S., Renfro A. L., Niu S., Quattrocchi C. C., Antalffy B. A., Sheldon M., Armstrong D. D., et al. Interaction of reelin signaling and Lis1 in brain development. Nat. Genet. 2003;35:270–276. doi: 10.1038/ng1257. [DOI] [PubMed] [Google Scholar]
- 16.Howell B. W., Hawkes R., Soriano P., Cooper J. A. Neuronal position in the developing brain is regulated by mouse disabled – 1. Nature (London) 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- 17.Howell B. W., Herrick T. M., Cooper J. A. Reelin-induced tryosine phosphorylation of disabled 1 during neuronal positioning. Genes Dev. 1999;13:643–648. doi: 10.1101/gad.13.6.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keshvara L., Magdaleno S., Benhayon D., Curran T. Cyclin-dependent kinase 5 phosphorylates disabled 1 independently of Reelin signaling. J. Neurosci. 2002;22:4869–4877. doi: 10.1523/JNEUROSCI.22-12-04869.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice D. S., Sheldon M., D'Arcangelo G., Nakajima K., Goldowitz D., Curran T. Disabled-1 acts downstream of Reelin in a signaling pathway that controls laminar organization in the mammalian brain. Development. 1998;125:3719–3729. doi: 10.1242/dev.125.18.3719. [DOI] [PubMed] [Google Scholar]
- 20.Ohshima T., Ward J. M., Huh C. G., Longenecker G., Veeranna, Pant H. C., Brady R. O., Martin L. J., Kulkarni A. B. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc. Natl. Acad. Sci. U.S.A. 1996;93:11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chae T., Kwon Y. T., Bronson R., Dikkes P., Li E., Tsai L. H. Mice lacking p35, a neuronal specific activator of Cdk5, display cortical lamination defects, seizures, and adult lethality. Neuron. 1997;18:29–42. doi: 10.1016/s0896-6273(01)80044-1. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y., Walsh C. A. Protein–protein interactions, cytoskeletal regulation and neuronal migration. Nat. Rev. Neurosci. 2001;2:408–416. doi: 10.1038/35077559. [DOI] [PubMed] [Google Scholar]
- 23.Becker A. J., Klein H., Baden T., Aigner L., Normann S., Elger C. E., Schramm J., Wiestler O. D., Blumcke I. Mutational and expression analysis of the reelin pathway components CDK5 and doublecortin in gangliogliomas. Acta Neuropathol. (Berl) 2002;104:403–408. doi: 10.1007/s00401-002-0570-4. [DOI] [PubMed] [Google Scholar]
- 24.Gupta A., Tsai L. H., Wynshaw-Boris A. Life is a journey: a genetic look at neocortical development. Nat. Rev. Genet. 2002;3:342–355. doi: 10.1038/nrg799. [DOI] [PubMed] [Google Scholar]
- 25.Walsh C. A., Goffinet A. M. Potential mechanisms of mutations that affect neuronal migration in man and mouse. Curr. Opin. Genet. Dev. 2000;10:270–274. doi: 10.1016/s0959-437x(00)00076-9. [DOI] [PubMed] [Google Scholar]
- 26.Schaar B. T., Kinoshita K., McConnell S. K. Doublecortin microtubule affinity is regulated by a balance of kinase and phosphatase activity at the leading edge of migrating neurons. Neuron. 2004;41:203–213. doi: 10.1016/s0896-6273(03)00843-2. [DOI] [PubMed] [Google Scholar]
- 27.Gdalyahu A., Ghosh I., Levy T., Sapir T., Sapoznik S., Fishler Y., Azoulai D., Reiner O. DCX, a new mediator of the JNK pathway. EMBO J. 2004;23:823–832. doi: 10.1038/sj.emboj.7600079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka T., Serneo F. F., Tseng H. C., Kulkarni A. B., Tsai L. H., Gleeson J. G. Cdk5 phosphorylation of doublecortin ser297 regulates its effect on neuronal migration. Neuron. 2004;41:215–227. doi: 10.1016/s0896-6273(03)00852-3. [DOI] [PubMed] [Google Scholar]
- 29.Craig A. G., Hoeger C. A., Miller C. L., Goedken T., Rivier J. E., Fischer W. H. Monitoring protein kinase and phosphatase reactions with matrix-assisted laser desorption/ionization mass spectrometry and capillary zone electrophoresis: comparison of the detection efficiency of peptide–phosphopeptide mixtures. Biol. Mass Spectrom. 1994;23:519–528. doi: 10.1002/bms.1200230810. [DOI] [PubMed] [Google Scholar]
- 30.Cech N. B., Enke C. G. Practical implications of some recent studies in electrospray ionization fundamentals. Mass Spectrom. Rev. 2001;20:362–387. doi: 10.1002/mas.10008. [DOI] [PubMed] [Google Scholar]
- 31.Nuhse T. S., Stensballe A., Jensen O. N., Peck S. C. Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteom. 2003;2:1234–1243. doi: 10.1074/mcp.T300006-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Arnott D., Gawinowicz M. A., Grant R. A., Neubert T. A., Packman L. C., Speicher K. D., Stone K., Turck C. W. ABRF-PRG03: phosphorylation site determination. J. Biomol. Tech. 2003;14:205–215. [PMC free article] [PubMed] [Google Scholar]
- 33.Wettenhall R. E., Aebersold R. H., Hood L. E. Solid-phase sequencing of 32P-labeled phosphopeptides at picomole and subpicomole levels. Methods Enzymol. 1991;201:186–199. doi: 10.1016/0076-6879(91)01017-v. [DOI] [PubMed] [Google Scholar]
- 34.Czupalla C., Nurnberg B., Krause E. Analysis of class I phosphoinositide 3-kinase autophosphorylation sites by mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:690–696. doi: 10.1002/rcm.967. [DOI] [PubMed] [Google Scholar]
- 35.Tan T. C., Valova V. A., Malladi C. S., Graham M. E., Berven L. A., Jupp O. J., Hansra G., McClure S. J., Sarcevic B., Boadle R. A., et al. Cdk5 is essential for synaptic vesicle endocytosis. Nat. Cell Biol. 2003;5:701–710. doi: 10.1038/ncb1020. [DOI] [PubMed] [Google Scholar]
- 36.Yasukawa T., Kanei-Ishii C., Maekawa T., Fujimoto J., Yamamoto T., Ishii S. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J. Biol. Chem. 1995;270:25328–25331. doi: 10.1074/jbc.270.43.25328. [DOI] [PubMed] [Google Scholar]
- 37.Fernandez-Patron C., Calero M., Collazo P. R., Garcia J. R., Madrazo J., Musacchio A., Soriano F., Estrada R., Frank R., Castellanos-Serra L. R., et al. Protein reverse staining: high-efficiency microanalysis of unmodified proteins detected on electrophoresis gels. Anal. Biochem. 1995;224:203–211. doi: 10.1006/abio.1995.1031. [DOI] [PubMed] [Google Scholar]
- 38.Gobom J., Nordhoff E., Mirgorodskaya E., Ekman R., Roepstorff P. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 1999;34:105–116. doi: 10.1002/(SICI)1096-9888(199902)34:2<105::AID-JMS768>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 39.Larsen M. R., Cordwell S. J., Roepstorff P. Graphite powder as an alternative or supplement to reversed-phase material for desalting and concentration of peptide mixtures prior to matrix-assisted laser desorption/ionization-mass spectrometry. Proteomics. 2002;2:1277–1287. doi: 10.1002/1615-9861(200209)2:9<1277::AID-PROT1277>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 40.Robinson P. J., Sontag J. M., Liu J. P., Fykse E. M., Slaughter C., McMahon H., Sudhof T. C. Dynamin GTPase regulated by protein kinase C phosphorylation in nerve terminals. Nature (London) 1993;365:163–166. doi: 10.1038/365163a0. [DOI] [PubMed] [Google Scholar]
- 41.Songyang Z., Lu K. P., Kwon Y. T., Tsai L. H., Filhol O., Cochet C., Brickey D. A., Soderling T. R., Bartleson C., Graves D. J., et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol. Cell. Biol. 1996;16:6486–6493. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright H. T. Nonenzymatic deamidation of asparaginyl and glutaminyl residues in proteins. Crit. Rev. Biochem. Mol. Biol. 1991;26:1–52. doi: 10.3109/10409239109081719. [DOI] [PubMed] [Google Scholar]
- 43.Lagerwerf F. M., van de Weert M., Heerma W., Haverkamp J. Identification of oxidized methionine in peptides. Rapid Commun. Mass Spectrom. 1996;10:1905–1910. doi: 10.1002/(SICI)1097-0231(199612)10:15<1905::AID-RCM755>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 44.Jiang X., Smith J. B., Abraham E. C. Identification of a MS–MS fragment diagnostic for methionine sulfoxide. J. Mass Spectrom. 1996;31:1309–1310. [Google Scholar]
- 45.Xie M., Vander Velde D., Morton M., Borchardt R. T., Schowen R. L. pH-induced change in the rate-determining step for the hydrolysis of the Asp/Asn-derived cyclic-imide intermediate in protein degradation. J. Am. Chem. Soc. 1996;118:8955–8956. [Google Scholar]
- 46.Camp H. S., Tafuri S. R., Leff T. c-Jun N-terminal kinase phosphorylates peroxisome proliferator-activated receptor-γ1 and negatively regulates its transcriptional activity. Endocrinology. 1999;140:392–397. doi: 10.1210/endo.140.1.6457. [DOI] [PubMed] [Google Scholar]
- 47.Burgess H. A., Martinez S., Reiner O. KIAA0369, doublecortin-like kinase, is expressed during brain development. J. Neurosci. Res. 1999;58:567–575. [PubMed] [Google Scholar]
- 48.Lin P. T., Gleeson J. G., Corbo J. C., Flanagan L., Walsh C. A. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J. Neurosci. 2000;20:9152–9161. doi: 10.1523/JNEUROSCI.20-24-09152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgess H. A., Reiner O. Doublecortin-like kinase is associated with microtubules in neuronal growth cones. Mol. Cell. Neurosci. 2000;16:529–541. doi: 10.1006/mcne.2000.0891. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi S., Saito T., Hisanaga S., Pant H. C., Kulkarni A. B. Tau phosphorylation by cyclin-dependent kinase 5/p39 during brain development reduces its affinity for microtubules. J. Biol. Chem. 2003;278:10506–10515. doi: 10.1074/jbc.M211964200. [DOI] [PubMed] [Google Scholar]
- 51.des Portes V., Francis F., Pinard J. M., Desguerre I., Moutard M. L., Snoeck I., Meiners L. C., Capron F., Cusmai R., Ricci S., et al. Doublecortin is the major gene causing X-linked subcortical laminar heterotopia (SCLH) Hum. Mol. Genet. 1998;7:1063–1070. doi: 10.1093/hmg/7.7.1063. [DOI] [PubMed] [Google Scholar]
- 52.Pilz D. T., Matsumoto N., Minnerath S., Mills P., Gleeson J. G., Allen K. M., Walsh C. A., Barkovich A. J., Dobyns W. B., Ledbetter D. H., et al. LIS1 and XLIS (DCX) mutations cause most classical lissencephaly, but different patterns of malformation. Hum. Mol. Genet. 1998;7:2029–2037. doi: 10.1093/hmg/7.13.2029. [DOI] [PubMed] [Google Scholar]