Abstract
IL-1F7b, a novel homologue of the IL-1 (interleukin 1) family, was discovered by computational cloning. We demonstrated that IL-1F7b shares critical amino acid residues with IL-18 and binds to the IL-18-binding protein enhancing its ability to inhibit IL-18-induced interferon-γ. We also showed that low levels of IL-1F7b are constitutively present intracellularly in human blood monocytes. In this study, we demonstrate that similar to IL-18, both mRNA and intracellular protein expression of IL-1F7b are up-regulated by LPS (lipopolysaccharide) in human monocytes. In stable transfectants of murine RAW264.7 macrophage cells, there was no IL-1F7b protein expression despite a highly active CMV promoter. We found that IL-1F7b-specific mRNA was rapidly degraded in transfected cells, via a 3′-UTR (untranslated region)-independent control of IL-1F7b transcript stability. After LPS stimulation, there was a rapid transient increase in IL-1F7b-specific mRNA and concomitant protein levels. Using sequence alignment, we found a conserved ten-nucleotide homology box within the open reading frame of IL-F7b, which is flanking the coding region instability elements of some selective genes. In-frame deletion of downstream exon 5 from the full-length IL-1F7b cDNA markedly increased the levels of IL-1F7b mRNA. A similar coding region element is located in IL-18. When transfected into RAW264.7 macrophages, IL-18 mRNA was also unstable unless treated with LPS. These results indicate that both IL-1F7b and IL-18 mRNA contain functional instability determinants within their coding region, which influence mRNA decay as a novel mechanism to regulate the expression of IL-1 family members.
Keywords: cytokine, gene regulation, interleukin 1, lipopolysaccharide, monocyte, macrophage
Abbreviations: CHO, Chinese-hamster ovary; IL-1, interleukin 1; IL-18BP, IL-18-binding protein; LPS, lipopolysaccharide; mAb, monoclonal antibody; ORF, open reading frame; PBMC, peripheral blood mononuclear cells; UTR, untranslated region
INTRODUCTION
The IL-1 (interleukin 1) family is an expanding family of cytokines sharing a similar all β-barrel structure consisting of 12 β-strands [1–3]. Six additional members of the IL-1 gene family have been discovered from the expressed sequence tag database searches expanding the total number to ten members [4–11]. The novel members derive from a common ancestor as does IL-1 and IL-18 [12,13]. Except for IL-18, which is found on chromosome 11, the IL-1 family members map to the same cluster on human chromosome 2 [5,12–14].
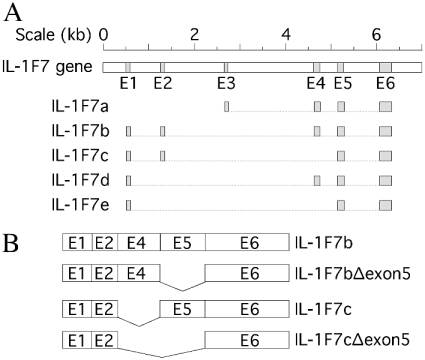
In a previous study, we reported that the IL-1 homologue IL-1F7b shares critical amino acid residues with IL-18 [15] and that IL-1F7b binds to the IL-18 receptor α chain [10]. Five different splice variants of IL-1F7 have been described (IL-1F7a–e) [5,7,10,11,13]. Isoform a has a unique N-terminus encoded by exon 3 of the IL-1F7 gene that is not expressed in the other isoforms [13]. The short isoforms IL-1F7c, IL-1F7d and IL-1F7e lack exon 4, 2 or both, respectively. None of the IL-1F7 variants expresses a typical signal peptide but the N-terminal sequence encoded by exon 1 contains a prodomain that may be processed by caspase-1 [16]. Despite extensive database searches and sequencing of the IL-1-gene locus, no murine homologue of IL-1F7 has yet been found.
The low affinity binding of IL-1F7b to the IL-18Rα was demonstrated using surface plasmon resonance [16] or a receptor pull-down assay [10]; however, despite binding to the IL-18Rα, no direct IL-18-agonistic or -antagonistic effect was observed. We found that IL-1F7b binds to the IL-18BP (IL-18 binding protein) [15], the natural inhibitor of IL-18 activity [17], and, unexpectedly, we observed that IL-1F7b enhanced the ability of IL-18BP to inhibit IL-18-induced interferon-γ. Accordingly, it was proposed that IL-1F7b functions as the receptor antagonist for IL-18 by first binding to the IL-18BP followed by binding to the IL-18Rβ chain, thus depriving this chain from participating in IL-18 signal transduction. IL-1F7b has also been proposed to possess Th1 anti-tumour properties but the underlying mechanism was not unveiled [18].
Transcripts of IL-1F7b were detected by real-time PCR in several tissues and were most abundant in testis, thymus and uterus [10]. Up-regulation of transcripts by PMA was shown for human PBMC (peripheral blood mononuclear cells) and dendritic cells [10]. IL-1F7b protein is expressed in human monocytes [15], tonsil plasma cells as well as primary breast carcinoma cells [16].
The gene expression of IL-1 or IL-18 is regulated at different levels (reviewed in [19,20]): (i) regulation of promoter activity; (ii) dissociation of transcription from translation through degradation of mRNA; and (iii) posttranslational regulation through processing of the inactive precursor to an active cytokine via limited proteolysis. In the present study, we used peripheral human monocytes to analyse the regulation of IL-1F7b by LPS (lipopolysaccharide) at the transcriptional and protein level in comparison with IL-18. Using stable transfectants, we obtained evidence that there was a 3′-UTR (untranslated region)-independent control of IL-1F7b transcript stability. Sequence alignment showed that IL-1F7b contains an A-rich homology box, which flanks instability elements of the functional coding region in a variety of mRNAs. Therefore, we evaluated whether coding region determinants can regulate the turnover of both IL-1F7b and IL-18 mRNAs representing a novel mechanism for gene regulation of IL-1 family members.
MATERIALS AND METHODS
Reagents, cells and antibodies
All reagents were purchased from Sigma Chemical Co. (St. Louis, MO, U.S.A.) unless indicated otherwise. RAW264.7, a murine macrophage cell line, COS7, a simian virus 40-transformed monkey kidney carcinoma cell line, and CHO (Chinese-hamster ovary) cells were purchased from A.T.C.C. (Rockville, MD, U.S.A.). The monoclonal antibodies against human IL-18 (MAB318) and against His6-tagged proteins (MAB050) were purchased from R&D Systems (Minneapolis, MN, U.S.A.). For the generation of polyclonal antibodies against IL-1F7b, a rabbit was immunized with recombinant IL-1F7b produced in Escherichia coli as described in [15]. Restriction enzymes, primers, DNA ligase, DNaseI and reverse transcriptase were purchased from Invitrogen (Carlsbad, CA, U.S.A.). Biolase DNA polymerase was obtained from Bioline USA (Randolph, MA, U.S.A.).
Generation of monoclonal antibodies against IL-1F7b
Recombinant IL-1F7b was produced using pMAL™ protein expression and purification system (New England Biolabs, Beverly, MA, U.S.A.) as described in [15]. After affinity chromatography with amylose-coupled resin (New England Biolabs), the maltose-binding protein–IL-1F7b fusion protein was cleaved with Factor Xa (New England Biolabs) for 2 h. The mixture of cleaved and non-cleaved proteins was separated using a Superdex 75HR 10/30 gel filtration column connected to an ÄKTA-FPLC apparatus (Amersham Biosciences, Piscataway, NJ, U.S.A.). Peak proteins corresponding to IL-1F7b were pooled and used to immunize 6-week-old female BALB/c mice. After five injections, the mouse with the highest serum titre was injected with a boost of 25 μg of IL-1F7b in PBS intraperitonally 1 week before fusion. Recombinant IL-1F7b with a His6 tag (IL-1F7b-his6) expressed in E. coli using pPROEX HTa (Invitrogen) [15] was used to screen clones. Positive hybridomas were subcloned and expanded for antibody production using stirred tank fermentation (Bioexpress, West Lebanon, NH, U.S.A.). IgG was purified from the cell culture supernatant using Protein A–Sepharose according to standard procedures and dialysed against PBS.
Isolation of monocytes
These studies were approved by the Combined Colorado Investigational Review Board and each subject gave informed consent. PBMC were purified from heparinized blood of healthy donors [21]. The monocytes were isolated from PBMC using MACS® monocyte isolation kit (Miltenyi Biotec, Auburn, CA, U.S.A.) following the manufacturers’ recommendation and remained on ice until stimulation with LPS.
Western blot
PAGE was performed using standard 10% SDS gels or 4–15% Tris/HCl gradient gels (Bio-Rad Laboratories, Hercules, CA, U.S.A.) and separated proteins were blotted on to nitrocellulose (Hybond™ ECL™, Amersham Biosciences, Piscataway, NJ, U.S.A.). Non-specific binding sites were blocked with 5% (w/v) non-fat milk in PBS/0.05% Tween 20. For detection of His6-tagged IL-1F7b in the lysate of transfected cells, an antibody raised against His6-tagged proteins was used at a concentration of 1 μg/ml. For the detection of IL-1F7b in human monocytes, a mAb (monoclonal antibody)) against IL-1F7b (clone 222) was used at 5 μg/ml. Western blots were developed with enhanced chemiluminescence (SuperSignal®, Pierce, Rockford, IL, U.S.A.).
ELISA specific for IL-1F7b-His6
Maxisorp® 96-well microtitre plates (Nalge Nunc International, Rochester, NY, U.S.A.) were coated with anti-His6-tag mAb at 1 μg/ml in carbonate buffer (pH 9.6) overnight. After blocking non-specific sites with 1% BSA in PBS/0.05% Tween 20 for 1 h, the lysate of RAW264.7/IL-1F7b-His6 transfectants was applied together with recombinant E. coli-produced IL-1F7b-His6 as a standard. Bound proteins were detected using a rabbit anti-IL-1F7b serum at a dilution of 1:500 in PBS/Tween/1%BSA and peroxidase-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Bar Harbor, ME, U.S.A.).
Cloning of IL-1F7b and IL-1F7 isoforms c
The IL-1F7b cDNA was cloned from a human spleen library as described in [15]. IL-1F7c was generated by a two-step PCR [22] using IL-1F7b cDNA as a template. The following oligonucleotide primers were used: primer 1 (sense, 5′-ATATCTCGAGCCACCATGTCCTTTGTGGGGGAG) corresponds to nucleotides 1–18 of IL-1F7b cDNA, extended at its 5′-end with the Kozak sequence [23] for optimal expression and a XhoI site; primer 2 (antisense, 5′-GGCTAATGCAAAGAAGATCTTTGTGTGAACAAAATTCATGGCG) contains nucleotides 165–123 and primer 3 (sense, 5′-CGCCATGAATTTTGTTCACACAAAGATCTTCTTTGCATTAGCC) contains 123–165 of IL-1F7c ORF (open reading frame); primer 4 (antisense, 5′-TATAGCGGCCGCCTAATCGCTGACCTCACTG) corresponds to 658–645 of human IL-1H4 ORF plus a NotI site. Products of a PCR with primers 1 and 2 and IL-1F7b as template revealed the 5′-half of IL-1F7c cDNA. Another PCR with primers 3 and 4 revealed the 3′-region of the IL-1F7c cDNA. Both fragments were purified by electrophoresis on 1% agarose gel and eluted using a gel extraction system (Promega, Madison, WI, U.S.A.). The two cDNA segments were mixed at a 1:1 ratio and added as templates to the second PCR reaction using primers 1 and 4. The resulting IL-1F7c cDNA was sequenced and cloned into pTarget expression plasmid (Promega) using XhoI and NotI restriction sites.
Mutagenesis of IL-1F7b and IL-1F7c
Mutants of IL-1F7b or IL-1F7c lacking exon 5 (see Figure 6B) were generated by the two-step PCR technique described above. The following mutagenic primers were used: primer 5 (sense, 5′-GATAAAAACTACATACGCCCAGAGGAGAAACTGATGAAGCTGG) contains nucleotides 244–265 and 410–430 of human IL-1F7b ORF and primer 6 (antisense, 5′-CCAGCTTCATCAGTTTCTCCTCTGGGCGTATGTAGTTTTTATC) which corresponds to nucleotides 430–410 and 265–244 of human IL-1F7b ORF; primer 7 (sense, 5′-GCCATGAATTTTGTTCACACAAAGGAGAAACTGATGAAGCTGG) contains nucleotides 124–145 and 290–310 of human IL-1F7c ORF and primer 8 (antisense, 5′-CCAGCTTCATCAGTTTCTCCTTTGTGTGAACAAAATTCATGGC), which corresponds to nucleotides 310–290 and 145–124 of human IL-1F7c. All constructs were sequenced and ligated into pTarget using XhoI and NotI restriction sites.
Figure 6. A homology region within sequences containing instability elements is conserved in IL-1F7b.
A region flanking the instability site in human plasminogen activator inhibitor type 2 mRNA [25] is also present in human IL-1 family members as IL-1F7b, IL-1β IL-18 and IL-1Ra. Alignment was generated by using LALIGN-program (www.ch.embnet.org) to compare two DNA sequences for local similarity.
Generation of stable RAW264.7 transfectants
The murine macrophage cell line RAW264.7 was maintained in RPMI 1640 containing 10% (v/v) heat-inactivated foetal calf serum (Invitrogen), 2 mM glutamine, 100 μg/ml streptomycin and 100 units/ml penicillin (Cellgro Mediatech, Herndon, VA, U.S.A.) in a humidified atmosphere at 37 °C with 5% CO2. RAW264.7 cells were transfected with different forms of IL-1F7b or IL-1F7c in pTarget using calcium phosphate [24]. The CMV promoter of pTarget is constitutively active and allows high expression of the inserted gene. Transfected cells were selected in a medium supplemented with 200 μg/ml neomycin. Resistant cells were subcloned and the cell lysate was tested for IL-1F7b expression using Western blotting, stained with rabbit IL-1F7b antiserum or RT (reverse transcriptase)–PCR for IL-1F7c. RAW264.7 cells were transfected with IL-1F7b or IL-1F7c mutants lacking exon 5 or IL-18 in pTarget using LIPOFECTAMINE™ (Invitrogen). Neomycin-resistant clones (>200) were pooled and tested for transgene expression using RT–PCR. COS7 cells and CHO cells were transfected with IL-1F7b in pTarget using DEAE-dextran [24].
mRNA stabilization experiments, RNA isolation and quantification
For mRNA stabilization experiments, 2.5×105 transfected RAW264.7 cells/well were seeded in six-well plates overnight before stimulation with 10 ng/ml of LPS (E. coli 055:B5). Alternatively, human PBMC (5×106 cells) were stimulated with 10 μg/ml of LPS. Cells were harvested after the indicated period of time and washed in 0.9% NaCl. Total RNA was purified using TRI reagent (Sigma) according to the manufacturer's recommendation. Aliquots of 2 μg of total RNA were digested with RNase-free DNaseI to remove any remaining plasmid DNA after transfection. Then 1 μg of DNaseI-treated RNA was reverse-transcribed using Superscript-RT (Invitrogen) in a total volume of 20 μl. For the subsequent PCR, 2 μl of reverse-transcription product was added to a final volume of 25 μl. The following pair of internal primers was used to detect transfected IL-1F7: primer 9 (sense, 5′-GGGAGAACTCAGGAGTGAAAAT) and primer 10 (antisense, 5′-TCCTTTCTCCGCAGAGGCTGA) used for PBMC; primer 11 (antisense, 5′-GGGAGAACTCAGGAGTGAAAAT) used for RAW264.7/IL-1F7 transfectants. Primer 12 (sense, 5′-CAGTAGAAGACAATTGCATCAA) and primer 13 (antisense, 5′-GTGAACATTATAGATCTATCCC) were used to amplify human IL-18 transfected into RAW264.7 cells. PCR reaction consisted of 35 or 27 cycles for PBMC or RAW264.7 transfectants respectively, followed by a final extension phase at 72 °C for 10 min. The number of cycles was established in separate experiments to obtain semi-quantitative results. RAW264.7 cells transfected with an antisense construct of murine ribosomal protein S3 in pTarget (S3rev) were used as a control for transcription. PCR using GAPDH-specific primers was performed as an internal control for each RNA sample. An equal volume of each reaction was applied to a 1% agarose gel containing ethidium bromide and visualized under UV light. Signals were analysed by densitometry using BioRad Quantity One® Software package.
Immunohistochemistry and confocal microscopy
Freshly isolated human monocytes or RAW264.7/IL-1F7b transfectants were washed in PBS and resuspended in freshly prepared 4% paraformaldehyde in PBS. After fixation for 15 min at room temperature (20 °C), the cells were spread on charged glass slides (Superfrost®/Plus; Fisher Scientific, Pittsburgh, PA, U.S.A.). Co-staining was performed using a monoclonal antibody against human IL-18 (1 μg/ml) or a preparation of rabbit anti-IL-1F7b IgG (2 μg/ml) in PBS containing 1% BSA. The corresponding concentration of non-immune mouse or rabbit IgG was used as a negative control. The background signal of the individual negative control was subtracted from the signal obtained by the specific antibody. A goat anti-mouse antibody, conjugated to Cy3 (dilution 1:300; Jackson ImmunoResearch Laboratories, West Grove, PA, U.S.A.) and a goat anti-rabbit antibody conjugated to Alexa488 (dilution 1:100; Molecular Probes, Eugene, OR, U.S.A.), were used for detection. Nuclei were stained blue using 1 μg/100 ml bisbenzimide (Sigma). Digital confocal imaging was performed using a Leica DM RXA microscope equipped with SlideBook Software for MacIntosh (Intelligent Imaging Innovations, Denver, CO, U.S.A.).
Statistical analysis
Results are expressed as means±S.E.M. Differences between treated and non-treated groups were compared by ANOVA and Bonferroni–Dunn post hoc tests. Statistical analyses were performed with the statistical package Statview™ 512+ (Brain-Power, Calabasas, CA, U.S.A.).
RESULTS
IL-1F7b is up-regulated in human monocytes by LPS
We have shown recently that IL-1F7b is expressed constitutively in human monocytes [15]. In the present study, we investigated whether the IL-1F7b protein expression is up-regulated by treatment with LPS as shown for IL-1β, IL-1Ra and IL-18. IL-1F7b shares critical amino acid sequence homology with IL-18 and binds to IL-18Rα [10,16]. IL-1F7b also binds to IL-18BP [15]. We therefore compared the expression of IL-1F7b and IL-18 in human monocytes using co-staining of the two cytokines. As observed in our previous study, low expression of IL-1F7b was readily detected in resting monocytes before stimulation with LPS (Figure 1A, upper panel). After stimulation for 4 h with LPS, both IL-1F7b and IL-18 were markedly increased intracellularly (Figure 1A, lower panel). Up-regulation of IL-1F7b was 2–3-fold when assessed by analysing the fluorescence signal of at least 70 individual cells (Figure 1B). The staining for IL-1F7b was similar to that for IL-18, revealing an intracellular granular-like pattern surrounding the nuclear membrane and the inner surface of the plasma membrane.
Figure 1. IL-1F7b is up-regulated in human monocytes by LPS and co-localizes with IL-18.
(A) Freshly isolated human monocytes were stimulated or not with LPS (10 μg/ml) for 4 h. After co-staining against IL-1F7b and Il-18, the cells were visualized using confocal digital microscopy. Red dye, anti-IL-1F7b; green dye, anti-IL-18; blue dye, nuclear stain. (B) Fluorescence was recorded for single cells and the mean counts of intensity (±S.E.M.) for IL-1F7b or IL-18 were calculated by analysing at least 70 individual cells. Statistical differences were calculated by ANOVA; ***P<0.0001.
Co-localization of IL-1F7b with IL-18
On the basis of similar intracellular distributions, we determined whether IL-1F7b and IL-18 are expressed in the same compartment of the cell. Digital cross-sections of 0.5 μm were analysed for pixel-based co-localization of IL-1F7b and IL-18. We observed that 60–80% of IL-1F7b appeared to co-localize with IL-18 positive staining in monocytes. The percentage of co-localization was similar before and after stimulation with LPS (results not shown).
IL-1F7b is up-regulated by LPS in stable transfectants
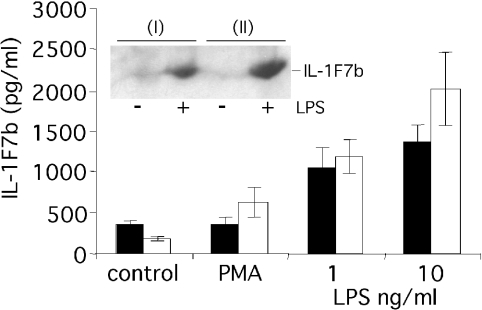
Despite a constitutively active CMV promoter, murine RAW264.7 cells stably transfected with IL-1F7b cDNA in pTarget only expressed minor levels of IL-1F7b protein intracellularly as assessed by ELISA, Western blot (Figure 2) or immunohistochemistry (results not shown). No IL-1F7b was detected in the cell supernatant. Transient transfection of COS7 or CHO cells with IL-1F7b in pTarget also did not result in detectable levels of soluble or intracellular IL-1F7b despite strong expression of a control gene (murine S3 protein) (results not shown). However, IL-1F7b was strongly expressed in RAW264.7 transfectants after stimulation with LPS at a low concentration of 1 ng/ml. Compared with LPS, stimulation with PMA was less efficient to induce IL-1F7b expression (Figure 2).
Figure 2. IL-1F7b protein expression in stably transfected murine RAW264.7 macrophages is up-regulated after stimulation.
Two different clones of RAW264.7 cells (0.5×106 cells) stably transfected with IL-1F7b-His6 were stimulated with LPS or PMA (10 ng/ml) for 12 h. Mean levels of IL-1F7b-His6 (±S.E.M.) in the lysate of transfected cells of three independent experiments were detected using the ELISA specific for IL-1F7b-His6 or by Western-blot analysis (10% SDS/PAGE) using an anti-His6-tag-specific antibody (inset).
LPS induces stabilization of IL-1F7b-specific mRNA in transfected RAW264.7 cells
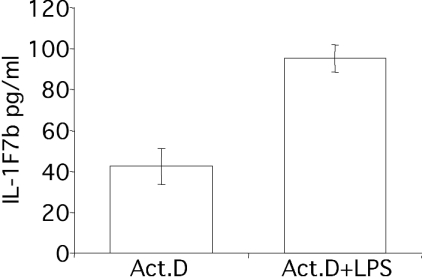
Enhanced transcription was unlikely to explain the increased expression of IL-1F7b after LPS treatment, since a control gene in pTarget was constitutively expressed in a variety of transfected cells. In addition, LPS also induced IL-1F7b protein expression in transfected RAW264.7 cells in the presence of actinomycin D, where stimulation of transcription was excluded (Figure 3). Therefore, we hypothesized that in transfected RAW264.7 cells, increased levels of IL-1F7b protein after LPS treatment are quite probably due to mRNA stabilization. RAW264.7/IL-1F7b transfectants were stimulated with LPS, total RNA was isolated and reverse-transcribed and semi-quantitative PCR analysis was performed to evaluate steady-state levels of IL-1F7b-specific mRNA. As shown in Figure 4(A), expression of IL-1F7b-specific mRNA was markedly induced by LPS compared with near absence before stimulation. Up-regulation of IL-1F7b mRNA was rapid and readily observed after 20 min. Maximum levels of IL-1F7b-specific mRNA were detected 60 min after LPS treatment. As a control, the cDNA for murine ribosomal protein S3 in antisense orientation (S3rev) in pTarget was used to generate stable transfectants of RAW264.7 cells. As shown in Figure 4(B), S3rev-specific mRNA was constitutively expressed in transfected RAW264.7 cells and not increased by LPS, reconfirming that the activity of the CMV promoter in pTarget does not depend on LPS treatment. These results therefore indicate that IL-1F7b-specific mRNA is rapidly degraded in non-stimulated cells but stabilized after LPS treatment.
Figure 3. LPS stimulates IL-1F7b protein in transfected RAW264.7/IL-1F7b cells in the presence of actinomycin D.
RAW264.7 cells (0.5×106 cells) stably transfected with IL-1F7b-His6 were pretreated with actinomycin D (1 μg/μl) for 1 h and subsequently stimulated with LPS (10 ng/ml) for 12 h. Mean levels of IL-1F7b-His6 (±S.E.M.) in the lysate of transfected cells of three independent experiments were detected using the ELISA specific for IL-1F7b-His6.
Figure 4. LPS induces stabilization of IL-1F7b mRNA in transfected RAW264.7/IL-1F7b cells.
Subclones of RAW264.7 cells stably transfected with IL-1F7b (A) or a control gene (S3rev) (B) were stimulated with LPS (10 ng/ml). Total RNA was isolated after the indicated time and reverse-transcribed as described. Semi-quantitative PCR was performed and the product was applied to a 1% agarose gel. Results from a single experiment out of at least three representative experiments are shown.
IL-1F7b mRNA in human PBMC is induced by LPS
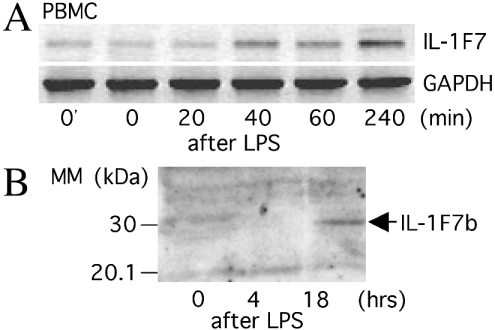
Next, we determined whether LPS treatment also up-regulates IL-1F7b-specific mRNA in primary human cells. Similar to RAW264.7/IL-1F7b transfectants, human PBMC were stimulated with LPS and total RNA was isolated at several time points. First-strand cDNA was synthesized and analysed by semi-quantitative PCR. The number of PCR cycles was adjusted to obtain semi-quantitative results. As demonstrated in RAW264.7/IL-1F7b transfectants, IL-1F7-specific mRNA was induced in PBMC within 20–40 min after LPS treatment (Figure 5A). Amplification of the indicated band and subsequent sequencing proved the amplification of IL-1F7b-specific mRNA. IL-1F7b mRNA levels increased for more than 240 min until the end of the incubation period. IL-1F7b protein within the lysate of human monocytes was detected 18 h after LPS stimulation on a Western blot (Figure 5B). The molecular size of naturally expressed IL-1F7b in human monocytes is 30 kDa and corresponds to the size of the recombinant protein expressed in RAW264.7 cells (Figure 2, inset).
Figure 5. Up-regulation of IL-1F7b mRNA and protein in human PBMC or monocytes after LPS treatment.
(A) Human PBMC (5×106 cells/sample) were stimulated with LPS (10 μg/ml). Total RNA was isolated after the indicated time, reverse-transcribed and analysed as described in Figure 3. 0′indicates RNA isolated from PBMC lysed immediately after separation. (B) Isolated human monocytes (5×106 cells/sample) were stimulated with LPS (10 μg/ml) in the presence of 1% autologous serum. After the indicated time, cells were washed and lysed in PBS/1% Triton X-100. The cell lysate was separated on a 4–15% gradient SDS/PAGE and proteins were blotted on nitrocellulose. The blot was stained using a monoclonal antibody against IL-1F7b. Results from a single experiment out of at least three representative experiments are shown.
A known mRNA instability sequence is conserved in IL-1F7b
A significant degree of homology has been described for a 10-nucleotide A-rich coding region, which was found flanking the known instability determinants of transcripts in human plasminogen activator inhibitor type 2, yeast MATα1, c-Myc proto-onkogen, human urokinase-type plasminogen activator receptor and vascular endothelial growth factor [25]. This region partially overlaps with known or putative binding sites for proteins associated with mRNA instability. We found a similar homology box within the coding sequence of IL-1F7b, IL-18, IL-1β and IL-1Ra (Figure 6). The homology box in IL-1F7b is located at the 3′-end of exon 4 (see Figure 6A).
Deletion of exon 5 stabilizes IL-1F7b mRNA
The deletion of sequences within the coding region containing instability elements was shown to increase the steady-state half-life of the corresponding mRNA [26–28]. Since the homology box in IL-1F7b is found at the 3′-end of exon 4, adjacent instability elements were quite probably located in exon 4 or exon 5. To localize the instability element within the coding region of IL-1F7b mRNA, we cloned both an isoform of IL-1F7b lacking exon 4 (IL-1F7c) as well as mutants of IL-1F7b and IL-1F7c lacking exon 5 (Figure 7B). Each plasmid was transfected into RAW264.7 cells and stable transfectants were isolated. Steady-state levels of mRNA before and after LPS treatment were analysed by semi-quantitative PCR. As shown in Figure 8(A), basal mRNA levels for both IL-1F7b and isoform IL-1F7c were rapidly degraded in RAW264.7 transfectants unless they were stimulated with LPS. The densitometric analysis illustrated in Figure 8(C) shows higher basal levels for IL-1F7c mRNA compared with IL-1F7b mRNA, but a more rapid increase after stimulation with LPS. In contrast, the transcripts of deletion mutants of IL-1F7b and IL-1F7c lacking exon 5 were constitutively expressed at significantly higher levels under the same experimental conditions (Figures 8B and 8C). In fact, the decay of mRNA in IL-1F7b and IL-1F7c deletion mutants was significantly reduced after LPS treatment during the incubation period of 4 h. These results demonstrate that the predominant instability element of IL-1F7b mRNA is located within exon 5.
Figure 7. Exon–intron structure of the IL-1F7b gene.
(A) The exon–intron structure of the IL-1F7 gene was created using the genomic BAC clone RP11-67L14 from chromosome 2 (GenBank® accession no. AC079753) applying the pairwise BLAST algorithm with IL-1F7b cDNA (GenBank® accession no. AF200496). The structure shown is in accordance with recently published results [12,13]. (B) Mutants of IL-1F7b and IL-1F7c isoforms lacking exon 5 were generated as illustrated.
Figure 8. Exon 5 confers instability to IL-1F7 mRNA.
RAW264.7 cells were stably transfected with wild-type or mutants of IL-1F7b and IL-1F7c lacking exon 5. Three different subclones transfected with IL-1F7b or IL-1F7c (A) or the bulk culture of cells transfected with IL-1F7b or IL-1F7c lacking exon 5 (B) were stimulated with LPS (10 ng/ml) for the indicated periods of time. Semi-quantitative PCR was performed on isolated RNA and the products were applied to a 1% agarose gel. The corresponding bands were analysed by densitometry and are expressed as percentage of maximum signal. (A, B) One representative gel for each experiment is shown. (C) The densitometric analysis of three to four independent experiments for each construct is shown (mean±S.E.M.).
LPS induces stabilization of IL-18 mRNA and protein expression in transfected RAW264.7 cells
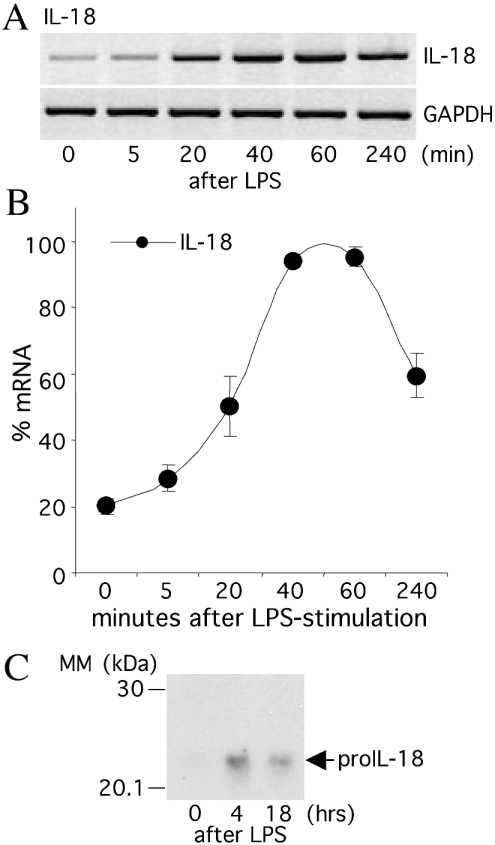
Steady-state IL-18 mRNA is readily expressed in human unfractionated blood and PBMC before stimulation, but further induced by LPS [21]. Using sequence alignment similar to IL-1F7b, the ten-nucleotide homology box flanking known instability determinants was observed at two different sites within the mRNA of IL-18 (Figure 6). Therefore, we speculated whether mRNA stabilization via coding region determinants also contributes to the regulation of IL-18. RAW264.7 cells were transfected with human IL-18 cDNA in pTarget and the bulk culture of stable transfectants was assessed for mRNA expression. Similar to IL-1F7b and IL-1F7c, basal levels of IL-18 mRNA were relatively low but markedly increased by LPS (Figures 9A and 9B). IL-18 mRNA levels rapidly increased and reached maximal levels after 1 h. A significant decrease was seen after an additional 3 h. The increase in mRNA expression is concomitant with an increase of intracellular IL-18 protein expression (Figure 9C). There is less intracellular IL-18 after 18 h incubation, suggesting secretion of IL-18. Since the activity of the CMV promoter in pTarget does not critically depend on co-stimulatory signals as LPS, these results indicated that functional instability elements are also present in the coding region of IL-18.
Figure 9. LPS induces stabilization of IL-18 mRNA and protein expression in transfected RAW264.7 cells.
RAW264.7 cells stably transfected with human IL-18 cDNA were stimulated with LPS (10 ng/ml). IL-18 mRNA and intracellular protein levels were analysed at the indicated times. (A) Semi-quantitative PCR of one representative experiment. (B) Densitometric analysis of six independent experiments (means±S.E.M.). (C) The lysate of transfected RAW264.7/IL-18 cells before and after treatment with LPS (10 ng/ml) was separated by SDS/PAGE (10% gel) and blotted on to nitrocellulose. The blot was stained using a mAb against human IL-18.
DISCUSSION
We have recently reported that the IL-1 homologue IL-1F7b is constitutively present intracellularly at low levels in blood monocytes from healthy human donors [15]. Low levels of steady-state mRNA for IL-1F7 were also detected in resting human PBMC [10] or in monocytes [11]. In the present study, we show that both mRNA and protein expression of IL-1F7b are up-regulated by LPS. Unexpectedly, murine RAW264.7 macrophage cells stably transfected with IL-1F7b cDNA under the control of a constitutively active CMV promoter were almost negative for IL-1F7b protein expression. We were able to demonstrate that IL-1F7b-specific mRNA is degraded in these transfectants and rapidly stabilized after LPS treatment resulting in an increase in IL-1F7b protein. A similar mechanism of regulation was observed in RAW cells stably transfected with human IL-18.
IL-1F7b and IL-18 share critical amino acids [15]. The significance of this homology is underscored by the observation that IL-1F7b binds to the IL-18Rα [10,16] and also to the IL-18BP [15], the natural inhibitor of IL-18. Since IL-1F7b is expressed at low levels in resting monocytes [15], we tested whether there is up-regulation during inflammation and compared the expression of IL-1F7b and IL-18 in LPS-treated and untreated monocytes using co-staining of each cytokine. Intracellular levels of both proteins were markedly increased after LPS stimulation within 4 h as assessed by digital laser microscopy. Moreover, 60–80% of IL-1F7b appears to be expressed in the same cellular compartment as IL-18. In fact, IL-18 accumulates in secretory lysosomes and is eventually secreted after stimulation [29]. Therefore, we speculate that IL-1F7b might be secreted via the same pathway as IL-18. Although this mechanism still needs to be established, evidence has been published that IL-1F7b is indeed secreted from adenovirus-transfected cells [16,18].
There was no apparent explanation why RAW264.7 cells stably transfected with IL-1F7b in the vector pTarget rarely contained significant levels of IL-1F7b protein unless stimulated with LPS. The CMV promoter of pTarget is constitutively active and does not depend on additional stimuli as shown for a control gene. Moreover, LPS also induced IL-1F7b protein expression in stable transfectants treated with actinomycin D when transcription is not contributing. Therefore, stimulation of transcription is unlikely to explain the increase of IL-1F7b mRNA and protein and we hypothesized that LPS treatment induced stabilization of IL-1F7b-specific mRNA. Indeed, we observed that steady-state levels of IL-1F7b-specific mRNA are rapidly increased in transfected RAW264.7 cells after LPS treatment. Since only the ORF was inserted into the pTarget for transfection, sequences within the 3′- or 5′-UTR of the IL-1F7b gene did not contribute to mRNA stability. Consequently, coding region determinants will probably act in a cis-dominant fashion to destabilize IL-1F7b-specific mRNA. These coding-region-instability elements are highly efficient in inducing degradation of IL-1F7b-specific mRNA. In fact, virtually no IL-1F7b transcripts were detected in transfected RAW264.7 cells before LPS treatment. Accordingly, significant IL-1F7b protein levels were only detected after stimulation. This observation was not restricted to RAW264.7 transfectants, since COS7 or CHO cells transiently transfected with IL-1F7b in pTarget also did not express detectable levels of IL-1F7b protein despite strong expression of a control gene.
In mammalian cells, the half-life of a particular mRNA can change severalfold without change in transcription [30]. Stability of mRNA is an important mechanism to control cytokine production. For example, regulatory motifs for IL-1β gene expression were found in the AU-rich 3′-UTR, which account for LPS-mediated mRNA stabilization (reviewed in [19]). Functional mRNA instability elements within the coding region have been shown to be involved in the control of mRNA turnover of an increasing number of transcripts such as human plasminogen activator inhibitor type 2 [25], yeast MATα1 [31], c-Myc proto-oncogene [27], human urokinase-type plasminogen activator receptor [28] and vascular endothelial growth factor [32]. Coding-region determinants were also shown to be involved in the regulation of IL-2 [33] as well as IL-11 mRNA [34]. Earlier to the present study, there was no evidence for the existence of functional mRNA stability determinants within the coding sequence of cytokines of the IL-1 family.
Tierney and Medcalf [25] described an A-rich homology box of ten nucleotides, which flanks known or suspected coding-region-instability elements in various genes suggesting that a common motif might play a broad role in the control of mRNA turnover. Notably, this homology box need not necessarily overlap the binding sites for proteins to prevent mRNA decay and its functional relevance is speculative [25]. We found a similar degree of homology in the coding region of members of the IL-1 family, e.g. IL-1β, IL-18, IL-1Ra as well as IL-1F7b. In IL-1F7b, this homology box is located at the 3′-end of exon 4. The discovery of the ten-nucleotide homology sequence initiated the present study of deletion mutants of IL-1F7b lacking exon 4 and/or exon 5 in RAW264.7 cells for mRNA stability. IL-1F7c is an isoform lacking exon 4. We were able to show that exon 5 contains a critical instability region in IL-1F7b, since significantly higher steady-state mRNA levels of IL-1F7b or IL-1F7c mutants lacking exon 5 were observed in RAW264.7 transfectants before stimulation with LPS. More importantly, the decay of specific mRNA after transient stabilization by LPS treatment was significantly reduced in the mutants of IL-1F7b or IL-1F7c lacking exon 5. A slightly higher basal level of steady-state mRNA was observed in IL-1F7c lacking exon 4, suggesting a less significant role of sequences within exon 4 to mediate IL-1F7 mRNA instability. The identification of the precise coding-region-instability sequence is the subject of ongoing studies. This work will also include the destabilization of otherwise stable mRNAs by sequences of IL-1F7b.
In human PBMC, a different kinetic pattern of IL-1F7b mRNA expression was observed after LPS treatment. IL-1F7b-specific mRNA levels increased during the incubation period of 4 h after LPS treatment, whereas IL-1F7b mRNA in RAW264.7 transfectants consistently showed a maximum level after 40–60 min. This may represent the difference between primary cells and transfected cell lines. Trans-acting mechanisms like endogenous IL-1F7 promoter activity or sequences within the AU-rich 3′-UTR for mRNA stability, as shown for IL-1β, probably participate in regulating IL-1F7b mRNA and protein expression in vivo. In addition to protein levels assessed by immunohistochemistry and mRNA expression, we found up-regulation of naturally expressed IL-1F7b in the lysates of human PBMC after stimulation with LPS by Western blotting. The molecular size of naturally expressed IL-1F7b is similar to the size of the recombinant IL-1F7b expressed in eukaryotic cells.
Gene expression of IL-18 appears to be different compared with other cytokines. It is regulated by at least two different promoters, one constitutively active and one inducible by LPS. In addition, unlike IL-1β, the human IL-18 mRNA does not contain AU-rich destabilizing elements within the 3′-UTR, implying that IL-18 mRNA may have a longer half-life [35]. Our previous studies have shown that IL-18 mRNA is constitutively expressed at significant levels in resting human PBMC or whole blood but increased by treatment with LPS [21]. As discussed above, the ORF of IL-18 also contains the ten-nucleotide A-rich homology box flanking known instability determinants at two different sites. In fact, using the same experimental design as applied for IL-1F7b, we found that IL-18 mRNA expressed under the control of a constitutively active CMV promoter in stable RAW264.7 transfectants was also rapidly degraded. After stimulation with LPS, markedly increased levels of IL-18 mRNA and protein were observed. These observations indicate that coding region instability elements are also involved in IL-18 mRNA turnover showing an additional mechanism for IL-18 gene regulation.
The expression of pro-inflammatory cytokines is a tightly controlled event to keep balance between the beneficial host defence functions and adverse effects. For example, an imbalance of cytokine effects of the host as described for IL-1 or TNFα in rheumatoid arthritis or inflammatory bowel disease, may be due to a failure to down-regulate the production of these cytokines. In conclusion, the rapid degradation of mRNA for IL-1F7b and IL-18 through instability elements within the coding region is a novel mechanism to regulate the expression of IL-1 family members in addition to the involvement of the 3′- and 5′-UTR, which controls mRNA stability of other cytokines.
Acknowledgments
This study was supported by NIH grants AI-15614 and HL-68743 (to C.A.D.) and the Deutsche Forschungsgemeinschaft BU-1222/2-1 (to P.B.).
References
- 1.Priestle J. P., Schar H. P., Grutter M. G. Crystallographic refinement of interleukin 1 β at 2.0 Å resolution. Proc. Natl. Acad. Sci. U.S.A. 1989;86:9667–9671. doi: 10.1073/pnas.86.24.9667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vigers G. P., Anderson L. J., Caffes P., Brandhuber B. J. Crystal structure of the type-I interleukin-1 receptor complexed with interleukin-1β. Nature (London) 1997;386:190–194. doi: 10.1038/386190a0. [DOI] [PubMed] [Google Scholar]
- 3.Schreuder H., Tardif C., Trump-Kallmeyer S., Soffientini A., Sarubbi E., Akeson A., Bowlin T., Yanofsky S., Barrett R. W. A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist. Nature (London) 1997;386:194–200. doi: 10.1038/386194a0. [DOI] [PubMed] [Google Scholar]
- 4.Barton J. L., Herbst R., Bosisio D., Higgins L., Nicklin M. J. A tissue specific IL-1 receptor antagonist homolog from the IL-1 cluster lacks IL-1, IL-1ra, IL-18 and IL-18 antagonist activities. Eur. J. Immunol. 2000;30:3299–3308. doi: 10.1002/1521-4141(200011)30:11<3299::AID-IMMU3299>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 5.Busfield S. J., Comrack C. A., Yu G., Chickering T. W., Smutko J. S., Zhou H. Leiby K. R., Holmgren L. M., Gearing D. P., Pan Y. Identification and gene organization of three novel members of the IL-1 family on human chromosome 2. Genomics. 2000;66:213–216. doi: 10.1006/geno.2000.6184. [DOI] [PubMed] [Google Scholar]
- 6.Debets R., Timans J. C., Homey B., Zurawski S., Sana T. R., Lo S., Wagner J., Edwards G., Clifford T., Menon S., et al. Two novel IL-1 family members, IL-1 δ and IL-1 ε, function as an antagonist and agonist of NF-κ B activation through the orphan IL-1 receptor-related protein 2. J. Immunol. 2001;167:1440–1446. doi: 10.4049/jimmunol.167.3.1440. [DOI] [PubMed] [Google Scholar]
- 7.Kumar S., McDonnell P. C., Lehr R., Tierney L., Tzimas M. N., Griswold D. E. Capper E. A., Tal-Singer R., Wells G. I., Doyle M. L., et al. Identification and initial characterization of four novel members of the interleukin-1 family. J. Biol. Chem. 2000;275:10308–10314. doi: 10.1074/jbc.275.14.10308. [DOI] [PubMed] [Google Scholar]
- 8.Lin H., Ho A. S., Haley-Vicente D., Zhang J., Bernal-Fussell J., Pace A. M. Hansen D., Schweighofer K., Mize N. K., Ford J. E. Cloning and characterization of IL-1HY2, a novel interleukin-1 family member. J. Biol. Chem. 2001;276:20597–20602. doi: 10.1074/jbc.M010095200. [DOI] [PubMed] [Google Scholar]
- 9.Mulero J. J., Pace A. M., Nelken S. T., Loeb D. B., Correa T. R., Drmanac R., Ford J. E. IL1HY1: a novel interleukin-1 receptor antagonist gene. Biochem. Biophys. Res. Commun. 1999;263:702–706. doi: 10.1006/bbrc.1999.1440. [DOI] [PubMed] [Google Scholar]
- 10.Pan G., Risser P., Mao W., Baldwin D. T., Zhong A. W., Filvaroff E., Yansura D. Lewis L., Eigenbrot C., Henzel W. J., et al. IL-1H, an interleukin 1-related protein that binds IL-18 receptor/IL-1Rrp. Cytokine. 2001;13:1–7. doi: 10.1006/cyto.2000.0799. [DOI] [PubMed] [Google Scholar]
- 11.Smith D. E., Renshaw B. R., Ketchem R. R., Kubin M., Garka K. E., Sims J. E. Four new members expand the interleukin-1 superfamily. J. Biol. Chem. 2000;275:1169–1175. doi: 10.1074/jbc.275.2.1169. [DOI] [PubMed] [Google Scholar]
- 12.Nicklin M. J., Barton J. L., Nguyen M., FitzGerald M. G., Duff G. W., Kornman K. A sequence-based map of the nine genes of the human interleukin-1 cluster. Genomics. 2002;79:718–725. doi: 10.1006/geno.2002.6751. [DOI] [PubMed] [Google Scholar]
- 13.Taylor S. L., Renshaw B. R., Garka K. E., Smith D. E., Sims J. E. Genomic organization of the interleukin-1 locus. Genomics. 2002;79:726–733. doi: 10.1006/geno.2002.6752. [DOI] [PubMed] [Google Scholar]
- 14.Mulero J. J., Nelken S. T., Ford J. E. Organization of the human interleukin-1 receptor antagonist gene IL1HY1. Immunogenetics. 2000;51:425–428. doi: 10.1007/s002510050640. [DOI] [PubMed] [Google Scholar]
- 15.Bufler P., Azam T., Gamboni-Robertson F., Reznikov L. L., Kumar S., Dinarello C. A., Kim S. H. A complex of the IL-1 homologue IL-1F7b and IL-18-binding protein reduces IL-18 activity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13723–13728. doi: 10.1073/pnas.212519099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S., Hanning C. R., Brigham-Burke M. R., Rieman D. J., Lehr R., Khandekar S., Kirkpatrick R. B., Scott G. F., Lee J. C., Lynch F. J., et al. Interleukin-1F7B (IL-1H4/IL-1F7) is processed by caspase-1 and mature IL-1F7B binds to the IL-18 receptor but does not induce IFN-γ production. Cytokine. 2002;18:61–71. doi: 10.1006/cyto.2002.0873. [DOI] [PubMed] [Google Scholar]
- 17.Novick D., Kim S. H., Fantuzzi G., Reznikov L. L., Dinarello C. A., Rubinstein M. Interleukin-18 binding protein: a novel modulator of the Th1 cytokine response. Immunity. 1999;10:127–136. doi: 10.1016/s1074-7613(00)80013-8. [DOI] [PubMed] [Google Scholar]
- 18.Gao W., Kumar S., Lotze M. T., Hanning C., Robbins P. D., Gambotto A. Innate immunity mediated by the cytokine IL-1 homologue 4 (IL-1H4/IL-1F7) induces IL-12-dependent adaptive and profound antitumor immunity. J. Immunol. 2003;170:107–113. doi: 10.4049/jimmunol.170.1.107. [DOI] [PubMed] [Google Scholar]
- 19.Dinarello C. A. Biologic basis for interleukin-1 in disease. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 20.Nakanishi K., Yoshimoto T., Tsutsui H., Okamura H. Interleukin-18 regulates both Th1 and Th2 responses. Annu. Rev. Immunol. 2001;19:423–474. doi: 10.1146/annurev.immunol.19.1.423. [DOI] [PubMed] [Google Scholar]
- 21.Puren A. J., Fantuzzi G., Gu Y., Su M. S., Dinarello C. A. Interleukin-18 (IFNγ-inducing factor) induces IL-8 and IL-1β via TNFα production from non-CD14+human blood mononuclear cells. J. Clin. Invest. 1998;101:711–721. doi: 10.1172/JCI1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S. H., Azam T., Novick D., Yoon D. Y., Reznikov L. L., Bufler P., Rubinstein M., Dinarello C. A. Identification of amino acid residues critical for biological activity in human interleukin-18. J. Biol. Chem. 2002;277:10998–11003. doi: 10.1074/jbc.M108311200. [DOI] [PubMed] [Google Scholar]
- 23.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell (Cambridge, Mass.) 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 24.Sambruck J., Fritsch E. F., Maniatis T. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 25.Tierney M. J., Medcalf R. L. Plasminogen activator inhibitor type 2 contains mRNA instability elements within exon 4 of the coding region. Sequence homology to coding region instability determinants in other mRNAs. J. Biol. Chem. 2001;276:13675–13684. doi: 10.1074/jbc.M010627200. [DOI] [PubMed] [Google Scholar]
- 26.Parker R., Jacobson A. Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MAT α 1 gene are involved in promoting rapid mRNA decay in yeast. Proc. Natl. Acad. Sci. U.S.A. 1990;87:2780–2784. doi: 10.1073/pnas.87.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeilding N. M., Lee W. M. Coding elements in exons 2 and 3 target c-myc mRNA downregulation during myogenic differentiation. Mol. Cell. Biol. 1997;17:2698–2707. doi: 10.1128/mcb.17.5.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shetty S., Kumar A., Idell S. Posttranscriptional regulation of urokinase receptor mRNA: identification of a novel urokinase receptor mRNA binding protein in human mesothelioma cells. Mol. Cell. Biol. 1997;17:1075–1083. doi: 10.1128/mcb.17.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardella S., Andrei C., Poggi A., Zocchi M. R., Rubartelli A. Control of interleukin-18 secretion by dendritic cells: role of calcium influxes. FEBS Lett. 2000;481:245–248. doi: 10.1016/s0014-5793(00)02015-9. [DOI] [PubMed] [Google Scholar]
- 30.Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hennigan A. N., Jacobson A. Functional mapping of the translation-dependent instability element of yeast MATα1 mRNA. Mol. Cell. Biol. 1996;16:3833–3843. doi: 10.1128/mcb.16.7.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dibbens J. A., Miller D. L., Damert A., Risau W., Vadas M. A., Goodall G. J. Hypoxic regulation of vascular endothelial growth factor mRNA stability requires the co-operation of multiple RNA elements. Mol. Biol. Cell. 1999;10:907–919. doi: 10.1091/mbc.10.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragheb J. A., Deen M., Schwartz R. H. CD28-Mediated regulation of mRNA stability requires sequences within the coding region of the IL-2 mRNA. J. Immunol. 1999;163:120–129. [PubMed] [Google Scholar]
- 34.Yang L., Steussy C. N., Fuhrer D. K., Hamilton J., Yang Y. C. Interleukin-11 mRNA stabilization in phorbol ester-stimulated primate bone marrow stromal cells. Mol. Cell. Biol. 1996;16:3300–3307. doi: 10.1128/mcb.16.7.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tone M., Thompson S. A., Tone Y., Fairchild P. J., Waldmann H. Regulation of IL-18 (IFN-γ-inducing factor) gene expression. J. Immunol. 1997;159:6156–6163. [PubMed] [Google Scholar]