Abstract
Human LAMAN (lysosomal α-mannosidase) was synthesized as a 120 kDa precursor in transfected COS cells [African-green-monkey kidney cells], which was partly secreted as a single-chain form and partly sorted to the lysosomes being subsequently cleaved into three peptides of 70, 40 and 15 kDa respectively. Both the secreted and the lysosomal forms contained endo H (endoglucosidase H)-resistant glycans, suggesting a common pathway through the trans-Golgi network. A fraction of LAMAN was retained intracellularly as a single-chain endo H-sensitive form, probably in the ER (endoplasmic reticulum). The inherited lack of LAMAN causes the autosomal recessive storage disease α-mannosidosis. To understand the biochemical consequences of the disease-causing mutations, 11 missense mutations and two in-frame deletions were introduced into human LAMAN cDNA by in vitro mutagenesis and the resulting proteins were expressed in COS cells. Some selected mutants were also expressed in Chinese-hamster ovary cells. T355P (Thr355→Pro), P356R, W714R, R750W and L809P LAMANs as well as both deletion mutants were misfolded and arrested in the ER as inactive single-chain forms. Six of the mutants were transported to the lysosomes, either with less than 5% of normal specific activity (H72L, D196E/N and R220H LAMANs) or with more than 30% of normal specific activity (E402K LAMAN). F320L LAMAN resulted in much lower activity in Chinese-hamster ovary cells when compared with COS cells. Modelling into the three-dimensional structure revealed that the mutants with highly reduced specific activities contained substitutions of amino acids involved in the catalysis, either co-ordinating Zn2+ (His72 and Asp196), stabilizing the active-site nucleophile (Arg220) or positioning the active-site residue Asp319 (Phe320).
Keywords: inherited disease, intracellular transport, lysosomal α-mannosidase, lysosome, α-mannosidosis
Abbreviations: CHO cell, Chinese-hamster ovary cell; COS cell, African-green-monkey kidney cell; DMEM, Dulbecco's modified Eagle's medium; endo H, endoglucosidase H; ER, endoplasmic reticulum; GH38, glycosylhydrolase family 38; LAMAN, lysosomal α-mannosidase; LAMP-1, lysosome-associated membrane protein 1; PDI, protein disulphide-isomerase; PNGase F, peptide N-glycosidase F
INTRODUCTION
Lysosomal α-mannosidase (LAMAN; EC 3.2.1.24) belongs to GH38 (glycosylhydrolase family 38) [1]. It is an acidic exoglycosidase that cleaves α-1,2, α-1,3 and α-1,6 mannosidic linkages [2] during the ordered degradation of N-linked oligosaccharides [3]. The three-dimensional structures of two GH38 enzymes, Golgi α-mannosidase II from Drosophila melanogaster [4] and LAMAN from bovine kidney [5], are known. These enzymes have a distorted seven-stranded α/β barrel active-site region, and the base of the active site is well preserved, including a Zn2+-binding site and the reaction nucleophile [6] following strand 4. The substrate-binding residues have been predicted based on comparison with related enzymes [5] and inhibitor binding [4].
The human enzyme is synthesized as a polypeptide of 1011 amino acids that is post-translationally modified by N-glycosylation, disulphide bridge formation, proteolysis, zinc binding and homodimer formation [7]. We refer to the resulting major proteolytic peptides as peptides a–e starting from the N-terminus [7,8]. The enzyme is probably transported to the lysosomes via the mannose 6-phosphate-dependent pathway, since the extracellular uptake of recombinant human LAMAN produced in CHO cells (Chinese-hamster ovary cells) is inhibited by mannose 6-phosphate [9]. The gene is located on chromosome 19q13.1 [10] and consists of 24 exons spanning 21.5 kb [11].
Deficiency of LAMAN causes the autosomal recessive disorder α-mannosidosis. Affected cells accumulate mannose-containing oligosaccharides in the lysosomes, resulting in the formation of large vacuoles containing storage material. Typical clinical symptoms are mental retardation, skeletal deformities, hearing loss and immunodeficiency [12], with varying degrees of severity. The variation in the clinical expression may be partly due to environmental factors and partly due to genetic factors other than the LAMAN gene, since complete lack of enzymic activity is found in patients with different clinical manifestations [13]. This is consistent with the observation of similar patterns and levels of oligomannoses in the urine of mildly and severely affected patients [14] and the fact that affected siblings can exhibit different clinical presentations [15]. α-Mannosidosis has also been found, and the respective mutations are reported, in cat [16], cow [8] and guinea-pig [17]. A mouse knockout model has also been established [18]. In affected cows and cats, the clinical symptoms are generally severe central nervous system disorder and premature death, whereas guinea-pigs and mice express a mild form of the disease.
We reported previously a spectrum of disease-causing mutations in the LAMAN gene. These include eight splicing, nine missense and three nonsense mutations, as well as two small insertions and three deletions in humans [7,13] and two missense mutations in cattle [8]. Only three human mutants, H72L (His72→Leu), P356R and R750W, have been confirmed as disease-causing based on activity measurements after transfection of mutant constructs into COS cells [African-green-monkey (Cercopithecus aethiops sabaeus) kidney cells] [19], but the consequences to intracellular transport were not investigated. In the present study, we have characterized the intracellular transport of human LAMAN in transfected COS cells and studied the intracellular transport of 11 α-mannosidosis-related LAMAN mutants, nine with amino acid substitutions and two with small deletions.
MATERIALS AND METHODS
Site-directed mutagenesis and DNA preparations
The human LAMAN cDNA inserted into the expression vector pcDNA 3.1 [9] was used as the template for site-directed mutagenesis using the QuikChange™ Site-Directed Mutagenesis kit (Stratagene) and primers as described in Table 1. The presence of mutations was verified by DNA sequencing. The complete LAMAN-coding sequences were sequenced for each mutant to exclude the introduction of other mutations caused by the PCR [ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit or Thermo Sequenase Terminator Cycle Sequencing kit (Amersham Biosciences)]. Plasmids were purified using the NucleoBond® Plasmid Purification kit (Clontech Laboratories) or the Concert Purification System (Life Technologies; Invitrogen).
Table 1. Primers used for the construction of mutants.
The substituted nucleotides are indicated in boldface and dashes indicate the positions at which deletions were introduced to mimic the effect of the splice site mutations [2].
| Primer | Direction | Sequence |
|---|---|---|
| H72L | Forward | 5′-GCCTCACACACTTGATGACGTG-3′ |
| Reverse | 5′-CACGTCATCAAGTGTGTGAGGC-3′ | |
| D196E | Forward | 5′-GTGGCCTGGCACATTGAGCCCTTCGGCCAC-3′ |
| Reverse | 5′-GTGGCCGAAGGGCTCAATGTGCCAGGCCAC-3′ | |
| D196N | Forward | 5′-GTGGCCTGGCACATTAACCCCTTCGGCCAC-3′ |
| Reverse | 5′-GTGGCCGAAGGGGTTAATGTGCCAGGCCAC-3′ | |
| R220H | Forward | 5′-GACGGCTTCTTCTTTGGGCACCTTGATTATCAAG-3′ |
| Reverse | 5′-CTTGATAATCAAGGTGCCCAAAGAAGAAGCCGTC-3′ | |
| Del(256–260) (IVS5−1G-C) | Forward | 5′-GACCTCTTCAC_TGGTTACAACCCGCC-3′ |
| Reverse | 5′-GGCGGGTTGTAACCA_GTGAAGAGGTC-3′ | |
| F320L | Forward | 5′-CATGGGCTCGGACCTCCAATATGAGAATGC-3′ |
| Reverse | 5′-GCATTCTCATATTGGAGGTCCGAGCCCATG-3′ | |
| Del(339–342) (IVS7+2T-G) | Forward | 5′-CTCATCCGGCT_GCAGGCAAAAGGAAG-3′ |
| Reverse | 5′-CTTCCTTTTGCCTGC_AGCCGGATGAG-3′ | |
| T355P | Forward | 5′-GTTCTCTACTCCCCCCCCGCTTGTTAC-3′ |
| Reverse | 5′-GTAACAAGCGGGGGGGGAGTAGAGAAC-3′ | |
| P356R | Forward | 5′-CTCTACTCCACCCGCGCTTGTTACCTC-3′ |
| Reverse | 5′-GAGGTAACAAGCGCGGGTGGAGTAGAG-3′ | |
| E402K | Forward | 5′-CCCTCAAACGCTACAAGCGCCTCAGC-3′ |
| Reverse | 5′-GCTGAGGCGCTTGTAGCGTTTGAGGG-3′ | |
| W714R | Forward | 5′-CCTGGAGCTAGAGCGGTCGGTGGGG-3′ |
| Reverse | 5′-CCCCACCGACCGCTCTAGCTCCAGG-3′ | |
| R750W | Forward | 5′-GACAGCAATGGCTGGGAGATCCTGGAG-3′ |
| Reverse | 5′-CTCCAGGATCTCCCAGCCATTGCTGTC-3′ | |
| L809P | Forward | 5′-GAGATGGCTCGCCGGAGCTCATGGTGC-3′ |
| Reverse | 5′-GCACCATGAGCTCCGGCGAGCCATCTC-3′ |
Cell culturing, transient transfection and metabolic labelling
COS-7 cells (CRL-1651; A.T.C.C., Manassas, VA, U.S.A.) were cultured in DMEM (Dulbecco's modified Eagle's medium), supplemented with 10% (v/v) foetal calf serum and antibiotics. For transfection, 350–400×103 cells/well were seeded in 3 cm culture plates (Nunc Brand Products; Nalge Nunc International, Rochester, NY, U.S.A.) 1 day before transfection. The next day, cells were transfected with 2.0 μg of the respective DNA constructs using LIPOFECTAMINE™ 2000 (Life Technologies). As negative control, cells were transfected with pcDNA 3.1 lacking the LAMAN cDNA insert (mock construct). The medium was removed 2 days post-transfection and replaced by DMEM without methionine/cysteine (BioWhittaker, Wokingham, Berks., U.K.). After incubation for 60 min, the medium was replaced by 200 μl of the same medium and added to 100–123 μCi/ml 35S-labelled methionine/cysteine (Amersham Biosciences). After 30 min pulse labelling, the cells were harvested or washed twice with complete DMEM and chased for different time periods. CHO K1-cells (CCL-61; A.T.C.C.) were transfected as described above, except that, on the day after transfection, they were transferred to a T-25 culture flask containing 0.4 μg/ml G418 (Sigma). After approx. 10 days, colonies containing stably transformed cells were transferred to T-75 culture flasks.
Cell harvest and activity analysis
Cells from 3 cm plates were harvested by trypsinization and lysed in PBS containing 1% Triton X-100 (ICN Biomedicals, Aurora, OH, U.S.A.), 0.5% deoxycholic acid (Sigma) and a protease inhibitor mixture (Sigma) consisting of 2 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 mM EDTA and 100 μg/ml PMSF. Enzymic activity was measured in the cell lysate and medium by incubation with 4 mM p-nitrophenyl α-D-mannopyranoside (Sigma) in 0.1 M acetic acid (pH 4.5) at 37 °C. The reaction was stopped by the addition of an equal volume of 13 mM glycine, 67 mM NaCl and 83 mM Na2CO3 (pH 10.7). The absorbance was recorded at 405 nm; 1 unit of enzymic activity was defined as the amount of enzyme that released 1 μmol of p-nitrophenol/min. The activity was correlated with the total amount of protein in the sample, which was measured by the DC protein assay kit (Bio-Rad Laboratories) according to the supplier's instructions.
Immunoprecipitation and SDS/PAGE
Immunoprecipitation of LAMAN from cell lysate and medium was performed using 1.5 μl of antiserum raised against native human placenta LAMAN [15] in the presence of 0.05% BSA and PBS. After overnight incubation, 1 mg of the cellular fraction of Staphylococcus aureus (Sigma) was used to precipitate the immunocomplexes. Immunoprecipitated LAMAN was either incubated with p-nitrophenyl α-D-mannopyranoside to measure α-mannosidase activity or analysed by SDS/PAGE (8–25% gel) under reducing conditions using the Phast system (Amersham Biosciences), followed by autoradiography. The 14C-labelled molecular-mass standards for autoradiography were obtained from Amersham Biosciences.
Deglycosylation
Immunoprecipitated LAMAN was added to 5 μl of 0.5% SDS and incubated at 100 °C for 3 min. Then, 3 μl of 6% Nonidet P40 and 5 μl of PBS were added, followed by the addition of either 2 μl of 5 units/ml endo H (endoglucosidase H; EC 3.2.1.96; Roche Biochemicals, Basel, Switzerland) or 2 μl of 1 mg/ml PNGase F [peptide N-glycosidase F or peptide-N4(N-acetyl-β-glucosaminyl) asparagine amidase F; EC 3.5.1.52; Hampton Research, Laguna Niguel, CA, U.S.A.] and incubated overnight at 37 °C.
Western blotting
Cell lysate or medium, prepared as described above, was subjected to SDS/PAGE using an 8–25% gradient and electroblotted on to a PVDF membrane (Immobilon™-P; Millipore) according to the manufacturer's instructions (PhastSystem; Amersham Biosciences). The membrane was incubated in 1:3000-diluted antiserum against native recombinant human LAMAN (gift from HemeBiotech, Hillerød, Denmark) overnight at 4 °C in PBS containing 3% (w/v) BSA (Sigma). Alkaline phosphatase-conjugated goat anti-rabbit (IgG) serum (1:3000 dilution) was used as a secondary antibody (Bio-Rad Laboratories). Immunocomplexes were visualized by chemiluminescence using the CDP-STAR substrate (Roche Biochemicals).
Immunofluorescence
COS-1 cells (A.T.C.C.) were cultured as described above: 2×105 cells/well were plated on poly-D-lysine (Sigma)-pretreated coverslips in six-well culture plates (Nunc) 1 day before transfections. Transfections were performed using 1.5 μg of plasmid DNA and LIPOFECTAMINE™ (Life Technologies) according to the manufacturer's instructions. Protein synthesis was arrested 36–42 h post-transfection with serum-free DMEM containing 50 μg/ml cycloheximide (Sigma) for 2 h at 37 °C, and the cells were washed with PBS and fixed in ice-cold methanol for 6 min at −20 °C. Unspecific staining was blocked by incubation for 30 min at room temperature (22 °C) in blocking buffer [0.5% BSA/0.2% saponin (Sigma) in PBS]. Coverslips were incubated for 45 min at room temperature in primary antibody diluted in blocking buffer. The following antibodies were used: rabbit anti-native human LAMAN [15] (1:1000 dilution), rabbit anti-bovine LAMAN peptide abc [10] (1:500 dilution), mouse anti-PDI (where PDI stands for protein disulphide-isomerase, EC 5.3.4.1) (1:50 dilution; StressGen, Victoria, BC, Canada), mouse anti-LAMP-1 (where LAMP-1 stands for lysosome-associated membrane protein 1) (1:50 dilution; antibody H4A3, developed by Thomas August, John Hopkins University, Baltimore, MD, U.S.A.). Mouse anti-LAMP-1, under the auspices of the National Institute of Child Health and Human Development (NICHD), was obtained from Developmental Studies Hybridoma Bank and maintained by the Department of Biological Sciences at the University of Iowa (Iowa City, IA, U.S.A.). Cells were washed with blocking buffer and incubated with 40 μl of secondary antibody [FITC- and TRITC (tetramethylrhodamine β-isothiocyanate)-conjugated mouse anti-rabbit and rat anti-mouse antibodies (Sigma) diluted to 1:200] in blocking buffer for 45 min at room temperature. After washing with PBS, the coverslips were mounted in glycerol and analysed by a DMR immunofluorescence microscope (Leica Microscope and Scientific Instruments Group, Solms, Germany), using the Quips fluorescence in situ hybridization image capture system (Applied Imaging, Santa Clara, CA, U.S.A.). The experiments were repeated at least twice.
Structural analysis
The missense mutations were modelled into the structure of bovine LAMAN (PDB code 1o7d) [5]. The Figures were obtained with Molscript and Raster3D [20,21] and edited with Photoshop.
RESULTS AND DISCUSSION
Intracellular transport of wild-type LAMAN in transfected COS cells
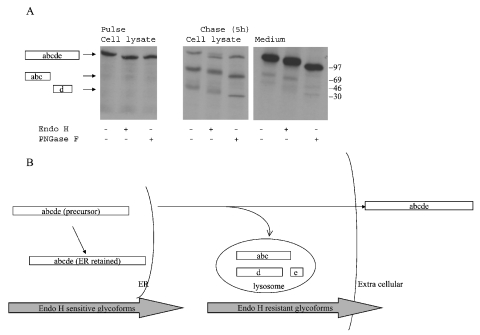
Wild-type LAMAN was synthesized as a 120 kDa single-chain precursor as judged from autoradiography after 30 min of pulse labelling (Figure 1A, left panel). This precursor contained only endo H-sensitive N-glycans (Figure 1A, left panel), indicating that LAMAN had not yet reached the complex glycan-synthesizing enzymes of the trans-Golgi network. After 5 h of chase, the intracellular level of the single-chain form had decreased and two peptides of 70 and 40 kDa had appeared (Figure 1A, middle panel), corresponding in molecular masses to peptides abc and d respectively of human placenta LAMAN [7]. The 15 kDa peptide e [7] was not observed. Probably, its small number of methionine residues and lack of cysteine residues caused too weak a signal in autoradiography. The 70 and 40 kDa peptides were partially endo H-resistant (Figure 1A, middle panel), indicating that they had been transported through the trans-Golgi network. The extracellular form of LAMAN also contained endo H-resistant N-glycans (Figure 1A, right panel), indicating that the lysosomal and extracellular forms followed the same route through the trans-Golgi network. Since the secreted form was of the single-chain form, proteolysis into peptides abc and d must have taken place after sorting from the secretory pathway and entry of LAMAN into the hydrolytic environments of the endosomal/lysosomal system. The physiological importance of this cleavage is unclear, since the single-chain form is enzymically active [9]. The presence of endo H-sensitive N-glycans in mature lysosomal and secreted LAMAN (Figure 1A, middle and right panels) was consistent with the phosphorylation of certain N-glycans during intracellular transport, since this modification prevents synthesis of complex type structures. This conforms to the mannose 6-phosphate-inhibited uptake of recombinant human LAMAN into fibroblasts [9] and the presence of endo H-sensitive N-glycans in human placenta LAMAN [7], and is consistent with nascent human LAMAN utilizing the mannose 6-phosphate-dependent pathway in the COS cells. Overexpression studies on other lysosomal proteins in COS cells have also shown that the secreted forms are mannose-6-phosphorylated, and this is probably due to an oversaturation of the mannose-6-phosphate receptor [12]. Some of the LAMAN remained as an intracellular single-chain form after 5 h chase (Figure 1A, middle panel). Since all of its N-glycans were of endo H-sensitive type (Figure 1A, middle panel), this was probably a form that was retained in the ER (endoplasmic reticulum). It may have been folded incorrectly and thus trapped in the ER by the folding control system. Surprisingly, the quantity of this form was not reduced even after 12 h chase, indicating a very slow turnover. A schematic model of the intracellular transport of wild-type LAMAN in COS cells is presented in Figure 1(B).
Figure 1. Intracellular transport of human LAMAN.
(A) COS-7 cells were transfected with pcDNA constructs containing wild-type human LAMAN cDNA using LIPOFECTAMINE™ 2000. The cells were pulsed 48 h later with [35S]methionine/cysteine for 30 min and the cells were subsequently harvested or chased for 5 h. Human LAMAN was precipitated from the cell lysates or the media with a polyclonal antiserum [7]. The resulting immunoprecipitates were treated with endo H or PNGase F, separated by SDS/PAGE and analysed by autoradiography. The positions of the respective LAMAN peptide fragments are indicated on the left. (B) A model of the intracellular transport of human LAMAN in COS cells, based on the results shown in (A).
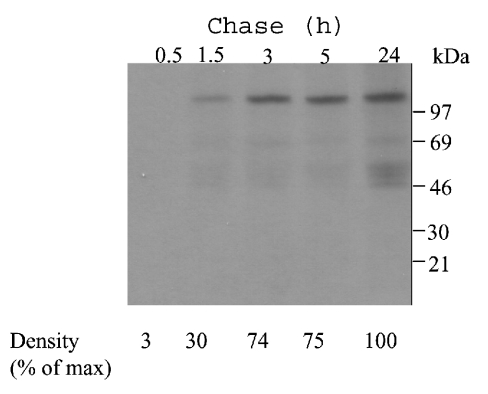
As determined from the autoradiograph in Figure 2, approx. 30% of the secreted LAMAN had reached the medium after 1.5 h chase, and the level of LAMAN protein in the medium reached a plateau 3 h after the end of labelling (Figure 2). Two weak bands of approx. 70 and 50 kDa appeared in the medium, as seen in both Figures 1(A) and 2. These may be the result of a slow proteolysis between peptides c and d, as observed previously in overexpressing CHO cells [9]. Also, the intracellular forms had reached their peak levels after 3 h chase (results not shown), suggesting that the forms obtained after 5 h chase (Figure 1A, middle and right panels) corresponded to the mature forms. This transport kinetics is similar to that for other lysosomal proteins, e.g. human sulphamidase [22]. The proteolytic maturation of sulphamidase in transfected cells begins only after 8 h of chase [22] and is thus a late lysosomal maturation. In contrast, the proteolytic processing of LAMAN into peptides abc, d and e is an early lysosomal maturation, since it occurred shortly after entry into the lysosomes. Probably, further partial processing of peptide abc into peptides a, b and c, as seen in purified human placenta LAMAN [7], is a late lysosomal processing event. The level of radioactive LAMAN appearing in the medium after 5 h chase was higher than the intracellular level (Figure 1A), and it correlated with a larger increase in LAMAN activity in the medium compared with that in the cell lysate 48 h after transfection of the COS cells (results not shown), suggesting that most of the nascent recombinant LAMAN protein was secreted. Secretion of lysosomal proteins in high-producer cells has also been reported in [23] and may be caused by saturation of the sorting system in the trans-Golgi network.
Figure 2. Secretion kinetics of human LAMAN.
COS-7 cells were transfected with pcDNA constructs containing a human LAMAN cDNA construct as described in Figure 1. Subsequent to the pulse period of 30 min with [35S]methionine/cysteine, the cells were chased for the indicated times. Human LAMAN was immunoprecipitated from the medium and analysed by autoradiography. The bands were scanned and the intensity of the respective bands was correlated with that of the band representing the 24 h chase.
Intracellular transport of LAMAN mutants in COS cells
Eleven putative disease-causing mutations (Tables 1 and 2) were introduced into human LAMAN cDNA. In addition, the D196E and D196N mutants affecting the active-site reaction nucleophile [5,6] were constructed to demonstrate the biochemical consequences of an active-site substitution. The constructs were transfected into COS-7 cells, and the cell extracts and culture media were assayed for α-mannosidase activity both on cell lysates and immunoprecipitates. Only COS cells transfected with the following LAMAN constructs expressed activities higher than the background: pcDNA-E402K, pcDNA-R220H and pcDNA-F320L (Table 3).
Table 2. Summary of the LAMAN mutations presented.
| Type of variation | |||
|---|---|---|---|
| cDNA level | Protein level | Exon/peptide | Reference |
| Human mutations | |||
| 215A-T | H72L | 2/a | [7] |
| IVS5−1G-C | Del(256–260) | 6/a | [13] |
| IVS7+2T-G* | Del(339–342) | 7/a | [13] |
| 1063A-C | T355P | 8/b | [13] |
| 1067C-G | P356R | 8/b | [13] |
| 1204G-A* | E402K | 9/b | [13] |
| 2140T-C | W714R | 17/d | [13] |
| 2248C-T | R750W | 18/d | [13] |
| 2426T-C | L809P | 20/d | [13] |
| Bovine mutations | |||
| 662G-A | R220H† | 2/a | [8] |
| 961T-C | F320L† | 3/a | [8] |
| Substitutions of the active-site nucleophile | |||
| 588C-G | D196E | 4/a | This study |
| 586G-A | D196N | 4/a | This study |
* These two mutations are located on the same allele.
† Correspond to R221H and F321L respectively in bovine LAMAN.
Table 3. Biochemical characterization of the variants expressed in COS cells.
| Peptide forms | ||||||
|---|---|---|---|---|---|---|
| Location | ||||||
| Variant | Intracellular proteolysed | Extracellular single-chain | ER | Lysosomes | Enzyme activity* (fold increase) | Group† |
| Wild-type | + | + | − | + | 8 (14)‡ | 1 |
| H72L | + | + | − | + | 1 (1)‡ | 1 |
| D196E§ | + | + | − | + | 1 | 1 |
| D196N§ | + | + | − | + | 1 | 1 |
| R220H∥ | + | + | − | + | 1.5 (1.5–2)‡ | 1 |
| F320L∥ | + | + | − | + | 3–4 (1)‡ | 1 |
| T335P | − | − | + | − | 1 | 2 |
| P356R | − | − | + | − | 1 | 2 |
| E402K | + | + | − | + | 3–4 | 1 |
| W714R | − | − | + | − | 1 | 2 |
| R750W | − | − | + | − | 1 | 2 |
| L809P | − | − | + | − | 1 | 2 |
| Del(256–260) | − | − | + | − | 1 | 2 |
| Del(339–342) | − | − | + | − | 1 | 2 |
* Activity relative to mock-transfected cells.
† Group 1 is defined as being transported to the lysosomes, whereas group 2 is accumulating in the ER.
‡ In CHO cells.
§ Substitution of the active-site nucleophile.
∥ Bovine mutations introduced into human LAMAN.
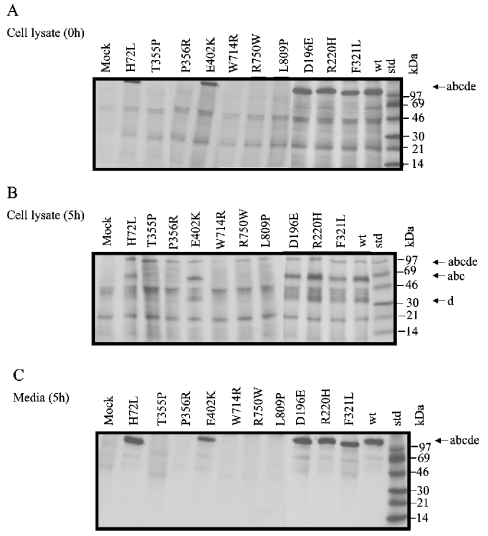
To study the expression and proteolytic maturation, COS cells transfected with different mutated pcDNA-LAMAN constructs were metabolically labelled with [35S]methionine/cysteine for 30 min and chased for 5 h. After the pulse period, five of the labelled mutants (H72L, E402K, D196E, R220H and F320L) were immunoprecipitated from the cell lysates as single-chain forms (Figure 3A). Subsequently, these mutants were partly transported to the lysosomes as judged from proteolytic processing into the peptides abc and d (Figure 3B) and partly secreted as single-chain forms (Figure 3C). Thus it appeared that these mutants exhibited an intracellular transport similar to wild-type LAMAN. In contrast, other mutants (T355P, P356R, W714R, R750W and L809P) could not be immunoprecipitated after the pulse period (Figure 3A) and were precipitated neither as the proteolytically processed lysosomal form (Figure 3B) nor as the secreted single-chain form (Figure 3C). This result indicates that these mutants did not reach beyond the Golgi apparatus and were either accumulating in the ER in a form that was not recognized by the antibody or quickly degraded shortly after synthesis or subjected to a decreased translation rate.
Figure 3. Intracellular transport of mutant human LAMANs.
COS-7 cells were transfected with pcDNA constructs containing missense mutations obtained by site-directed mutagenesis (Table 1) and metabolically labelled as described in Figure 1. Subsequent to the pulse period of 30 min with [35S]methionine/cysteine, the cells were either harvested (A) or subjected to 5 h of chase (B, C). Human LAMAN was immunoprecipitated from the cell extracts (A, B) or the media (C) and analysed by autoradiography.
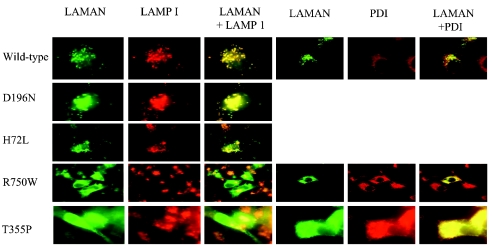
To study the levels of LAMAN protein expression, the proteins were denatured before binding to the primary antibody, thus excluding any folding-dependent binding. When the LAMAN expression was studied by immunofluorescence, the proteins were denatured during fixation of the COS cells and, consequently, exhibited affinities towards antibodies against denatured LAMAN (Figure 4). As seen in the upper panels of Figure 4, the wild-type, H72L and D196N LAMANs were localized to vesicular structures that co-localized with the lysosomal marker LAMP-1. Similar co-localization was also observed for the D196E, R220H, F320L and E402K LAMANs (results not shown). These mutants exhibited lysosomal processing similar to that of wild-type LAMAN (Figure 3B), thus confirming the normal sorting of these mutants. T355P and R750W LAMANs were expressed at significant levels as evidenced by immunofluorescence studies (Figure 4, bottom panels). This indicated that the apparently low expression levels of these mutants as judged by metabolic labelling (Figures 3A and 3B) were due to a low affinity of the folded mutants for the antibody raised against native LAMAN. Probably, these mutants folded abnormally, preventing the normal expression of antigenic epitopes. Such misfolding was consistent with the localization of these mutants to structures that co-localized with the ER marker PDI, but not with LAMP-1 (Figure 4, bottom panel), indicating that the T355P and R750W LAMANs were retained in the ER by the folding control system. Similar co-localizations were also found for the P356R, W714R and L809P LAMANs as well as the two deletion mutants, del(256–259) and del(339–342) LAMANs (results not shown).
Figure 4. Intracellular distribution of human LAMAN in COS cells observed with immunofluorescence microscopy.
Double immunofluorescence staining was performed using antibodies against LAMAN, LAMP-1 and PDI. Yellow indicates overlap of the LAMAN peptides (green) and LAMP-1 (red) or PDI (red).
Western blotting was performed to investigate in more detail the expression levels and structures of the ER-retained forms. All the seven mutants that accumulated in the ER as judged from immunofluorescence (Figure 4) existed as intracellular 120 kDa single-chain forms (Figure 5A). These mutants were neither proteolytically processed nor secreted (Figures 5A and 5B), and this is consistent with their ER retention. The other mutants were proteolytically processed as judged from the appearance of the 40 kDa peptide d (Figure 5A) and were secreted as single-chain forms (Figure 5B), suggesting that their intracellular pathways were similar to the wild-type. This result was confirmed for a selected group of mutants expressed in CHO cells (results not shown).
Figure 5. Western-blot analyses of mutant human LAMANs.
Cell lysate and medium from COS cells transfected with various mutant LAMAN constructs were subjected to SDS/PAGE and electroblotted on to a PVDF membrane. The membrane was incubated in diluted antiserum against native recombinant human LAMAN as described in the Materials and methods section.
Results obtained from the expression of LAMAN mutants in COS cells and the selected mutants in CHO cells are summarized in Table 3. We divided the mutants into two main groups according to their intracellular routes: (i) lysosomal sorting and secretion and (ii) transport arrest in the ER. The phenotype of the group 1 mutants is characteristic for proteins that fold sufficiently to pass the ER folding control system, although some group 1 mutants may possess subtle alterations in their structures, which decrease their activities compared with wild-type LAMAN in transfected COS cells. Five of the mutants in group 1 exhibited <5% of the increase in activity caused by the wild-type construct D196N, D196E, H72L and R220H, whereas the activities caused by the F320L and E402K mutants were 30–40% of the wild-type activity (Table 3). In CHO cells, two of the group 1 mutants, H72L and R220H, had similar activity as in COS cells, whereas F320L had much lower activity in CHO cells (Table 3). The phenotype of the group 2 mutants is characteristic for proteins that are not folded correctly and are thus retained by the folding control system of ER. In agreement with this model, none of the group 2 mutants exhibited any significant activity (Table 3).
Modelling of the mutations into the three-dimensional structure of LAMAN
To understand the structural consequences of the disease-causing missense mutations, we modelled each of the amino acid replacements into the three-dimensional structure of bovine LAMAN.
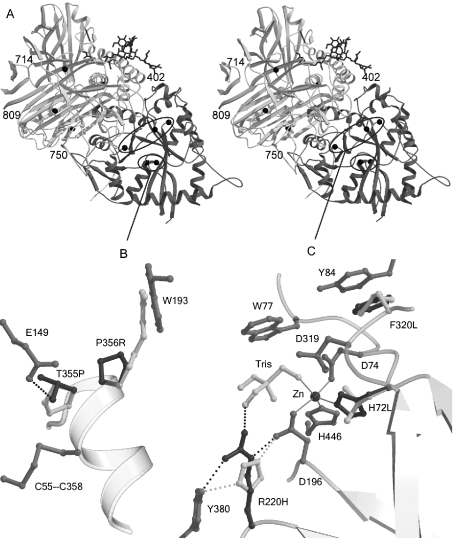
Group 2 mutants contained mutations in either the active-site domain (Thr355, Pro356) or on a domain interface (Arg750) or interior of the domain formed by peptide d (Trp714, Leu809). Two examples of substitutions in this group are shown in Figure 6(A). In the LAMAN three-dimensional structure, Thr355 is hydrogen-bonded to Glu149, an interaction that the mutated Pro355 cannot make. During the folding, Thr355 probably also hydrogen-bonds to the free cysteine residue Cys358, which forms a disulphide bridge with Cys55 in the mature structure [5]. Pro356 is in an optimal position to initialize the formation of the following α-helix and to affect the rate of LAMAN folding [5]. The P356R substitution also introduces a residue that does not fit the folded structure well. A human LAMAN mutant lacking the Cys55-Cys358 disulphide bridge also exhibited a group 2 phenotype (T. Berg, unpublished work), suggesting that any mutation affecting the formation of this disulphide bridge causes misfolding and ER retention. Probably, the T355P and P356R LAMANs are two examples of such mutations, since they were both predicted to affect the positioning of Cys358.
Figure 6. Amino acid substitutions modelled in the bovine LAMAN structure.
(A) Stereographic representation of the complete structure. The active-site domain is darker than the rest of the structure. The affected residues are indicated with black balls. Mutations outside the active-site domain are labelled, and mutations in the N-terminal domain are shown in detail in (B, C). (B) Mutations affecting the folding of LAMAN. The mutated wild-type residues are shown in dark grey and the modelled mutations in white. Hydrogen bonds are displayed with broken lines and metal co-ordination with solid lines. The T355P mutation disrupts a hydrogen bond to Glu149 and, probably, to the free cysteine residue Cys358, which in the mature enzyme participates in a disulphide bridge. Pro356 is in optimal position to initialize helix formation and is important for the rate of folding of LAMAN. (C) Mutations that inactivate the enzyme, but allow folding, are located close to the active site: although His220 hydrogen-bonds to the nucleophile Asp196, it cannot co-ordinate the substrate as Arg220 probably does. His72 is involved in the metal binding, whereas Phe320 follows the active-site residue Asp319 and makes a stacking interaction with Tyr84. Probably, the hydrophobic stacking also stabilizes Trp77, which is involved in substrate binding.
Figure 6(B) depicts a model of those group 1 mutants that are located in the active site. The D196N/E changes were not modelled, as the function of the Asp196 active-site nucleophile is well established [5,6], and the biochemical phenotype of the substitutions (Table 3) was consistent with this function. His72 co-ordinates the active-site Zn2+, and the H72L mutation abolishes this interaction (Figure 6B). The OD1 oxygen of the reaction nucleophile Asp196 directly co-ordinates Zn2+, indicating that the Zn2+ positions Asp196 and stabilizes the protonation states of the reacting partners. It has been suggested that Zn2+ is directly involved in the catalysis by Golgi α-mannosidase II in co-ordinating the O2 and O3 oxygens of the mannose substrate and facilitating the deglycosylation step by moving from a less favoured hexavalent co-ordination on the substrate to a quinquivalent co-ordination after product release [4]. These functions of Zn2+ fit well with the complete loss of activity after substitution of the Zn2+-co-ordinating His72. Furthermore, the functional importance of His72 is shown by its conservation among all the GH38 enzymes (results not shown). The R220H mutant possessed less than 5% of the wild-type specific activity (Table 3). This conforms to the activity measured in livers from cattle affected with the R220H mutation [8], indicating that the activity phenotype was caused mainly by a decreased specific activity and was not due to abnormal folding or increased turnover in the lysosomes. In the bovine LAMAN structure, Arg220 is hydrogen-bonded to both the reaction nucleophile Asp196 and a Tris molecule (Figure 6B). This suggests a dual role for Arg220 in both the activation and orientation of the nucleophile Asp196 as well as in substrate recognition. The modelled residue His220 can also position Asp196 correctly (Figure 6B); however, due to the substantially lower pKa value of a histidine side chain, it is not as effective in activation of the nucleophile. In the GH38 family, Arg220 can be substituted only with a lysine residue (results not shown), which has similar chemical properties to an arginine residue. Hydrogen-bonding to the Tris molecule in the bovine LAMAN structure and superimposition with α-amylases [5] also suggest a role for Arg220 in substrate binding. Both arginine and lysine residues at position 220 have unoccupied co-ordination sites towards the substrate. Histidine, on the other hand, is substantially shorter and has no unco-ordinated N atoms according to our model as shown in Figure 6(B).
Phe320 follows a potential catalytic acid–base Asp319 [4,5] and, thus, its function could be to anchor Asp319, probably by making a stacking interaction with Tyr84 (Figure 6B). In our measurements in COS cells, the F320L mutant appeared to have 30–40% of the wild-type activity. The activity was, however, much lower when the mutant was expressed in CHO cells (Table 3). In livers from cattle affected with the F320L mutation, the measured activity was less than 1% [8,24], which was accompanied by very low levels of LAMAN protein as judged by Western blotting [8]. Assuming normal levels of LAMAN synthesis, this indicated that the endogenous F321L mutant folded abnormally in bovine cells or was unstable and quickly degraded. Phe320 is not conserved among GH38 enzymes. The high activity of the F320L mutant in COS cells relative to CHO cells and cells from cattle affected with the F321L mutation, may be due to the overproduction system. Overproduction may overload the folding control or the lysosomal transport and turnover systems in COS cells, as observed earlier with disease-causing mutants of β-glucuronidase and lysosomal β-galactosidase [24,25].
Genotype–phenotype correlation
Eight of the mutants associated with human α-mannosidosis exhibited no detectable activity in COS cells, including five group 2 mutants, one group 1 mutant (H72L) and two deletion mutants (Table 3). Thus the corresponding DNA mutations are probably disease-causing. One mutant E402K caused a 3–4-fold increase in activity in transfected COS cells (Table 3), which is nearly half of the increase caused by a wild-type construct. However, in the patient known to carry the E402K substitution, the mutation causing the del(339–342) variation is located on the same allele [13]. Since del(339–342) LAMAN is a group 2 mutant and possesses no activity (Table 3), it is conceivable that the E402K substitution is a rare variant that is not disease-causing. Molecular modelling showed that Glu402 forms a hydrogen bond to Gln444 (results not shown), which may be important in the anchoring of the nearby active-site residues His446 (Figure 6B) and Asp447 [5]. Lys402 may form an alternative hydrogen bond to Gln444, suggesting that this variation is tolerated. In LAMAN from D. melanogaster, position 402 is occupied by histidine (results not shown), indicating that changes are tolerated at this position.
In contrast with the COS cells transfected with mutants containing human disease-causing mutations, the cells transfected with constructs containing either of the two bovine disease-causing mutations exhibited significant LAMAN activities (Table 3). This is consistent with activity measurements in human fibroblasts [7] and in bovine tissues and cells [8,26]. However, the enzymic activities in cattle do not coincide with mild clinical phenotypes, since the affected cattle are often stillborn or show early severe neurological disorders [26]. It thus appears that the minimum amount of LAMAN activity required to prevent the disease is higher in cattle than in humans. This may be due to the nature of the respective glycans that accumulate within the lysosomes of human and bovine cells in the absence of LAMAN activity. The major human lysosomal storage product is the trisaccharide Manα1,3Manβ1,4GlcNAc, whereas, in cattle, the branched pentasaccharide Manα1,3(Manα1,6)Manβ1,4GlcNAcβ1,4GlcNAc accumulates in the complete absence of LAMAN [2], due to the lack of lysosomal chitobiase in cattle [3]. Since linear glycans are better substrates for LAMAN compared with branched glycans [27] and α1,3-linkages are cleaved faster than α1,6-linkages [28], a higher LAMAN activity is required to cleave the bovine storage product compared with the human. This may explain why both the bovine models of α-mannosidosis can possess significant LAMAN activity in cells and tissues despite the severe clinical phenotypes [8,26]. Consistent with this hypothesis, the only other example of significant LAMAN activity connected to α-mannosidosis is in affected long-haired cats [16,29]. Similar to cattle, cats also lack chitobiase activity [3]. Thus the function of chitobiase is probably to increase the rate of N-glycan degradation in the lysosomes.
In the present study, we have found that all the previously reported mutations suggested to cause human α-mannosidosis resulted in no significant activity in transfected COS cells, confirming that they are disease-causing. Most of the missense mutations resulted in misfolding and ER retention. This is consistent with the limited number of active-site residues in a protein compared with residues that are important for the folding process. The biochemical characterization of the various mutations has given new insight into the folding mechanism and catalysis of LAMAN.
Acknowledgments
This project was supported by the European Commission contract no. QLK3-CT2001-02458. The opinions expressed in this paper do not necessarily represent the opinion of the European Commission and the Commission is not responsible for any use that might be made of the data presented here. We thank Jouni Vesa for helpful advice on the immunofluorescence analyses.
References
- 1.Henrissat B., Bairoch A. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeGasperi R., al Daher S., Daniel P. F., Winchester B. G., Jeanloz R. W., Warren C. D. The substrate specificity of bovine and feline lysosomal α-D-mannosidases in relation to α-mannosidosis. J. Biol. Chem. 1991;266:16556–16563. [PubMed] [Google Scholar]
- 3.Aronson N. N., Kuranda M. J. Lysosomal degradation of Asn-linked glycoproteins. FASEB J. 1989;3:2615–2622. doi: 10.1096/fasebj.3.14.2531691. [DOI] [PubMed] [Google Scholar]
- 4.Van den Elsen J. M., Kuntz D. A., Rose D. R. Structure of Golgi α-mannosidase II: a target for inhibition of growth and metastasis of cancer cells. EMBO J. 2000;20:3008–3017. doi: 10.1093/emboj/20.12.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heikinheimo P., Helland R., Leiros H.-K., Karlsen S., Evjen G., Ravelli R., Schoen G., Ruigrok R., Tollersrud O. K., McSweeney S., et al. The structure of bovine lysosomal α-mannosidase suggests a novel mechanism for low pH activation. J. Mol. Biol. 2003;327:631–644. doi: 10.1016/s0022-2836(03)00172-4. [DOI] [PubMed] [Google Scholar]
- 6.Numao S., He S., Evjen G., Howard S., Tollersrud O. K., Withers S. G. Identification of Asp197 as the catalytic nucleophile in the family 38 α-mannosidase from bovine kidney lysosomes. FEBS Lett. 2000;484:175–178. doi: 10.1016/s0014-5793(00)02148-7. [DOI] [PubMed] [Google Scholar]
- 7.Nilssen O., Berg T., Riise H. M., Ramachandran U., Evjen G., Hansen G. M., Malm D., Tranebjaerg L., Tollersrud O. K. α-Mannosidosis: functional cloning of the lysosomal α-mannosidase cDNA and identification of a mutation in two affected siblings. Hum. Mol. Genet. 1997;6:717–726. doi: 10.1093/hmg/6.5.717. [DOI] [PubMed] [Google Scholar]
- 8.Tollersrud O. K., Berg T., Healy P., Evjen G., Ramachandran U., Nilssen Ø. Purification of bovine lysosomal α-mannosidase, characterization of its gene and determination of two mutations that cause α-mannosidosis. Eur. J. Biochem. 1997;246:410–419. doi: 10.1111/j.1432-1033.1997.00410.x. [DOI] [PubMed] [Google Scholar]
- 9.Berg T., King B., Meikle P. J., Nilssen O., Tollersrud O. K., Hopwood J. J. Purification and characterization of recombinant human lysosomal α-mannosidase. Mol. Genet. Metab. 2001;73:18–29. doi: 10.1006/mgme.2001.3173. [DOI] [PubMed] [Google Scholar]
- 10.Nebes V. L., Schmidt M. C. Human lysosomal α-mannosidase: isolation and nucleotide sequence of the full-length cDNA. Biochem. Biophys. Res. Commun. 1994;200:239–245. doi: 10.1006/bbrc.1994.1440. [DOI] [PubMed] [Google Scholar]
- 11.Riise H. M., Berg T., Nilssen O., Romeo G., Tollersrud O. K., Ceccherini I. Genomic structure of the human lysosomal α-mannosidase gene (MANB) Genomics. 1997;42:200–207. doi: 10.1006/geno.1997.4668. [DOI] [PubMed] [Google Scholar]
- 12.Thomas G. H. Disorders of glycoprotein degradation and structure: α-mannosidosis, β-mannosidosis, fucosidosis, sialidosis and aspartylglucosaminuria, carbohydrate-deficient glycoprotein syndrome. In: Schriver C. R., Beaudet A. L., Sly W. S., Valle D., editors. The Metabolic and Molecular Bases of Inherited Disease. 8th edn. New York: McGraw-Hill; 2001. pp. 3507–3534. [Google Scholar]
- 13.Berg T., Riise H. M., Hansen G. M., Malm D., Tranebjaerg L., Tollersrud O. K., Nilssen O. Spectrum of mutations in α-mannosidosis. Am. J. Hum. Genet. 1999;64:77–88. doi: 10.1086/302183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warner T. G., Mock A. K., Nyhan W. L., O'Brien J. S. α-Mannosidosis: analysis of urinary oligosaccharides with high performance liquid chromatography and diagnosis of a case with unusually mild presentation. Clin. Genet. 1984;25:248–255. doi: 10.1111/j.1399-0004.1984.tb01985.x. [DOI] [PubMed] [Google Scholar]
- 15.Michelakakis H., Dimitriou E., Mylona-Karayanni C., Bartsocas C. S. Phenotypic variability of mannosidosis type II: report of two Greek siblings. Genet. Couns. 1992;3:195–199. [PubMed] [Google Scholar]
- 16.Berg T., Tollersrud O. K., Walkley S. U., Siegel D., Nilssen Ø. Purification of feline lysosomal α-mannosidase, determination of its cDNA sequence and identification of a mutation causing α-mannosidosis in Persian cats. Biochem. J. 1997;328:863–870. doi: 10.1042/bj3280863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berg T., Hopwood J. J. α-Mannosidosis in the guinea pig: cloning of the lysosomal α-mannosidase cDNA and identification of a missense mutation causing α-mannosidosis. Biochim. Biophys. Acta. 2002;1586:169–176. doi: 10.1016/s0925-4439(01)00081-3. [DOI] [PubMed] [Google Scholar]
- 18.Stinchi S., Lullmann-Rauch R., Hartmann D., Coenen R., Beccari T., Orlacchio A., von Figura K., Saftig P. Targeted disruption of the lysosomal α-mannosidase gene results in mice resembling a mild form of human α-mannosidosis. Hum. Mol. Genet. 1999;8:1365–1372. doi: 10.1093/hmg/8.8.1365. [DOI] [PubMed] [Google Scholar]
- 19.Gotoda Y., Wakamatsu N., Kawai H., Nishida Y., Matsumoto T. Missense and nonsense mutations in the lysosomal α-mannosidase gene (MANB) in severe and mild forms of α-mannosidosis. Am. J. Hum. Genet. 1998;63:1015–1024. doi: 10.1086/302048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraulis P. J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 21.Merritt E. A., Murphy M. E. P. Raster3D Version 2.0: a program for photorealistic molecular graphics. Acta Crystallogr. D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 22.Perkins K. J., Byers S., Yogalingam G., Weber B., Hopwood J. J. Expression and characterization of wild type and mutant recombinant human sulfamidase. Implications for Sanfilippo (mucopolysaccharidosis IIIA) syndrome. J. Biol. Chem. 1999;274:37193–37199. doi: 10.1074/jbc.274.52.37193. [DOI] [PubMed] [Google Scholar]
- 23.Ling P., Roberts R. M. Overexpression of uteroferrin, a lysosomal acid phosphatase found in porcine uterine secretions, results in its high rate of secretion from transfected fibroblasts. Biol. Reprod. 1993;49:1317–1327. doi: 10.1095/biolreprod49.6.1317. [DOI] [PubMed] [Google Scholar]
- 24.Wu B. M., Tomatsu S., Fukuda S., Sukegawa K., Orii T., Sly W. S. Overexpression rescues the mutant phenotype of L176F mutation causing β-glucuronidase deficiency mucopolysaccharidosis in two Mennonite siblings. J. Biol. Chem. 1994;269:23681–23688. [PubMed] [Google Scholar]
- 25.Zhang S., Bagshaw R., Hilson W., Oho Y., Hinek A., Clarke J. T. R., Hinek A., Callahan W. Characterisation of β-galactosidase mutations Asp332→Asn and Arg148→Ser and a polymorphism Ser532→Gly in a case of GM1 gangliosidosis. Biochem. J. 2000;348:621–632. [PMC free article] [PubMed] [Google Scholar]
- 26.Healy P. J., Harper P. A., Dennis J. A. Phenotypic variation in bovine α-mannosidosis. Res. Vet. Sci. 1990;49:82–84. [PubMed] [Google Scholar]
- 27.Tulsiani D. R., Touster O. Substrate specificities of rat kidney lysosomal and cytosolic α-D-mannosidases and effects of swainsonine suggest a role of the cytosolic enzyme in glycoprotein catabolism. J. Biol. Chem. 1987;262:6506–6514. [PubMed] [Google Scholar]
- 28.Michalski J. C., Haeuw J. F., Wieruszeski J. M., Montreuil J., Strecker G. In vitro hydrolysis of oligomannosyl oligosaccharides by the lysosomal α-D-mannosidases. Eur. J. Biochem. 1990;189:369–379. doi: 10.1111/j.1432-1033.1990.tb15498.x. [DOI] [PubMed] [Google Scholar]
- 29.Cummings J. F., Wood P. A., de Lahunta A., Walkley S. U., Le Boeuf L. The clinical and pathologic heterogeneity of feline α-mannosidosis. J. Vet. Intern. Med. 1988;2:163–170. doi: 10.1111/j.1939-1676.1988.tb00311.x. [DOI] [PubMed] [Google Scholar]