Abstract
Muskelin is an intracellular protein with a C-terminal kelch-repeat domain that was initially characterized as having functional involvement in cell spreading on the extracellular matrix glycoprotein thrombospondin-1. As one approach to understanding the functional properties of muskelin, we have combined bioinformatic and biochemical studies. Through analysis of a new dataset of eight animal muskelins, we showed that the N-terminal region of the polypeptide corresponds to a predicted discoidin-like domain. This domain architecture is conserved in fungal muskelins and reveals a structural parallel between the muskelins and certain extracellular fungal galactose oxidases, although the phylogeny of the two groups appears distinct. In view of the fact that a number of kelch-repeat proteins have been shown to self-associate, co-immunoprecipitation, protein pull-down assays and studies of cellular localization were carried out with wild-type, deletion mutant and point mutant muskelins to investigate the roles of the discoidin-like and kelch-repeat domains. We obtained evidence for cis- and trans-interactions between the two domains. These studies provide evidence that muskelin self-associates through a head-to-tail mechanism involving the discoidin-like domain.
Keywords: discoidin domain, fungi, galactose oxidase, kelch repeat, muskelin, β-propeller
Abbreviations: BTB/POZ domain, bric-a-brac, tramtrack, broad-complex/poxvirus and zinc finger domain; CDD, conserved domain database; CTLH motif, C-terminal to LisH motif; DTT, dithiothreitol; ECM, extracellular matrix; EGFP, enhanced green fluorescent protein; EGFP–MK, EGPF fused to muskelin; EGFP–MKKC, EGPF fused to amino acids 244–735 of muskelin; EGFP–MKDD and GST-MKDD, EGFP and GST respectively fused to the muskelin discoidin-like domain; EST, expressed sequence tag; FGI, Fungal Genome Initiative; GST, glutathione S-transferase; LisH motif, Lissencephaly-1 homology motif; ORF, open reading frame; PSI, proteasome inhibitor I [benzyloxycarbonyl-Ile-Glu(OtBu)-Ala-Leu-CHO]; SMART, simple modular architecture research tool; SMC, smooth muscle cell; WICGR, Whitehead Institute Center for Genome Research; Z-LLF-CHO, benzyloxycarbonyl-Leu-Leu-phenylalaninal
INTRODUCTION
Muskelin is an intracellular protein that was initially characterized as having functional involvement in cell spreading on the ECM (extracellular matrix) glycoprotein thrombospondin-1 [1]. Mouse C2C12 skeletal myoblasts selected for stable overexpression of muskelin showed qualitative alterations in cytoskeletal organization when adherent to thrombospondin-1, that involved decreased numbers of fascin microspikes and the assembly of focal adhesions. These effects were not apparent in cells adherent to fibronectin or after antisense-mediated depletion of muskelin [1]. This specificity of response suggested that muskelin is linked to cell-signalling processes that are differentially regulated on the basis of ECM context. To elucidate the underlying mechanisms, this laboratory is building an understanding of muskelin, its cellular interactions, biological roles and regulation.
Muskelin is encoded by a single gene in the mouse, and highly conserved orthologues have been identified in Homo sapiens [2], Rattus norvegicus (GenBank NM_031359) and Drosophila melanogaster [3]. Initial bioinformatic analysis of these orthologues showed that muskelin is a multidomain protein [3]. The central region of the protein is predicted to have α-helical secondary structure and contains two sequence motifs, the LisH motif [Lissencephaly-1 homology motif; SMART (simple modular architecture research tool) #SM0667] and the CTLH motif (C-terminal to LisH motif; SMART #SM0668). The LisH motif is an α-helical region of approx. 60 amino acids at the N-terminus of Lis1 (Lissencephaly-1) that contributes to its binding interactions with the α-subunit of platelet-activating factor acetylhydrolase [4] and with the microtubule motor protein dynein [5]. The motif is also found in other microtubule-binding proteins and in proteins that are not associated with microtubules [6]. The CTLH motif is another α-helical segment of unknown function that is present in several LisH motif-containing proteins [6].
The C-terminal half of muskelin contains six repeated kelch motifs and a short C-terminal region. The kelch motif was first described in Drosophila kelch [7], and is now known to be present as five, six or seven repeats in a plethora of proteins in diverse organisms from all domains of life and in poxviruses [8–11]. The crystal structure of fungal galactose oxidase, which contains seven kelch motifs, revealed that each kelch motif forms a four-stranded antiparallel β-sheet that corresponds to a blade within a β-propeller structure [8,12,13] (PDB 1GOF). Many kelch proteins of animals contain a BTB/POZ domain (bric-a-brac, tramtrack, broad-complex/poz virus zinc finger domain) N-terminal to the kelch repeats [11]. In some of these proteins, including kelch, mayven, MIPP [mouse IAP (inhibitor of apoptosis protein)-promoted placental gene], actinfilin and Nd-1, the kelch-repeat β-propeller binds to actin, and the proteins self-associate into dimers or oligomers through trans-interactions of the BTB/POZ domain [14–17]. Multimerization is necessary for the physiological function of kelch in mediating actin filament cross-linking within egg chamber ring canals [14]. In other BTB/kelch-repeat proteins, for example Keap-1 and gigaxonin, the kelch β-propellers interact with individual protein partners, and it is not known if self-association through the BTB domain takes place [18,19].
The presence of the LisH and CTLH motifs distinguishes muskelin from other kelch-repeat proteins and suggests potential for multiple protein–protein interaction sites. To fully understand the domain organization of muskelin, we carried out bioinformatic analyses with a new dataset of species orthologues of muskelin that we had developed. We report that the N-terminal portion of the polypeptide is predicted to fold as a Factor 5, Factor 8_type_C domain, also known as the discoidin domain (Conserved Domain Database entry 24469). This finding revealed a structural commonality between the intracellular muskelins and extracellular fungal galactose oxidases that led us to evaluate the molecular phylogeny of muskelins and their relationship with galactose oxidases. We also obtained experimental evidence for self-association of muskelin, dependent on the discoidin-like domain and the kelch-repeat β-propeller.
MATERIALS AND METHODS
Cell culture and materials
C2C12 mouse skeletal myoblasts were grown in Dulbecco's modified Eagle's medium (Gibco) containing 20% (v/v) heat-inactivated fetal calf serum. Rat aortic vascular SMCs (smooth muscle cells), human embryonic kidney 293T cells and COS-7 green monkey kidney cells were grown in Dulbecco's modified Eagle's medium containing 10% (v/v) fetal calf serum. All cell lines were maintained at 37 °C in a water-saturated incubator with a 5% CO2 atmosphere, and were routinely replated every third day in standard tissue culture plastic products. pEGFP-C and pEGFP-N1 plasmids were purchased from Clontech.
Bioinformatic and phylogenetic analysis of muskelin and galactose oxidase orthologues
To identify additional muskelin orthologues, TBLASTN and BLASTP searches of the non-redundant database and dbest were carried out at EMBL/NCBI/DDJN with mouse or Drosophila melanogaster muskelin as the query sequences. The complete nucleotide sequencing of a full-length Danio rerio muskelin cDNA from IMAGE clone BF158025 (Reseach Genetics) was managed by Alexia Zaromytidou, and the sequence was deposited in GenBank under accession number AF418017. Multiple sequence alignment of amino acids 1–172 of muskelins from Homo sapiens (AF047489), Mus muscularis (U72194), Rattus norvegicus (NM_031359), Danio rerio (AF418017), Drosophila melanogaster (AF049333), Anopheles gambiae (EAA097111 and BM606168), Ciona intestinalis {BW24484 and other ESTs (expressed sequence tags) in Cluster 05014v1; [20]} and Gallus gallus (AJ451619 and BW210776) was performed in CLUSTAL-W [21] and is presented in Boxshade 3.2. The amino acid sequences and the derived 50% consensus sequence were examined further against the PROSITE [22], SMART [23] and PFAM [24] databases, as well as the CDD (conserved domain database) [25], at default parameters.
Choanoflagellate ESTs were searched by TBLASTX at http://projects.bocklabs.wisc.edu/carroll/choano [26]. Fungal genomes were searched through the BLAST genomes interface at NCBI [27] and through the FGI (Fungal Genome Initiative) site at the WICGR (Whitehead Institute Center for Genome Research; http://www-genome.wi.mit.edu). The complete genomes queried included the Fusarium graminearum and Coprinus cinereus sequencing projects (Whitehead Institute/MIT Center for Genome Research), the Magnaporthe grisae sequencing project [Ralph Dean, Fungal Genomics Laboratory at North Carolina State University (http://www.fungalgenomics.ncsu.edu) and Whitehead Institute/MIT Center for Genome Research], and the Ustilago maydis sequencing project [10 × genome assembly by WICGR and Bayer WGS of strain 521; Ustilago maydis Sequencing Project, Center for Genome Research (http://www.broad.mit.edu)]. Galactose oxidase sequences were identified by use of Hypomyces rocellus/Dactylium dendroides galactose oxidase (GenBank accession no. A38084) as a BLAST query sequence against GenBank and the available complete and partial fungal genome sequences at NCBI BLAST genomes and FGI/WICGR. These included the complete genomes of the ascomycotes Saccharomyces cerevisiae [28], Schizosaccharomyces pombe [29], Neurospora crassa [30] and Encephalitizoon cuniculi [31] and the basidomycotes Ustilago maydis, Coprinus cinereus and Cryptococcus neoformans. Identified ORFs (open reading frames) were aligned in CLUSTALW, and putative active-site motifs identified by comparison with Hypomyces rocellus galactose oxidase. ORFs were identified by a combination of the BLAST search matches with muskelin or galactose oxidase, translation of genomic DNA by the NCBI ORF-finder program, and CLUSTALW alignment and manual inspection of translated regions against known muskelins or galactose oxidases. These searches were carried out between July and December 2003. Secondary structure predictions were made using the Jpred program (http://www.compbio.dundee.ac.uk/jpred).
Six kelch repeats from muskelins and galactose oxidases were used as a basis for phylogenetic comparison. For Encephalitizoon cuniculi muskelin and the galactose oxidases, which all have seven kelch repeats, the six kelch repeats were selected on the basis of CLUSTALW alignments which showed that kelch repeats 2–7 of galactose oxidase aligned with repeats 1–6 of mouse muskelin, and also that Encephalitizoon cuniculi kelch-repeat 4 did not align with repeats 1–6 of the other muskelins. Phylogenetic analyses were performed at San Diego Supercomputer Biology Workbench with the PROTPARS, DRAWTREE and CLUSTALTREE methods and are displayed in DRAWTREE.
Preparation of muskelin expression constructs
The untagged mouse muskelin in pcDNA3 was described previously [1]. A V5His6-tagged version was prepared by PCR of mouse muskelin cDNA with the primer pair 5′-GGTGCTGACAAGATGGCG and 5′-CAGTGTGATCAGGTCTAC and TOPO cloning of the PCR product into the pcDNA3.1V5His vector according to the manufacturer's procedures (Invitrogen). To generate fusions of muskelin with EGFP (enhanced green fluorescent protein), mouse muskelin cDNA was digested from pBluescript-SK [1] with EcoRI and ligated into pEGFP-C2 (Clontech) using T4 DNA ligase (NEB) according to the manufacturer's protocol. A C-terminal EGFP fusion protein was prepared by PCR using the oligonucleotides 5′-GATCATGAATTCGGGTGCTGACAAGATGGCG and 5′-GACTGCAGAATTCGCAGTGTGATCAGGTC. The 2245 bp PCR product was digested with EcoRI and ligated into pEGFP-N1. A mutant muskelin, MK-Y488A/V495A, was prepared by site-directed mutagenesis of mouse muskelin in the pcDNA3.1 vector, using the oligonucleotide pair 5′-ATTTCTTTAGTGCTGATGTGGACTCAGATCATGCAGACATAATTT and 5′-AAATTATGTCTGCATGATCTGAGTCCACATCAGCACTAAAGAAAT and the Quickchange site-directed mutagenesis kit (Stratagene). EGFP–MK-Y488A/V495A was prepared by digestion of muskelin cDNA from pcDNA3.1-MK-Y488A/V495A with EcoRI and ligation into pEGFP-C3. The muskelin cDNAs in all of these constructs were manually DNA sequenced across the 5′ ligation sites to determine that the reading frames were correct. To prepare EGFP fused to the muskelin discoidin-like domain (EGFP–MKDD), plasmid pEGFP-MK was digested with EcoRV and SmaI and religated with T4 DNA ligase. EGPF fused to amino acids 244–735 of muskelin (EGFP–MKKC) was prepared by PCR of muskelin cDNA with the primer pair 5′-GGCTTAGAATTCCAGGAGTATAAGCCA and 5′-CGTGACGGTACCTACAGTGTGATCAGGTC, which included EcoRI and KpnI restrictions sites respectively. The PCR product was digested accordingly for ligation into pEGFP-C2.
A GST (glutathione S-transferase) fusion protein encoding the N-terminal portion of mouse muskelin (amino acids 1–158; discoidin-like domain) was prepared by amplifying the corresponding cDNA by PCR using the oligonucleotides 5′-GCTACCTTAGGATCCATGGCGGCTGGC and 5′-ATGCATGGATCCTAGATATCAGGATCATCAATGC. The 501 bp reaction product was digested with BamHI and ligated into the pGEX-1 plasmid (Pharmacia), in-frame with the N-terminal GST tag (construct termed GST–MKDD). The GST–MKDD construct was confirmed by manual sequencing. Expression of GST–MKDD fusion protein in Escherichia coli was induced by treating mid-exponential liquid cultures with 0.1 mM isopropyl β-D-thiogalactoside for 3 h at 30 °C before lysis by sonication. The fusion protein was purified by standard procedures using glutathione–agarose beads (Sigma) and eluted with 10 mM GSH [32]. GST protein was purified by the same protocol and used as a control.
Transient transfection
Cells for transient transfection were plated at 30–40% confluency and transfected with 2 μg of plasmid DNA by either the SuperFect or the PolyFect method according to the manufacturer's protocols (Qiagen). Cells were incubated for 36 h before use in experiments. Pilot experiments established that muskelin expression was maximal at this time.
Pull-down experiments and immunoblotting
293T human embryonic kidney cells were transiently transfected as described. For muskelin pull-down assays, confluent 100 mm plates of cells were lysed in 1 ml of buffer containing 1% (v/v) Triton X-100, 1% (v/v) glycerol, 150 mM NaCl, 50 mM Tris/HCl, pH 7.5, and protease inhibitor cocktail (Roche). Lysates were centrifuged for 10 min at 13000 g, and 10 μg of either GST–MKDD or GST was added together with 50 μl of 60% (w/v) glutathione–Sepharose 4B beads (Amersham) in lysis buffer. Samples were incubated with rotation overnight at 4 °C. Beads were pelleted by centrifugation and washed five times in 1 ml of lysis buffer. Proteins were resuspended in 40 μl of SDS/PAGE sample buffer containing 100 mM DTT (dithiothreitol) and resolved on SDS/12.5%-polyacrylamide gels.
In experiments to determine protein expression levels, whole-cell lysates were prepared from transfected cells by lysis in 2×SDS/PAGE sample buffer containing 100 mM DTT, 100 mM Tris/HCl, pH 6.8, 4% (w/v) SDS and 20% (v/v) glycerol. An aliquot of 20 μg of each sample was separated on 10% (w/v) polyacrylamide gels and transferred electrophoretically to a PVDF membrane (Millipore) in buffer containing 25 mM Tris/HCl, pH 9.4, 40 mM glycine and 10% (v/v) methanol by semi-dry blotting (Owl Separation Systems). Blots were blocked in 0.5% (w/v) casein or 2.5% (w/v) non-fat dried milk before being probed with JOD-2 rabbit antibodies against muskelin [1], mouse monoclonal antibody against GFP (Roche), or mouse monoclonal antibody against γ-tubulin (clone GTU-88; Sigma), followed by ECL® detection using alkaline phosphatase-conjugated secondary antibodies (Tropix) and Hybond® ECL film (Amersham).
Blot overlay assay
The blot overlay method was developed from that described by Li et al. [33]. Cell extracts or purified proteins were resolved on SDS/10%-polyacrylamide gels under reducing conditions and transferred to nitrocellulose. Blots were rinsed in buffer A (10 mM Hepes/KOH, pH 7.5, 60 mM KCl, 1 mM EDTA, 1 mM 2-mercaptoethanol) for 5 min at 4 °C and then incubated in buffer A supplemented with decreasing concentrations of guanidine hydrochloride (Sigma) (6, 3, 1.5, 0.75, 0.38, 0.19, 0.1 and 0 M) for 10 min at each step, to renature the immobilized proteins. The membranes were blocked by incubation in buffer A containing 5% (w/v) milk powder and 0.05% Nonidet-40 for 60 min, and then in buffer A containing 1% milk powder and 0.05% Nonidet-40 for 60 min. [35S]Methionine-labelled muskelin was prepared by coupled in vitro transcription/translation reactions using the TNT T7 transcription/translation kit (Promega) as described [3], and aggregates were removed by centrifugation at 100000 g for 30 min. Membranes were overlaid for 12 h with 3–5 ml of buffer A supplemented with 50 μl of in vitro-translated muskelin (approx. 100 ng of muskelin). Membranes were then rinsed with buffer A for 5 min and exposed to Kodak Biomax film at −70 °C.
Actin co-sedimentation
Purified rabbit skeletal actin (Cytoskeleton Inc.) was suspended at 2 mg/ml in G-buffer (10 mM Tris, pH 7.5, 130 mM KCl, 20 mM NaCl, 2 mM MgCl2, 1 mM 2-mercaptoethanol, 0.1 mM EGTA) and cleared of aggregates by centrifugation at 100000 g for 30 min at 4 °C. The supernatant was transferred to a fresh tube and ATP added to a concentration of 0.2 μM in the presence or absence of predetermined amounts of in vitro-translated muskelin. The mixture was incubated at 25 °C for 30 min. Polymerized F-actin, or muskelin alone, was then pelleted by centrifugation at 100000 g for 30 min at 25 °C. Equal portions of supernatant and the resuspended pellet were mixed with SDS/PAGE sample buffer containing 10 mM DTT for SDS/PAGE analysis followed by immunoblotting. Blots were probed for actin (clone AC-40; Sigma) or exposed to autoradiographic film to detect radiolabelled muskelin.
Microtubule co-sedimentation
Purified bovine brain tubulin (ICN) was diluted in ice-cold suspension buffer (80 mM Mes, pH 6.8, 5 mM DTT, 1 mM EGTA, 1 mM MgCl2) to a final concentration of 2 mg/ml, and aggregates were removed by centrifugation at 100000 g for 30 min at 4 °C. The supernatant was transferred to a freshly chilled tube and polymerization initiated by the addition of 1 mM GTP and 10 mg/ml taxol (final concentrations). The mixture was incubated at 37 °C for 45 min and then diluted 1:1 (v/v) to give a final tubulin concentration of 1 mg/ml. Equal aliquots were incubated for 30 min at 37 °C with or without in vitro-translated muskelin. Microtubules, or muskelin alone, were then pelleted at 100000 g for 30 min at 37 °C underlaid by a cushion of 25% (w/v) sucrose in polymerization buffer containing taxol. Equal aliquots of the supernatant and pelleted microtubules were separated into equal volumes of SDS/PAGE sample buffer containing 100 mM DTT and analysed by SDS/PAGE and immunoblotting for tubulin. Muskelin was detected by autoradiography of the immunoblots.
Fluorescence microscopy
Cells were plated on coverslips at 4×104 cells/cm2 in 6-well plates and transfected with muskelin expression vectors using PolyFect as transfection agent as described above. For proteasome inhibition, transfected cells were treated 36 h later with 100 μM PSI [proteasome inhibitor I; benzyloxycarbonyl-Ile-Glu(OtBu)-Ala-Leu-CHO; Calbiochem] or 5 μM Z-LLF-CHO (benzyloxycarbonyl-Leu-Leu-phenylalaninal; Calbiochem) for 1, 6 or 12 h at 37 °C. After washing in PBS, cells expressing GFP constructs were fixed in 3.7% (v/v) formaldehyde in PBS for 10 min at room temperature, washed in PBS and mounted with VectaShield mounting medium (Vector Laboratories). Samples were examined by epifluorescence using a Leica DM LB microscope equipped with 100×/1.30 PH3 oil objective. Images were captured using a cooled charged-coupled device camera (Optronics) and MagnaFire digital imaging capture software (Optronics) at 1280×1024 pixels resolution and processed using Adobe PhotoShop 7.0.
Confocal images in XY and Z were obtained using a Bio-Rad MRC-1000 laser confocal microscope equipped with a 63× oil objective. From each transient transfection, >100 transfected cells were counted, and the percentage of cells containing muskelin particles was quantified. For fluorescent co-staining, cells were fixed with 3.7% (v/v) paraformaldehyde in PBS for 10 min and permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature, then incubated with TRITC (tetramethylrhodamine β-isothiocyanate)–phalloidin for 60 min at room temperature. Cells expressing endogenous muskelin were stained as described in [1], and cells expressing V5-tagged muskelin were stained with FITC-conjugated goat antibody to the V5-epitope tag (Abcam). Coverslips were washed in PBS and water before mounting with VectaShield mounting medium (Vector Laboratories).
RESULTS
Identification of the N-terminal region of muskelin as a predicted discoidin domain
The N-terminus of muskelin (amino acids 1–160 in human and mouse muskelins) is a highly conserved region of the protein, with 52% identity between the human and D. melanogaster orthologues [3]. We studied this region closely by multiple sequence alignment of a new dataset of eight animal muskelins that included previously unexamined insect and chordate orthologues (identified as described in the Materials and methods section) (Figure 1). The 50% identity consensus sequence derived from the CLUSTALW alignment was then used to query the CDD at NCBI [25]. This revealed that the N-terminal region (amino acids 1–145) of mouse muskelin is predicted to fold as an F5/F8_type_C domain, also known as the discoidin domain (CDD24469, Pfam 00754, PDB 1CZT) (e value of 7×10−6 against a threshold of e=0.01). The discoidin domain is an evolutionarily widespread (present in bacteria and eukaryotes; Pfam 00754 species tree), β-barrel fold that mediates specific protein–protein interactions in a number of extracellular proteins such as neuropilin. In clotting factors V and VIII, the domain is required for binding to membrane phospholipids [34]. More recently, discoidin domain folds have been identified in two intracellular proteins: the N-terminal domain of XRCC1 (X-ray cross-complementing group 1; PDB 1XNA) and APC10 (anaphase-promoting complex subunit 10; PDB 1JHJ). These domains mediate specific binding interactions within large multimolecular complexes [35,36].
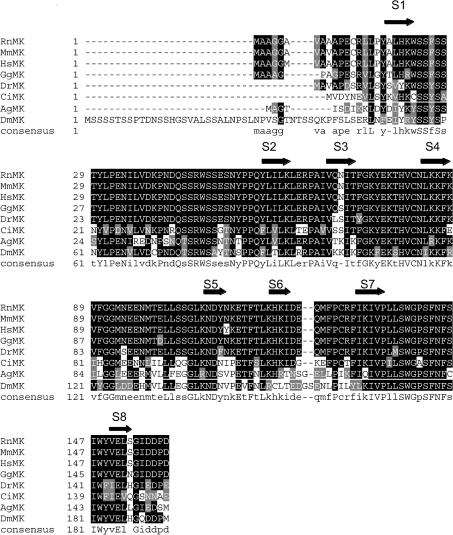
Figure 1. CLUSTALW alignment of the N-terminal regions of eight animal muskelins.
The N-terminal sequences of Anopheles gambiae (Ag; GenBank XP_314298 and BM606168), Ciona intestinalis (Ci; BW210776 and other ESTs in cluster 05014r1), Danio rerio (Dr; AF418017), Drosophila melanogaster (Dm; AF049333), Gallus gallus (Gg; AJ451691), Homo sapiens (Hs; NP_037387), Mus muscularis (Mm; NP_038819) and Rattus norvegicus (Rn; NM_031359) muskelins (MKs) were aligned in CLUSTALW and are presented in Boxshade. Black shading indicates identical residues, grey shading indicates conservative substitutions, and no shading indicates unrelated amino acids. The 50% identity consensus sequence is shown below the alignment. Blocks corresponding to the eight β-strands of a discoidin domain β-barrel, S1–S8, are indicated by arrows (see also Figure 2).
Inspection of the mouse muskelin N-terminal domain sequence and the muskelin consensus sequence in alignment with three discoidin domain structures from the PDB revealed strong conservation of residues and predicted β-strands in muskelin at positions corresponding to the eight antiparallel β-strands that make up the β-barrel of a discoidin domain structure (Figures 1 and 2). A very highly conserved 21 aa motif, the MIND motif, was identified previously within the N-terminus of muskelin (aa 132–152 of mouse muskelin [3]). The ends of this motif mapped on to the predicted seventh and eighth β-strands of the β-barrel, and thus would be predicted to be necessary for domain structure, through hydrophobic contacts with adjacent β-strands (S7 and S8; highlighted in red in Figures 2A and 2B). In conjunction with our previous findings, this analysis supports our view of muskelin as a multidomain protein with two major globular domains, the discoidin-like domain and the kelch-repeat β-propeller, that are linked together by a central α-helical region (Figure 3).
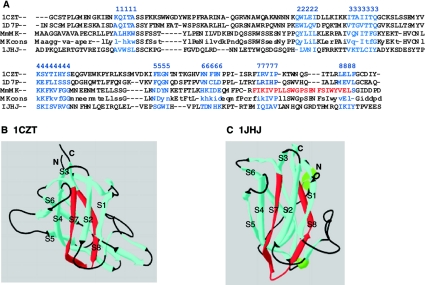
Figure 2. The muskelin N-terminal region is predicted to fold as a discoidin domain.
(A) Alignment by CLUSTALW of the N-terminal region of mouse muskelin (MmMK) and the 50% identity consensus sequence (MKcons) with three discoidin domain structures from PDB. 1CZT, C2 domain of coagulation factor V; 1D7P, C2 domain of coagulation factor VIII; 1JHJ, anaphase-promoting complex subunit 10. The eight β-strands of the β-barrel (numbered here 1–8) were assigned from the CDD and by inspection of the primary sequences against the structures in PDB, and are marked in blue. The MIND motif in muskelin is marked in red. (B, C) Ribbon diagrams of the discoidin domains of 1CZT (B) and 1JHJ (C). In each structure, β-strands are in blue (with the strands of the β-barrel labelled S1–S8), loops are in black, and the regions aligned with the muskelin MIND motif (as shown in A) are in red.
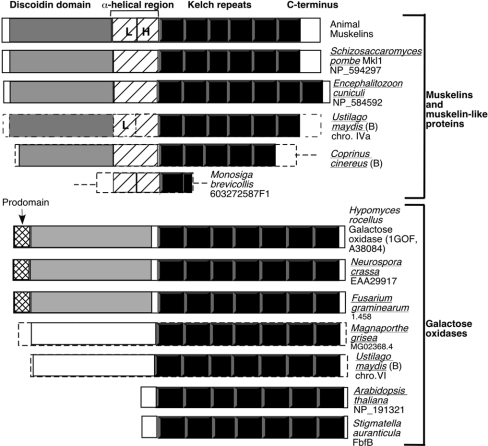
Figure 3. Domain architecture of muskelins and extracellular galactose oxidases.
In each stick model, the predicted discoidin-like domain is shown in grey, the kelch repeats in black, the central α-helical region of muskelin with diagonal lines, the prodomain of galactose oxidase as cross-hatching, and unrelated portions of the polypeptides in white. L, LisH motif; H, CTLH motif (as identified in SMART). The broken lines for Ustilago maydis, Coprinus cinereus, Magnaporthe grisea and Monosiga brevicollis N- and C-terminal ends indicate that the ends of the ORFs are not definitively assigned. The seven kelch repeats in NP_191321 and AAL25195 were identified by multiple sequence alignment with A38084 and assignment of the kelch repeats according to the 1GOF structure. NP_191321 and AAL25195 contain predicted signal peptides. B, basidiomycote. Organisms for which the complete genome sequence is available are underlined.
A limited number of eukaryotic proteins have the discoidin domain/kelch-repeat domain architecture
TBLASTX sequence searches of muskelin against the predicted proteomes of human, mouse, D. melanogaster, Anopheles gambiae, Caenorhabditis elegans and Arabidopsis thaliana has shown that muskelin is the only kelch-repeat protein of these organisms that has the predicted domain architecture of a discoidin-like domain and kelch repeats [11]. Because the discoidin domain and kelch repeats are present individually in numerous proteins from the bacterial and eukaryotic domains of life (Pfam 00754 and 01344 species trees), we were interested to establish whether proteins from any other organisms shared this domain architecture or would correspond to muskelin homologues. First, we carried out BLASTP sequence similarity searches of the GenBank non-redundant protein database with mouse muskelin as a query. In addition to re-identifying the muskelin-like protein of Schizosaccharomyces pombe (Mkl1, SPAC15.10 [3]; Figure 3), the hypothetical protein NP_584592 of Encephalitizoon cuniculi [31] was also found to contain a predicted N-terminal discoidin domain (e value=2×10−4) and kelch repeats. Encephalitizoon cuniculi is an intracellular eukaryotic parasite of animals that is considered to be a highly evolved fungus [37]. Further sequence analysis of NP_584592 in comparison with muskelin showed that it contains seven kelch repeats, but does not contain recognizable LisH or CTHL motifs, although the central region of the polypeptide was predicted to have α-helical secondary structure (Figure 3). Choanoflagellate protozoa are considered to have a close phylogenetic relationship with metazoan animals [38]. Searches of choanoflagellate EST databases identified a partial coding sequence for a muskelin-like protein in Monosiga brevicollis, including the α-helical region and a portion of the kelch repeats (EST 603272587F1; Figure 3). Thus muskelin-like proteins were identified in three out of the five eukaryotic kingdoms.
Because proteins of low sequence identity can have significant structural similarity, we also queried the CDD with muskelin. Interestingly, the single kelch-repeat protein for which a crystal structure has been determined, the extracellular galactose oxidase of the fungus Hypomyces rocellus/Dactylium dendroides, contains a discoidin domain and a kelch-repeat β-propeller domain (PDB 1GOF) [8,12,13]. The primary structures of muskelin and galactose oxidase do not have significant sequence similarity: in a multiple sequence alignment, mouse muskelin and 1GOF had 16% identity and 26% similarity, yet similarity is apparent at the level of domain architecture [Figure 3, CDART (conserved domain architecture retrieval tool) and results not shown]. Galactose oxidase is a member of a group of evolutionarily conserved copper-tyrosyl radical enzymes. Its function is to oxidize primary alcohols to aldehydes [39]. Related, predicted copper-tyrosyl oxidases have been identified in Neurospora crassa (EAA29917 [30]), Arabidopsis thaliana (NP_191321, pirT06758 [40]) and the bacteria Stigmatella aurantiaca (FbfB; CAA77680 and AAL25195 [41]) and Gloeobacter violaceus (NP_927197 [42]); however, of these, only certain fungal enzymes have the architecture of a discoidin domain and a kelch-repeat domain (Figure 3). In 1GOF, the discoidin domain acts as a substrate-binding site and the kelch β-propeller is the catalytic domain [8,13]. Glyoxyl oxidase is a related enzyme that lacks a discoidin domain and has a different substrate specificity, but shares the same active-site motifs [43,44]. In this family of copper-tyrosyl radical enzymes, two conserved sequence motifs, RXYHS and RXYXSS, are critical for co-ordination of the copper ion and for catalysis [44]. These motifs are not present in any of the muskelins that we have identified.
We noticed that the fungal kingdom is apparently the only one in which both muskelin-like proteins and extracellular galactose oxidases are present, yet from this first analysis of complete genomes, individual species encoded either a muskelin-like protein (Schizosaccharomyces pombe, Encephalitizoon cuniculi) or a galactose oxidase (Neurospora crassa), or neither (Saccharomyces cerevisiae). Searches of additional complete genomes of ascomycote fungi, available through the FGI/WICGR, identified further examples of galactose oxidase-like predicted proteins in the complete genomes of Fusarium graminearum (ORF on contig 1.458, scaffold 8, with 98% identity to IGOF; ORF on contig 1.10, scaffold 1, with 65% identity to 1GOF, and ORF on contig 1.160, scaffold 2, with 59% identity to 1GOF) and Magnaporthe grisea (one ORF on contig 2.473 with 44% identity to the 1GOF catalytic domain; MG02368.4), but did not identify muskelin-like proteins. Searches of the available genomes of basidiomycote fungi (one incomplete and three whole genome shotgun sequences at NCBI, and two datasets for the complete Ustilago maydis genome at FGI/WICGR, as of December 2003) for muskelin and galactose oxidase showed that no muskelin-like or galactose oxidase-like protein was encoded in the genome of Cryptococcus neoformans. The Coprinus cinereus genome encoded a muskelin-like protein (ORF in Cc1.52, scaffold 5; 32% identity with mouse muskelin over 550 aa). Importantly, the Ustilago maydis genome encoded both a galactose oxidase-like protein (ORF in Um1.94 scaffold 5 and Bayer contig 1020931, chromosome VI; 40% identity to 1GOF) and a muskelin-like protein (ORF in Um1.190, scaffold 16 and Bayer contig 0.7, chromosome IVa; 34% identity to mouse muskelin) (Figure 3).
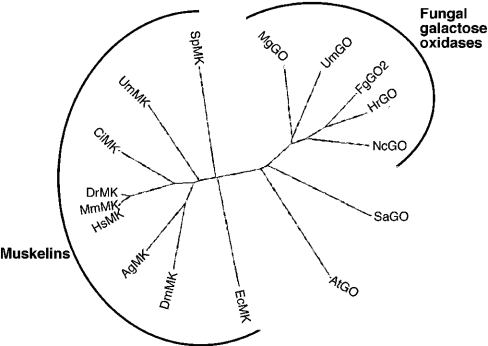
The overall amino acid identity between the fungal muskelins was approx. 30%, similar to their relatedness to animal muskelins (Table 1). The sequence identities between fungal galactose oxidases were between 38% and 52%, distinctly higher than the approx. 16% identity with mouse muskelin (Table 2). The overall identity between the muskelin-like and galactose oxidase-like proteins of Ustilago maydis was 18%. Thus the two protein sequences are as remote from one another when encoded within the same genome as when compared across eukaryotic kingdoms. The phylogenetic relationship between muskelins and galactose oxidases was examined further using unrooted dendrograms prepared in PROTPARS and DRAWTREE. Both sequence analysis methods produced similar results, with the muskelins grouping separately from the galactose oxidases (Figure 4). However, the relatively low identity between the fungal muskelins resulted in program-dependent differences in their exact placement relative to animal muskelins (results not shown). In conclusion, muskelins appear to have arisen within the opistokont lineage and to constitute a unique group of proteins. The two major domains (discoidin-like domain and kelch-repeats) have been conserved through their evolution.
Table 1. Amino acid sequence identity of muskelins.
The percentage sequence identities of muskelins from the indicated species are shown.
| Identity (%) | ||||||
|---|---|---|---|---|---|---|
| M. muscularis | D. melanogaster | S. pombe | E. cuniculi | U. maydis | C. cinereus | |
| Mus muscularis | 100 | 52 | 28 | 26 | 34 | 31 |
| Drosophila melanogaster | 100 | 28 | 33 | 27 | 30 | |
| Schizosaccharomyces pombe | 100 | 27 | 27 | 26 | ||
| Encephalitizoon cuniculi | 100 | 30 | 27 | |||
| Ustilago maydis | 100 | 26 | ||||
| Coprinus cinereus | 100 | |||||
Table 2. Amino acid sequence identity of fungal galactose oxidases.
The percentage sequence identities of galactose oxidases from the indicated species of fungi are shown.
| Identity (%) | ||||
|---|---|---|---|---|
| H. rocellus | N. crassa | U. maydis | M. grisea | |
| Hypomyces rocellus | 100 | 52 | 40 | 44 |
| Neurospora crassa | 100 | 38 | 43 | |
| Ustilago maydis | 100 | 40 | ||
| Magnaporthe grisea | 100 | |||
Figure 4. Phylogenetic relationships of muskelins and galactose oxidases.
Six kelch repeats from each of the indicated species of muskelin (MK) or galactose oxidase (GO) were selected for analysis as described in the Materials and methods section and aligned in CLUSTALW, and are presented as an unrooted dendrogram in DRAWTREE. The incomplete Coprinus cinereus muskelin sequence was not included. Key: Ag, Anopheles gambiae; At, Arabidopsis thaliana; Ci, Ciona intestinalis; Dr, Danio rerio; Dm, Drosophila melanogaster; Ec, Encephalitizoon cuniculi; Fg, Fusarium graminearum; Hr, Hypomyces rocellus; Hs, Homo sapiens; Mg, Magnaporthe grisea; Mm, Mus muscularis; Nc, Neurospora crassa; Sa, Stigmatella aurantiaca; Sp, Schizosaccharomyces pombe; Um, Ustilago maydis.
Biochemical analysis of muskelin self-association: roles for the discoidin-like domain and the kelch-repeat β-propeller
On the basis of this knowledge of muskelin domain architecture, and the fact that other kelch-repeat proteins oligomerize, we tested muskelin for self-association capacity. For this analysis, we developed several tagged versions of wild-type muskelin and two deletion mutants that contained either the discoidin-like domain or the C-terminal half containing the kelch-repeat β-propeller and C-terminus (Figure 5A). We also prepared predicted function-perturbing point mutations within the β-propeller. In each kelch motif, eight highly conserved residues define a consensus motif [7,8]. These residues are positioned on or closely adjacent to the four β-strands that form the core of each propeller blade, because the major structural requirement for assembly of a β-propeller is for hydrophobic interactions that enable correct packing of each propeller blade [45]. A number of natural disease-causing mutations in the kelch-repeat proteins RAG-2 and gigaxonin involve non-conservative substitutions of residues within the consensus motif that disrupt the hydrophobic packing of the blade and are deleterious to the circular closure and stabilization of the β-propeller [46–48]. To mimic these mutations, we mutated key consensus residues in kelch repeat 4 of muskelin (Figure 5A). Two mutants were prepared, EGFP–MK-Y488S/V495S and EGFP–MK-G474A/G475A. Both gave the same results in our experiments, and only the data for EGFP–MK-Y488A/V495A are presented.
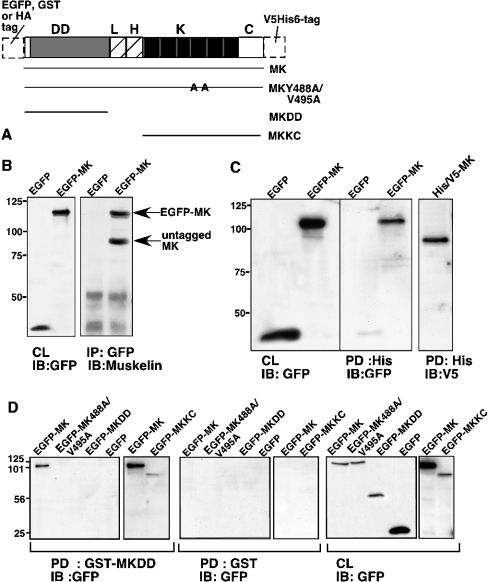
Figure 5. Biochemical analysis of muskelin self-association.
(A) Schematic diagram of muskelin constructs, their designations and the various N- and C-terminal tags utilized. HA, haemagglutinin; DD, discoidin-like domain; L, LisH motif; H, CTLH motif; K, kelch repeats; C, C-terminal region. (B) Co-immunoprecipitation of muskelin with EGFP–MK. Lysates from 293T cells expressing EGFP or EGFP–MK were mixed 1:1 (v/v) with SMC lysates and immunoprecipitated (IP) for GFP. Bound proteins were resolved on 10% (w/v) polyacrylamide gels, transferred to a PVDF membrane and probed with antibodies to muskelin. Expression of the transiently expressed proteins was monitored by immunoblot (IB) of cell lysates with anti-GFP antibody (CL). (C) Specific pull-down (PD) of EGFP–MK with MK–V5His6. Lysates of 293T cells transiently expressing MK–V5His6 were mixed 1:1 (v/v) with lysates from cells expressing EGFP or EGFP–MK and incubated with nickel beads. Bead-bound proteins were resolved on 10% (w/v) polyacrylamide gels, transferred to a PVDF membrane and probed with antibodies to polyhistidine or GFP. Expression of the transiently expressed proteins was monitored by immunoblotting (IB) of cell lysates (CL). (D) Mapping requirements for pull-down (PD) by GST–MKDD. Aliquots of 400 μg of detergent lysates of 293T cells transiently transfected with the indicated EGFP–MK proteins were incubated with either GST–MKDD or GST-loaded glutathione–agarose beads. Aliquots of 45 μg of the lysates were also taken to monitor protein levels (CL). The bound proteins were resolved on SDS/10%-polyacrylamide gels, transferred to a PVDF membrane and probed with antibody to GFP. Only wild-type EGFP–MK and EGFP–MKKC bound to GST–MKDD, and there was no binding to GST. The results are representative of three independent experiments.
We first tested co-immunoprecipitation of the full-length wild-type protein. Muskelin was specifically co-immunoprecipitated with EGFP–MK when mixed with lysates of EGFP- or EGFP–MK-expressing cells (Figure 5B). Next, MK–V5His6 was purified by binding to nickel beads and mixed with extracts from cells expressing EGFP or EGFP–MK. EGFP–MK was specifically captured by MK–V5His6 beads (Figure 5C). These results provide evidence for trans-interactions of the full-length protein. To investigate the roles of separate domains, we prepared a GST-fusion protein encoding the discoidin-like domain of muskelin (termed GST–MKDD) and used this as an affinity probe after purification on glutathione–agarose beads to pull down various forms of muskelin from cell extracts. EGFP–MK bound specifically to GST–MKDD beads, whereas EGFP alone did not interact (Figure 5D). The role of the kelch β-propeller in self-association was studied by use of the mutant EGFP–MK-Y488A/V495A. This protein did not bind to GST–MKDD, although it was expressed equivalently to EGFP–MK (Figure 5D). The same procedure was carried out with the MKDD and MKKC deletion constructs. EGFP–MKDD did not bind to GST–MKDD, whereas EGFP–MKKC was bound (Figure 5D), demonstrating a trans-interaction between the two domains in this assay.
Because of the known associations of other kelch-repeat proteins with actin or tubulin, and of the LisH motif with microtubules [6,9], we also tested whether muskelin bound to actin or to tubulin in vitro, under the hypothesis that such interactions might contribute to the stabilization of muskelin oligomers in intact cells. In blot overlay assays, untagged muskelin did not bind to monomeric actin or tubulin, presented in microgram quantities (Figure 6, and G. D. M. Collett and J. C. Adams, unpublished work). As a positive control, interaction of the known actin-binding protein, fascin, with G-actin was detectable with a 5 μg load of G-actin (Figure 6A). To determine whether muskelin bound to F-actin or to microtubules, co-sedimentation assays were carried out under the appropriate polymerization conditions, using protein samples precleared of aggregates by high-speed centrifugation. In four independent experiments, muskelin did not affect the proportion of either microtubules (Figure 6C) or F-actin (Figure 6D) that was pelleted. Although small amounts of muskelin co-sedimented with F-actin or microtubules, there was no consistent or significant enhancement over the amount that was pelleted independently of the presence of F-actin or microtubules (Figure 6). Thus muskelin self-association in vitro did not depend on co-association with actin or tubulin, either as monomers or polymers.
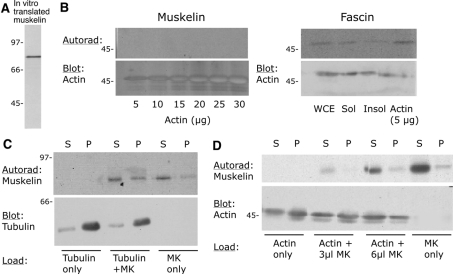
Figure 6. Muskelin does not bind actin or tubulin.
(A) Analysis by SDS/PAGE and autoradiography of [35S]methionine-labelled in vitro-translated muskelin, as used for the experiments shown in (B)–(D). (B) Analysis of the binding of muskelin or fascin to G-actin, detected by blot overlay. In the left-hand panel, the indicated concentrations of actin were resolved on an SDS/12.5%-PAGE gel, transferred to nitrocellulose, renatured and incubated with radiolabelled muskelin. In the right-hand panel, the same procedure was followed using 50 μg of whole-cell extract (WCE), or 50 μg of detergent-soluble (Sol) or -insoluble (Insol) fractions, or 5 μg of G-actin, and the blot was incubated with radiolabelled in vitro-translated fascin. (C, D) Results of co-sedimentation assays carried out as described in the Materials and methods section for microtubule assembly (C) or F-actin polymerization (D). Equal aliquots of supernatant (S) or pellet (P) were resolved on SDS/12.5%-polyacrylamide gels, transferred to nitrocellulose and probed with antibodies to tubulin or actin as appropriate. Muskelin was detected by autoradiographic exposure of the same blots (Autorad). The results are representative of three independent experiments. In all panels, molecular mass markers are in kDa.
Self-association of muskelin in cells
We next wished to determine whether the protein self-association properties that we had detected biochemically applied to muskelin in intact cells. The cells chosen for study were C2C12 cells and SMCs, which have abundant endogenous muskelin, and Cos-7 and 293T cells, in which endogenous muskelin was not detectable (Figure 7A) [1]. The wild-type and mutant EGFP-tagged proteins were expressed equivalently in Cos-7 cells (Figure 7B). First, the distribution of the wild-type protein was studied. By indirect immunofluorescent staining, endogenous muskelin has a predominantly cytoplasmic distribution, with some concentration in membrane ruffles [1,9]. By confocal microscopy, EGFP–MK expressed transiently in Cos-7 cells was found to be cytoplasmic (Figures 7C and 7D). It was noted that the expression of EGFP–MK also resulted in the formation of small cytoplasmic particles which varied in number and in size between cells (arrowed in Figure 7C). By 36 h after transfection of Cos-7 cells with EGFP–MK, 22.4±1.1% of the transfected cells contained particles, with 25% of these cells containing 2–10 particles (n=300 cells, from a total of three experiments). The diameter of the particles varied between 0.44 and 1.33 μm, with a mean (±S.E.M.) diameter of 0.76±0.03 μm (n=46). These distributions appeared to be specific to muskelin, because unfused EGFP, below the size limit for diffusion through the nuclear pore complex [49], was distributed between the nucleus and cytoplasm, and no cytoplasmic particles were observed (Figure 7E).
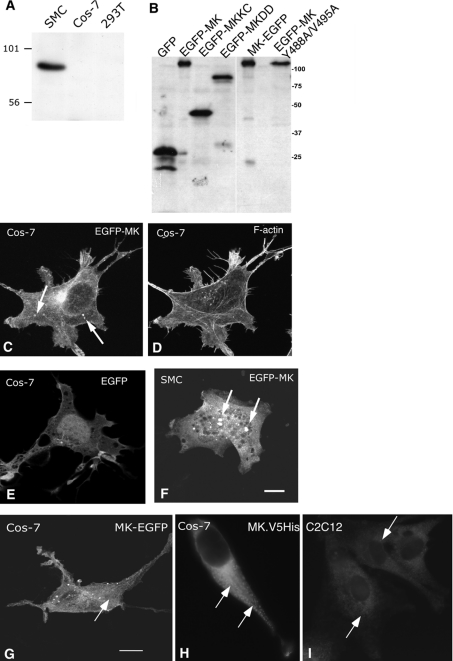
Figure 7. Expression and localization of muskelin in cells.
(A) Western blot of an SDS/10%-polyacrylamide gel loaded with 40 μg of 1% Triton X-100 cell extracts from SMCs, Cos-7 cells and 293T cells, probed with anti-muskelin antibodies. (B) Immunoblot showing relative expression of EGFP–MK proteins, detected by anti-GFP antibody. Protein designations are as in (A). Molecular mass markers are in kDa. (C)–(I) Confocal images of the distribution of transiently expressed EGFP–MK (C, D), MK–EGFP (G) or MK–V5His6 (H) in Cos-7 cells, or EGFP–MK in SMCs (F). Panels (C) and (D) show a Cos-7 cell co-stained for F-actin. (E) Distribution of EGFP in Cos-7 cells. (I) Endogenous muskelin of C2C12 cells (as XY section from Z stack). Arrows in panels (C) and (F)–(I) indicate the muskelin particles of various sizes observed in the cells. Bars=10 μm.
To determine whether the formation of the particles was a feature of expression in Cos-7 cells, EGFP–MK was transiently expressed in SMCs. In these cells, EGFP–MK also had a cytoplasmic distribution, and in some cases formed small particles (arrowed in Figure 7F). Similar results were obtained in C2C12 cells and 293T cells (not shown). To verify that formation of the EGFP–MK particles was not an artifact of the fusion of muskelin C-terminal to EGFP, the subcellular localizations of other forms of the protein were examined. A construct in which muskelin was fused N-terminal to EGFP (termed MK–EGFP) was expressed equivalently to EGFP–MK, as demonstrated by Western blot for EGFP on whole-cell lysates (Figure 7B). Cos-7 cells expressing MK–EGFP contained similar numbers of MK–EGFP particles to cells expressing EGFP–MK (Figure 7G), indicating that the formation of these bodies was independent of the design of the fusion protein. Furthermore, expression of muskelin with a C-terminal V5 epitope tag also resulted in the development of muskelin particles, as well as the general cytoplasmic staining (Figure 7H). C2C12 cells stained with affinity-purified antibodies to muskelin to reveal endogenous muskelin also showed small particles within the cytoplasm, although these were not as abundant as in the transfected cells (Figure 7I).
Aggresomes are bodies of approx. 5 μm, located at the microtubule-organizing centre, in which misfolded protein oligomers become sequestered when in excess of the capacity of the proteosome degradation pathway [50,51]. To clarify that the muskelin particles did not correspond to aggresomes, cells that had expressed EGFP–MK or EGFP–MK-Y488A/V495A for 24 h were incubated with the membrane-permeable proteosome inhibitors PSI or Z-LLF-CHO for 12 h. For both proteins and inhibitors, we observed a loss of the dispersed cytoplasmic EGFP signal and accumulation of EGFP–MK or EGFP–MK-Y488A/V495A in large, amorphous, juxtanuclear aggregates that appeared to correspond to aggresomes (Figures 8A and 8B; shown for PSI treatment only). Thus the small particles formed by endogenous or expressed forms of muskelin were clearly distinct from the large aggregates that accumulated under conditions of impaired protein catabolism.
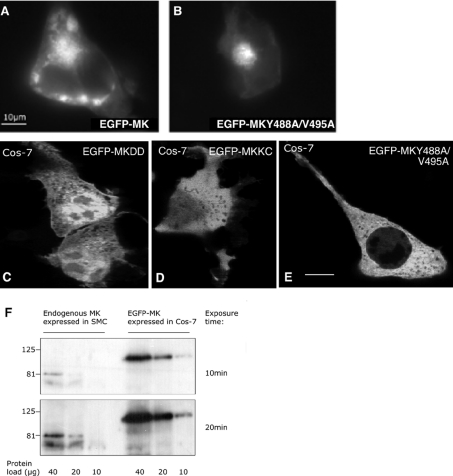
Figure 8. Roles of the discoidin-like domain and the β-propeller in particle formation in cells.
(A, B) Cos-7 cells expressing EGFP–MK (A) or EGFP–MK488A/V495A (B) were treated with 100 μM PSI for 12 h before fixation. Large muskelin aggregates were formed in the cells. (C)–(E) Localization of muskelin mutants. Confocal XY sections of Cos-7 cells transiently expressing EGFP–MKDD (C), EGFP–MKKC (D) or EGFP–MK-Y488A/V495A (E) are shown. None of these proteins formed particles. Bars=10 μm. (F) Expression of EGFP–MK in Cos-7 cells relative to endogenous muskelin in SMCs. The indicated protein loads of whole-cell extracts were resolved on an SDS/10%-polyacrylamide gel, transferred to a PVDF membrane and probed with anti-muskelin antibodies. Two ECL® exposure times were taken for quantification of expression using Scion Image (NIH). Pixel intensity per area was quantified at each exposure time (EGFP–MK signal from a 10 μg protein load compared with muskelin signal from 40 and 20 μg protein loads at 20 min exposure time) and mean values calculated.
The discoidin-like domain and kelch-repeat domain both contribute to the assembly of muskelin particles in cells
We intepret the muskelin particles to result from the self-association capacity of the protein. To determine the contributions of the discoidin-like domain and the kelch-repeat domain to particle formation, the localizations of the N- and C-terminal deletion mutant proteins were examined. EGFP–MKDD, with a molecular mass of 44 kDa, was distributed between the cytoplasm and the nucleus, and no muskelin particles were detected (Figure 8C). The molecular mass of this protein is below the size limit for diffusion through the nuclear pore complex [49]. The same distribution was observed for a haemagglutinin-tagged MKDD protein of 30 kDa (S. Prag and J. C. Adams, unpublished work). The EGFP–MKKC protein, of molecular mass 83 kDa, was located mainly in the cytoplasm, and no muskelin particles were detected. In some cells, nuclear muskelin was detected at low levels (Figure 8D). EGFP–MK-Y488A/V495A also had a diffuse cytoplas-mic distribution and did not form any particles (Figure 8E). Thus both a wild-type β-propeller and the discoidin-like domain are needed for particle formation. Localization within the cytoplasm also depends on the full-length protein.
In our studies of muskelin, we have found that SMCs and C2C12 cells contain high levels relative to other cell lines (S. Prag, G. D. M. Collett and J. C. Adams, unpublished work). To relate particle formation to the level of expression, we quantified the level of EGFP–MK relative to the endogenous level in SMCs, by immunoblotting matched protein loads from SMCs and transiently transfected Cos-7 cells with an antibody to muskelin. The level of expression achieved was at most 4-fold over that of SMCs (Figure 8F).
DISCUSSION
Muskelin is a unique, intracellular member of the kelch-repeat superfamily. Here we have combined bioinformatic approaches that identify the N-terminal region as a predicted discoidin-like domain with experimental data that demonstrate that the discoidin-like domain contributes to protein self-association in conjunction with the C-terminal kelch-repeat β-propeller. The predicted domain architecture of muskelin highlighted an interesting architectural parallel between muskelins and the extracellular galactose oxidases of fungi that led us to trace the molecular phylogeny of the two groups. These studies extend knowledge of muskelin as a multidomain protein, situate muskelin within the domains of life, and uncover new directions for future studies of protein function.
Muskelin was orginally identified in connection with cell responses to thrombospondin-1. The new phylogenetic data indicate that muskelins originated earlier in evolution than thrombospondins, which are present in the ECM of complex multicellular animals, including insects and chordates, but not Caenorhabditis elegans [52]. Animal muskelins have five conserved consensus motifs for phosphorylation by protein kinase C and two putative CK2 phosphorylation sites that are not shared by Schizosaccharomyces pombe Mkl1 [3]. By multiple sequence alignment, the fifth potential protein kinase C phosphorylation motif, TQR [3], is present in Ustilago maydis and Coprinus cinereus muskelins, but not in Encephalitizoon cuniculi muskelin, and Ustilago maydis also contains a potential protein kinase C phosphorylation motif, SPR, at the second site. The other putative protein kinase C or CK2 motifs are not present in these fungal muskelins. However, the clear conservation of muskelins in animal, fungal and prospectively protozoal kingdoms leads us to suggest that muskelins may have ancestral functions distinct from thrombospondin pathways, or that they were co-opted into thrombospondin regulation as a consequence of the evolution of the ECM in multicellular animals. The identification of muskelin in several well studied fungi and animals opens broad scope to address this question in the future. Interestingly, the relevant fungi have very diverse lifestyles that range from obligate intracellular parasitism of animals (Encephalitizoon cuniculi [31]), to a multicellular plant parasite (Ustilago maydis [53]), to free-living single-cell and multicellular forms (Schizosaccharomyces pombe and Coprinus cinereus respectively). In Schizosaccharomyces pombe, muskelin has been documented as a non-essential gene [54].
An obvious question arising from the domain architecture parallels between muskelin and galactose oxidase is whether the two groups of proteins represent the results of divergent or convergent evolution. Structural similarity in the absence of a distinct sequence relationship (i.e. below 20% identity) is generally believed to indicate either extreme divergence from a remote common ancestor or random evolutionary convergence to a very stable fold suited to the properties of both proteins. The predicted structural similarity between muskelins and extracellular galactose oxidase is based on a low, statistically insignificant, level of sequence identity (Table 2). Furthermore, only certain fungal galactose oxidases within the large galactose oxidase family contain a discoidin domain. The presence of galactose oxidase-like encoding sequences in the genomes of bacteria as well as plants and fungi indicates a last common ancestor of very early evolutionary origin. In contrast, the most parsimonious phylogenetic model for muskelins is to postulate a last common ancestor corresponding to an ancestor of opisthokont eukaryotes. Thus, on the basis of currently available genomic information, the two sets of proteins appear to have distinct evolutionary origins and to have undergone convergent evolution at the level of domain architecture within the fungal kingdom.
Because other kelch-repeat proteins have been shown to oligomerize through their N-terminal domains, we investigated whether the discoidin-like domain of muskelin is involved in self-association. We demonstrated by co-immunoprecipitation, protein pull-downs and cell-localization studies that muskelin undergoes self-association, and that both the discoidin-like domain and an intact kelch β-propeller are necessary for the interaction. Our cellular and biochemical data implicate kelch repeat 4 in the mechanism of muskelin self-association; however, the results do not rule out the possibility that this repeat binds to a bridging molecule that is necessary for multimerization. We established that bridging does not depend on binding to actin or tubulin. In our experiments, intact wild-type muskelin was necessary for the formation of particles in cells, whereas the C-terminal region was sufficient to bind the discoidin-like domain when presented as an affinity matrix. A possible explanation is that cis-interactions are needed to seed particle formation in cells, whereas the very high concentration of discoidin-like domain presented on the affinity matrix bypasses this requirement. A need for cis-interactions for particle formation in cells was corroborated by the finding that EGFP–MK-Y488A/V495A did not form the particles.
Both cis- and trans-interactions have been documented for other kelch-superfamily proteins. The kinase activity of the actin-fragmin kinase N-terminal domain is enhanced by the C-terminal kelch-repeat β-propeller when present in cis [55]. For HCF-1 (host cell factor-1), interactions of the C-terminal region with the N-terminal kelch-repeat β-propeller contribute to the formation of a complex with VP16 and DNA [56]. In addition, trans-interactions through dimerization of BTB domains have been demonstrated for several BTB/kelch proteins and confer actin cross-linking activity (see Introduction). Several BTB/kelch proteins have been noted to form cellular particles when overexpressed [17,57]. We postulate that physiologically regulated alterations in the ability of muskelin to self-associate may serve as a mechanism to regulate its molecular interactions and activities in the diverse organisms in which it is found.
Acknowledgments
We thank Nina Kureishy for preparing affinity-purified anti-muskelin IgG, Alexia Zaramytidou for managing the sequencing of zebrafish muskelin, Crystal Bykowski for purification of GST–MKDD fusion protein, Raymond Monk for assistance with molecular biology, and Mark Shipman (LMCB) for assistance with confocal microscopy. We thank the CCF Molecular Biotechnology core for DNA sequencing. We acknowledge the public database resources established by the Fungal Genome Initiative. This research was supported by the Wellcome Trust (Prize Studentship 046077 to G.D.M.C., and Senior Fellowship in Basic Biomedical Research 038284 to J.C.A.) and by a Danish Reseach Academy studentship to S.P.
References
- 1.Adams J. C., Seed B., Lawler J. Muskelin, a novel intracellular mediator of cell adhesive and cytoskeletal responses to thrombospondin-1. EMBO J. 1998;17:4964–4974. doi: 10.1093/emboj/17.17.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams J. C., Zhang L. cDNA cloning of human muskelin and localization of the muskelin (MKLN1) gene to human chromosome 7q32 and mouse chromosome 6 B1/B2 by physical mapping and FISH. Cytogenet. Cell Genet. 1999;87:19–21. doi: 10.1159/000015385. [DOI] [PubMed] [Google Scholar]
- 3.Adams J. Characterization of a Drosophila melanogaster orthologue of muskelin. Gene. 2002;297:69–78. doi: 10.1016/s0378-1119(02)00887-9. [DOI] [PubMed] [Google Scholar]
- 4.Cahana A., Escamez T., Nowakowski R. S., Hayes N. L., Giacobini M., von Holst A., Shmueli O., Sapir T., McConnell S. K., Wurst W., et al. Targeted mutagenesis of Lis1 disrupts cortical development and LIS1 homodimerization. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6429–6434. doi: 10.1073/pnas.101122598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith D. S., Niethammer M., Ayala R., Zhou Y., Gambello M. J., Wynshaw-Boris A., Tsai L. H. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- 6.Emes R. D., Ponting C. P. A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum. Mol. Genet. 2001;10:2813–2820. doi: 10.1093/hmg/10.24.2813. [DOI] [PubMed] [Google Scholar]
- 7.Xue F., Cooley L. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 8.Bork P., Doolittle R. F. Drosophila kelch motif is derived from a common enzyme fold. J. Mol. Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 9.Adams J., Kelso R., Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/s0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 10.Shchelkunov S., Totmenin A., Kolosova I. Species-specific differences in organization of orthopoxvirus kelch-like proteins. Virus Genes. 2002;24:157–162. doi: 10.1023/a:1014524717271. [DOI] [PubMed] [Google Scholar]
- 11.Prag S., Adams J. C. Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals. BMC Bioinformatics. 2003. 2003;4:42. doi: 10.1186/1471-2105-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito N., Phillips S. E., Stevens C., Ogel Z. B., McPherson M. J., Keen J. N., Yadav K. D., Knowles P. F. Novel thioether bond revealed by a 1.7 A crystal structure of galactose oxidase. Nature (London) 1991;350:87–90. doi: 10.1038/350087a0. [DOI] [PubMed] [Google Scholar]
- 13.Ito N., Phillips S. E., Yadav K. D., Knowles P. F. Crystal structure of a free radical enzyme, galactose oxidase. J. Mol. Biol. 1994;238:794–814. doi: 10.1006/jmbi.1994.1335. [DOI] [PubMed] [Google Scholar]
- 14.Robinson D. N., Cooley L. Drosophila kelch is an oligomeric ring canal actin organizer. J. Cell Biol. 1997;138:799–810. doi: 10.1083/jcb.138.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim I. F., Mohammadi E., Huang R. C. Isolation and characterization of IPP, a novel human gene encoding an actin-binding, kelch-like protein. Gene. 1999;228:73–83. doi: 10.1016/s0378-1119(99)00006-2. [DOI] [PubMed] [Google Scholar]
- 16.Soltysik-Espanola M., Rogers R. A., Jiang S., Kim T. A., Gaedigk R., White R. A., Avraham H., Avraham S. Characterization of Mayven, a novel actin-binding protein predominantly expressed in brain. Mol. Biol. Cell. 1999;10:2361–2375. doi: 10.1091/mbc.10.7.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Derin R., Petralia R. S., Li M. Actinfilin, a brain-specific actin-binding protein in postsynaptic density. J. Biol. Chem. 2002;277:30495–30501. doi: 10.1074/jbc.M202076200. [DOI] [PubMed] [Google Scholar]
- 18.Itoh K., Wakabayashi N., Katoh Y., Ishii Y., Igarashi K., Engel J. D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding J., Liu J. J., Kowal A. S., Nardine T., Bhattacharya P., Lee A., Yang Y. Microtubule-associated protein 1B: a neuronal binding partner for gigaxonin. J. Cell Biol. 2002;158:427–433. doi: 10.1083/jcb.200202055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satou Y., Yamada L., Mochizuki Y., Takatori N., Kawashima T., Sasaki A., Hamaguchi M., Awazu S., Yagi K., Sasakura Y., et al. A cDNA resource for the basal chordate Ciona intestinalis. Genesis. 2002;33:153–154. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- 21.Higgins D., Thompson J., Gibson T., Thompson J. D., Higgins D. G., Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falquet L., Pagni M., Bucher P., Hulo N., Sigrist C. J., Hofmann K., Bairoch A. The PROSITE database, its status in 2002. Nucleic Acids Res. 2002;30:235–238. doi: 10.1093/nar/30.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz J., Milpetz F., Bork P., Ponting C. P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonnhammer E. L., Eddy S. R., Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 25.Marchler-Bauer A., Panchenko A. R., Shoemaker B. A., Thiessen P. A., Geer L. Y., Bryant S. H. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King N., Hittinger C. T., Carroll S. B. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- 27.Cummings L., Riley L., Black L., Souvorov S. A., Resenchuk S., Dondoshansky I., Tatusova T. Genomic BLAST: custom-defined virtual databases for complete and unfinished genomes. FEMS Microbiol. Lett. 2002;216:133–138. doi: 10.1111/j.1574-6968.2002.tb11426.x. [DOI] [PubMed] [Google Scholar]
- 28.Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., Galibert F., Hoheisel J. D., Jacq C., Johnston, et al. Life with 6000 genes. Science. 1996;274:546–549. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 29.Wood V., Gwilliam R., Rajandream M. A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., et al. The genome sequence of Schizosaccharomyces pombe. Nature (London) 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 30.Galagan J. E., Calvo S. E., Borkovich K. A., Selker E. U., Read N. D., Jaffe D., FitzHugh W., Ma L. J., Smirnov S., Purcell S., et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature (London) 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 31.Katinka M. D., Duprat S., Cornillot E., Metenier G., Thomarat F., Prensier G., Barbe V., Peyretaillade E., Brottier P., Wincker P., et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature (London) 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J., Russel D. W. 3rd edn. Cold Spring Habour, NY: CSHL Press; 2001. Molecular Cloning, A Laboratory Manual. [Google Scholar]
- 33.Li M., Jan Y. N., Jan L. Y. Specification of subunit assembly by the hydrophilic amino-terminal domain of the Shaker potassium channel. Science. 1992;257:1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- 34.Baumgartner S., Hofmann K., Chiquet-Ehrismann R., Bucher P. The discoidin domain family revisited: new members from prokaryotes and a homology-based fold prediction. Protein Sci. 1998;7:1626–1631. doi: 10.1002/pro.5560070717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marintchev A., Mullen M. A., Maciejewski M. W., Pan B., Gryk M. R., Mullen G. P. Solution structure of the single-strand break repair protein XRCC1 N-terminal domain. Nat. Struct. Biol. 1999;6:884–893. doi: 10.1038/12347. [DOI] [PubMed] [Google Scholar]
- 36.Wendt K. S., Vodermaier H. C., Jacob U., Gieffers C., Gmachl M., Peters J.-M., Huber R., Sondermann P. Crystal structure of the APC/DOC1 subunit of the human anaphase-promoting complex. Nat. Struct. Biol. 2001;8:784–788. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- 37.Keeling P. J., McFadden G. I. Origins of microsporidia. Trends Microbiol. 1998;6:19–23. doi: 10.1016/S0966-842X(97)01185-2. [DOI] [PubMed] [Google Scholar]
- 38.Baldorf S. L. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 39.Rogers M. S., Dooley D. M. Copper-tyrosyl radical enzymes. Curr. Opin. Struct. Biol. 2003;7:189–196. doi: 10.1016/s1367-5931(03)00024-3. [DOI] [PubMed] [Google Scholar]
- 40.Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 41.Silakowski B., Ehret H., Schairer H. U. fbfB, a gene encoding a putative galactose oxidase, is involved in Stigmatella auranticiana fruiting body formation. J. Bacteriol. 1998;180:1241–1247. doi: 10.1128/jb.180.5.1241-1247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura Y., Kaneko T., Sato S., Mimuro M., Miyashita H., Tsuchiya T., Sasamoto S., Watanabe A., Kawashima K., Kishida Y., et al. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 2003;10:137–145. doi: 10.1093/dnares/10.4.137. [DOI] [PubMed] [Google Scholar]
- 43.Whittaker M. M., Kersten P. J., Nakamura N., Sanders-Loehr J., Schweizer E. S., Whittaker J. W. Glyoxyl oxidase from Phanerochaete chrysosporium is a new radical-copper oxidase. J. Biol. Chem. 1996;271:681–687. doi: 10.1074/jbc.271.2.681. [DOI] [PubMed] [Google Scholar]
- 44.Whittaker M. M., Kersten P. J., Cullen D., Whittaker J. W. Identification of catalytic residues in glyoxal oxidase by targeted mutagenesis. J. Biol. Chem. 1999;274:36226–36232. doi: 10.1074/jbc.274.51.36226. [DOI] [PubMed] [Google Scholar]
- 45.Fulop V., Jones D. T. Beta propellers: structural rigidity and functional diversity. Curr. Opin. Struct. Biol. 1999;9:715–721. doi: 10.1016/s0959-440x(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz K., Gauss G. H., Ludwig L., Pannicke U., Li Z., Linder D., Friedrich W., Seger R. A., Hansen-Hagge T. E., Desiderio S., et al. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 47.Callebaut I., Mornon J. P. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol. Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bomont P., Cavalier L., Blondeau F., Ben-Hamida C., Belal S., Tazir M., Demir E., Topaloglu H., Korinthenberg R., Tuysuz B., et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat. Genet. 2000;26:370–374. doi: 10.1038/81701. [DOI] [PubMed] [Google Scholar]
- 49.Johnston J. A., Ward C. L., Kopito R. R. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia-Mata R., Bebok Z., Sorscher E. J., Sztul E. S. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1999;146:1239–1254. doi: 10.1083/jcb.146.6.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters R. Nucleo-cytoplasmic flux and intracellular mobility in single hepatocytes measured by fluorescence microphotolysis. EMBO J. 1984;3:1831–1836. doi: 10.1002/j.1460-2075.1984.tb02055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams J. C., Monk R., Taylor A. L., Ozbek S., Fascetti N., Baumgartner S., Engel J. Characterization of Drosophila thrombospondin defines an early origin of pentameric thrombospondins. J. Mol. Biol. 2003;328:479–484. doi: 10.1016/s0022-2836(03)00248-1. [DOI] [PubMed] [Google Scholar]
- 53.Bolker M. Ustilago maydis – a valuable model system for the study of fungal dimorphism and virulence. Microbiology. 2001;147:1395–1401. doi: 10.1099/00221287-147-6-1395. [DOI] [PubMed] [Google Scholar]
- 54.Decottignies A., Sanchez-Perez I., Nurse P. Schizosaccharomyces pombe essential genes: a pilot study. Genome Res. 2003;13:399–406. doi: 10.1101/gr.636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eichinger L., Bomblies L., Vandekerckhove J., Schleicher M., Gettemans J. A novel type of protein kinase phosphorylates actin in the actin-fragmin complex. EMBO J. 1996;15:5547–5556. [PMC free article] [PubMed] [Google Scholar]
- 56.Hughes T. A., La Boissiere S., O'Hare P. Analysis of functional domains of the host cell factor involved in VP16 complex formation. J. Biol. Chem. 1999;274:16437–16443. doi: 10.1074/jbc.274.23.16437. [DOI] [PubMed] [Google Scholar]
- 57.Sasagawa K., Matsudo Y., Kang M., Fujimura L., Iitsuka Y., Okada S., Ochiai T., Tokuhisa T., Hatano M. Identification of Nd1, a novel murine kelch family protein involved in stabilization of actin filaments. J. Biol. Chem. 2003;277:44140–44146. doi: 10.1074/jbc.M202596200. [DOI] [PubMed] [Google Scholar]