Abstract
mAKAP (muscle-selective A-kinase-anchoring protein) co-ordinates a cAMP-sensitive negative-feedback loop comprising PKA (cAMP-dependent protein kinase) and the cAMP-selective PDE4D3 (phosphodiesterase 4D3). In vitro and cellular experiments demonstrate that PKA-phosphorylation of PDE4D3 on Ser-13 increases the affinity of PDE4D3 for mAKAP. Our data suggest that activation of mAKAP-anchored PKA enhances the recruitment of PDE4D3, allowing for quicker signal termination.
Keywords: A-kinase-anchoring protein (AKAP), cAMP-dependent protein kinase (PKA), increased affinity, phosphodiesterase, serine-13, signalling
Abbreviations: AEBSF, 4-(2-aminoethyl)benzenesulphonyl fluoride; AKAP, A-kinase-anchoring protein; mAKAP, muscle-selective AKAP; β2AR, β2-adrenergic receptor; FP, fluorescence polarization; HRP, horseradish peroxidase; IBMX, 3-isobutyl-1-methylxanthine; PDE, phosphodiesterase; PKA, cAMP-dependent protein kinase; PP, protein phosphatase; VSV, vesicular stomatis virus
INTRODUCTION
mAKAP (muscle-selective A-kinase-anchoring protein) is part of a large family of functionally related proteins called AKAPs that serve to sequester PKA (cAMP-dependent protein kinase) to distinct subcellular compartments for local activation [1,2]. This method of regulation ensures that the broad specificity of PKA is focused only on closely situated substrates. A second layer of anchored PKA regulation involves the dissociation of the catalytic subunit upon neighbouring increases of cAMP concentration. Thus the enzymes that generate and degrade cAMP, adenylate cyclases and PDEs (phosphodiesterases) respectively, control PKA activity tightly.
PDEs are encoded by 11 gene families, and within each family are various alternatively spliced forms [3]. PDEs are believed to play important roles in the integration of signalling pathways by controlling the spatial gradient of cyclic nucleotides [4,5]. Specifically hydrolysing cAMP, the PDE4 family is currently a target for small-molecule inhibitors to treat diseases such as asthma, COPD (chronic obstructive pulmonary disease) and cancer [6]. One mechanism used by this family to achieve local PDE activity is compartmentalization to specific areas in the cell by AKAPs. AKAP450 and mAKAP are both capable of scaffolding the signal-termination enzyme PDE4D3 within the same complex as PKA [5,7]. This provides a mechanism to control cAMP levels and thus PKA activity. In fact, functional and biochemical studies have shown that PDE4D3 is a substrate of PKA [8–11]. In the case of mAKAP, hormonal stimulation of cAMP levels causes the release of the catalytic subunit of mAKAP-anchored PKA and results in phosphorylation of PDE4D3 on Ser-54 (PDE4D3 Ser-54). This enhances PDE4D3 activity 2-fold, results in the breakdown of cAMP to basal levels and causes the re-formation of the PKA holoenzyme [5,8,9]. This negative-feedback loop is active at the nuclear membrane of cardiomyocytes where mAKAP is localized via three spectrin repeats [5,12].
Although biochemical studies have shown that PKA phosphorylates all the long isoforms of the PDE4D family for enzyme activation, PDE4D3 is unique in that it can be phosphorylated on a second site, Ser-13 (PDE4D3 Ser-13) [6,8]. However, the functional consequences of Ser-13-phosphorylation on PDE4D3 were unknown until now. mAKAP binds to the 15 unique amino acids that make up the N-terminus of PDE4D3, and, reciprocally, PDE4D3 binds to the central portion of mAKAP containing residues 1286–1831 [5]. Intrigued by the observation that the mAKAP-interaction site for PDE4D3 contains an established PKA-phosphorylation site, we sought to understand the molecular basis by which these two proteins interact. Using several different in vitro and cellular binding methods, our results demonstrate that PKA-phosphorylation of PDE4D3 on Ser-13 increases the affinity of PDE4D3 for mAKAP. Hence this is the first study to assign a functional consequence to PKA-phosphorylation of PDE4D3 Ser-13.
EXPERIMENTAL
Phosphorylation of PDE4D3 peptide and pull-down assay
Biotin-labelled PDE4D3 peptide 1–16 (Biotin-MMHVNNFPFRRHSTIC, synthesized by Cell Essentials, Boston, MA, U.S.A.) was used for phosphorylation by PKA. Peptide (5 mM) was incubated in PKA kinase buffer (50 mM Tris/HCl, pH 7.5, and 5 mM MgCl2) containing 100 μM ATP, 5 μM [γ-32P]ATP and 0.3 mM cAMP with or without the PKA catalytic subunit for 15 min at 30 °C. The reaction mixture was spotted on to p81 phosphocellulose paper, washed five times in 75 mM phosphoric acid and once in 95% ethanol. Filters were air-dried and were counted for radioactivity by liquid-scintillation counting.
For the pull-down assay, the same peptide (5 mM) was incubated in PKA kinase buffer containing 100 μM ATP and 0.3 mM cAMP with or without the PKA catalytic subunit for 2 h at 30 °C. To each sample, 15 μl of Neutravidin beads (Pierce) was added and then rocked for 1 h at room temperature (22 °C). The precipitates were washed three times with HSE buffer {20 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1% (v/v) Triton X-100, 1 mM DTT (dithiothreitol), 1 mM sodium orthovanadate, 50 mM sodium fluoride, 1 mM benzamidine, 0.2 μg/ml leupeptin, 0.2 μg/ml pepstatin and 1 mM AEBSF [4-(2-aminoethyl)benzenesulphonyl fluoride]} and resuspended in 500 μl of HSE buffer containing 5 μM recombinant mAKAP fragments. Reactions were incubated overnight at 4 °C with rocking. Precipitates were washed three times with HSE buffer, and bound proteins were subjected to SDS/7.5% PAGE, followed by immunoblotting with anti-His antibody (1:5000, Amersham Biosciences). For densitometry, the amount of mAKAP 1286–1831 fragment co-immunoprecipitated with the peptides was quantified using NIH Image software and expressed as arbitrary units.
In vitro peptide-binding assay
Biotin-labelled PDE4D3 peptide 1–16 and biotin-labelled PDE4D3 phospho-Ser-13 peptide 1–20 [Biotin-MMHVNNFPFRRHS(PO4)TICFNVN, synthesized by Cell Essentials] were resuspended in water containing 20% (v/v) DMSO to a working concentration of 50 μM. The mAKAP 1286–1831–His protein was generated by infecting H9C2 cells for 2 days with adenovirus directed to express the fragment and then lysing cells in HSE buffer. The extract was cleared by centrifugation at 16000 g for 10 min, and soluble extract was used as the input. For the binding assay, 25 μl of mAKAP 1286–1831–His-containing extract was added to peptides that were diluted to various concentrations in HSE lysis buffer containing 10% (v/v) DMSO in a final volume of 1 ml. To each sample, 20 μl of Neutravidin beads was added and incubated overnight with rocking at 4 °C. Precipitates were washed three times in HSE buffer, resolved by SDS/7.5% PAGE and analysed by anti-His immunoblotting. Quantifying bands using NIH Image, dividing each value by the largest value within an experiment and multiplying by 100 determined the normalized percentage bound. The reported values were generated from the average of three independent experiments.
FP (fluorescence polarization)
Peptides used for FP studies, PDE4D3 1–20 (MMHVNNFPFRRHSTICFNVN) and PDE4D3 phospho-Ser-13 1–20 [MMHVNNFPFRRHS(PO4)TICFNVN, both synthesized by Cell Essentials], were labelled N-terminally with FITC using the Fluorescein-C6-Amine Labeling kit (Panvera, Madison, WI, U.S.A.) following the manufacturer's instructions. Peptides (1 nM) were resuspended in PBS containing 5 mg/ml BSA, pH 7.0. Increasing concentrations of bacterially produced recombinant mAKAP 1286–1831–His were mixed with each FITC-labelled peptide. Each sample was incubated for 10 min. FP was measured on a Beacon 2000 (Panvera) following the manufacturer's instructions. Saturation-binding curves were generated with Prism graphing software (GraphPad, San Diego, CA, U.S.A.). Dissociation constants (Kd) were calculated from non-linear regression curves generated from the average of five independent experiments.
Plasmids, transfection and immunoprecipitation–Western analysis
The pcDNA3.1-mAKAP-MYC-HIS and pcDNA3.1-mAKAP-(Ile2062Pro)-MYC-HIS plasmids were described previously [12]. The pcI-NEO-PDE4D3-VSV (where VSV is vesicular stomatis virus) plasmid was a gift from Professor Miles Houslay (University of Glasgow, Glasgow, Scotland, U.K.). PDE4D3 phospho-mutants were created to mimic the non-phosphorylated form (pcI-NEO-PDE4D3-Ser13Ala-VSV) or the phosphorylated form (pcI-NEO-PDE4D3 Ser13Glu-VSV) by site-directed mutagenesis (Stratagene) following the manufacturer's instructions.
DNA (10 μg total) was transfected into 10-cm-diameter dishes of 50% confluent HEK-293 cells. At 2 days after transfection, cells were lysed with 1 ml of lysis buffer [10 mM Na2PO4, pH 7.4, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1% (v/v) Triton X-100, 1 mM benzamidine, 0.2 μg/ml leupeptin, 0.2 μg/ml pepstatin and 1 mM AEBSF], and extracts were cleared by centrifugation at 16000 g for 10 min. Equal amounts of protein were immunoprecipitated with 4 μg of anti-VSV antibody (Sigma). Immunoprecipitates were washed twice with lysis buffer, twice with lysis buffer containing 0.6 M NaCl and twice with lysis buffer without detergent, then resolved by SDS/7% PAGE and transferred on to nitrocellulose. Bound proteins were detected by blotting with anti-mAKAP-VO145–HRP (where HRP is horseradish peroxidase) (1:5000) and anti-VSV antibodies (1:150000). Anti-VO145 was generated in rabbits against the mAKAP fusion protein 1446–2312–His. Conjugation of anti-VO145 to HRP was performed according to the manufacturer's instructions (Alpha Diagnostic International, San Antonio, TX, U.S.A.).
Cell-stimulation studies were performed before lysis as follows. Cells were serum starved for 5 h, then stimulated with 10 μM forskolin and 50 μM IBMX (3-isobutyl-1-methylxanthine) or DMSO control for 15 min. For densitometry, bands were quantified using NIH Image software. The amount of mAKAP bound was normalized to the amount of PDE4D3 immunoprecipitated.
RESULTS AND DISCUSSION
mAKAP requires anchored PKA to interact with PDE4D3 efficiently in cells
PDE4D3 contains two PKA-phosphorylation sites, Ser-13 and Ser-54. PKA-phosphorylation of Ser-54 increases mAKAP-bound PDE4D3 activity 2-fold [8–10]. However, no known functional consequence has been ascribed to phosphorylation of Ser-13. Since the mAKAP-binding domain of PDE4D3 contains Ser-13, we hypothesized that PKA-phosphorylation of this site regulates PDE4D3 binding to mAKAP.
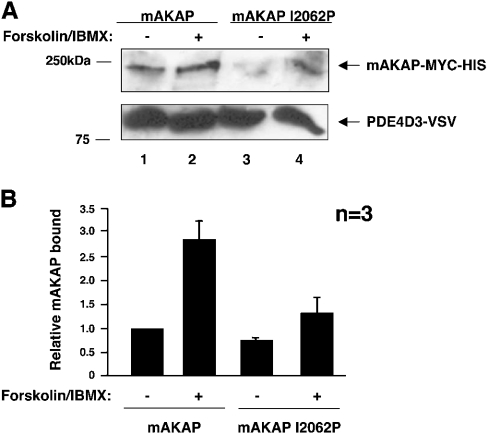
We tested this hypothesis in two ways with a heterologous expression system (Figure 1). First, we tested if the interaction between mAKAP and PDE4D3 increased when mAKAP-anchored PKA was fully activated by forskolin (Figure 1A, lanes 1 and 2). Secondly, we tested whether PDE4D3 preferentially associated with PKA-bound mAKAP using an mAKAP mutant that abolishes its ability to interact with the PKA holoenzyme (Ile2062Pro) (Figure 1A, lanes 3 and 4). Mutant and wild-type mAKAP were co-transfected into HEK-293 cells with PDE4D3–VSV. Before lysis, cells were serum-starved and then stimulated with forskolin and IBMX to fully activate cellular PKA. Immunoprecipitation of PDE4D3–VSV was performed with anti-VSV antibody, samples were washed, and bound proteins were resolved by SDS/PAGE. Equal levels of PDE4D3–VSV were immunoprecipitated as determined by anti-VSV Western blot (Figure 1A, lower panel). Co-immunoprecipitation of mAKAP was analysed by Western blot with anti-mAKAP–HRP antibody. Wild-type mAKAP clearly co-immunoprecipitated with PDE4D3 as expected (Figure 1A, upper panel, lane 1). Stimulation of the cells with forskolin and IBMX caused a 2.85±0.40-fold increase in mAKAP association (Figure 1A, upper panel, lane 2, and Figure 1B). Thus maximal activation of mAKAP-anchored PKA enhanced the association of mAKAP with PDE4D3. These data clearly support the idea that the catalytic function of mAKAP-anchored PKA is required for the high-affinity PDE4D3–mAKAP interaction in cells. Furthermore, association of PDE4D3–VSV with the mAKAP Ile2062Pro mutant was diminished to 0.75±0.04-fold as compared with wild-type mAKAP (Figure 1A, upper panel, lane 3, and Figure 1B). This demonstrates that displacing the PKA holoenzyme from mAKAP decreases the level of mAKAP associated with PDE4D3. Finally, when cells transfected with mAKAP Ile2062Pro were stimulated with forskolin and IBMX, the amount of associated mAKAP mutant increased to 1.32±0.32-fold (Figure 1A, upper panel, lane 4, and Figure 1B). However, this modest increase was still significantly less than that seen with stimulated wild-type mAKAP (Figure 1A, upper panel, lane 2, and Figure 1B), and it most likely reflects the global activation of un-anchored PKA. These data demonstrate that phosphorylation of PDE4D3 by mAKAP-anchored PKA, and not other kinases or unanchored PKA, regulates the strength of the interaction between the PDE and mAKAP.
Figure 1. mAKAP requires anchored PKA to interact strongly with PDE4D3 in cells.
(A) Wild-type mAKAP (lanes 1 and 2) and a mutant form of mAKAP that cannot anchor PKA (mAKAP I2062P) (lanes 3 and 4) were transfected into HEK-293 cells with PDE4D3–VSV, as indicated. At 2 days after transfection, cells were serum-starved for 5 h and stimulated with 10 μM forskolin and 50 μM IBMX for 15 min, as indicated. PDE4D3–VSV protein was immunoprecipitated with anti-VSV antibody, resolved by SDS/7% PAGE and transferred on to nitrocellulose. Immunoprecipitates were probed with anti-mAKAP–HRP to detect mAKAP (upper panel). Anti-VSV antibody was used to detect PDE4D3–VSV to show equivalent levels of immunoprecipitated PDE4D3 (lower panel). These blots are representative of three independent experiments. (B) Bands from (A) were quantified using NIH Image software. The amount of mAKAP bound to PDE4D3–VSV was normalized to the amount of PDE4D3–VSV immunoprecipitated. Results are means±S.E.M. for three independent experiments. The columns are labelled with transfection and stimulation conditions.
mAKAP 1286–1831 preferentially associates with phosphorylated PDE4D3 peptide
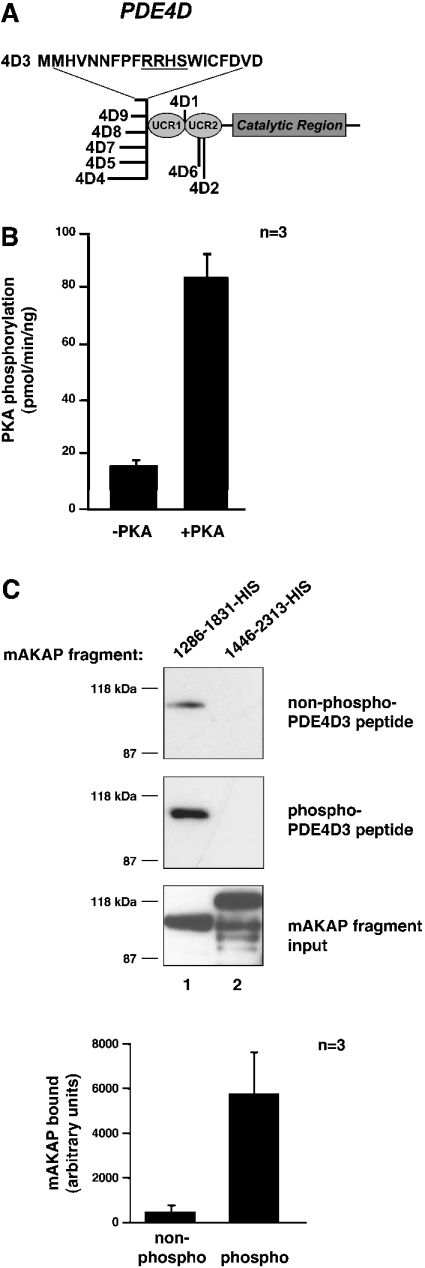
The PDE4D locus gives rise to nine different isoforms by alternative splicing. These nine forms contain an identical catalytic region, but unique N-termini (Figure 2A) [6,13,14]. The 15 unique N-terminal amino acid residues of PDE4D3 are sufficient to bind to mAKAP residues 1286–1831 [5]. Within this region is an established PKA-phosphorylation site, Ser-13 [8–11]. To determine whether phosphorylation of this residue affects mAKAP binding, we performed in vitro pull-down assays with phosphorylated and non-phosphorylated peptide containing this unique region (Figure 2C). Peptide was initially subjected to a kinase assay with and without the PKA catalytic subunit to confirm that the peptide could be sufficiently phosphorylated by PKA (Figure 2B). Phosphorylation was then carried out without radiolabel to generate phosphorylated peptide for the pull-down assay. PKA-phosphorylated and non-phosphorylated peptide were precipitated, incubated with bacterially produced His-tagged mAKAP fragments, washed, resolved by SDS/PAGE and analysed by Western blot. Only the mAKAP fragment 1286–1831–His bound significantly to the peptide (Figure 2C, top and middle blots, lane 1) even though similar amounts of mAKAP fragment 1446–2313–His were tested (Figure 2C, bottom blot). However, pull-down of the PKA-phosphorylated peptide resulted in more co-precipitation of mAKAP 1286–1831–His than non-phosphorylated peptide (Figure 2C, lane 1, compare top and middle blots). The difference in band density, 517±258 units for non-phosphorylated peptide and 5807±1794 units for PKA-phosphorylated peptide, is at least 5-fold as demonstrated in the graph of Figure 2(C). This experiment provides in vitro evidence that phosphorylation of PDE4D3 peptide by PKA increases its affinity for the mAKAP fragment. In combination with the previous experiment, this strengthens the notion that PKA-phosphorylation of the PDE positively affects association with mAKAP.
Figure 2. mAKAP 1286–1831 binds PKA-phosphorylated PDE4D3 peptide better than non-phosphorylated peptide in vitro.
(A) The structure of the PDE4D family of PDEs. The conserved catalytic core and upstream conserved regions (UCRs) are indicated. The locations of divergent sequences for each isoform are indicated, and the 20 unique amino acid residues of PDE4D3 are denoted by the one-letter amino acid code. The PKA-phosphorylation consensus site is underlined. (B) Phosphorylation of PDE4D3 peptide by PKA. PDE4D3 peptide was incubated in PKA kinase buffer containing 100 μM ATP, 5 μM [γ-32P]ATP and 0.3 mM cAMP with or without the PKA catalytic subunit for 15 min at 30 °C. The reaction mixture was spotted on to p81 phosphocellulose paper, washed five times in 75 mM phosphoric acid and once in 95% ethanol. Filters were air-dried and were counted by liquid-scintillation counting. Results are averages of three independent experiments. (C) Pull-down assay of mAKAP fragments with PDE4D3 peptide. Biotin–PDE4D3 peptide was incubated in PKA kinase buffer containing 100 μM ATP and 0.3 mM cAMP with or without the PKA catalytic subunit for 2 h at 30 °C. Neutravidin beads were added to collect the peptide and then rocked for 1 h at room temperature. The precipitates were washed three times and resuspended in 500 μl of HSE buffer containing 5 μM recombinant mAKAP fragments. Reactions were incubated overnight at 4 °C with rocking. Precipitates were washed three times with HSE buffer, and bound proteins were subjected to SDS/7% PAGE followed by immunoblotting with anti-His antibody. The top blot tests co-precipitation of mAKAP fragments with nonphosphorylated PDE4D3 peptide. The middle blot tests co-precipitation of mAKAP fragments with phosphorylated PDE4D3 peptide. The bottom blot shows that equal amounts of mAKAP fragments (lane 1, mAKAP 1286–1831–His; lane 2, mAKAP 1446–2313–His) were used in the assay. The 1286–1831–His bands from the top and middle blots were quantified using NIH Image software. The amount of mAKAP fragment bound to non-phosphorylated (non-phospho) and PKA-phosphorylated (phospho) peptides was graphed in arbitrary units. Results are means±S.E.M. for three independent experiments.
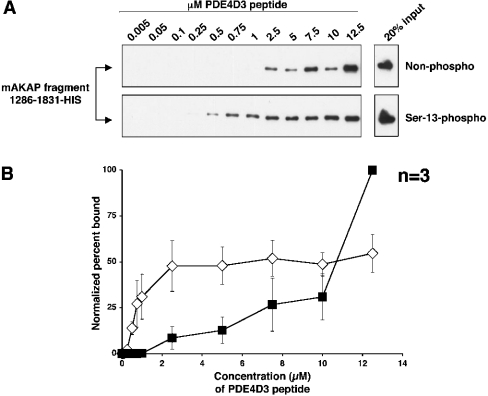
Phosphorylation of PDE4D3 peptide on Ser-13 increases its affinity for mAKAP 1286–1831
To quantitatively determine the effect of PDE4D3 Ser-13-phosphorylation on mAKAP 1286–1831 binding, an in vitro binding curve was performed using known concentrations of biotintagged peptide synthesized to contain phospho-Ser-13 and non-phospho-Ser-13 (Figure 3A). Known concentrations for each peptide were mixed with a fixed amount of cell extract containing mAKAP fragment 1286–1831–His. Pull-down of the peptide with Neutravidin beads demonstrated that the mAKAP fragment bound to lower concentrations of Ser-13-phosphorylated peptide than non-phosphorylated peptide. Non-phosphorylated peptide bound to the mAKAP fragment at a concentration of 2.5 μM, while Ser-13-phosphorylated peptide bound to the mAKAP fragment at least 5-fold better at a concentration of 0.5 μM. Bands were quantified by densitometry, and the normalized percentage bound was plotted against the concentration of peptide used (Figure 3B). The graph illustrates that the PDE4D3 peptide has a higher affinity for the mAKAP fragment when it is phosphorylated on Ser-13. One peculiar phenomenon observed in this experiment is that the non-phosphorylated peptide bound significantly more mAKAP than the Ser-13-phosphorylated peptide when 12.5 μM and higher peptide concentrations were tested. We cannot explain this unusual, but reproducible, effect that was observed in three independent experiments.
Figure 3. The mAKAP 1286–1831 fragment binds to lower concentrations of Ser-13-phosphorylated PDE4D3 peptide than non-phosphorylated peptide in vitro.
(A) Biotin–PDE4D3 peptides that were either non-phosphorylated (upper panel) or phosphorylated on Ser-13 (lower panel) were mixed at various concentrations (indicated in μM) with a fixed amount of mAKAP fragment 1286–1831–His. Biotin–PDE4D3 peptides were collected by Neutravidin beads, resolved by SDS/7% PAGE and subjected to Western analysis using anti-His antibody to detect the mAKAP fragment. The upper and lower right-hand panels represent 20% of the mAKAP fragment added to the samples. The blots shown are representative of three independent experiments. (B) Bands from (A) were quantified using NIH Image for each experiment, and the percentage of mAKAP 1286–1831–His bound to peptide for each concentration point was determined (see the Experimental section). To create the graph, the normalized percentage of mAKAP 1286–1831–His bound to biotin–Ser-13-phosphorylated PDE4D3 peptide (⋄) or biotin–non-phosphorylated PDE4D3 peptide (▪) was plotted against peptide concentration. Results are the means±S.E.M. for three independent experiments.
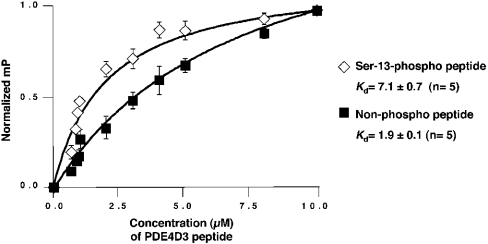
To obtain kinetic data describing the PDE4D3 peptide 1–20 and mAKAP fragment 1286–1831–His interaction, FP measurements were taken (Figure 4). Ser-13-phosphorylated and non-phosphorylated peptides were conjugated to FITC, and known concentrations were mixed with a fixed amount of bacterially expressed mAKAP 1286–1831–His protein. FP measurements were taken, and a saturation-binding curve for each peptide was generated as shown in Figure 4. From this information, we calculated the Kds for each experiment. The averaged results from five independent experiments showed a Kd of 7.1±0.7 μM for Ser-13-phosphorylated peptide and a Kd of 1.9±0.1 μM for non-phosphorylated peptide. Thus phosphorylation of the PDE4D3 peptide on Ser-13 increases the Kd for the mAKAP fragment at least 3-fold.
Figure 4. FP of the mAKAP 1286–1831–PDE4D3 peptide interaction.
FITC–Ser-13-phosphorylated PDE4D3 peptide (⋄) or FITC–non-phosphorylated PDE4D3 peptide (▪) was resuspended in PBS containing 5 mg/ml BSA, pH 7.0. Increasing concentrations of recombinant mAKAP 1286–1831–His were mixed with each FITC-labelled peptide. Each sample was incubated for 10 min. Saturation-binding curves were generated with Prism graphing software. Milli-polarization (mP)=FP/1000. Kds were calculated from non-linear regression curves generated from the average of five individual experiments.
Mimicking phosphorylation of PDE4D3 on Ser-13 increases its affinity for mAKAP
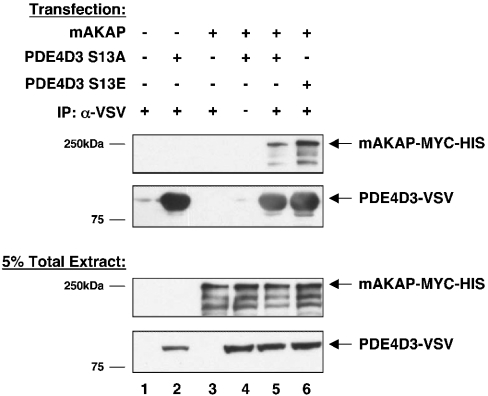
To understand if there is a consequence when PDE4D3 is phosphorylated on Ser-13 in cells, Ser-13 mutants were generated and used for immunoprecipitation–Western blotting experiments (Figure 5). A mutant that abolishes the phosphorylation site, Ser13Ala, and a mutant that mimics the phosphorylation site, Ser13Glu, were transfected into HEK-293 cells with full-length mAKAP. Immunoprecipitation of PDE4D3–VSV was performed using anti-VSV antibody, samples were washed, and bound proteins were resolved by SDS/PAGE. Co-immunoprecipitation of mAKAP was analysed by Western blot using anti-mAKAP–HRP antibody. More mAKAP bound to the PDE4D3 Ser13Glu mutant than to the PDE4D3 Ser13Ala mutant (Figure 5, top panel, compare lanes 5 and 6), even though equal levels of mAKAP and PDE4D3 mutants were expressed (Figure 5, bottom panels) and equal levels of PDE4D3 mutants were immunoprecipitated (Figure 5, second panel). These data demonstrate that in cells, full-length mAKAP preferentially binds to PDE4D3 when it carries a negative charge on Ser-13, thus highlighting the functional significance of PKA-phosphorylation of PDE4D3 on Ser-13.
Figure 5. mAKAP binds more strongly to PDE4D3 Ser13Glu than to PDE4D3 Ser13Ala in cells.
PDE4D3–VSV phospho-mutants were created to mimic the non-phosphorylated form (PDE4D3–VSV Ser13Ala) or the phosphorylated form (PDE4D3–VSV Ser13Glu). PDE4D3–VSV phosphomutants were co-transfected with mAKAP into HEK-293 cells as indicated. At 2 days after transfection, PDE4D3–VSV protein was immunoprecipitated with anti-VSV antibody. Immunoprecipitates were probed with anti-mAKAP–HRP to detect mAKAP (top panel) and anti-VSV to detect PDE4D3 phospho-mutants (second panel). The input (5% of total extract) was immunoblotted with anti-mAKAP–HRP (third panel) or anti-VSV (bottom panel) antibodies to demonstrate equivalent expression levels of mAKAP and PDE4D3 phospho-mutants in all transfection conditions. These blots are representative of five independent experiments.
In the present paper, we show evidence that PKA-phosphorylation of PDE4D3 on Ser-13 results in a molecular change with the ability to affect signal transduction potential. Cell-based experiments comparing wild-type mAKAP and a mutant unable to anchor the PKA holoenzyme demonstrated that phosphorylation of PDE4D3 by fully activated mAKAP-anchored PKA is important for the high-affinity interaction between PDE4D3 and mAKAP. In vitro binding assays and FP using PDE4D3 peptide and mAKAP fragments clearly established that phosphorylation of PDE4D3 on Ser-13 enhances the recruitment of mAKAP. Further cellular studies using PDE4D3 mutants that abolish or mimic Ser-13-phosphorylation confirm this observation. Our data demonstrating the dependence upon mAKAP to anchor PKA for this effect give credence to the anchoring hypothesis that enzymes sequestered into signal transduction units are discretely activated for local activation and regulate local effects such as binding affinity. In addition to the present study, a study of gravin/AKAP250 demonstrated that phosphorylation of gravin by gravin-anchored PKA regulates its interaction with the β2AR (β2-adrenergic receptor) [15]. The authors demonstrated that activation of the β2AR with isoprenaline (isoproterenol) stimulated phosphorylation of gravin by PKA. Much like the present study, disruption of the amphipathic helix in gravin to abolish the binding site of the PKA RII subunit resulted in ablation of phosphorylation on gravin and, in turn, binding of gravin to β2AR. Hence, in several instances, increasing the binding affinity through anchored PKA-phosphorylation regulates the binding of an AKAP to another signalling enzyme.
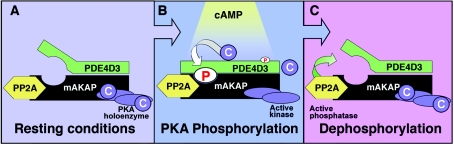
The increase in binding affinity due to phosphorylation of PDE4D3 on Ser-13 provides another level to the tightly regulated negative-feedback model depicted in Figure 6. That is, upon hormonal stimulation of a cell, cAMP increases and activates mAKAP-bound PKA. The catalytic subunits then phosphorylate local substrates, including PDE4D3. Robust phosphorylation of PDE4D3 on Ser-54 increases PDE activity 2-fold and causes cAMP breakdown. Phosphorylation of PDE4D3 on Ser-13 serves to enhance the binding affinity of PDE4D3 for mAKAP. As a result of PDE4D3 binding more tightly to mAKAP, PDE4D3 does not diffuse away from the local cAMP gradient. The significance of this effect is that it leads to localized PDE activity and quicker termination of the cAMP signal. Consequently, the return of basal cAMP levels allows for the re-formation of the PKA holoenzyme and resets the system.
Figure 6. Model for interaction of PDE4D3 with mAKAP in vivo.
(A) Under resting conditions, PDE4D3 binds to the mAKAP signalling complex. (B) Upon hormonal stimulation of a cell, cAMP increases and activates mAKAP-bound PKA. The catalytic subunits phosphorylate local substrates, including PDE4D3. Phosphorylation of PDE4D3 on Ser-13 enhances the binding affinity of PDE4D3 for mAKAP. Phosphorylation of PDE4D3 on Ser-54 increases PDE activity 2-fold and causes cAMP breakdown. (C) With the return of basal cAMP levels, the PKA holoenzyme reforms. Phosphatase activity dephosphorylates PDE4D3, and the signalling system is reset.
Although we now know that mAKAP-anchored PKA-phosphorylation of PDE4D3 on Ser-13 and on Ser-54 each have mutually exclusive signalling effects, we still do not know if the phosphorylation events have an effect on one another. For example, does phosphorylation of one residue enhance the phosphorylation of the other in vivo? Evidence from MacKenzie et al. [16] suggests that Ser-54 is the first residue phosphorylated in cells. Using anti-phospho-Ser-13 and anti-phospho-Ser-54 antibodies, their study demonstrated that in COS7 cells, two populations of PDE4D3 phosphorylated on Ser-54 existed after 1 min of exposure to forskolin and IBMX. This is in contrast with only a single species detected for PDE4D3 phosphorylated on Ser-13. The authors describe the anti-phospho-Ser-54 doublet as two species of phosphoproteins: one that is phosphorylated only on Ser-54, and one that is doubly phosphorylated on Ser-54 and on Ser-13. This suggests that phosphorylation of Ser-54 precedes that of Ser-13 in this experiment. However, it is unlikely that phosphorylation of Ser-54 is a requirement for Ser-13-phosphorylation in vivo because Sette and Conti [8] demonstrated that PDE4D3 mutants Ser13Ala and Ser54Ala could each be phosphorylated to similar degrees using a cell-free phosphorylation system [8]. Finally, as shown by our data, mAKAP can bind to PDE4D3 that is not phosphorylated on Ser-13, but phosphorylation of this residue enhances the affinity of the interaction. While it is uncertain whether phosphorylation of one site influences the rate of phosphorylation at the second site, the current evidence suggests that the PDE4D3-phosphorylation events do not depend upon one another.
Finally, implicated in the regulation of the mAKAP-anchored negative-feedback loop is the requirement for a phosphatase to dephosphorylate Ser-13 and Ser-54. Co-immunoprecipitation experiments from rat heart extract demonstrated that PP (protein phosphatase) 2A subunits A and C are associated with the mAKAP complex [17]. PP1 and mAKAP were shown to co-sediment with the calcium-activated calcium release channel RyR (ryanodine receptor) prepared from cardiac sarcoplasmic reticulum [18]. Whether one of these two candidates resets the PDE or whether another phosphatase is responsible is not known. Future studies are planned to determine if an anchored phosphatase plays a role in this feedback loop and to determine if other signalling components, such as adenylate cyclases, associate with mAKAP at the nuclear envelope.
Acknowledgments
We thank Professor Miles Houslay (University of Glasgow, Glasgow, Scotland, U.K.) for the gift of the PDE4D3–VSV mammalian expression plasmid. This work was supported by NIH (National Institutes of Health) NRSA (National Research Service Award) grant F32HL68480 (to J.J.C.M.) and NIH grant DK54441 (to J.D.S.).
References
- 1.Colledge M., Scott J. D. AKAPs: from structure to function. Trends Cell Biol. 1999;9:216–221. doi: 10.1016/s0962-8924(99)01558-5. [DOI] [PubMed] [Google Scholar]
- 2.Tasken K., Aandahl E. M. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol. Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 3.Francis S. H., Turko I. V., Corbin J. D. Cyclic nucleotide phosphodiesterases: relating structure and function. Prog. Nucleic Acid Res. Mol. Biol. 2001;65:1–52. doi: 10.1016/s0079-6603(00)65001-8. [DOI] [PubMed] [Google Scholar]
- 4.Yarwood S. J., Steele M. R., Scotland G., Houslay M. D., Bolger G. B. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J. Biol. Chem. 1999;274:14909–14917. doi: 10.1074/jbc.274.21.14909. [DOI] [PubMed] [Google Scholar]
- 5.Dodge K. L., Khouangsathiene S., Kapiloff M. S., Mouton R., Hill E. V., Houslay M. D., Langeberg L. K., Scott J. D. mAKAP assembles a protein kinase A/PDE4 phosphodiesterase cAMP signaling module. EMBO J. 2001;20:1921–1930. doi: 10.1093/emboj/20.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houslay M. D., Adams D. R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tasken K. A., Collas P., Kemmner W. A., Witczak O., Conti M., Tasken K. Phosphodiesterase 4D and protein kinase a type II constitute a signaling unit in the centrosomal area. J. Biol. Chem. 2001;276:21999–22002. doi: 10.1074/jbc.C000911200. [DOI] [PubMed] [Google Scholar]
- 8.Sette C., Conti M. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. J. Biol. Chem. 1996;271:16526–16534. doi: 10.1074/jbc.271.28.16526. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann R., Wilkinson I. R., McCallum J. F., Engels P., Houslay M. D. cAMP-specific phosphodiesterase HSPDE4D3 mutants which mimic activation and changes in rolipram inhibition triggered by protein kinase A phosphorylation of Ser-54: generation of a molecular model. Biochem. J. 1998;333:139–149. doi: 10.1042/bj3330139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oki N., Takahashi S. I., Hidaka H., Conti M. Short term feedback regulation of cAMP in FRTL-5 thyroid cells: role of PDE4D3 phosphodiesterase activation. J. Biol. Chem. 2000;275:10831–10837. doi: 10.1074/jbc.275.15.10831. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez R., Sette C., Yang D., Eglen R. M., Wilhelm R., Shelton E. R., Conti M. Activation and selective inhibition of a cyclic AMP-specific phosphodiesterase, PDE-4D3. Mol. Pharmacol. 1995;48:616–622. [PubMed] [Google Scholar]
- 12.Kapiloff M. S., Schillace R. V., Westphal A. M., Scott J. D. mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J. Cell Sci. 1999;112:2725–2736. doi: 10.1242/jcs.112.16.2725. [DOI] [PubMed] [Google Scholar]
- 13.Wang D., Deng C., Bugaj-Gaweda B., Kwan M., Gunwaldsen C., Leonard C., Xin X., Hu Y., Unterbeck A., De Vivo M. Cloning and characterization of novel PDE4D isoforms PDE4D6 and PDE4D7. Cell. Signalling. 2003;15:883–891. doi: 10.1016/s0898-6568(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 14.Gretarsdottir S., Thorleifsson G., Reynisdottir S. T., Manolescu A., Jonsdottir S., Jonsdottir T., Gudmundsdottir T., Bjarnadottir S. M., Einarsson O. B., Gudjonsdottir H. M., et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 15.Tao J., Wang H. Y., Malbon C. C. Protein kinase A regulates AKAP250 (gravin) scaffold binding to the β2-adrenergic receptor. EMBO J. 2003;22:6419–6429. doi: 10.1093/emboj/cdg628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacKenzie S. J., Baillie G. S., McPhee I., MacKenzie C., Seamons R., McSorley T., Millen J., Beard M. B., van Heeke G., Houslay M. D. Long PDE4 cAMP specific phosphodiesterases are activated by protein kinase A-mediated phosphorylation of a single serine residue in Upstream Conserved Region 1 (UCR1) Br. J. Pharmacol. 2002;136:421–433. doi: 10.1038/sj.bjp.0704743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapiloff M. S., Jackson N., Airhart N. mAKAP and the ryanodine receptor are part of a multi-component signaling complex on the cardiomyocyte nuclear envelope. J. Cell Sci. 2001;114:3167–3176. doi: 10.1242/jcs.114.17.3167. [DOI] [PubMed] [Google Scholar]
- 18.Marx S. O., Reiken S., Hisamatsu Y., Jayaraman T., Burkhoff D., Rosemblit N., Marks A. R. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 2000;101:365–376. doi: 10.1016/s0092-8674(00)80847-8. [DOI] [PubMed] [Google Scholar]