Abstract
Considering the fundamental mechanism causing singularity phenomena, we performed the following abduction: Assuming that a multicellular system is driven by spontaneous fluctuation of each cell and dynamic interaction of the cells, state transition of the system would be experimentally predictable from cellular heterogeneity. This study evaluates the abductive hypothesis by analyzing cellular heterogeneity to distinguish pre-state of state transition of differentiating cells with Raman spectroscopy and human induced pluripotent stem cells (hiPSCs) technique. Herein, we investigated the time development of cellular heterogeneity in Raman spectra during cardiomyogenesis of six hiPSC lines and tested two types of analyses for heterogeneity. As expected, some spectral peaks, possibly attributed to glycogen, correctively exhibited higher heterogeneity, prior to intensity changes of the spectrum in the both analyses in the all cell-lines tested. The combination of spectral data and heterogeneity-based analysis will be an approach to the arrival of biology that uses not only signal intensity but also heterogeneity as a biological index.
Keywords: DNB, spectroscopy, new biomarker, iPS, differentiation
Significance
This study experimentally proved that cellular heterogeneity in Raman spectrum enables to detect the pre-state transition of hiPSCs faster than its signal intensity changes. To achieve this, a Raman spectral method with small experimental deviations was newly developed. Cellular heterogeneity of Raman peaks attributed glycogen, showed significant increase at the 2nd day after induction of cardiomyogenesis, which is significantly earlier than intensity changes at the 4th day. In addition, there are peaks that exhibited a common change in heterogeneity between cell lines. The results provide the concept of heterogeneity-based markers to biophysics.
Introduction
Singularity phenomenon in a multicellular system is caused by avalanche of cell-cell interaction triggered by sudden state transition of a single cell, here called singularity cell. A basic mechanism at work here is that interactions between individual cells form a field that influences the cells. This has been discussed in the bioholonics theory proposed in the 1980s, which states that life phenomena are autonomous cooperative systems whose constituent elements are “holons”, which are relationships between individuals and fields, regardless of layer [1]. The driving force of state transition of cell in a biological system is spontaneous fluctuation, often called Yuragi [2,3], which has been experimentally confirmed in muscle contraction, signal transduction in a cell, and recognition by brain [4]. We previously found that, although ESCs does not have any oscillators or circadian rhythm, it exhibits a collective differentiation only with state transition driven by fluctuation and slaving by cell-cell interaction: the cell-cell interaction plays a role in maintaining cells in a stable state by mutually restricting spontaneous fluctuations with neighboring cells, and appearance-disappearance of cell-cell interactions causes collective state transition of cells in a colony [5]. We think that the mechanism for cells to synchronize in a multicellular system by collaboration of the fluctuation and the cell-cell interaction is the bottom foundation of Singularity phenomenon.
Abductively considering a biological event, cell differentiation in this study, state fluctuation of an individual cell or the heterogeneity of the cells should increase prior to a collective differentiation of a cell colony. Cell differentiation can be represented by a ball rolling on a sloping surface of ridges and valleys that are dynamically formed by the destruction and reconstruction of complicated cross-interplay of elements composing epigenetic, genetic network, etc., as described in Waddington’s epigenetic landscape [6]. The population distribution of the cell state is larger in the valley, which is called an attractor. The cell state stochastically fluctuates even in a stable attractor, and its transition is observed as a stochastic switching between different cell states by experimenters [7]. In such a dynamic complex system, the fluctuation of an observable/measurable parameter slows down as the switching event approaches, resulting in greater variance, which is called critical slowing down [8,9]. According to the above picture, an observable/measurable parameter that exhibits increased fluctuation when focusing on time course or an increased cellular heterogeneity when focusing on cell population should be present prior to cell state transition, including escape from pluripotency. Many researchers may have qualitatively noticed this phenomenon. For example, transcriptome analysis has shown that the expression of certain genes was more heterogeneous in mouse embryonic stem cells (ESCs) immediately after losing pluripotency [10]. However, it has not yet been quantitatively proven.
In this study, we demonstrate a quantitative proof of the above-mentioned abductive hypothesis based on a Raman fingerprinting technology and human induced pluripotent stem cells (hiPSCs). Even if the hypothesis is true, it is unclear what and when will they exhibit fluctuation/heterogeneity. Therefore, measurement and analysis in the proof require comprehensiveness. The Raman spectrum emitted from a cell includes many spectral peaks derived from the comprehensive chemical status of molecular-bonds vibrations in a cell, and can be predictably linked to transcriptomic data [11,12]. So far, we have experimentally demonstrated that the spectral feature in Raman spectroscopy can be used to discriminate between cell states, similar to fingerprinting [13–15]. Moreover, even if the proof is achieved with only in an experimental model cell, it does not necessarily mean that our proof is universal. It is preferable to prove the hypothesis using cells in the same state with different genetic backgrounds. Induced pluripotent cells technology it is suitable for this purpose. Some research groups have previously reported on the use of Raman fingerprinting for identifying differentiating states of human induced pluripotent stem cells (hiPSCs) [16–20]. Naturally, the time course of differentiation of many single cells should be measured using multiple cell line types. Simplicity of observation is another reason for the use of Raman spectroscopy due to its unnecessity of any staining procedure.
Here, we prepared six hiPSC-lines derived from different origins, collected Raman spectra of single-cells during cardiomyogenesis of them, and finally found that some of Raman spectra peaks exhibit heterogeneous features just before a hiPSC loses pluripotency, meaning the proof of our abductive hypothesis. Experimentally, the cellular heterogeneity was quantitated by coefficient of variation (CV) as the index of the degree of Yuragi, and the CV in the specific spectral peaks, most likely attributed to glycogen, increased more rapidly than signal intensity changes upon loss of pluripotency, meaning that the CV can be used as a biomarker for future cell state transition. Thus, our results demonstrated such analysis based on the Yuragi concept can detect state transition of a multicellular system prior to the conventional intensity-based analysis. Collective differentiation observed in this study is one of the simplest singularity phenomena. We believe that the Yuragi-based analysis promises to be a standard research tool in Singularity biology.
Materials and Methods
Raman Spectroscopy
We acquired Raman spectra from cells using a home-built line-scan Raman microscope based on inverted microscope (IX81: Olympus, Tokyo, Japan) equipped with spectrometer (MK-300: Bunkoukeiki, Tokyo, Japan) and a two-dimensional detector (PIX-IS400, Princeton Instruments, Trenton, NJ), as described in the previous reports [13–15]. A 532 nm laser (Ventus532: Laser Quantum, Stockport, UK) was used for illumination. The laser was shaped into a line with a cylindrical lens and directed onto the cells on a glass substrate. The laser intensity was 2.4 mW/μm2. The backward-scattering light was collected by an objective and captured on a 400×1340 pixels detector after being passed through a polychromator. The Raman spectrum was acquired over a range of 160 cm–1 to 2200 cm–1 on the 1340 pixels (1.52 cm–1/pixel). By scanning the line on an axis perpendicular to it, we acquired a hyperspectral image (Figure 1A). The impact of photodamage due to the strong laser irradiation is one of the main factors behind the low-reproducibility of Raman spectroscopic measurements. To minimize photodamage, we limited the exposure time to 15 s and scanned only 10 steps at 3.35 μm/step. Total time to acquire a single hyperspectral image was only 150 s. Number of cells measured for Raman spectra is summarized in Supplementary Table S1.
Figure 1 .
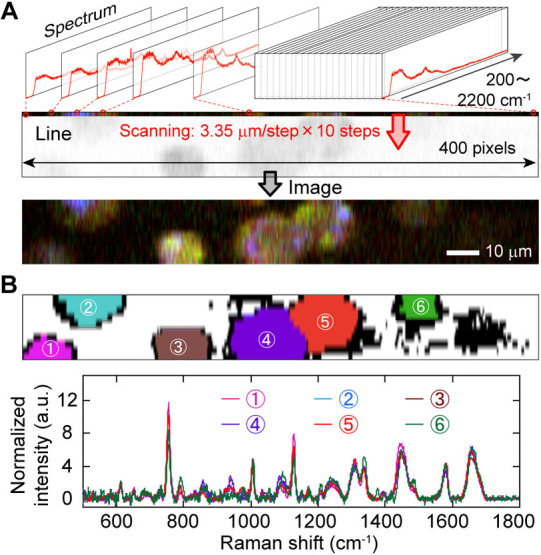
The present single-cell Raman spectroscopy. (A) Explanative cartoon of the procedure to acquire hyper spectral image. (B) Raman spectra of six cells of 253G1 hiPSC. The upper panel is the segmentation results and the lower is the spectra. The numbers and colors match to the upper and lower.
Cell Culture
The hiPSC-lines used in the study were purchased from Riken Cell Bank (Ibaraki, Japan). Cells were cultured on iMatrix-511 (Takara Bio, Tokyo, Japan) coated dishes in StemFit AK02N medium (Ajinomoto, Tokyo, Japan) at 37°C and 5% CO2. The medium was changed daily. For cardiomyocyte differentiation, cells were cultured until 70% confluency and induced to differentiate using a PSC Cardiomyocyte Differentiation Kit (Gibco, Waltham, MA) following the manufacturer’s instructions. For Raman measurements, cells were harvested using TrypLE Select (Gibco) and washed with PBS and centrifuged at 150×g for 5 min before being resuspended in FluoroBrite DMEM Media (Gibco).
Transcriptomic Analysis
To analyze the transcriptome of human iPSCs and their differentiated derivatives, SurePrint G3 Human GE microarray 8×60K Ver.3.0 (Agilent Technologies) was used. The cells were harvested from the same culture dish used for the Raman measurement. The RNA was isolated using an RNeasy mini kit (Qiagen, Venlo, Netherlands) and labeled with Cy3-CTP using a Low Input Quick Amp Labeling Kit (Agilent Technologies). Microarray hybridization was performed, and the hybridized slides were scanned using a microarray scanner (Agilent Technologies) following the supplier’s protocol. The fluorescent signals were processed with the Feature Extraction software (Agilent Technologies) and the processed signal data were normalized and analyzed using GeneSpring GX software Ver. 14.9 (Agilent Technologies). The microarray expression analysis was performed using biologically triplicate samples.
Spectral Analysis
For spectral analysis, the broad peaks from the glass substrate and auto-fluorescence was subtracted using rolling-circle algorithm, as described in supplementary text. For automated identification of spectral peaks, we applied the zero-crossing algorism to an averaged spectrum. All processes were performed using a custom-made software programmed in C++ (Visual Studio 2008, Microsoft, USA) and image processing was carried out using the OpenCV library (ver. 2.4.3). All analyses were performed with blinding information about the data, such as the differentiating days and the cell line, for the analyst. The principal component analysis (PCA) was performed using a function in the OpenCV library. Before PCA processing, the data were standardized with Z-score normalization.
Dynamic Network Biomarkers (DNB) Analysis
The procedure for DNB analysis in this study is based on a previous report [21,22]. The first step involved assigning each Raman shift to a component of the complex network. Significantly fluctuating components (Raman shifts) were identified using the F-test (with a significance level of alpha=0.05) against Raman intensities of the experimental group (each day after differentiation) versus the control group (Day-0) and multiple-test correction was performed using the Benjamini-Hochberg method. Next, the Pearson’s correlation coefficient (rij) was calculated between the fluctuating components with i and j representing different indices of the components. Hierarchical clustering was then performed using the average linkage method with for the distance (d) of 1 - abs(r). A maximum cluster was specified using a distance threshold of 0.2 in this study. The average standard deviation and average correlation strength of the components in this cluster were then analyzed. If both increased and showed a peak, it indicated the transition state of the system, and the components in this cluster were identified as DNB Raman shifts.
Results
Pre-investigation of Reproducibility in the Biological Replicate
The greatest concern in Raman spectroscopy of living cells is the potential for cell damage due to intense light exposure, as the Raman scattering signal is weak. Exposure to radiation, including laser irradiation, can stimulate collagen and glycogen production in cells, which may alter the shape of the acquired Raman spectra, as shown in previous studies [23,24]. Therefore, in this study, a line-confocal Raman spectroscopy system was used instead of a point-focus system to minimize exposure time and reduce the risk of cell damage [13,25].
We acquired a hyperspectral Raman image of 400×10 pixels (corresponding to 125 μm×33.5 μm) including one to six hiPSCs of the 253G1 cell-line. Each pixel had a spectrum from 160 to 2200 cm–1 (Figure 1A), and we obtained an average Raman spectrum within each segmented cell area in the image (Figure 1B). However, the low-reproducibility of Raman spectrum measurement of living hiPSCs is often due to slight deviations in the optical adjustment, contamination from undesired background signal in the obtained spectrum, photodamage due to prolonged or intense laser irradiation, among other factors. This disturbance hinders the quantification of cellular heterogeneity. Therefore, we established an analysis protocol to realize technical-deviation-free Raman finger-printing by mathematically predicting background signals on cell-existing areas (Supplementary Text S1–S4, and Figures S1–S6).
Even if the technical reproducibility of the measurement system is high enough, reproducibility in biological replicates must also be ensured in biological assays/experiments. In many cases, cells are removed from the deep freezer for each data set, thawed, and then processed for proliferation culture. During the interval time between experimental replicates, optical aberrations could occur, and/or the cell condition could vary depending on the analyst’s experimental skills. This can also affect the quantification of cellular heterogeneity.
We collected single-cell spectra in total of seven culture dishes in two independent experiments, with an interval of three months, and investigated the difference in the average Raman spectrum in each dish (Figure 2A). At first glance, there was no apparent difference in the mean spectra in the dishes (Figure 2B), indicating high reproducibility. Nevertheless, a heatmap of differences from the total averaged spectrum highlighted the slight differences in biological replication (Figure 2C). To investigate this slight difference in more detail, we performed a principal component analysis (PCA) of all single-cell Raman spectra. The difference was represented on the second principal component (PC2; contribution rate=20%), not on the first principal component (PC1; contribution rate=34%) (Figure 2D). Thus, the variation in biological replicates was not the primary principal component in single-cell Raman spectra of hiPSC-lines. In other words, the heterogeneity in single-cell Raman spectrum was greater than the deviation in the biological replicates as shown in the PCA score plot because deviation between single cells (each dot) is larger in PC1 axis within the dish whereas deviation is larger in PC2 axis when compared between dishes (Figure 2D).
Figure 2 .
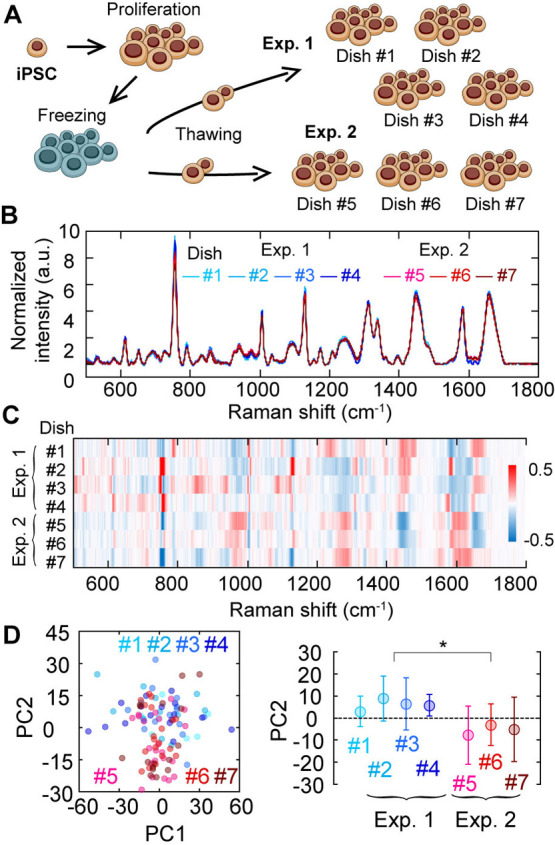
Pre-investigation of reproducibility in biological replicate. (A) Explanative cartoon of the investigation of biological-duplicate reproducibility. (B) Average Raman spectra of 253G1 hiPSC of seven dishes obtained in two distinct sample preparations. Different color, different dish. Blueness, the first preparation (Exp.1); Redness, the second preparation (Exp.2). (C) A heatmap of the differences from the total average spectrum among seven dishes. Total average spectrum calculated from all 7 dishes were subtracted from average spectrum of each dish (#1~#7). The color indicates the difference of mean spectrum of each dish and the average spectrum of all seven dishes. (D) Principal component analysis of Raman spectra of single cells in seven dishes. Color indicates each dish number. The left panel is PC2 vs PC1 plot and the right is the dish dependence of the PC2 value. Error bars are standard deviations. Asterisks indicate less than 0.05 of the p-value in the Student’s t-test.
Comparison Among iPSC-lines in Raman Spectrum
Supposing that the Raman spectrum obtained from a cell corresponds to gene-expression profile [12], it had been expected that the Raman spectrum emitted from hiPSCs would vary depending on human individuality. To test this possibility, we compared the average Raman spectra emitted from six hiPSC-lines derived from different origins: two controls (253G1 and 648A1), two mitochondrial disease-derived (HPS0458 and HPS0461), and two Fabry disease-derived (HPS2108 and HPS3284).
Interestingly, we could hardly detect apparent difference in Raman spectrum between them (Figure 3A), whereas the genetic profile exhibited obvious difference between the hiPSC-lines (Supplementary Figure S7). The tiny difference in the hiPSC-lines was visualized on the heatmap of differences from the total average spectrum, exhibiting that the difference between two independent experiments was comparable to those between the hiPSC-lines (Figure 3B). Performing PCA, the hiPSC-line dependency was represented onto the PC2, similar to the difference seen in biological replication (Figure 3C). Comparing the average PC2 values, while the feature of healthy, mitochondrial disease and Fabry disease could be statistically discriminated on the PC2 axis, there was little difference between cell lines in the same axis (Figure 3D). In short, the differences among the hiPSC-lines in pluripotent state were not large enough to be distinguishable though there were indeed small differences on the average.
Figure 3 .
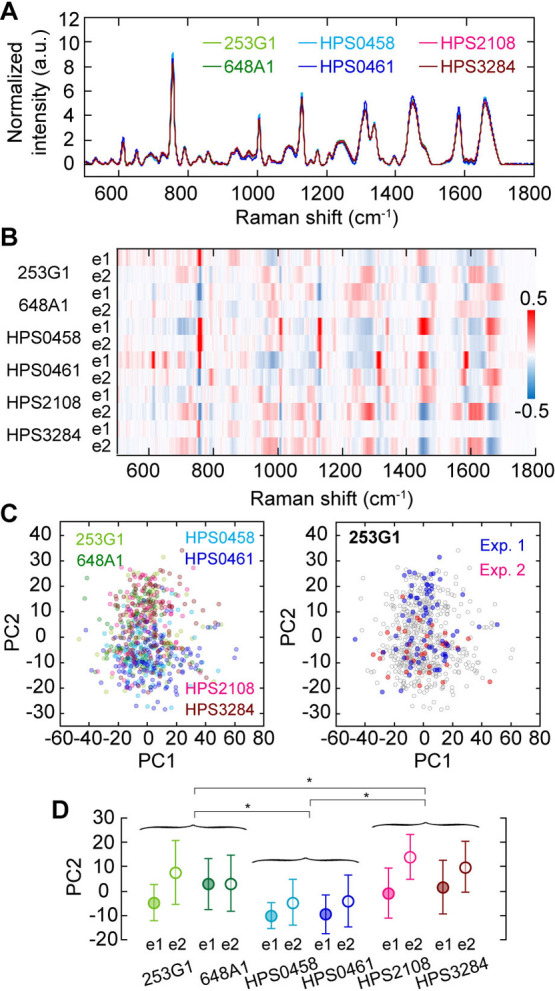
Comparison among average Raman spectra of six different hiPSC-lines. (A) Mean Raman spectra of six hiPSC-lines. Different color, different hiPSC-line. (B) A heatmap of the differences from the total averaged spectrum among six hiPSC-lines and two distinct sample preparations. The color indicates the difference of mean spectrum of each hiPSC-line and each preparation (e1 or e2) and the average spectrum of all 12 collections. (C) Principal component analysis of Raman spectra of single cells in six hiPSC-lines (left) and in two sample preparations of 253G1 (right). Different color, different hiPSC-line in the left panel. The gray plots in the right panel are superimpose of the left panel. (D) hiPSC-line and sample preparation dependences of the PC2 value in C. Error bars are standard deviations. Asterisks indicate less than 0.05 of the p-value in the Student’s t-test.
Coefficient of Variation Analysis in Cellular Raman Spectroscopy
Based on the above results, we concluded that the present Raman spectroscopic analysis is highly reproducible in technical replicates and, thus, suitable to quantitate cellular heterogeneity in a dish and explore a common behavior independent of the origin’s individuality. The next step was to explore the specific Raman peaks whose intensity become more fluctuating/heterogeneous prior to state transition of cell in hiPSC early differentiation.
We induced cardiomyogenesis to the six different hiPSC lines using a common protocol and collected single-cell spectra from a total of seven culture dishes into two independent experiments, with an interval time of three months, at 0th, 1st, 2nd, 4th, 7th and 10th days after the differentiation induction (Figure 4A). Intensity changes of the 38 main Raman peaks in the mean intensity spectrum in a dish were clearly visualized during myocardial differentiation of the six different hiPSC-lines (Figure 4B, upper, and Supplementary Figure S8). Some of the peaks showed common changes regardless of hiPSC-line. For example, the peak indicating cytochrome-C (755 cm–1) decreased (Figure 4B, upper, yellow arrow) and that of lipids (1448 cm–1) increased (Figure 4B, upper, green arrow), respectively. Changes in these peaks, which were mostly common among the cell lines, were observed on the 2nd or the 4th day after the differentiation induction (Supplementary Figure S9A–C). On the other hand, in the spectrum of standard deviation in a dish, peaks showing large change were observed at earlier days than the intensity change after the induction of differentiation (Figure 4B, lower).
Figure 4 .
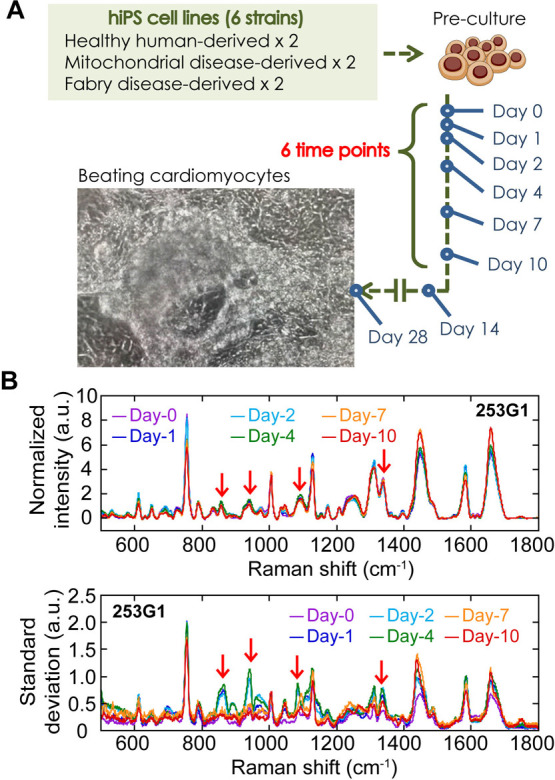
Raman spectra measurement during myocardial differentiation of six hiPSC-lines. (A) Explanative cartoon of the Raman observation in cardiomyogenesis of distinct 6 hiPSC strains. (B) Mean intensity (upper) and mean standard deviation (lower) of Raman spectra of 253G1 before (Day-0, magenta) and on 1st (Day-1, blue), 2nd (Day-2, green), 4th (Day-4, green), 7th (Day-7, orange) and 10th (Day-10, red) day after differentiation induction. Red arrows show peaks that was identified as early differentiation marker in CV analysis. Combined data of two experimental batches was used.
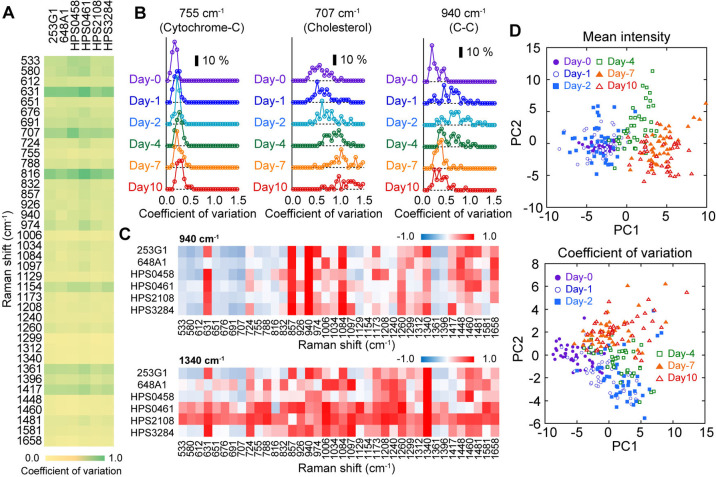
As a metric for quantifying cellular heterogeneity in a dish, we used the coefficient of variation (CV), which is the standard deviation over the mean intensity. Similar to the intensity spectrum, there was no obvious hiPSC-line dependence in the CV spectrum before the myocardial differentiation (Figure 5A). Cell-to-cell variation was small in most of the peaks in all cell-lines tested (Figure 5A) Using the CV metric, we investigated the time development of cellular heterogeneity with the differentiation progression (Supplementary Figure S9B). While 755 cm–1 peaks did not largely change throughout the progression of differentiation (Figure 5B, left), 707 cm–1 peak, attributed to cholesterol [26], increased from the 4th day after the induction of differentiation (Figure 5B, middle). High correlations between the hiPSC-lines in the time development of the CV were observed for almost all peaks (Supplementary Figure S9D). Among them, we identified peaks at 857, 940, 1084, and 1340 cm–1 as early differentiation markers that increased more than two-fold immediately after the induction and decreased in the latter half in all hiPSC-lines (Figure 5B, right and Supplementary Figure S9B, red characters). The time courses of the CV at 857, 940, and 1084 cm–1 peaks during the 10 days of early differentiation were commonly correlated in all cell lines (Figure 5C, upper), which is reasonable because those peaks derive from C-C bond-related vibration modes [27,28]. That of the 1340 cm–1 peak, attributing CH deformation [11], was loosely linked to those of various peaks such as 940, 1006, 1084, 1208, and 1260 cm–1 peaks (Figure 5C, lower). By performing principal component analysis using all spectra data of all cell lines, distribution shift in the PC space could be detected on the 4th day after the induction in the conventional intensity-based analysis (Figure 5D, upper) and on the 1st day in the present heterogeneity-based analysis (Figure 5D, lower). In the gene expression profile, we confirmed the expression changes of some genes prior to those of core transcription factors characterized to pluripotent state (Supplementary Figure S7, CXCL1, HESX1, WNT3A, etc.).
Figure 5 .
Coefficient of variation analysis of Raman spectra during myocardial differentiation of six hiPSC-lines before induction of differentiation (Day-0). (A) Spectra of coefficient of variation (CV) of six hiPSC-lines. Color from yellow to green correspond to value from 0 to 1.0. (B) Time developments of histogram of CV at the peaks of 755 cm–1 peaks (left), 707 cm–1 peak (middle) and 940 cm–1 peak (right). Scale bar indicates 10% frequency. All six cell-lines were used for analysis. (C) A heatmap of the correlations of time development of CV Correlation of CV time development between 940 cm–1 (upper) or 1340 cm–1 (lower) was calculated against other peaks and visualized. Color from blue to red correspond to value from –1.0 to 1.0. (D) Principal component analysis of mean intensity of Raman spectra in a dish (upper) and that of CV (lower). Each plot indicates each dish. Color is the same to Figure 4B.
Dynamical Network Biomarkers (DNB) Analysis in Cellular Raman Spectroscopy
The DNB theory proposes a more theoretical method for the use of the early-warning signals [29,30], and it has been successful in identifying a group of genes whose expression fluctuates prior to the onset of metabolic syndrome in mice [21]. The DNB theory is based on the idea that there should be elements that collectively fluctuate just before the critical slowing down [29,30]. Assuming that a cell is a system consisting of a large number of elements and their interactions, the DNB theory has the potential to be applied to predict state transitions of the cell system. In an analysis based on the DNB theory, “fluctuating” variables that are correlated and fluctuate collectively in measured multivariate data can be statistically identified. Evaluating the significantly fluctuating and highly correlated components of a biological system provides an early signal of the state transitions in the system (earlier than ordinary intensity-based biomarkers) and simultaneously identifies groups of signature components [29,30]. Corresponding to experiments in cell biology such as this study, the analysis based on DNB theory can automatically identify a group of Raman peak spectra with cooperative transitions in the degree of single-cell heterogeneity along a time development before the state transition of a cell. Recently, we reported that analysis based on DNB theory could detect early-state transition of immune T cells in antigen-presenting cell (APC) activation in Raman spectrum measurement [21]. Finally, we investigated the feasibility of the DNB analysis to screen the peaks identified shown in Figure 5 in a human-independent manner.
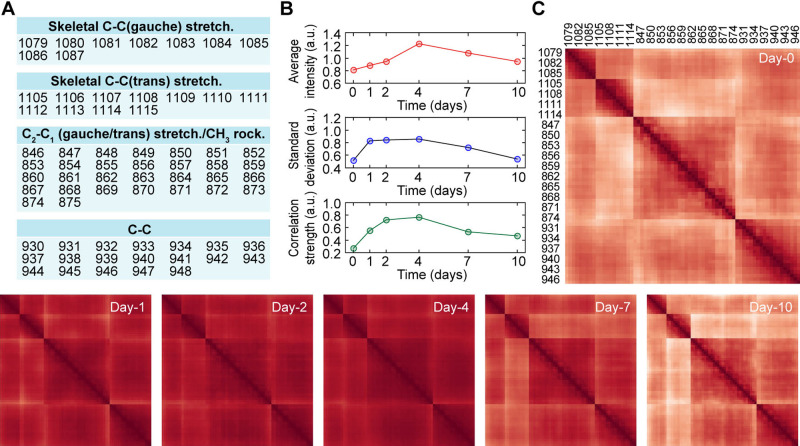
The DNB analysis was applied to the present dataset to investigate the intercorrelation of Raman peak heterogeneity. The DNB analysis identified mainly four groups as fluctuating elements on the 2nd and 4th day after induction, which are attributed to C-C bond related modes (Figure 6A and Supplementary Figure S9), consistent with the result of the CV analysis. The average intensity of these peaks reached the maximum on the 4th day and then monotonically decreased (Figure 6B, top). The average standard deviation immediately increased to the maximum on the 1st day and then decreased after being maintained until the 4th day (Figure 6B, middle). The correlation strength of those peaks behaved in a manner between the maximum intensity and the standard deviation (Figure 6B, bottom). Notably, on the 10th day, the intensity and the correlation stopped decreasing to approximately half of the maximum values, while the standard deviation returned to the initial value. To visualize the time development of the correlations among the identified marker groups, heatmaps were displayed showing the correlation strength between each two markers at each elapsed day (Figure 6C). Correlation strength was derived by first calculating the correlation coefficient for all combinations of Raman shifts extracted in DNB analysis (Figure 6A). By taking the sum of absolute values of the correlation coefficients and dividing it by the number of Raman shifts, average correlation strength can be calculated. Before the myocardial differentiation, the heatmap exhibited each cluster of four groups shown in Figure 6A (Figure 6C, Day-0). The correlation between groups increased from the 1st day to the 4th day after the myocardial induction and then returned (Figure 6C, Day-1 to Day-10). Thus, the correlation among four groups increased before the 4th day in the conventional intensity-based analysis.
Figure 6 .
DNB analyses of Raman spectra during myocardial differentiation of six hiPSC-lines. (A) A table of dynamic network biomarker identified by the DNB analysis. The number indicates the wavenumber identified as a marker. (B) Time developments of average intensity (top), standard deviation (middle), and correlation strength (bottom) of the peak groups identified as the marker. The correlation strength does not include auto-correlation. (C) Time developments of heatmap of correlation between the identified markers. Darker, higher; lighter, lower.
Discussion
In summary, we have clearly demonstrated the increased single-cell heterogeneity in the Raman spectrum before the escape from the pluripotent state, defined by the intensity profile with the development of a highly-reproducible single-cell Raman spectroscopic protocol. This study experimentally proves the abductive hypothesis that cellular heterogeneity represented in Raman spectrum of hiPSCs increases faster than signal intensity changes immediately after losing pluripotency, independent of hiPSC-lines tested.
If the transient increase in standard deviation observed in Figure 4B was derived from deviations and/or vibrations in the measurement system, the increase in standard deviation should be confirmed not only for specific peaks but also for all peaks or the entire spectrum. Therefore, we concluded that the single-cell heterogeneity observed here was derived from the fluctuation of cellular state. Then, the Yuragi-based analysis such as CV analysis and DNB analysis can detect the state transition prior to the conventional intensity-based analysis. Those Raman peaks identified by CV analysis and DNB analysis can be used as markers to define a cell state corresponding to pre-transition state defined by intensity-based analysis. In retrospect, upon reviewing our previous data, we found that cellular heterogeneity in Raman spectrum increased before the cells entered a stable state, in all cases including muscle differentiation, early embryonic stem cell, and T cell differentiation in mouse cell lines [11–15]. Increased heterogeneity of Raman spectra before the change in Raman peak intensity demonstrates that although intracellular concentration of chemical component became heterogeneous before the change in cell state, averaged concentration within the population does not change, indicating the difficulty in detecting the pre-transition state by traditional assay methods which measure the average concentration of the cell component. Interestingly, another group previously reported that Raman spectral signals collected from fetal mouse heart tissue were noisier and more heterogeneous than those obtained from adult mice [31]. Given this study and the previous results, we here propose the use of heterogeneity as an indicator to define pre-transition state of a cell.
The heterogeneity-based analyses, including CV analysis and DNB analysis, identified some peaks correctively fluctuated between the 1st and 2nd day after myocardial differentiation induction: 857, 940, 1084, and 1340 cm–1 in the CV analysis (Figure 5A), and 857, 940, 1084, and 1110 cm–1 in the DNB analysis (Figure 6A). The intracellular component with characteristic peaks at these wavelengths is glycogen [32,33]. This result was consistent with the previous report that standard deviation of Raman peaks attributed to glycogen (852 cm–1 and 938 cm–1 in this report) of human ESCs (hESCs) was greater than that of hiPSCs [16]. In general, it is experimentally difficult to stably maintain human cells in a fully-pluripotent state, called naïve state, so reprogrammed stem cells called “iPS cells” are typically in a partially–pluripotent, or primed state. We hypothesize that glycogen expression fluctuates when cells exit the stable primed state, regardless of whether they are undergoing reprogramming or differentiating.
The CV analysis identified an interesting heterogeneity-based biomarker of 707 cm–1 attributed to cholesterol. The CV of 707 cm–1 constantly increased from the 0th day to the 10th day after differentiation induction (Figure 5B, middle) whereas the peak intensity decreased (Supplementary Figure S9A). Transcriptomic analysis previously demonstrated that gene expressions related to cholesterol efflux and transport are upregulated from the 0th day to the 15th day in hiPSC cardiomyogenesis [34]. We speculate that changes in metabolic activity can be detected as cellular heterogeneity at the Raman spectral peak reflecting cholesterol. On the other hand, another group reported an increase in cholesterol expression before and after cardiomyogenesis [31]. However, they used hESCs instead of hiPSCs and a different differentiation protocol. Furthermore, they cultured the cells using ultra-low attachment dishes and collagen-coated dishes for differentiating culture and harvested them prior to Raman measurements. In contrast, we used uncoated glass bottom dishes to prevent changes in cell state caused by mechanical stimulation during cell reseeding over differentiation progression and measured Raman scattering spectra without reseeding. These experimental differences may have resulted in differences in the absolute amount of cholesterol expression in cells. Although further validation experiments are needed, it is possible that cellular heterogeneity provides a more accurate reflection of gene network activation than the increase or decrease in intensity.
The result of DNB analysis can be interpreted as followed: just after the differentiation induction, the low collectiveness allowed independent fluctuation of each element. With the progression of differentiation, these elements interacted with each other, resulting in a positive feedback loop that increased their intensities. Once the interaction network was established on the 4th day, the fluctuation was restricted, and the intensity and correlation transitioned to a constant due to the construction of a new stable network. The network reconstruction was clearly highlighted by the time-series changes in the correlation heatmap (Figure 6C). The results of DNB analysis indicate that the intermolecular network of C-C bond-related vibrations, most likely responsible for glycogen, is a key factor in driving hiPSCs out of the pluripotent state.
The transition of cellular Raman spectrum during hiPSC differentiation has been previously investigated [16–20]. All of these studies have demonstrated the clear separation and discrimination of iPSCs and other cells based on unsupervised machine learning methods such as PCA and t-distributed stochastic neighbor embedding (tSNE), and/or deep learning methods using intensity information. However, to the author’s knowledge, this study is the first example of an analysis that actively uses information on cellular heterogeneity in Raman spectrum. Therefore, the results in this study cannot simply compare to those previously reported, and the above discussions and hypotheses do not have any previous evidences and need to be newly proven in the future. In other words, by considering and quantitating cellular heterogeneity based on concept of singularity biology, this study generated a new biological question that has not been discussed so far.
The concept of Yuragi-based analysis might open the next door in presymptomatic medicine because markers identified by the Yuragi-based analysis can detect the state transition of a multi-cellular system in advance, meaning a possibility of on-set prediction of disease. This study is also the first comparative investigation of the Raman spectral signatures of hiPSCs derived from different individuals. Our findings demonstrate that the differences in origin were minimal compared to the cellular heterogeneity in the Raman spectrum, highlighting the advantages of using Raman spectroscopy as a quality evaluation tool for hiPSCs. This is because the marker threshold can be determined without depending on individual differences. The present method, using Raman spectrum and Yuragi-based analysis, has the potential to become a standard technique in hiPSC quality control.
In this article, we described the new method, the results and the finding, and the proposal of the new biological markers to define pre-transition state of cells. In general, Yuragi-concept is basically stands for temporal fluctuations where state of the element fluctuates by time. However, assuming that ergodicity is valid within the system, random temporal fluctuation of the elements appears as heterogeneity in the population. This assumption that ergodicity is valid in the system, however, have to be proven in the future study. Notably, while the abductive hypothesis proven in this study is based on the concepts of holons and Yuragi, the idea of the experiments we preformed arose from the first principle of Singularity biology, “see the forest for the trees.” The concept of Singularity biology experimentally summarized the two fundamental concepts debated for several decades, leading to the new biological question, tool and hypothesis in this study.
Conclusion
In conclusion, we propose a new analytical concept that cellular heterogeneity in Raman spectrum could detect pre-state transition of cells. The Yuragi-based biomarker is perhaps applicable to any kinds of single-cell omics technologies, not just in hiPSC quality evaluation and Raman spectroscopy. We hope for the emergence of biology that uses not only signal intensity but also fluctuation/heterogeneity as a biological index. This study demonstrated the possibility that analysis based on “Singularity biology” contribute to not only basic science but also future medicines, including presymptomatic medicine and regenerative medicine.
Conflict of Interest
The authors declare no competing interests.
Author Contributions
All experiments, H.F.; analytical development, T.M.W.; Genetic analyses, K.S., Y.K. and K.A; hiPSC lines establish and culture, K.S. and K.A.; DNB analysis, Y.Y., T.H., and K.K.; project administration, T.M.W.; All authors have read and agreed to the published version of the manuscript.
Data Availability
The evidence data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary Materials
Acknowledgements
We would like to thank Professor Tamiki Komatsuzaki (Hokkaido University) for his critical discussions on data analysis, Keiko Yoshizawa (RIKEN) for assistance in preparing hiPSCs, and Arno Germond and Yasuhiro Maeda for their contribution to the construction of the Raman microscope. We also thank Editage (www.editage.com) for providing English language editing services and Mr. Steven Robbins for English correction/proof. This work was primarily supported by Secom Science and Technology Foundation (Specific Field Research grant FY 2017), Japan Agency for Medical Research and Development under grant number 17bm0804008, and MEXT Grant-in-Aid for Scientific Research on Innovative Areas “Singularity Biology”, Grant Number JP18H05409. Part of the fluctuation analysis was supported by JST Moonshot R&D Grant Number JPMJMS2021, JSPS KAKENHI Grant Number JP 17KT0050, and a Grant-in-Aid for the Cooperative Research Project from the Institute of Natural Medicine, University of Toyama in FY2019 and FY2020. Finally, we greatly appreciate Prof. Toshio Yanagida (Osaka University) for providing us the basic concept and discussion of “Yuragi”.
References
- [1].Shimizu, H. A general approach to complex systems in bioholonics. in Lasers and synergetics: A colloquium on coherence and self-organization in nature. (Graham, R., Wunderlin, A. eds.), pp. 204–223 (Springer Berlin Heidelberg, Berlin, Heidelberg, 1987). [Google Scholar]
- [2].Yanagida, T., Murata, T. Functional roles of yuragi in biosystems. in Fluctuation-induced network control and learning: Applying the yuragi principle of brain and biological systems. (Murata, M., Leibnitz, K. eds.), pp. 31–47 (Springer Singapore, Singapore, 2021). [Google Scholar]
- [3].Leibnitz, K. Introduction to yuragi theory and yuragi control. in Fluctuation-induced network control and learning: Applying the yuragi principle of brain and biological systems. (Murata, M., Leibnitz, K. eds.), pp. 3–30 (Springer Singapore, Singapore, 2021). [Google Scholar]
- [4].Yanagida, T., Ueda, M., Murata, T., Esaki, S., Ishii, Y.. Brownian motion, fluctuation and life. Biosystems 88, 228–242 (2007). https://doi.org/10.1016/j.biosystems.2006.08.012 [DOI] [PubMed] [Google Scholar]
- [5].Okamoto, K., Germond, A., Fujita, H., Furusawa, C., Okada, Y., Watanabe, T. M.. Single cell analysis reveals a biophysical aspect of collective cell-state transition in embryonic stem cell differentiation. Sci. Rep. 8, 11965 (2018). https://doi.org/10.1038/s41598-018-30461-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Waddington, C. H. The strategy of the genes; a discussion of some aspects of theoretical biology (Allen & Unwin, London, 1957). [Google Scholar]
- [7].Chalancon, G., Ravarani, C. N. J., Balaji, S., Martinez-Arias, A., Aravind, L., Jothi, R., et al. . Interplay between gene expression noise and regulatory network architecture. Trends Genet. 28, 221–232 (2012). https://doi.org/10.1016/j.tig.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Carpenter, S. R., Brock, W. A.. Rising variance: A leading indicator of ecological transition. Ecol. Lett. 9, 311–318 (2006). https://doi.org/10.1111/j.1461-0248.2005.00877.x [DOI] [PubMed] [Google Scholar]
- [9].Scheffer, M., Bascompte, J., Brock, W. A., Brovkin, V., Carpenter, S. R., Dakos, V., et al. . Early-warning signals for critical transitions. Nature 461, 53–59 (2009). https://doi.org/10.1038/nature08227 [DOI] [PubMed] [Google Scholar]
- [10].Böttcher, M., Tada, Y., Moody, J., Kondo, M., Ura, H., Abugessaisa, I., et al. Single-cell transcriptomics, scrna-seq and c1 cage discovered distinct phases of pluripotency during naïve-to-primed conversion in mice. bioRxiv (2020). https://doi.org/10.1101/2020.09.25.313239
- [11].Germond, A., Ichimura, T., Horinouchi, T., Fujita, H., Furusawa, C., Watanabe, T. M.. Raman spectral signature reflects transcriptomic features of antibiotic resistance in escherichia coli. Commun. Biol. 1, 85 (2018). https://doi.org/10.1038/s42003-018-0093-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kobayashi-Kirschvink, K. J., Gaddam, S., James-Sorenson, T., Grody, E., Ounadjela, J. R., Ge, B., et al. Raman2rna: Live-cell label-free prediction of single-cell rna expression profiles by raman microscopy. bioRxiv (2021). https://doi.org/10.1101/2021.11.30.470655 [DOI] [PMC free article] [PubMed]
- [13].Ichimura, T., Chiu, L. D., Fujita, K., Kawata, S., Watanabe, T. M., Yanagida, T., et al. . Visualizing cell state transition using raman spectroscopy. PLoS One 9, e84478 (2014). https://doi.org/10.1371/journal.pone.0084478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ichimura, T., Chiu, L. D., Fujita, K., Machiyama, H., Kawata, S., Watanabe, T. M., et al. . Visualizing the appearance and disappearance of the attractor of differentiation using raman spectral imaging. Sci. Rep. 5, 11358 (2015). https://doi.org/10.1038/srep11358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ichimura, T., Chiu, L. D., Fujita, K., Machiyama, H., Yamaguchi, T., Watanabe, T. M., et al. . Non-label immune cell state prediction using raman spectroscopy. Sci. Rep. 6, 37562 (2016). https://doi.org/10.1038/srep37562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tan, Y., Konorov, S. O., Schulze, H. G., Piret, J. M., Blades, M. W., Turner, R. F.. Comparative study using raman microspectroscopy reveals spectral signatures of human induced pluripotent cells more closely resemble those from human embryonic stem cells than those from differentiated cells. Analyst 137, 4509–4515 (2012). https://doi.org/10.1039/c2an35507h [DOI] [PubMed] [Google Scholar]
- [17].Parrotta, E., De Angelis, M. T., Scalise, S., Candeloro, P., Santamaria, G., Paonessa, M., et al. . Two sides of the same coin? Unraveling subtle differences between human embryonic and induced pluripotent stem cells by raman spectroscopy. Stem Cell Res. Ther. 8, 271 (2017). https://doi.org/10.1186/s13287-017-0720-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hsu, C. C., Xu, J., Brinkhof, B., Wang, H., Cui, Z., Huang, W. E., et al. . A single-cell raman-based platform to identify developmental stages of human pluripotent stem cell-derived neurons. Proc. Natl. Acad. Sci. U.S.A. 117, 18412–18423 (2020). https://doi.org/10.1073/pnas.2001906117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li, R., Walsh, P., Truong, V., Petersen, A., Dutton, J. R., Hubel, A.. Differentiation of human ips cells into sensory neurons exhibits developmental stage-specific cryopreservation challenges. Front. Cell Dev. Biol. 9, 796960 (2021). https://doi.org/10.3389/fcell.2021.796960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ishigaki, M., Hitomi, H., Ozaki, Y., Nishiyama, A.. Exposing intracellular molecular changes during the differentiation of human-induced pluripotent stem cells into erythropoietin-producing cells using raman spectroscopy and imaging. Sci. Rep. 12, 20454 (2022). https://doi.org/10.1038/s41598-022-24725-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Koizumi, K., Oku, M., Hayashi, S., Inujima, A., Shibahara, N., Chen, L. N., et al. . Identifying pre-disease signals before metabolic syndrome in mice by dynamical network biomarkers. Sci. Rep. 9, 8767 (2019). https://doi.org/10.1038/s41598-019-45119-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Haruki, T., Yonezawa, S., Koizumi, K., Yoshida, Y., Watanabe, T. M., Fujita, H., et al. . Application of the dynamical network biomarker theory to raman spectra. Biomolecules 12, 1730 (2022). https://doi.org/10.3390/biom12121730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].de Oliveira, M. A. S., Smith, Z. J., Knorr, F., de Araujo, R. E., Wachsmann-Hogiu, S.. Long term raman spectral study of power-dependent photodamage in red blood cells. Appl. Phys. Lett. 104, 103702 (2014). https://doi.org/10.1063/1.4868253 [Google Scholar]
- [24].Paidi, S. K., Diaz, P. M., Dadgar, S., Jenkins, S. V., Quick, C. M., Griffin, R. J., et al. . Label-free raman spectroscopy reveals signatures of radiation resistance in the tumor microenvironment. Cancer Res. 79, 2054–2064 (2019). https://doi.org/10.1158/0008-5472.CAN-18-2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Palonpon, A. F., Ando, J., Yamakoshi, H., Dodo, K., Sodeoka, M., Kawata, S., et al. . Raman and sers microscopy for molecular imaging of live cells. Nat. Protoc. 8, 677–692 (2013). https://doi.org/10.1038/nprot.2013.030 [DOI] [PubMed] [Google Scholar]
- [26].Krafft, C., Neudert, L., Simat, T., Salzer, R.. Near infrared raman spectra of human brain lipids. Spectrochim. Acta A Mol. Biomol. Spectrosc. 61, 1529–1535 (2005). https://doi.org/10.1016/j.saa.2004.11.017 [DOI] [PubMed] [Google Scholar]
- [27].Qiu, S., Huang, Q., Huang, L., Lin, J., Lu, J., Lin, D., et al. . Label-free discrimination of different stage nasopharyngeal carcinoma tissue based on raman spectroscopy. Oncol. Lett. 11, 2590–2594 (2016). https://doi.org/10.3892/ol.2016.4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hu, Z., Wang, X., Wang, W., Zhang, Z., Gao, H., Mao, Y.. Raman spectroscopy for detecting supported planar lipid bilayers composed of ganglioside-gm1/sphingomyelin/cholesterol in the presence of amyloid-beta. Phys. Chem. Chem. Phys. 17, 22711–22720 (2015). https://doi.org/10.1039/c5cp02366a [DOI] [PubMed] [Google Scholar]
- [29].Chen, L., Liu, R., Liu, Z. P., Li, M., Aihara, K.. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci. Rep. 2, 342 (2012). https://doi.org/10.1038/srep00342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Aihara, K., Liu, R., Koizumi, K., Liu, X., Chen, L.. Dynamical network biomarkers: Theory and applications. Gene 808, 145997 (2022). https://doi.org/10.1016/j.gene.2021.145997 [DOI] [PubMed] [Google Scholar]
- [31].Brauchle, E., Knopf, A., Bauer, H., Shen, N., Linder, S., Monaghan, M. G., et al. . Non-invasive chamber-specific identification of cardiomyocytes in differentiating pluripotent stem cells. Stem Cell Reports 6, 188–199 (2016). https://doi.org/10.1016/j.stemcr.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Duraipandian, S., Mo, J., Zheng, W., Huang, Z.. Near-infrared raman spectroscopy for assessing biochemical changes of cervical tissue associated with precarcinogenic transformation. Analyst 139, 5379–5386 (2014). https://doi.org/10.1039/c4an00795f [DOI] [PubMed] [Google Scholar]
- [33].Konorov, S. O., Schulze, H. G., Piret, J. M., Aparicio, S. A., Turner, R. F., Blades, M. W.. Raman microscopy-based cytochemical investigations of potential niche-forming inhomogeneities present in human embryonic stem cell colonies. Appl. Spectrosc. 65, 1009–1016 (2011). https://doi.org/10.1366/11-06312 [DOI] [PubMed] [Google Scholar]
- [34].Barison, M. J., Pereira, I. T., Waloski Robert, A., Dallagiovanna, B.. Reorganization of metabolism during cardiomyogenesis implies time-specific signaling pathway regulation. Int. J. Mol. Sci. 22, 1330 (2021). https://doi.org/10.3390/ijms22031330 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The evidence data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.