Abstract
MAPK (mitogen-activated protein kinase) pathways constitute major regulators of cellular transcriptional programmes. We analysed the ERK1,2 (extracellular-signal-regulated kinase 1,2) transcriptome in a non-transformed MEC (mammary epithelial cell) line, MCF-12A, utilizing rAd MEK1EE, a recombinant adenovirus encoding constitutively active MEK1 (MAPK/ERK kinase 1). rAd MEK1EE infection induced morphological changes and DNA synthesis which were inhibited by the MEK1,2 inhibitor PD184352. Hierarchical clustering of data derived from seven time points over 24 h identified 430 and 305 co-ordinately up-regulated and down-regulated genes respectively. c-Myc binding sites were identified in the promoters of most of these up-regulated genes. A total of 46 candidate effectors of the Raf/MEK/ERK1,2 pathway in MECs were identified by comparing our dataset with previously reported Raf-1-regulated genes. These analyses led to the identification of a suite of growth factors co-ordinately induced by MEK1EE, including multiple ErbB ligands, vascular endothelial growth factor and PHRP (parathyroid hormone-related protein). PHRP is the primary mediator of humoral hypercalcaemia of malignancy, and has been implicated in metastasis to bone. We demonstrate that PHRP is secreted by MEK1EE-expressing cells. This secretion is inhibited by PD184352, but not by ErbB inhibitors. Our results suggest that, in addition to anti-proliferative properties, MEK1,2 inhibitors may be anti-angiogenic and possess therapeutic utility in the treatment of PHRP-positive tumours.
Keywords: extracellular-signal-regulated kinase 1,2 (ERK1,2); expression profiling; mammary epithelial cell; microarray; mitogen-activated protein kinase (MAPK); parathyroid hormonerelated protein (PHRP)
Abbreviations: AP-1, activator protein-1; CDK, cyclin-dependent kinase; delta-Raf-ER, Raf/oestrogen receptor/green fluorescent protein fusion protein; DUSP, dual-specificity phosphatase; EGF, epidermal growth factor; ERK, extracellular-signal-regulated kinase; HB-EGF, heparin-binding EGF; HHM, hypercalcaemia of malignancy; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; MEC, mammary epithelial cell; MEK, MAPK/ERK kinase; MEK1EE, constitutively active MEK1; MKP, MAPK phosphatase; PHRP, parathyroid hormone-related protein; rAd MEK1EE, recombinant adenovirus encoding MEK1EE; rAd LacZ, recombinant adenovirus encoding LacZ; TGF, transforming growth factor; VEGF, vascular endothelial growth factor
INTRODUCTION
MAPK (mitogen-activated protein kinase) signalling cascades integrate and are major transducers of extracellular stimuli that result in diverse cellular responses [1]. MAPKs are organized into evolutionarily conserved modules composed of an upstream MEKK (MEK kinase) which phosphorylates and activates a MEK (MAPK/ERK kinase), which in turn phosphorylates and activates a MAPK [ERK (extracellular-signal-regulated kinase)] [1,2]. MEKs are dual-specificity protein kinases displaying stringent substrate selectivity that results in the activation of a limited number of closely related downstream MAPKs. The MEK1,2/ERK1,2 signalling module is the predominant MAPK pathway mediating responses to both proliferative and differentiative signals [1,2]. The only known substrates of MEK1,2 are ERK1,2 and the only known activators of ERK1,2 are MEK1,2.
Considerable interest in understanding the contribution of MEK1,2/ERK1,2 signalling to tumorigenesis is driven by this MAPK module residing both upstream and downstream of known oncogenes. ErbB tyrosine kinase receptors, Ras and Raf are upstream activators of MEK1,2 and ERK1,2. Recently, missense mutations in B-Raf were identified as occurring in 66% of malignant melanomas [3]. In addition, the proto-oncogene products c-Fos, c-Jun and c-Myc encode downstream transcription factor targets of activated ERK1,2 [4].
ERK1,2 activity is elevated in tumorigenic cell lines as well as primary tumour tissue [5]. Small-molecule inhibitors targeting both Raf and MEK1,2 are in clinical trials in oncology [5]. MEK1,2 inhibitors inhibit the growth of tumour xenografts, reverse cellular transformation, decrease cell proliferation, and in some instances induce apoptosis [5].
Consistent with their role as major effectors of Ras, dominant negative mutants of either MEK1 or ERK2 inhibit Ras- and Raf-mediated transformation. Likewise, expression of constitutively active forms of MEK1 can result in cellular transformation [1,2]. A constitutively activated ERK2–MEK1 fusion protein is sufficient to transform NIH3T3 cells when sequestered in the nucleus, but not in the cytoplasm [6]. This observation suggests that the nuclear substrates of ERK1,2 drive cellular transformation.
The global transcriptional profile induced by ERK1,2 activation remains largely undefined [4]. Previous expression profiling experiments have evaluated upstream activators such as Ras and Raf [7,8]. MEK1,2 are the most well characterized substrates and effectors of Raf; however, Raf-mediated, MEK1,2-independent, events have been reported [9]. The ERK1,2 transcriptome has been evaluated indirectly utilizing the MEK1,2 inhibitor PD98059 in Ras-transformed fibroblasts [7]. However, PD98059 also inhibits MEK5 [10]. Proteomic studies [11] as well as expression profiling experiments utilizing filter arrays of 1176 cDNAs [12] have yielded novel ERK1,2 targets. However, the number of regulated events identified was limited and suggested the potential for identifying additional novel ERK1,2-regulated genes. Using a direct method, we have evaluated the ERK1,2 transcriptome in an MEC (mammary epithelial cell) line. Our results demonstrate the strong co-ordinate induction of a suite of growth factors with known importance in multiple aspects of oncogenesis, including multiple ErbB ligands, VEGF (vascular endothelial growth factor) and parathyroid hormone-related protein (PHRP). PHRP is a peptide hormone responsible for HHM (hypercalcaemia of malignancy) and is implicated in metastasis to bone. We demonstrate that PHRP is secreted by MCF-12A cells after the expression of MEK1EE (constitutively active MEK1). PHRP secretion is inhibited by the MEK1,2 inhibitor PD184352, but not by ErbB inhibitors.
EXPERIMENTAL
Cell culture
MCF-12A cells (from A.T.C.C.) were cultured in Dulbecco's modified Eagle's medium/Ham's nutrient mixture F12 (1:1, v/v) supplemented with 20 ng/ml EGF (epidermal growth factor), 100 ng/ml cholera toxin (Calbiochem, La Jolla, CA, U.S.A.), 10 μg/ml insulin, 0.5 μg/ml cortisol and 5% (v/v) horse serum (complete medium).
Adenoviral infection
rAd MEK1EE and rAd LacZ (recombinant adenovirus encoding MEK1EE or LacZ respectively) were kindly provided by M. Cobb (University of Texas Southwestern Medical Center, Dallas, TX, U.S.A.). rAd MEK1EE encodes mutant (S218E and S222E), constitutively active MEK1 [13]. Adenoviruses were amplified in 293 cells, and the MEK1EE mutations were confirmed by sequence analysis. Viral titres were determined by HPLC. MCF-12A cells were plated at (4–5)×104 cells/cm2 in complete medium for 24 h and subsequently serum starved for 24 h in serum-free medium (Dulbecco's modified Eagle's medium/F12, supplemented with 0.5 μg/ml cortisol, 5.0 μg/ml insulin and 11 μg/ml bovine transferrin). Following serum starvation, cells were infected with 7.5×109 viral particles/ml of either rAd MEK1 EE or rAd LacZ in complete medium for 1 h at 37 °C and then returned to serum-free medium. At 0, 4, 6, 8, 12, 16 and 24 h after the 1 h infection, cells were rinsed in 37 °C in PBS, harvested using guanidine isothiocyanate, snap-frozen in liquid N2 and stored at −80 °C. Under these experimental conditions, 100% infectivity was achieved, while serum and non-specific adenoviral effects were minimized.
RNA isolation and cRNA probe synthesis
Total RNA was isolated by CsCl gradient centrifugation as described [14]. Total RNA from each sample was used to prepare biotinylated target RNA using the Genechip T7-Oligo(dT) Promoter Primer Kit (Affymetrix, #900375) and Invitrogen reagents in accordance with Affymetrix protocols (www.Affymetrix.com/support/technical/manual/expression_manual.affx). Briefly, 10 μg of total RNA was used to generate first-strand cDNA using a T7-linked oligo(dT) primer. Following second-strand synthesis, double-stranded cDNA was purified using a Qiagen RNeasy kit. The synthesis of biotin-labelled cRNA probes was carried out using biotin-labelled ribonucleotides and in vitro transcription reagents (Enzo Diagnostics; Affymetrix, #900182), resulting in approx. 100-fold amplification of RNA. The biotin-labelled cRNA probes were purified and fragmented in fragmentation buffer (Affymetrix, #900371) by incubation at 95 °C for 35 min.
Hybridization to Affymetrix U95A GeneChips
Spike controls were added to 10 μg of fragmented cRNA prior to overnight hybridization to human U95A oligonucleotide arrays for 16 h at 42 °C. Arrays were washed and stained with streptavidin/phycoerythrin, before scanning on an Affymetrix GeneChip scanner. Probe synthesis, hybridizations and scaling of raw data were performed by Expression Analysis Inc. (Durham, NC, U.S.A.) in accordance with Affymetrix protocols. Data were analysed using Microarray Suite 5.0.
Quality control
The amount of starting RNA was determined by absorption spectrophotometry, and the quality was confirmed using an Agilent Bioanalyser. Prior to hybridization to U95A GeneChips, MEK1EE and LacZ cRNA probe quality was assessed by hybridization to Affymetrix Test Microarray Chips (#900341). Following hybridization to U95A chips, array images were inspected visually to confirm scanner alignment and the absence of bubbles and scratches. 3′/5′ ratios for glyceraldehyde-3-phosphate dehydrogenase and β-actin were between 1.0 and 3.4. BioB spike controls were present on all chips, with BioC, BioD and CreX present in increasing intensities. Background fluorescence was less than 100 (50–80). Q-values ranged from 1.9 to 2.2. Global scaling of raw data using a target intensity of 500 produced scale factors that ranged from 3.6 to 9.5.
Quantitative real-time PCR
Quantitative real-time PCR was performed using an ABI Prism 7700 sequence detector (Applied Biosystems, Foster City, CA, U.S.A.). Probe and primers for quantitative real-time PCR were designed based on GenBank sequences using Primer Express software (Applied Biosystems) (see Table S1, at http://www.BiochemJ.org/bj/381/bj3810635add.htm). Sequence specificity was confirmed by BLAST analysis. Alternatively, Taqman™ (Applied Biosystems) probe/primer sets were used as indicated. Of five housekeeping genes tested for use as an endogenous control, the ribosomal protein S9 was determined to be the least variable across time points, and was chosen. The same total RNA utilized for hybridization to U95A arrays was reverse-transcribed with Superscript II (Gibco BRL). The resulting cDNA was used in quantitative real-time PCR experiments. Cycle threshold values were normalized by comparing cycle thresholds obtained for S9 in parallel reactions.
Data analysis
Probe-set signal intensities for all elements on the U95A array were subjected to global scaling as described above. Differential gene expression was determined by generating logarithmic ratios of scaled signal intensities of all elements on the arrays for MEK1EE relative to LacZ. A filter query was then applied to identify all genes that were differentially expressed by a minimum of 2.5-fold up or down relative to the LacZ control at any one time point. The filter criteria also stipulated that, in addition to a minimum 2.5-fold change in mRNA abundance, at least one of the probe sets had to be assigned a P value of ≤0.04, indicating that the transcript was present (‘P’ call) in that sample. Genes meeting these criteria were analysed further using a hierarchical clustering algorithm to identify groups of genes exhibiting similar expression profiles [15]. The clustering algorithm was implemented using S-PLUS (Mathsoft Inc., Seattle, WA, U.S.A.).
Transcription factor sites and promoter analysis
Of the 430 up-regulated genes contained within group A (see the Results section), 301 had genomic sequences that could be defined unambiguously as potential regulatory sequences by the transcript mapping approach using AAT [16], as described previously [17]. Sequences 2000 bp immediately upstream of the transcription start sites were then extracted as promoter sequences. These promoter region sequences were pooled and checked for transcription factor binding sites by running a match against the TRANSFAC transcription factor binding site Matrix libraries [18]. Similarly, a reference promoter database from all the genes contained within GenBank Refseq (Release 129), except those genes that did not have a continuous region upstream of the first exon start site containing at least 2000 bp, was also constructed. The constructed reference database contained 4221 genes.
A matrix similarity score of 0.8 was used as the cut-off value to calculate the observed number of each matrix in the sample list. The same analysis was applied on the reference database to calculate the expected frequency of each matrix. The expected number of each matrix in the sample list is equal to the expected frequency multiplied by the promoter number in the sample list. The discrepancies are evaluated by a statistical variable chi-square [19]. The chi-square values were converted into P values. P<0.005 was used as a cut-off in the present study.
Western analysis
Samples of 30 μg of total protein were resolved on 10% (w/v) polyacrylamide gels under reducing conditions. Antibodies recognizing dual-phophorylated ERK1,2 and pan ERK1,2, and MEK1,2 were from Biosource International (Camarillo, CA, U.S.A.) and Cell Signaling Technologies (Beverly, MA, U.S.A.) respectively.
Thymidine incorporation assays
Cells were plated on to Cytostar scintillating microplates (AmershamPharmacia, Piscataway, NJ, U.S.A.) and infected as described above. [14C]Thymidine incorporation was quantified by β-scintillation counting. PD184352, Iressa and GW2016 were synthesized as described in patent W09901426 [19a], patent W09633980 [19b] and Cockerill et al. [20] respectively.
PHRP RIA
Cells were plated on to 12-well plates and infected with rAd MEK1EE or rAd LacZ in triplicate as described above. After the 1 h infection, medium was replaced with 500 μl of serum-free medium with or without inhibitors. Conditioned medium was collected 48 h later, cleared by centrifugation at 20817 g for 15 min and stored at −80 °C. Cells were counted using a haemocytometer. PHRP in conditioned medium was detected using a two-site RIA (Nichols Diagnostics, San Juan Capistrano, CA, U.S.A.).
RESULTS
Expression of rAd MEK1EE and activation of ERK1,2
Total RNA was collected at seven time points (0 to 24 h) following adenoviral infection. Whole-cell lysates from parallel cultures were collected for evaluation of transgene expression and ERK1,2 activation. MEK1EE transgene mRNA and protein expression were confirmed using quantitative real-time PCR and Western analysis. MEK1EE mRNA was detectable by quantitative real-time PCR at time 0 (cycle threshold=27), which corresponded to the end of the 1 h infection period, and reached near maximal levels by 4 h post-infection (cycle threshold=18) (Table S2; http://www.BiochemJ.org/bj/381/bj3810635add.htm).
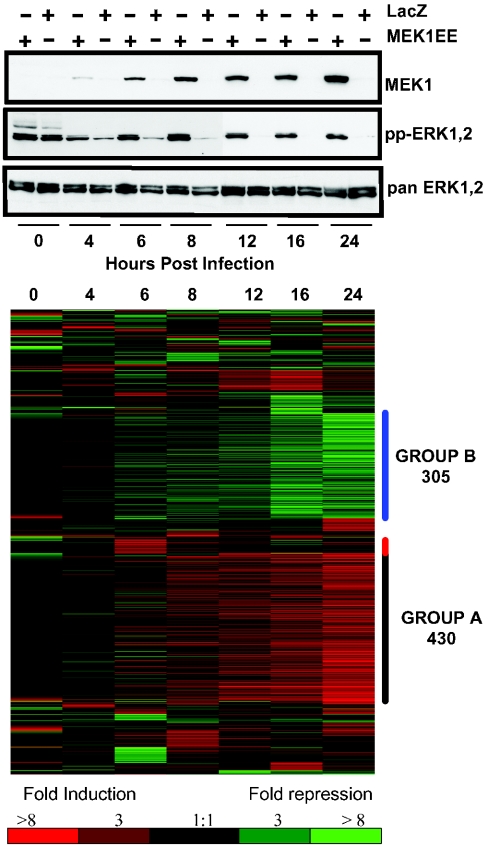
MEK1 protein expression was detected as early as 4 h post-infection (Figure 1A). Differences in ERK1,2 activation, attributable to MEK1EE, were observed by 4 h post-infection (Figure 1A). None of p38, JNK (c-Jun N-terminal kinase) and Akt were selectively activated by rAd MEK1EE, as assessed by Western analysis of parallel lysates utilizing phospho-specific antisera (results not shown).
Figure 1. Expression of MEK1EE and activation of ERK1,2.
(A) MCF-12A cells were infected and cell lysates harvested for Western analysis in parallel with cells infected for array analysis. Total MEK1 protein was detected by immunoblotting with an anti-MEK1 antibody (upper panel). Blots were stripped and reprobed with antibodies specific for ppERK1,2 (dual-phosphorylated ERK1,2; middle panel) and total ERK (lower panel). (B) Heat map depicting detected increases (red) and decreases (green) in mRNA expression of genes represented by 1347 elements analysed across the seven time points. Comparisons were made between MEK1EE and LacZ mRNA levels for each time point.
Analysis of changes in expression identified on the U95A GeneChip
cRNA derived from rAd MEK1EE- and rAd LacZ-infected cells was hybridized to Affymetrix U95A oligonucleotide GeneChips containing probe sets representing approx. 10000 human genes and ESTs (expressed sequence tags). An initial lenient filter query was established that required a ≥2.5-fold change in expression and a Present call for at least one time point. A total of 1347 elements met these criteria and were subjected to further statistical analysis (Table S3; http://www.BiochemJ.org/bj/381/bj3810635add.htm). Of these, 114 elements were determined to be redundant (95% identity over the overlapping sequences and 98% identity over a 100 bp window). Of the resultant 1233 non-redundant genes, 465 were differentially expressed by ≥2.5-fold at two or more time points.
Changes in relative mRNA abundance were confirmed for 26 genes by quantitative real-time PCR (Table S2; http://www.BiochemJ.org/bj/381/bj3810635add.htm). A subset of these genes is shown in Table 1. For all but one gene, i.e. that encoding ErbB3, the U95A array results were confirmed (Table S2). Results for genes in which no or very modest changes were detected on the array were also validated [c-Jun, MKP1 (MAPK phosphatase 1), EGF receptor]. Finally, changes in the expression of known transcriptional targets of ERK1,2, such as c-Fos, JunB and MKP3, were detected.
Table 1. Quantitative real-time PCR analysis of a subset of genes differentially regulated 10-fold or more by MEK1EE.
The fold change in mRNA abundance in MCF-12A cells infected with rAd MEK1EE relative to those infected with rAd LacZ is shown. Positive numbers indicate fold up-regulation and negative numbers indicate fold down-regulation. GenBank accession numbers correspond to elements on the Affymetrix U95A microarray and were used for probe/primer design for validation by quantitative real-time PCR.
| Up- or down-regulation (fold) | ||||||
|---|---|---|---|---|---|---|
| Gene product | Accession no. | 0 h | 6 h | 8 h | 12 h | 24 h |
| Amphiregulin | NM_001657 | 1.1 | 2.1 | 1.9 | 4.0 | 17.1 |
| BRCA1 | U14680 | 1.1 | 1.0 | 2.1 | 5.3 | 13.0 |
| BRCA2 | U43746 | 1.0 | 1.4 | 2.8 | 4.6 | 16.9 |
| Bub1 | AF046078 | 1.1 | −1.1 | 2.0 | 6.1 | 14.9 |
| cdc2 | NM_001786 | 1.1 | 1.2 | 1.6 | 5.2 | 9.7 |
| COX2 | NM_000963 | −1.1 | −1.2 | 1.3 | 7 | 97 |
| DUSP5 | NM_004419 | 1.3 | 6.1 | 5.6 | 7.4 | 36.7 |
| Epiregulin | NM_001432 | −1.5 | 4.2 | 4.6 | 4.6 | 46.5 |
| HB-EGF | M60278 | −1.1 | 3.2 | 4.2 | 3.4 | 45.2 |
| PHRP | M24351 | 1.1 | 2.6 | 6.1 | 17.1 | 90.5 |
| TGFα | NM_003236 | 1.1 | 2.8 | 6.1 | 4.9 | 9.8 |
| VEGF | NM_003376 | 1.2 | 2.3 | 2.3 | 2.8 | 14.9 |
To identify genes with similar expression profiles, a hierarchical clustering algorithm was employed [15]. Approx. 50% of genes fell within two groups. Group A and group B consist of 430 co-ordinately up-regulated and 305 down-regulated genes respectively (Figure 1B). Of the 465 genes differentially expressed by ≥2.5-fold at two or more time points, 344 were found in groups A (233) and B (111). Thus subsequent analyses focused on genes contained within groups A and B. Genes contained within these groups that were differentially expressed by ≥4.5-fold are shown in Table S4 (http://www.BiochemJ.org/bj/381/bj3810635add.htm).
MEK1EE induces the expression of multiple immediate early genes, including the MKPs
Activation of ERK1,2 led to the induction of multiple immediate early genes, including members of the DUSP (dual-specificity phosphatase) gene family, also known as the MKPs (Tables S2 and S4; http://www.BiochemJ.org/bj/381/bj3810635add.htm). These well established transcriptional targets of MAPKs dephosphorylate and down-regulate MAPK activity by removal of the activating phosphates within the Thr-Xaa-Tyr motif of the MAPK activation lip [21]. The 143-fold induction of MKP3 mRNA at 24 h was the largest change in gene expression detected across all time points. MKP2 (DUSP4) and MKP3 (DUSP6) mRNAs were induced by 10- and 16-fold respectively at 6 h post-infection. Levels of transcripts encoding the less well characterized MKP, DUSP5 (VH3), were also increased by 6 h post-infection. This result was confirmed by quantitative real-time PCR (Table 1). DUSP5 has been shown to inactivate ERK1,2 in vitro and in vivo, and may function as a tumour suppressor [22–24]. Inactivation of DUSP5 by retroviral insertion within the coding region of DUSP5 resulted in malignancy in CDKn2a−/− mice (where CDK is cyclin-dependent kinase) [24]. The regulation of DUSP5 mRNA by MEK1,2/ERK1,2 has not been reported previously.
Additional immediate early genes induced by ERK1,2 included numerous transcriptional regulators. Among these were IEX-1 (immediate early response gene X-1), which potentiates ERK1,2 activation in response to growth factors [25], and the AP-1 (activator protein-1) components Fos and Jun. Induction of c-Fos mRNA was detected at 6 h (3-fold) and reached a maximal level of 7-fold by 12 h. JunB mRNA was modestly up-regulated (3–4-fold; Table S2; http://www.BiochemJ.org/bj/381/bj3810635add.htm). In contrast, induction of the reported ERK1,2 transcriptional target c-Jun was near the limit of detection for quantitative real-time PCR (2-fold at both 16 and 24 h; Table S2).
Additional transcription factors induced by MEK1EE expression were members of the bHLH-Zip (basic helix–loop–helix) family (c-Myc, Max, E2F5), the Forkhead/Winged helix family (hepatocyte nuclear factor-3β) and the Ets family (Elk-1) (Table S3; http://www.BiochemJ.org/bj/381/bj3810635add.htm). Multiple binding partners of Myc were regulated, including Mad4 (down-regulated by 4.6-fold at 24 h), which is associated with cellular differentiation [26].
Transcript mapping and transcription factor site analysis
Binding sites for two transcription factors, Myc and E2F, occurred in a significantly greater number of the promoters of group A genes relative to a set of randomly selected promoter sequences (P<0.005). Of the 301 genes analysed, 238 contained c-Myc binding sequences in their promoter regions (Table S5; http://www.BiochemJ.org/bj/381/bj3810635add.htm). Of these genes, 18 also contained binding sites for E2F (Table S5).
MEK1EE expression regulates genes involved in the reorganization of cytoarchitecture
Some of the most substantial decreases in mRNA expression occurred among genes regulating cytoskeletal reorganization and cell attachment (Tables S3 and S4; http://www.BiochemJ.org/bj/381/bj3810635add.htm). Cytokeratins 1, 4, 10, 13, 15 and 16 were maximally decreased by 14-, 7-, 4-, 5-, 6- and 2.5-fold respectively. Cytokeratin 18 was the only cytokeratin to be induced (2.9-fold at 24 h). Similarly, cytokeratin 18 was induced, whereas cytokeratins 1 and 10 were repressed, by H-ras expression in mouse keratinocytes [27].
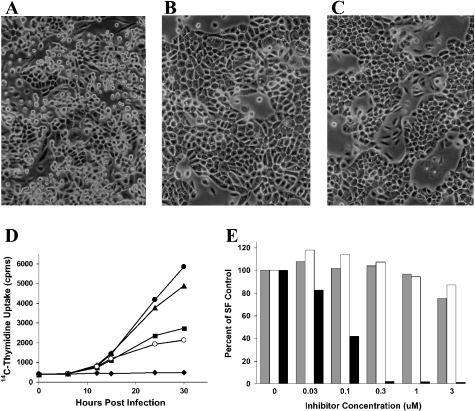
In contrast, substantial induction of laminin and integrin receptor mRNAs was detected (Tables S3 and S4). The heterotrimeric laminin proteins mediate cell adhesion and migration via interaction with specific integrin receptors [28]. MEK1EE expression altered cellular morphology, which is probably linked to the changes in expression detected in these cytoskeletal elements (Figures 2A and 2B). The MEK1,2 inhibitor PD184352 blocked the morphological changes in MEK1-expressing cells at a concentration that also inhibited DNA synthesis (Figure 2C). Changes in cell morphology and in the abundance of mRNAs encoding cytokeratins and integrins have been described previously in the murine MEC line EpH4 expressing constitutively active MEK1 [12]. Interestingly, these EpH4 cells also demonstrated a metastatic phenotype in vivo.
Figure 2. Expression of MEK1EE induces morphological changes and DNA synthesis in MCF-12A cells.
Phase-contrast images are shown of cells grown in serum-free medium 48 h post-infection with rAd MEK1EE (A), rAd LacZ (B) or rAd MEK1EE plus 1 μM PD184352 (C). (D) Thymidine incorporation is shown at the time points indicated after 24 h of serum starvation and a 1 h infection with rAd MEK1EE (▴) or rAd LacZ (▪), and in uninfected MCF-12A cells grown in serum (•) or serum-free medium (○). Counts in wells with medium but no cells are also shown (♦). (E) A MEK1 inhibitor, but not ErbB inhibitors, inhibit MEK1EE-induced thymidine incorporation. MCF-12A cells were treated with Iressa (grey bars), GW2016 (white bars) or PD184352 (black bars) at the concentrations indicated immediately following a 1 h infection with rAd MEK1EE. [14C]Thymidine incorporation was measured 30 h post-infection [serum-free (SF) medium] and is expressed as a percentage of that in DMSO-treated MCF-12A cells infected in parallel. The data are representative of at least two experiments performed in triplicate.
MEK1EE expression induces transcriptional changes that favour cell cycle progression
ERK1,2 activation resulted in a reciprocal pattern of expression of genes encoding cell cycle regulatory proteins that was suggestive of a pro-proliferative effect (Table S4; http://www.BiochemJ.org/bj/381/bj3810635add.htm). Ras and its downstream targets ERK1,2 are thought to regulate entry into the cell cycle primarily through induction of cyclin D1 mRNA expression via ERK1,2-regulated transcription factors, including AP-1 [29–31]. Cyclin D1 mRNA was induced 8-fold at 16 h, as determined by quantitative real-time PCR (Table S2). Other mRNAs encoding cyclins involved in the G/S-phase transition that were also induced included cyclin E2 (6–14-fold) and cyclin A (2.5–6-fold). DNA synthesis requires activation of cyclinE/A–CDK2 complexes by dephosphorylation of CDK2 by the phosphatase cdc25A. cdc25A, a known c-Myc target gene, was induced 6-fold beginning at 12 h. Transcripts encoding cyclins responsible for G2/M transition were also increased. mRNAs encoding cyclin B and its catalytic subunit cdc2 (cdk1) were co-ordinately induced, beginning at 12 h and increasing to 6.1-fold and 4.6-fold respectively by 24 h. Multiple genes involved in mitotic spindle assembly were up-regulated, including CENP-E (centromere protein E; 4.9–23.7-fold), mitotic kinesins and the protein kinases Bub1 (2–8-fold by array; 2–15-fold by quantitative real-time PCR), MAD2L (2.8–14.4-fold) and Aurora2 (2.7–4.9-fold).
MEK1EE expression led to the suppression of several negative regulators of cell cycle progression, such as the CDK inhibitor p57, also known as Kip2 (2.8-fold and 10-fold down-regulation at 8 h and 16 h respectively). The transcriptional regulation of p57/Kip2 by ERK1,2 has not been reported previously. In contrast, the expression of mRNA encoding the CDK2 inhibitor p21 (Cip1/WAF1) was modestly down-regulated by 2.8-fold at 8 h (confirmed by quantitative real-time PCR; Table S2). Quantitative real-time PCR analysis indicated no change in p53 transcript abundance. In addition, the anti-proliferative genes TGFβ (transforming growth factor β), BTG-1 (B-cell translocation gene-1) and gas-1 were negatively regulated by activated ERK1,2.
Consistent with these changes, MEK1EE expression resulted in an increased rate of [14C]thymidine incorporation relative to rAd LacZ-infected cells (Figure 2D). Thymidine incorporation was inhibited by the MEK1,2 inhibitor PD184352, but not by the ErbB inhibitors Iressa or GW2016 (Figure 2E). This suggests that entry into S phase is dependent on active MEK1, and is not due to stimulation of ErbB receptors via the induction of autocrine loops (see below).
Multiple genes involved in the DNA damage response are induced by MEK/ERK, including the tumour suppressor genes BRCA1 and BRCA2
mRNAs encoding the breast cancer susceptibility genes BRCA1 and BRCA2 were induced (Table S3; http://www.BiochemJ.org/bj/381/bj3810635add.htm). Quantitative real-time PCR analysis of BRCA1,2 mRNA expression confirmed the array results (2.0–2.8-fold at 8 h, increasing to 13–17-fold at 24 h; Table 1). mRNAs encoding multiple BRCA-interacting proteins were also up-regulated by MEK1EE, including the zinc finger protein BARD1 (BRCA1-associated ring domain 1; 2.8–4.6-fold increase), the RecQ helicase BLM (4.1–13.2-fold) and the mismatch-repair gene MSH2 (2.9–4.3-fold). MSH2 and BLM interact with BRCA1 as components of the BRCA1-associated surveillance complex, BASC, which is involved in DNA damage recognition and repair [32]. mRNAs encoding the tumour suppressor and DNA damage response gene GADD45α were also increased by MEK1EE. Increased GADD45α expression was confirmed by quantitative real-time PCR (Table S2).
Multiple apoptotic genes are induced in response to active MEK1/ERK1,2
MEK1EE expression induced multiple mRNAs encoding proteins involved in apoptosis. Increased expression of the pro-apoptotic Fas/Apo-1 receptor was detected by multiple elements on the array. In addition, increases were detected in mRNAs encoding the pro-apoptotic adaptor molecule CRADD (2.7-fold at 8 h) and caspase 8 (MACHα-1, FLICE) (3.3-fold at 8 h). In contrast, the pro-apoptotic cytokine TRAIL was down-regulated by 2.5–7.2-fold (Table S3; http://www.BiochemJ.org/bj/381/bj3810635add.htm).
Analysis of gene regulation events common to Raf-1 and MEK1
The only known cellular substrates and effectors of Raf-1 are MEK1,2. A careful analysis of the transcriptional output of Raf-1 in MCF-10A cells utilizing Affymetrix HuGene FL probe arrays has been reported previously [8]. Like MCF-12A cells, MCF-10A is an immortalized, non-transformed MEC line. An MCF-10A cell line expressing delta-Raf-ER (a Raf/oestrogen receptor/green fluorescent protein fusion protein) was treated with 4-hydroxytamoxifen to induce Raf-1 activity. The transcriptional changes induced by active MEK1EE and Raf-1 were compared. A total of 132 Raf-1-induced expression changes (114 up-regulated and 18 down-regulated) were reported [8]. BLAST analysis identified 111 elements representing genes common to both the Raf-1 and MEK1 datasets; 41 non-redundant genes were up-regulated and five were down-regulated in both datasets (Table 2). It is likely that these are genes regulated by Raf through activation of MEK1,2. This analysis revealed the induction of a suite of growth factors implicated in tumorigenesis.
Table 2. Gene expression induced by both constitutively active Raf-1 and MEK1.
The first two columns provide gene names and the corresponding annotation derived from the GeneCards database (from Schulze et al. [8]). Columns 3–5 are from Schulze et al. [8] and show GenBank accession numbers from the Affymetrix HuGeneFL chip and the fold change in expression at 8 and 72 h after delta-Raf-ER activation in MCF-10A cells. Column 6 lists the GenBank accession numbers from the Affymetrix U95A array matched by Blast analysis to the genes listed in column 3. Columns 7–13 list the fold changes in mRNA abundance after infection with rAd MEK1EE at the time points indicated. Positive and negative numbers represent fold up- and down-regulation respectively. IGF, insulin-like growth factor.
| Raf-1 time course | MEK1EE time course | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Change (fold) | Change (fold) | |||||||||||
| Gene name/function | Gene annotation | Accession no. | 8 h | 72 h | Matched gene | 0 h | 4 h | 6 h | 8 h | 12 h | 16 h | 24 h |
| Angiogenesis | ||||||||||||
| Follistatin | Antagonist of anti-angiogenic inhibin β A | M19481 | 2.1 | 12.7 | M19481 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 | 2.1 | 67.6 |
| VEGF | Vascular endothelial growth factor | M27281 | 3.9 | 4.5 | AF024710 | 1.4 | −1.3 | 1.0 | 1.8 | 1.4 | 1.1 | 5.8 |
| VEGF | Vascular endothelial growth factor | M27281 | 3.9 | 4.5 | AF022375 | 1.0 | −1.1 | 1.4 | 1.8 | 1.4 | 1.9 | 4.9 |
| Apoptosis/complement | ||||||||||||
| CD55/DAF | Decay-accelerating factor inhibitor of complement | M31516 | 4.4 | 10.4 | M31516 | −1.2 | 1.3 | 1.3 | 2.0 | 1.5 | 1.4 | 3.8 |
| Fas/Apo-1 | Death receptor | X83492 | 3.4 | 2.7 | X83492 | −1.9 | −1.5 | −2.2 | 2.5 | 1.0 | 3.1 | 1.8 |
| Fas/Apo-1 | Death receptor | X83492 | 3.4 | 2.7 | X83490 | −1.1 | −1.1 | 3.9 | 1.7 | 1.4 | 1.6 | 1.4 |
| Fas/Apo-1 | Death receptor | X83492 | 3.4 | 2.7 | X89101 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.7 | −1.1 |
| IPL/TSSC3 | Tumour-suppressing subtransferable candidate 3 | AF001294 | 5.1 | 4.6 | AF001294 | −1.3 | 1.5 | 1.2 | 1.7 | 2.3 | 2.7 | 1.8 |
| Calcium homoeostasis | ||||||||||||
| Calbindin 2 | Calcium-binding protein | X56667 | 2.7 | 8.2 | X56667 | 1.2 | 1.7 | 1.2 | 1.1 | 2.1 | −1.6 | 3.4 |
| PTHLH | Parathyroid hormone-related protein | M17183 | 2.1 | 3.3 | J03802 | 0.0 | 0.0 | −1.9 | 0.0 | 0.0 | 0.0 | 12.8 |
| PTHLH | Parathyroid hormone-related protein | M17183 | 2.1 | 3.3 | M24351 | 0.0 | 0.0 | 0.0 | 2.6 | 3.9 | 2.8 | 6.1 |
| PTHLH | Parathyroid hormone-related protein | M24351 | 2.5 | 3.2 | M24351 | 0.0 | 0.0 | 0.0 | 2.6 | 3.9 | 2.8 | 6.1 |
| Stanniocalcin 1 | Calcium homoeostasis | U25997 | 2.8 | 14.2 | U25997 | 0.0 | 0.0 | 0.0 | 0.0 | 2.7 | 2.5 | 2.1 |
| Cell surface/cell adhesion/extracellular matrix | ||||||||||||
| CD66a/biliary glycoprotein | CAECAM-1 cell adhesion molecule | X16354 | 5.7 | 13.1 | X16354 | 0.0 | 0.0 | 0.0 | 0.0 | 2.6 | 8.6 | 4.6 |
| CEACAM6 | Carcinoembryonic antigen-related cell adhesion molecule 6 | M18728 | 3.2 | 12 | M18728 | 0.0 | −1.3 | −1.2 | 1.3 | 3.6 | 1.9 | 4.0 |
| Integrin α5 | Fibronectin receptor | X06256 | 6.7 | 7.7 | X06256 | −1.2 | −1.2 | 0.0 | 0.0 | 0.0 | 1.6 | 3.7 |
| Integrin α6 | Cell adhesion protein | X53586 | 7.5 | 8.2 | S66213 | 1.4 | −1.1 | 1.1 | 2.2 | 2.0 | 2.5 | 3.4 |
| Laminin α3 | Extracellular matrix component | L34155 | 2 | 3.7 | L34155 | 1.5 | 1.1 | 1.1 | 1.5 | 1.9 | 1.8 | 2.7 |
| Laminin γ2 | Extracellular matrix component | U31201 | 6.9 | 9.4 | U31201 | 1.7 | −1.6 | 4.5 | 1.3 | 11.4 | 4.7 | 5.0 |
| Matrix metalloproteinase 1 | Interstitial collagenase | X54925 | −1.5 | 19.2 | M13509 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.8 |
| Growth factors and cytokines | ||||||||||||
| Amphiregulin | EGF-like growth factor | M30703 | 23.2 | 20.8 | M30704 | −1.0 | −1.3 | 2.3 | 1.5 | 2.3 | 3.9 | 11.0 |
| HB-EGF | Heparin-binding EGF-like growth factor | M60278 | 36.8 | 42.9 | M60278 | 1.3 | 0.0 | 0.0 | 1.8 | 2.3 | 2.8 | 33.9 |
| TGFα | EGF-like growth factor | X70340 | 2.9 | 3.3 | X70340 | −1.4 | −1.4 | 1.6 | 3.9 | 3.7 | 4.5 | 8.5 |
| Metabolism | ||||||||||||
| 5′-Nucleotidase (CD73) | Hydrolysis of extracellular nucleotides | X55740 | 6 | 8.4 | X55740 | 1.6 | 1.1 | 1.3 | 2.9 | 5.1 | 5.1 | 8.2 |
| Ornithine decarboxylase | Biosynthesis of polyamines | M33764 | 2.9 | 3.2 | M33764 | 1.3 | −1.1 | −1.1 | 2.4 | 1.9 | 2.3 | 4.0 |
| Ornithine decarboxylase | Biosynthesis of polyamines | M33764 | 2.9 | 3.2 | X16277 | 1.3 | −1.4 | 1.1 | 1.8 | 1.5 | 1.7 | 2.8 |
| SULT2B1a | Hydroxysteroid sulphotransferase | U92314 | 10.5 | 14.7 | U92315 | −1.4 | −1.3 | 1.1 | 1.2 | 1.7 | 1.4 | 2.8 |
| Uridine phosphorylase | Phospholysis of uridine | X90858 | 2.7 | 4.9 | X90858 | −1.0 | −1.2 | −1.1 | 1.4 | 2.5 | 4.6 | 28.2 |
| Signalling | ||||||||||||
| EphA2 | Ephrin receptor protein tyrosine kinase | M59371 | 2.3 | 3.2 | M59371 | 1.3 | −1.8 | 2.3 | 1.9 | 1.7 | 1.2 | 3.6 |
| Grb10 | Adaptor protein in insulin and IGF-1 signalling | D86962 | 3.1 | 2.3 | D86962 | 1.1 | 1.8 | −1.4 | 1.2 | 1.3 | 1.2 | 2.9 |
| Hydroxysteroid (11-β) DH2 | Cortisol-metabolizing enzyme | U26726 | −17.2 | −22.3 | U26726 | −1.2 | −1.0 | −1.7 | −2.3 | −3.4 | −11.3 | −7.1 |
| Jagged 1 | Notch receptor ligand | U73936 | 3.3 | 2.8 | U77914 | 1.2 | 1.2 | −1.3 | 1.4 | 1.7 | 1.6 | 2.9 |
| MKP2 | MAPK phosphatase 2 | U48807 | 3.4 | 3.5 | U48807 | 1.1 | −1.5 | 10.3 | 5.5 | 4.3 | 4.9 | 4.3 |
| MKP5 | MAPK phosphatase 5 | U15932 | 2.9 | 6.2 | U15932 | 1.1 | −1.1 | 4.1 | 4.3 | 3.2 | 4.9 | 24.0 |
| Transcription | ||||||||||||
| BRF2 | Tis11d butyrate/EGF-response factor 2 | X78992 | 10.3 | 11.9 | X78992 | −1.0 | −1.4 | 1.9 | 1.9 | 1.3 | 1.9 | 2.5 |
| BRF2 | Tis11d butyrate/EGF-response factor 2 | X78992 | 10.3 | 11.9 | U07802 | −1.3 | 1.3 | 1.5 | 2.1 | 2.6 | 3.6 | 2.3 |
| BRF2 | Tis11d butyrate/EGF-response factor 2 | U07802 | 3.8 | 4.3 | U07802 | −1.3 | 1.3 | 1.5 | 2.1 | 2.6 | 3.6 | 2.3 |
| c-Myc | Transcription factor | L00058 | 3.7 | 3.3 | V00568 | −1.1 | 1.1 | 3.5 | 1.9 | 1.6 | 1.6 | 4.1 |
| c-Myc | Transcription factor | M13929 | 3.6 | 3.0 | V00568 | −1.1 | 1.1 | 3.5 | 1.9 | 1.6 | 1.6 | 4.1 |
| EGR3 | Early growth response gene 3 | X63741 | 5.7 | 3.7 | X63741 | 1.2 | −1.2 | 0.0 | 4.1 | 32.3 | −1.0 | 0.0 |
| ETV5 | Ets-related transcription factor | X96381 | 2.9 | 6.2 | X96381 | 0.0 | 0.0 | 0.0 | 0.0 | 4.8 | 8.0 | 14.2 |
| HMGIC | High-mobility-group protein isoform C | U28749 | 3.5 | 6.5 | X92518 | 0.0 | 0.0 | 0.0 | 2.6 | 2.8 | 3.3 | 3.0 |
| JunB | Subunit of AP-1 | U20734 | 13.4 | 8.3 | X51345 | −1.1 | −1.1 | 2.8 | −1.1 | 1.1 | −1.4 | 3.6 |
| JunB | Subunit of AP-1 | U20734 | 13.4 | 8.3 | M29039 | 1.3 | 0.0 | 2.4 | 1.5 | 1.4 | 0.0 | 3.1 |
| Other functions | ||||||||||||
| ATP6B1 | Lysosomal proton pump subunit | M25809 | 1.4 | −3.0 | M25809 | −1.4 | 1.1 | 1.9 | −1.3 | −1.3 | −2.6 | −2.3 |
| Gu-protein | RNA helicase | U41387 | 3.2 | 2.2 | U41387 | −1.6 | 1.1 | 1.2 | 3.6 | 2.6 | 3.4 | 2.4 |
| KCNO1 | Potassium channel | U90065 | 4.6 | 3.6 | U33632 | 1.1 | 1.2 | −1.6 | 1.4 | 1.2 | 2.5 | 1.2 |
| PAI-3 | Plasminogen activator inhibitor III | M68516 | −1.5 | −3.5 | M68516 | −1.0 | −1.1 | −2.8 | −1.5 | −3.3 | −5.4 | −29.3 |
| SLC26A2 | Sulphate transporter | U14528 | 2.9 | 3.7 | U14528 | 1.1 | 1.4 | 1.1 | 2.5 | 1.8 | 1.8 | −1.5 |
| Unknown function | ||||||||||||
| GLVR1 | Leukaemia virus receptor 1 phosphate transporter | L20859 | 5.4 | 6.1 | L20859 | −1.2 | −1.0 | 3.3 | 4.0 | 3.5 | 4.1 | 5.4 |
| GS3786 | Human cancellous bone osteoblast mRNA | D87120 | 3.1 | 2.6 | D87120 | −1.2 | −1.1 | 2.1 | 3.7 | 2.6 | 3.0 | 3.9 |
| GS3955 | Osteoblast mRNA for GS3955 | D87119 | 9.7 | 13.3 | D87119 | 1.5 | −1.0 | 1.3 | 3.7 | 4.0 | 4.4 | 6.1 |
| hSBP1 | Selenium binding protein 1 | U29091 | −3.9 | −9.3 | U29091 | 1.7 | 1.0 | −1.4 | −1.6 | −1.9 | −4.0 | −2.5 |
| Ndr1 | RTP Drg1 potential target gene for N-myc | D87953 | 1.4 | 3.0 | D87953 | −1.1 | −1.0 | 1.1 | −1.1 | 2.9 | 1.8 | 1.5 |
| PHLDA1 | Pleckstrin homology-like domain protein | Z50194 | 17 | 27.5 | Z50194 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 42.6 |
| RIL | 37 kDa LIM domain protein | X93510 | 3.6 | −2.4 | X93510 | 1.3 | −1.4 | −1.4 | −2.2 | −2.2 | −3.3 | −2.3 |
Angiogenesis
MEK1EE expression induced several RNAs encoding proteins involved in angiogenesis. mRNAs encoding VEGF, a pro-angiogenic factor, were up-regulated by both delta-Raf-ER (3.9–4.5-fold at 72 h) and MEK1EE (4.9-fold at 24 h). Quantitative real-time PCR confirmed this result (Table 1). Follistatin was also strongly up-regulated by delta-Raf-ER (12.7-fold at 72 h) and by MEK1EE (67.6-fold at 24 h). Follistatin may play a role in angiogenesis by binding to and inhibiting the biological activities of activin [33]. mRNA encoding PAI3 (plasminogen activator inhibitor 3), an anti-angiogenic factor, was down-regulated by both Raf-1 (3.5-fold, 72 h) and MEK1 (3.3–29.3-fold, 24 h).
Ras/Raf/ERK ErbB ligand positive-feedback loop
Among the genes induced by both Raf-1 and MEK1EE were multiple ErbB ligands [8]. Confirmation by quantitative real-time PCR demonstrated that transcripts encoding HB-EGF (heparinbinding EGF), epiregulin and TGFα were co-ordinately up-regulated by MEK1EE, by 3–4-fold, beginning at 6 h post-infection (Table 1). HB-EGF and epiregulin reached maximal increases of 45- and 46-fold respectively by 24 h (Table 1). Amphiregulin mRNA was also induced (4-fold at 12 h; 17-fold at 24 h). EGF mRNA was not detected in rAd MEK1EE-infected MCF-12A cells by quantitative real-time PCR (Table S2; http://www.BiochemJ.org/bj/381/bj3810635add.htm).
Calcium and bone metabolism
Several genes involved in calcium metabolism and bone formation were regulated by both Raf and MEK1EE. mRNAs encoding calbindin 2 were increased 8.2-fold and 3.4-fold by Raf-1 and MEK1EE respectively. Stanniocalcin-2 mRNA was modestly up-regulated by MEK1EE expression (2.1–2.7-fold) at three time points, but was strongly up-regulated (14.2-fold, 72 h) by delta-Raf-ER. Stanniocalcin-2 is a putative calcium-regulating hormone and may have an anti-hypocalcaemic function [34]. Two genes associated with bone formation were down-regulated by MEK1EE, but were not reported in the delta-Raf-ER dataset. Bone morphogenetic protein 1 (decreased by 3.2–4.8-fold, Table S4; http://www.BiochemJ.org/bj/381/bj3810635add.htm) is a member of the TGFβ gene family and is a stimulator of bone formation. Oesteoblast-specific factor-2, which is negatively regulated by parathyroid hormone [35], was maximally down-regulated 41-fold at 6 h (Table S3).
A striking finding was the induction of PHRP mRNA by both activated Raf and MEK1. PHRP, a peptide hormone, is the major mediator of HHM, and may be involved in the process of skeletal metastases from primary breast tumours [36]. PHRP mRNA was modestly up-regulated (2–3-fold) by activated Raf-1. MEK1EE expression induced PHRP mRNA by up to 6-fold. Quantitative real-time PCR revealed strong induction of PHRP mRNA by MEK1EE (90.5-fold, 24 h) (Table 1).
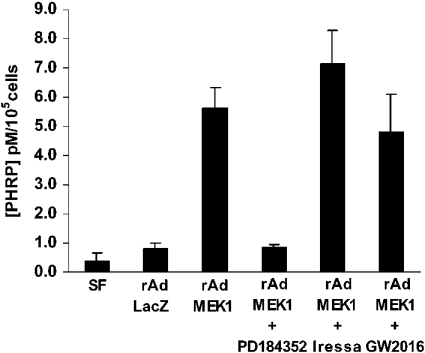
The induction of PHRP by MEK1EE was evaluated further by assessing PHRP peptide secretion into serum-free medium. PHRP levels were increased by MEK1EE by >7-fold compared with LacZ-infected control cells (5.6±0.7 and 0.8±0.2 pM/105 cells respectively) (Figure 3). Treatment of MEK1EE-expressing MCF-12A cells with the MEK1,2 inhibitor PD184352 completely abolished the increase in PHRP secretion. However, treatment with the ErbB inhibitors Iressa and GW2016 did not affect the secretion of PHRP induced by MEK1EE. Thus PHRP secretion in these MEK1EE-expressing cells appears to be due to activation of the MEK1,2/ERK1,2 pathway. The activation by ErbB ligands of additional signalling pathways does not appear to be required for PHRP secretion by these MEK1EE-expressing cells.
Figure 3. Induction of PHRP by MEK1EE.
PHRP secretion into conditioned medium is induced by active MEK1 and inhibited by the MEK inhibitor PD184352 (1 μM), but not by the ErbB inhibitors Iressa or GW2016 (3 μM). MCF-12A cells were plated and infected as described in the Experimental section. Concentrations were determined from a standard curve assayed in parallel, and the results are expressed as pM/105 cells. SF, serum-free medium.
DISCUSSION
We report here the direct evaluation of a MAPK-regulated transcriptional programme in a mammalian cell line, yielding the identification of a large number of co-ordinately induced or repressed genes. These results provide a molecular characterization of phenotypic changes induced in a MEC line upon ERK1,2 activation. These changes include increased DNA synthesis, changes in cytoarchitecture, and induction of mRNAs encoding autocrine growth factors. Increased cell cycle entry, morphological changes and growth factor independence are all hallmarks of epithelial cell transformation. Thus the expression profile described here provides a potential molecular characterization of the link between regulation of transcription by the ERK1,2 pathway and cellular transformation.
MCF-12A cells are immortal but non-transformed [37]. Therefore these cells must retain intact molecular mechanisms to prevent unbridled cellular proliferation. MKPs, pro-apototic proteins such as Fas, and DNA damage/cell cycle checkpoint proteins such as BRCA1,2 are probable components of this safety net. BRCA1,2 mutations predispose carriers to increased cancer risk, but the mechanisms remain unknown [38]. Multiple functions have been described for the BRCA proteins, including the detection and repair of DNA damage, cell cycle checkpoint control, and the regulation of transcription and apoptosis [38]. DNA-damaging agents, serum and cell cycle transitions induce BRCA1,2 transcription [38]. However, the specific signalling pathways regulating the transcription of BRCA1,2 have not been elucidated. Our results support a link between the MEK1,2/ERK1,2 pathway and BRCA1,2. Breast cancer cells lacking functional BRCA1 or BRCA2 may be disadvantaged with regard to controlling cellular growth driven by activation of the classical Ras/MAPK pathway.
In the present paper we report a significant enrichment of Myc binding sites in the promoters of genes co-ordinately up-regulated after MEK1EE expression. This promoter analysis does not address all of the possible transcription factors involved in the regulation of genes downstream of ERK1,2. Rather, we sought to determine if any transcription factors could be strongly and statistically associated with the gene expression pattern detected in response to MEK1EE expression. Thus we chose to limit our analysis to only those transcription factors detected as being up-regulated by MEK1EE expression, the promoters of the genes that were co-ordinately up-regulated (group A), and the transcription factor binding sites that were enriched in these promoters relative to their presence in the promoters of a randomly selected set of genes. The identification of c-Myc binding sites in the majority of genes induced by ERK1,2 is consistent with the known importance of c-Myc in mediating Ras/Raf/MEK1,2/ERK1,2 output [31]. Among the genes identified with c-Myc binding sites in their promoters were many of the most strongly regulated genes, including those encoding the MKPs, BRCA2, COX2 (cyclo-oxygenase 2), and the growth factors PHRP, VEGF, HB-EGFR and amphiregulin.
The most well characterized pathway from Ras to MEK1,2/ERK1,2 activation is through Raf-1. Thus the transcriptional programme regulated by both Raf and MEK1 is likely to be relevant to cellular transformation. We have identified 46 genes that are strong candidate downstream effectors of the Ras/Raf/MEK1,2/ERK1,2 pathway in MECs. Among these genes are those encoding hormones and growth factors (ErbB ligands, VEGF and PHRP) implicated in multiple facets of oncogenesis.
Up-regulation of VEGF in fibroblasts by expression of constitutively active MEK1 has been reported previously [39]. MEK1 regulation of VEGF may be cell type-specific, because in parallel experiments utilizing intestinal epithelial cells VEGF expression was not induced by constitutively active MEK1 [40]. The data presented here and by Schulze et al. [8] suggest direct regulation of VEGF expression in breast epithelial cells via the Raf-1/MEK1,2/ERK1,2 pathway. Thus inhibitors of Raf and MEK1 may have anti-angiogenic as well as anti-proliferative properties.
Induction of ErbB ligands by constitutively active Ras has been reported previously [41–43]. Both Raf-dependent and Raf-independent induction mechanisms have been reported [41–43]. Our data extend these findings and, for the first time, demonstrate induction of ErbB ligands by expression of constitutively active MEK1. Our data also strongly suggest that the induction of ErbB ligands by Raf-1 in MCF-10A cells occurs via MEK1,2/ERK1,2. This positive regulatory loop involving the ErbB ligands and receptors provides a demonstrated survival benefit to cells. It mediates the serum-independent proliferation of H-Ras-transformed fibroblasts [42] and protection of human MECs from Anoikas-induced apoptosis [8]. ErbB inhibitors did not inhibit thymidine incorporation in MCF-12A cells expressing MEK1EE. In contrast, proliferation and ERK1,2 activation were inhibited by treatment with the ErbB inhibitor Iressa in V12K-Ras-transformed MCF-12A cells (results not shown).
PHRP is expressed in normal breast, where it is thought to mobilize calcium for the production of milk. It is also produced by many tumours and is the primary mediator of HHM. This disorder occurs in approx. 40% of breast cancer patients and is the most prevalent life-threatening disorder of metabolism associated with cancer [44]. PHRP stimulates bone resorption by osteoclasts. A role for PHRP in metastasis to bone has also been proposed [36]. Approx. 92% of skeletal metastases from breast cancers express PHRP, as opposed to 20% of non-skeletal metastases [45]. Growth factors as well as Ras induce PHRP [44]. The pathways controlling PHRP expression have only been partially characterized. The involvement of the Ras/Raf/MEK, Rac/JNK and TGFβ/p38 pathways in PHRP induction has been inferred via dominant negative and inhibitor studies [46–48].
In summary, we present the first demonstration of PHRP mRNA and protein secretion due to expression of constitutively active MEK1. PHRP secretion was inhibited by the MEK1,2 inhibitor PD184352, but not by the ErbB inhibitors Iressa and GW2016. These findings suggest the direct regulation of PHRP by the MEK1,2 pathway. These results strongly support the rationale for investigating the potential positive impact of inhibitors of the MEK1,2 pathway on the treatment of PHRP-positive tumours.
Online data
Acknowledgments
We thank P. Kirschmeier and W.R. Bishop and members of the SPRI Oncology Dept for helpful discussions, and for their support and encouragement; R.A. Weinberg for comments regarding the manuscript; the SPRI Discovery Technology group for their support of this work; and E. Schaefer, W.L. Ling, G. Vellekamp, S. Ravidran, X.Y. Cai and D. Brassard for adenoviral amplification, determination of viral titres, and sequencing.
References
- 1.Pearson G., Robinson F., Beers Gibson T., Xu B.-e., Karandikar M., Berman K., Cobb M. H. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 2.Lewis T. S., Shapiro P. S., Ahn N. G. Signal transduction through MAP kinase cascades. Adv. Cancer Res. 1998;74:49–139. doi: 10.1016/s0065-230x(08)60765-4. [DOI] [PubMed] [Google Scholar]
- 3.Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M. J., Bottomley W., et al. Mutations of the BRAF gene in human cancer. Nature (London) 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 4.Treisman R. Regulation of transcription by MAP kinase cascades. Curr. Opin. Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 5.Allen L. F., Sebolt-Leopold J., Meyer M. B. CI-1040 (PD184352), a targeted signal transduction inhibitor of MEK (MAPKK) Semin. Oncol. 2003;30(Suppl. 16):105–116. doi: 10.1053/j.seminoncol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Robinson M. J., Stippec S. A., Goldsmith E., White M. A., Cobb M. H. A constitutively active and nuclear form of the kinase ERK2 is sufficient for neurite outgrowth and cell transformation. Curr. Biol. 1998;8:1141–1150. doi: 10.1016/s0960-9822(07)00485-x. [DOI] [PubMed] [Google Scholar]
- 7.Zuber J., Tchernitsa O. I., Hinzmann B., Schmitz A. C., Grips M., Hellriegel M., Sers C., Rosenthal A., Schafer R. A genome-wide survey of Ras transformation targets. Nat. Genet. 2000;24:144–152. doi: 10.1038/72799. [DOI] [PubMed] [Google Scholar]
- 8.Schulze A., Lehmann K., Jefferies H. B. J., McMahon M., Downward J. Analysis of the transcriptional program induced by Raf in epithelial cells. Genes Dev. 2001;15:981–994. doi: 10.1101/gad.191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearson G., Bumeister R., Henry D. O., Cobb M. H., White M. A. Uncoupling Raf1 from MEK1/2 impairs only a subset of cellular responses to Raf activation. J. Biol. Chem. 2000;275:37303–37306. doi: 10.1074/jbc.C000570200. [DOI] [PubMed] [Google Scholar]
- 10.Mody N., Leitch J., Armstrong C., Dixon J., Cohen P. Effects of kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 2001;502:21–24. doi: 10.1016/s0014-5793(01)02651-5. [DOI] [PubMed] [Google Scholar]
- 11.Lewis T. S., Hunt J. B., Aveline L. D., Jonscher K. R., Louie D. F., Yeh J. M., Nahreini T. S., Resing K. A., Ahn N. G. Identification of novel kinase pathway signaling targets by functional proteomics and mass spectrometry. Mol. Cell. 2000;6:1343–1354. doi: 10.1016/s1097-2765(00)00132-5. [DOI] [PubMed] [Google Scholar]
- 12.Pinkas J., Leder P. MEK1 signaling mediates transformation and metastasis of EpH4 mammary epithelial cells independent of an epithelial to mesenchymal transition. Cancer Res. 2002;62:4781–4790. [PubMed] [Google Scholar]
- 13.Klesse L. J., Meyers K. A., Marshall C. J., Parada L. F. Nerve growth factor induces survival and differentiation through two distinct signaling cascades in PC12 cells. Oncogene. 1999;18:2055–2068. doi: 10.1038/sj.onc.1202524. [DOI] [PubMed] [Google Scholar]
- 14.Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 15.Eisen M. B., Spellman P. T., Brown P. O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X., Adams M. D., Zhou H., Kerlavage A. R. A tool for analyzing and annotating genomic sequences. Genomics. 1997;46:37–45. doi: 10.1006/geno.1997.4984. [DOI] [PubMed] [Google Scholar]
- 17.Wang L., Wu Q., Qiu P., Mirza A., McGuirk M., Kirschmeier P., Greene J. R., Wang Y., Pickett C. B., Liu S. Analyses of p53 target genes in the human genome by bioinformatic and microarray approaches. J. Biol. Chem. 2001;276:43604–43610. doi: 10.1074/jbc.M106570200. [DOI] [PubMed] [Google Scholar]
- 18.Wingender E., Chen X., Hehl R., Karas H., Liebich I., Matys V., Meinhardt T., Pruss M., Reuter I., Schacherer F. Transfac: An integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu P., Ding W., Jiang Y., Greene J. R., Wang L. Computational analysis of composite regulatory elements. Mamm. Genome. 2002;13:327–332. doi: 10.1007/s00335-001-2141-8. [DOI] [PubMed] [Google Scholar]
- 19a.Barrett S., Bridges A., Doherty A., Dudley D., Saltiel A., Tecle H. 4-Bromo or 4-iodo phylamino benzhydroxamic acid derivatives and their use as MEK inhibitors. 1999 World Patent WO 99/01426.
- 19b.Gibson K. Quinazoline derivatives. 1996 World Patent WO 96/33980.
- 20.Cockerill S., Stubberfield C., Stables J., Carter M., Guntrip S., Smith K., McKeown S., Shaw R., Topley P., Thomsen L. Indazolylamino quinazolines and pyridopyrimidines as inhibitors of the EGFr and c-erbB-2. Bioorg. Med. Chem. Lett. 2001;11:1401–1405. doi: 10.1016/s0960-894x(01)00219-0. [DOI] [PubMed] [Google Scholar]
- 21.Theodosiou A., Ashworth A. MAP kinase phosphatases. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-reviews3009. REVIEWS3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwak S. P., Dixon J. E. Multiple dual specificity protein tyrosine phosphatases are expressed and regulated differentially in liver cell lines. J. Biol. Chem. 1995;270:1156–1160. doi: 10.1074/jbc.270.3.1156. [DOI] [PubMed] [Google Scholar]
- 23.Kovanen P. E., Rosenwald A., Fu J., Hurt E. M., Lam L. T., Giltnane J. M., Wright G., Staudt L. M., Leonard W. J. Analysis of gamma c-family cytokine target genes. Identification of dual-specificity phosphatase 5 (DUSP5) as a regulator of mitogen-activated protein kinase activity in interleukin-2 signaling. J. Biol. Chem. 2003;278:5205–5213. doi: 10.1074/jbc.M209015200. [DOI] [PubMed] [Google Scholar]
- 24.Lund A. H., Turner G., Trubetskoy A., Verhoeven E., Wientjens E., Hulsman D., Russell R., DePinho R. A., Lenz J., van Lohuizen M. Genome-wide retroviral insertional tagging of genes involved in cancer in cdkn2a-deficient mice. Nat. Genet. 2002;32:160–165. doi: 10.1038/ng956. [DOI] [PubMed] [Google Scholar]
- 25.Garcia J., Ye Y., Arranz V., Letourneux C., Pezeron G., Porteu F. IEX-1: A new ERK substrate involved in both ERK survival activity and ERK activation. EMBO J. 2002;21:5151–5163. doi: 10.1093/emboj/cdf488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kime L., Wright S. C. Mad4 is regulated by a transcriptional repressor complex that contains Miz-1 and c-Myc. Biochem. J. 2003;370:291–298. doi: 10.1042/BJ20021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng C., Kilkenny A. E., Roop D., Yuspa S. H. The v-Ras oncogene inhibits the expression of differentiation markers and facilitates expression of cytokeratins 8 and 18 in mouse keratinocytes. Mol. Carcinog. 1990;3:363–373. doi: 10.1002/mc.2940030608. [DOI] [PubMed] [Google Scholar]
- 28.Patarroyo M., Tryggvason K., Virtanen I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin. Cancer Biol. 2002;12:197–207. doi: 10.1016/S1044-579X(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 29.Roovers K., Assoian R. K. Integrating the kinase signal into the G1 phase of the cell cycle machinery. Bioessays. 2000;22:818–826. doi: 10.1002/1521-1878(200009)22:9<818::AID-BIES7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Shaulian E., Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20:2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 31.Sears R. C., Nevins J. R. Signaling networks that link cell proliferation and cell fate. J. Biol. Chem. 2002;277:11617–11620. doi: 10.1074/jbc.R100063200. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- 33.Kozian D. H., Ziche M., Augustin H. G. The activin-binding protein follistatin regulates autocrine endothelial cell activity and induces angiogenesis. Lab. Invest. 1997;76:267–276. [PubMed] [Google Scholar]
- 34.Honda S., Kashiwagi M., Ookata K., Tojo A., Hirose S. Regulation by 1[alpha],25-dihydroxyvitamin D3 of expression of stanniocalcin messages in the rat kidney and ovary. FEBS Lett. 1999;459:119–122. doi: 10.1016/s0014-5793(99)01225-9. [DOI] [PubMed] [Google Scholar]
- 35.Adams A. E., Rosenblatt M., Suva L. J. Identification of a novel parathyroid hormone-responsive gene in human osteoblastic cells. Bone. 1999;24:305–313. doi: 10.1016/s8756-3282(98)00188-4. [DOI] [PubMed] [Google Scholar]
- 36.Guise T. A. Parathyroid hormone-related protein and bone metastases. Cancer. 1997;80(Suppl. 8):1572–1580. doi: 10.1002/(sici)1097-0142(19971015)80:8+<1572::aid-cncr7>3.3.co;2-d. [DOI] [PubMed] [Google Scholar]
- 37.Paine T. M., Soule H. D., Pauley R. J., Dawson P. J. Characterization of epithelial phenotypes in mortal and immortal human breast cells. Int. J. Cancer. 1992;50:463–473. doi: 10.1002/ijc.2910500323. [DOI] [PubMed] [Google Scholar]
- 38.Venkitaraman A. R. Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J. Cell Sci. 2001;114:3591–3598. doi: 10.1242/jcs.114.20.3591. [DOI] [PubMed] [Google Scholar]
- 39.Pages A., Milanini J., Richard D. E., Berra E., Gothie E., Vinals F., Pouyssegur J. Signaling angiogenesis via p42/p44 kinase cascade. Ann. N.Y. Acad. Sci. 2000;902:187–200. doi: 10.1111/j.1749-6632.2000.tb06313.x. [DOI] [PubMed] [Google Scholar]
- 40.Rak J., Mitsuhashi Y., Sheehan C., Tamir A., Viloria-Petit A., Filmus J., Mansour S. J., Ahn N. G., Kerbel R. S. Oncogenes and tumor angiogenesis: Differential modes of vascular endothelial growth factor up-regulation in Ras-transformed epithelial cells and fibroblasts. Cancer Res. 2000;60:490–498. [PubMed] [Google Scholar]
- 41.McCarthy S. A., Samuels M. L., Pritchard C. A., Abraham J. A., McMahon M. Rapid induction of heparin-binding epidermal growth factor/diptheria toxin receptor expression by Raf and Ras oncogenes. Genes Dev. 1995;9:1953–1964. doi: 10.1101/gad.9.16.1953. [DOI] [PubMed] [Google Scholar]
- 42.Hamilton M., Wolfman A. Oncogenic Ha-Ras-dependent mitogen-activated protein kinase activity requires signaling through the epidermal growth factor receptor. J. Biol. Chem. 1998;273:28155–28162. doi: 10.1074/jbc.273.43.28155. [DOI] [PubMed] [Google Scholar]
- 43.Gangarosa L. M., Sizemore N., Graves-Deal R., Oldham S. M., Der C. J., Coffey R. J. A Raf-independent epidermal growth factor receptor autocrine loop is necessary for ras transformation of rat intestinal epithelial cells. J. Biol. Chem. 1997;272:18926–18931. doi: 10.1074/jbc.272.30.18926. [DOI] [PubMed] [Google Scholar]
- 44.Warrel R. P., DeVita V. T., Hellman J. S., Rosenberg S. A. Cancer, Principles and Practices in Oncology. Philadelphia: Lippincott-Raven; 1997. Metabolic emergencies; pp. 2486–2500. [Google Scholar]
- 45.Vargas S., Gillespie M., Powell G., Southby J., Dankas J., Moseley J., Martin T. Localization of parathyroid hormone-related protein mRNA expression in breast cancer and metastatic lesions by in situ hybridization. J. Bone Miner. Res. 1992;7:971–979. doi: 10.1002/jbmr.5650070814. [DOI] [PubMed] [Google Scholar]
- 46.Aklilu F., Park M., Goltzman D., Rabbani S. A. Induction of parathyroid hormone-related peptide by the Ras oncogene: Role of Ras farnesylation inhibitors as potential therapeutic agents for hypercalcemia of malignancy. Cancer Res. 1997;57:4517–4522. [PubMed] [Google Scholar]
- 47.Aklilu F., Gladu J., Goltzman D., Rabbani S. A. Role of mitogen-activated protein kinases in the induction of parathyroid hormone-related peptide. Cancer Res. 2000;60:1753–1760. [PubMed] [Google Scholar]
- 48.Kakonen S.-M., Selander K. S., Chirgwin J. M., Yin J. J., Burns S., Rankin W. A., Grubbs B. G., Dallas M., Cui Y., Guise T. A. Transforming growth factor-beta stimulates parathyroid hormone-related protein and osteolytic metastases via smad and mitogen-activated protein kinase signaling pathways. J. Biol. Chem. 2002;277:24571–24578. doi: 10.1074/jbc.M202561200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.