Abstract
Silicon is, besides oxygen, the most abundant element on earth. Only two taxa use this element as a major constituent of their skeleton, namely sponges (phylum Porifera) and unicellular diatoms. Results from combined cytobiological and molecularbiological techniques suggest that, in the demosponge Suberites domuncula, silicic acid is taken up by a transporter. Incubation of cells with the fluorescent silica tracer PDMPO [2-(4-pyridyl)-5-{[4-(2-dimethylaminoethylaminocarbamoyl)methoxy]phenyl}-oxazole] showed a response to silicic acid by an increase in fluorescence; this process is temperature-dependent and can be blocked by DIDS (4,4-di-isothiocyanatostilbene-2,2-disulphonic acid). The putative NBC (Na+/HCO3−) transporter was identified, cloned and analysed. The deduced protein comprises all signatures characteristic of those molecules, and phylogenetic analysis also classifies it to the NBC transporter family. This cDNA was used to demonstrate that the expression of the gene is strongly up-regulated after treatment of cells with silicic acid. In situ hybridization demonstrated that the expression of the sponge transporter occurs in those cells that are located adjacent to the spicules (the skeletal element of the animal) or in areas in which spicule formation occurs. We conclude that this transporter is involved in silica uptake and have therefore termed it the NBCSA {Na+/HCO3−[Si(OH)4]} co-transporter.
Keywords: Na+/HCO3−[Si(OH)4] (NBCSA)-related transporter, 2-(4-pyridyl)-5-{[4-(2-dimethylaminoethylaminocarbamoyl)methoxy]phenyl}oxazole (PDMPO), silica uptake, spicule formation, sponges, Suberites domuncula
Abbreviations: aa, amino acid; AEs, anion-exchangers; BTS, bicarbonate transporter superfamily; CMFSW, Ca2+- and Mg2+-free artificial seawater; CMFSW-E, CMFSW containing 2.5 mM EDTA; DIDS, 4,4-di-isothiocyanatostilbene-2,2-disulphonic acid; DIG, digoxigenin; HCO3− transporter, bicarbonate transporter; Myr, million years; Na+/HCO3−[Si(OH)4], NBCSA co-transporter; NDAE, Na+-dependent Cl−–HCO3− exchanger; PDMPO, 2-(4-pyridyl)-5-{[4-(2-dimethylaminoethylaminocarbamoyl)methoxy]phenyl}oxazole; PPR, pentatricopeptide repeat; RACE, rapid amplification of cDNA ends; SiO2, silica; SW, seawater; TM, transmembrane
INTRODUCTION
Sponges (phylum Porifera) are the phylogenetically oldest metazoan organisms which use silica for the formation of their skeleton. These animals evolved prior to the Cambrian explosion, approx. 525 Myr (million years) ago. On the basis of molecular data it is now established that the class of Hexactinellida is the oldest taxon of Porifera; they form, together with Demospongiae, the group of siliceous sponges, while the class of Calcarea, with their calcareous skeleton, is younger and represents a sister group to Cnidaria [1]. Also diatoms (Bacillariophyta) possess a siliceous skeleton. However, these organisms are evolutionarily younger (100 Myr [2]), belong to the Protista, and consequently have used an independent strategy to utilize silica for the construction of their skeletal elements. In the present study the mechanism of uptake of silicic acid by siliceous sponges has been examined at the molecular level for the first time.
On the basis of paleontological data it had been proposed that the major reason for the use of silica rather than of calcium carbonate can be seen in the environment from which the respective taxa evolved. The siliceous sponges emerged in the eutrophic milieu, while the calcareous sponges evolved approx. 20 Myr later in an oligotrophic environment [3]. The chemical composition of the skeletal elements, the spicules, in Demospongiae and in Hexactinellida is mostly SiO2 (silica) and water (reviewed in [4]). The spicules are surrounded by a membrane termed the silicalemma [5].
For Demospongiae it has been well established that the siliceous spicules have a central core and an axial filament composed of protein [6]. Important is the observation that these filaments are impregnated with silica [7]. Specialized cells, the sclerocytes, which are characterized by a high metabolic activity, secrete spicules [8]. Secretion of spicules appears to proceed either intracellularly or extracellularly, very likely depending on the size of the spicules (reviewed in [9]). It has been proposed that the axial filament is synthesized intracellularly and subsequently exocytosed into the mesohyl, the bulky extracellular matrix in which sponge cells are embedded [6,10]. The axial filament is composed of cathepsin-related molecules, the silicateins, e.g. forms α, β or γ [11–14]. Biochemical evidence has been presented suggesting that silicatein catalyses the polycondensation of silicon alkoxides.
In sponges, silica is present as amorphous silica with approx. 10–15% of water [15]. In calcareous sponges the spicules are composed of up to 90% of CaCO3 and up to about 2% of water [16]. The obvious similarity between silicon and carbon as the two basic components of the spicules in siliceous and calcareous sponges is corroborated by recent findings showing that the carbonic anhydrase-related silicase also isolated from a sponge can split the ester-related bond in polymerized silicic acid [17].
In contrast with what is known for diatoms, nothing is known at the enzymic level about the silica uptake in sponges. As reviewed recently [18], the uptake of silica in diatoms (specifically Cylindrotheca fusiformis) is storable, indicating a carrier-mediated process. The transporter acts as a sodium/silicic acid symporter, more specifically in an electrogenic transport with a Si(OH)4/Na+ ratio of 1:1 [19]. In addition, evidence has been presented that the diatom Phaeodactylum tricornutum can transport the ionized form, SiO(OH)3−, as well [20]. The transporter was cloned [21] and found to consist of ten predicted transmembrane (TM) segments. As expected, especially because of the long time and large taxonomical distance between diatoms and sponges, the deduced diatom polypeptide shows a sequence similarity neither to metazoan proteins in general nor to sponge proteins in particular (http://spongebase.genoserv.de/). Hence the evolutionary history of the silicic acid transporter in diatoms is independent of that expected to exist in Porifera. In these multicellular animals, silica uptake, very likely in the form of silicic acid, depends on the concentration of soluble silica in the surrounding medium and on temperature – parameters which favour an active transport mechanism [4,22,23]. In addition, it has to be postulated that the uptake of silica in sponges must be very efficient and energy-consuming, since the concentration of silicic acid in the surrounding milieu is low and crucial for the formation of spicules. The optimal silica concentration for the synthesis of spicules is between 5 and 100 μM [24]. Lastly, the growth rate of sponge spicules is fast and, for freshwater sponges, has been measured at 5 μm/h [25]. Hence it can be deduced that sponges have an efficient system to take up silica from the environment.
Our attempts to identify the silicic acid transporter in the demosponge Suberites domuncula, with degenerate primers that were suitable for the PCR-coupled cloning of the diatom transporter, or by choosing differential display of mRNA at different silica concentrations in the seawater in which the sponges were maintained, failed. We finally followed a PCR cloning strategy with primers directed against the conserved regions of the BTS [HCO3− (bicarbonate) transporter superfamily]. Several groups of HCO3− transporters exist in Metazoa; the BTS comprising the NBC (Na+/HCO3−) co-transporters and the Cl−/HCO3− AEs (anion-exchangers) (see [26]). Two further HCO3− transporters are noteworthy in this context, namely the K+/HCO3− cotransporter [27] and the Na+-dependent NDAE (Cl−–HCO3− exchanger) [26,28]. The latter transporter is – like all the transporters mentioned – involved in the pH/ionic homoeostasis and functionally active if the outside milieu is higher in Na+ (than inside), higher in pH and lower in Cl−. This cloning strategy was successful.
In the present study we demonstrate that the cells from S. domuncula react to exposure to silicic acid with a change of the fluorescence of PDMPO [2-(4-pyridyl)-5-{[4-(2-dimethylaminoethylaminocarbamoyl)methoxy]phenyl}oxazole], a known fluorescent silica tracer [29]. This reaction depends on temperature and is sensitive to the inhibitor DIDS. The suggestion that a transporter is involved in this reaction is corroborated by our success in identifying and cloning an Na+/HCO3− co-transporter. Evidence is presented that this transporter is involved in silicic acid transport and hence has been termed Na+/HCO3−[Si(OH)4] co-transporter or NBCSA-related transporter.
MATERIALS AND METHODS
Materials
The sources of chemicals and enzymes used have been given previously [1,13,30]. PDMPO LysoSensor yellow/blue was obtained from Molecular Probes (Leiden, The Netherlands); natural sterile filtered seawater (SW), DIDS and Na2SiO3 (sodium metasilicate) were from Sigma–Aldrich (Taufkirchen, Germany). The compositions of CMFSW (Ca2+- and Mg2+-free artificial seawater) and CMFSW containing 2.5 mM EDTA (CMFSW-E) were given previously [31].
Sponges and cells
Live specimens of S. domuncula (Porifera, Demospongiae, Hadromerida) were collected in the Adriatic Sea near Rovinj (Croatia) and kept in aquaria in Mainz, Germany, for more than 10 months at a temperature of 17 °C prior to their use.
Single cells were obtained as described in [32]. Briefly, tissue samples were cut into 3 mm3 cubes; these were transferred into Falcon tubes and dissociated into single cells with CMFSW-E. After incubation for 40 min with shaking, the supernatant was collected and filtered through a 40 μm-mesh nylon net. The sponge cells were obtained by centrifugation (500 g for 5 min), washed twice in CMFSW and the final pellet was resuspended in SW (seawater) to a density of 6×106 cells/ml. A 1 ml portion of this cell suspension was added to a chambered coverglass (Lab-Tek Chamber Slide System; Nunc, Wiesbaden, Germany) that had been coated with poly-L-lysine [10 μg/ml; Mr 70000–150000 (Sigma–Aldrich)] for at least 2 h at room temperature. The cultures were kept at 4° overnight.
Primmorph formation
The procedure for the formation of primmorphs, three-dimensional aggregates composed of proliferating cells, from single cells was applied as described previously [32]. Briefly, single cells obtained by dissociation were cultured in natural SW supplemented with 1% RPMI 1640 medium. Primmorphs of at least 1 mm in diameter were formed after 5 days. Then these three-dimensional cell cultures were transferred into RPMI 1640 medium enriched with silicic acid (60 μM) [33] and incubated for up to 5 days and used for the in-situ-hybridization analysis.
PDMPO fluorescence studies
The fluorescent dye PDMPO was initially applied to measure acidification in cells [34], and later it was also used as a silica tracer for biological silicification studies [29].
In our experiments, the following courses of incubation were chosen. First, sponge cells kept for 12 h at 4 °C in SW were exposed for 2 min to ambient medium (containing 2 μM silicon) or medium supplemented with 60 μM Na2SiO3 at 4 °C or 17 °C as indicated. After 2 min, 3 μM PDMPO was added and the measurement of fluorescence intensity was started immediately; measurement continued for 16 min. Where indicated the (non-)competitive inhibitor DIDS (see [35]) was added to the cells at a concentration of 20 μM, together with silicic acid.
Digital fluorescence microscopy was performed with sponge cells to monitor PDMPO fluorescence as described in [29]. Sponge cells were inspected with an inverted-stage Olympus IX70 microscope with apochromatic reflected light fluorescence lenses (UApo40X/340). For the determination of fluorescence intensity, cells were illuminated with 340 nm light from a mercury source (OSP-EXA, Olympus). The emitted light (445 nm) was passed into an image-intensified CCD (charge-coupled-device) camera (C2400-87; Hamamatsu, Herrsching, Germany). Images were digitized as 256×256 pixels by 8-bit arrays with a computerized imaging system (Argus-50; Hamamatsu).
Isolation of a cDNA for the NBCSA-co-transporter-related sequence from S. domuncula
A cDNA encoding the potential Na+/HCO3−[Si(OH)4] co-transporter (termed NBCSA_SUBDO) was isolated from a cDNA library [1]. PCR with degenerate primers designed against the conserved aa (amino acid) sequences of the TM segments 1, 7 and 8 was performed with this library. PCR conditions were: 4 min at 94 °C (1 cycle) and 30 s at 94 °C, 30 s at 42 °C, 20 s at 72 °C (35 cycles). Fragments of the expected sizes were obtained and sequenced using standard procedures. The full-length cDNA was amplified by RACE (rapid amplification of cDNA ends) applying the Invitrogen (Groningen, The Netherlands) GeneRacer Kit, the manufacturer's manual being followed. The complete 3959-nt-long sequence, termed SDNBCSA, was obtained as determined by Northern blotting. Computer analysis of the deduced protein and its phylogenetic-tree assessment was performed as described previously [13].
Northern blotting
RNA was extracted from liquid-nitrogen-pulverized tissue of S. domuncula with TRIzol® Reagent (Gibco BRL, Grand Island, NY, U.S.A.). Sponge specimens were incubated for up to 5 days in the presence of either 2 μM silicic acid (ambient concentration present in the natural SW) or SW adjusted to a silicic acid concentration of 60 μM. Then tissue samples were taken for RNA isolation; 5 μg of total RNA was electrophoresed through a 1% formaldehyde/agarose gel and blotted on to Hybond-N+ nylon membrane (Amersham, Little Chalfont, Bucks., U.K.), the manufacturer's instructions being followed. Hybridization was performed with a 450-bp large part of either SDNBCSA or the cDNA of the housekeeping sponge gene for β-tubulin, SDTUB (accession number AJ550806); tubulin was used as an internal standard. The probes were labelled with the PCR DIG Probe Synthesis Kit (Roche, Mannheim, Germany), the Instruction Manual of the manufacturer being followed. After washing, DIG (digoxigenin)-labelled nucleic acid was detected with anti-DIG Fab fragments (conjugated to alkaline phosphatase; dilution 1:10000) and visualized by the chemiluminescence technique using CDP-Star [disodium 4-chloro-3-(methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro) tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate] according to the manufacturer's (Roche) instructions. The screens were scanned with the GS-525 Molecular Imager (Bio-Rad, Hercules, CA, U.S.A.).
In situ localization studies
In order to identify which cells express SDNBCSA within S. domuncula, two series of experiments were performed. First, tissue samples (4 mm3 cubes) from sponge specimens were treated in CMFSW containing 2.5 mM EDTA for 40 min. Then the non-disintegrated tissue samples were removed by sedimentation of the suspension for 1 min. The cells and spicules in the resulting supernatant were additionally subjected to sedimentation for 3 min. The sediment was collected, washed twice with CMFSW and finally centrifuged (5 min, 2000 g) on to Silane-Prep-Slides (Sigma–Aldrich) and used for in situ hybridization.
Secondly, primmorphs which were kept either in the absence (i) or in the presence (ii) of additional silicic acid (60 μM) for 3 or 5 days were used.
The in situ hybridization method applied was based on the procedure described by Polak and McGee [36], with the modifications described recently [37]. Frozen 8-μm-thick sections from primmorphs were obtained at −30 °C using a Slee (Mainz, Germany) cryostat. Cryosections and cells were fixed with 4% paraformaldehyde and then washed twice with 1×PBS at room temperature. The samples were incubated with 1 μg/ml Proteinase K for 30 min and subsequently fixed again with paraformaldehyde. The colour resulting from the sponge was removed with ethanol/propan-2-ol as described in [37]. After rehydration with 1×PBS, DIG-labelled DNA probes were added to the hybridization solution. Hybridization was performed overnight in a glass chamber at 40 °C; the subsequent washes were performed at 45 °C as described in [37]. The probes were labelled with the PCR DIG Probe synthesis kit (Roche). DNA probes were constructed on the basis of the S. domuncula cDNA sequences. Both antisense and sense probes were prepared by PCR using the linearized cDNA. The probe spanned a segment within the open reading frame (nt427–nt869) of the NBC-related sequence with a length of 452 bp (SDNBCSA). After blocking, the sections were incubated with an anti-DIG antibody conjugated to alkaline phosphatase. NBT/X-phosphate (NitroBlue Tetrazolium/5-bromo-4-chloroindol-3-yl phosphate) was used for the visualization of the signals. Positive signals were obtained with the antisense probes, whereas the sense probes (controls) showed no staining.
RESULTS
Effect of silicic acid on the spectral properties of cells in the presence of PDMPO
As outlined in the Materials and methods section, dissociated sponge cells were pre-incubated in medium for 2 min at 4 °C with 60 μM Na2SiO3 in the absence or presence of 20 μM DIDS. Then incubation of the cells continued at 4 °C or 17 °C with the fluorescent dye PDMPO, and the change of the emission fluorescence at 445 nm was monitored using digital fluorescence microscopy.
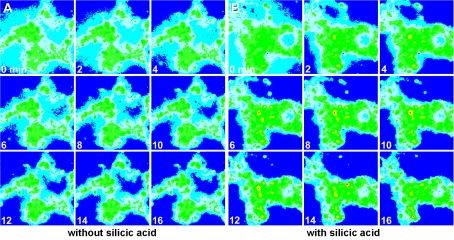
The change of the fluorescence intensity of the emission at 445 nm in response to silicic acid can be readily monitored by pseudo-colour image analysis (see the Materials and methods section). It is obvious that the change of the colour intensity from blue (low) to red (high intensity) is only marginal in the assays using cells and PDMPO without additional silicic acid at an incubation temperature of 17 °C (Figure 1A). However, if the cells were incubated with 60 μM silicic acid, a strong shift of the colour from blue to yellow and red is seen as quickly as 6 min after an incubation period with PDMPO (Figure 1B). From these data we conclude that the cells react to silicic acid with an intracellular change of the fluorescence properties of PDMPO.
Figure 1. Pseudo-colour ratio images of dissociated sponge cells incubated with 60 μM silicic acid as described in the Materials and methods section.
The cells were first incubated in SW with ambient (A) or 60 μM silicic acid (B) at 17 °C and after 2 min exposed to 3 μM PDMPO. Fluorescence intensity was recorded at 445 nm, and the images are shown at time zero and at 0, 2, 4, 6, 8, 10, 12, 14 and 16 min. Magnification ×30.
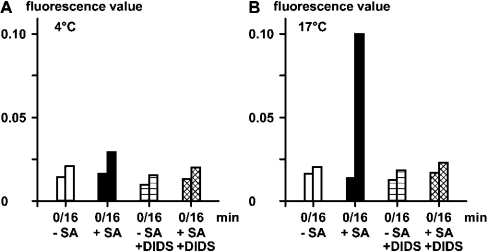
A quantitative analysis was performed to determine the effect of temperature and DIDS on the silicic acid-evoked change of the intracellular fluorescence using PDMPO. The measurements revealed that, at 4 °C, the fluorescence values were low and level at about 0.02 arbitrary unit (Figure 2A), irrespective of treatment of the cells with silicic acid and/or with DIDS. However, if the incubation with the fluorescence probe was performed at 17 °C, a strong effect of silicic acid was seen, which was dependent on the co-incubation with DIDS (Figure 2B). In the absence of silicic acid, a slight increase in fluorescence values from 0.020 to 0.025 was measured after a 16 min incubation period at 17 °C. If cells which had been preincubated with 60 μM silicic acid for 2 min (17 °C) were analysed, a strong increase of the fluorescence values resulted; after 16 min the value reached 0.10 (Figure 2B).
Figure 2. Effect of silicic acid (SA) on the florescence emission of PDMPO in sponge cells.
The cells were incubated in the absence (−SA; open bars) or presence of 60 μM silicic acid (+SA; Na2SiO3) (closed bars). In a parallel series of experiments the inhibitor DIDS was added at time zero in the absence (−SA/+DIDS; horizontally hatched) or presence of silicic acid (+SA/+DIDS; cross-hatched). The fluorescence probe PDMPO was added 2 min later and the measurements were taken for 16 min; the fluorescence values at 445 nm are given in arbitrary units. The studies were performed at either 4 °C (A) or 17 °C (B).
In order to determine if the fluorescence change caused by silicic acid can be blocked by the inhibitor DIDS, parallel experiments were performed with a 20 μM concentration of this (non-)competitive inhibitor. The experiments showed that, if DIDS is added together with silicic acid, only a marginal increase in the fluorescence values could be measured (Figure 2B). These data suggest that silicic acid is taken up by sponge cells via a transport mechanism which can be poisoned by DIDS. The most likely candidate transporter is a HCO3− transporter which contains sites known to be targets for DIDS. Until now, only one transporter could be identified in sponges, and that is the one described here.
NBCSA co-transporter: sequence data
The complete cDNA, SDNBCSA, was obtained by the PCR technique, followed by RACE completion. The 3959-nt long cDNA comprises one ORF (open reading frame) spanning nt128–130–nt3851–3853(stop). The 1241-aa-long deduced polypeptide, NBCSA_SUBDO, has a predicted Mr of 138264 and contains the highly conserved TM region (aa690–aa1209) (Figure 3A). The deduced protein is calculated to be an unstable protein with an instability index of 56.6, based upon the presence of certain dipeptides that frequently occur in metabolically unstable proteins [38].
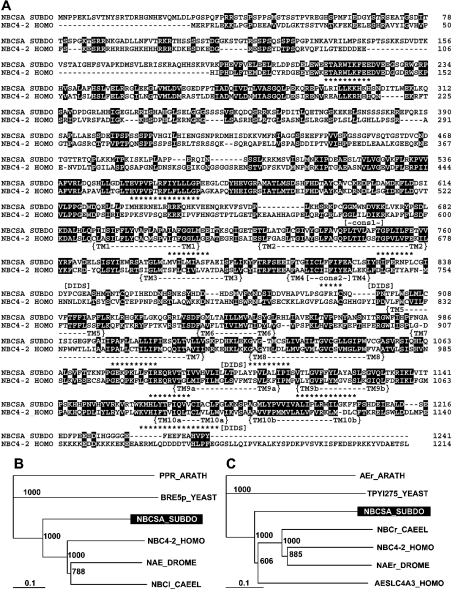
Figure 3. Na+/HCO3−[Si(OH)4] co-transporter (NBCSA)-related sequence from S. domuncula.
The sponge deduced protein (NBCSA_SUBDO) is aligned with the closest similar sequence from human, the solute carrier family 4, NBC co-transporter from human (NBC4–2_HOMO, accession number NP_003606). Residues conserved (similar or related with respect to their physico-chemical properties) in both sequences are shown in white on black. The TM region comprising ten helices (TM), the two AE consensus patterns (cons) and the putative DIDS motifs are marked. (B) The N-terminal part of these two sequences is compared with the regions present in the next similar sequences from the protostomians D. melanogaster, the Na+-driven AE 1 (NAE_DROME, NP_723264.1) and C. elegans, the probable electrogenic NBC co-transporter protein (NBCl_CAEEL, NP_492258.1) as well as the yeast, S. cerevisiae, protein Bre5p (BRE5p_YEAST, NP_014449.1) and the plant (A. thaliana) PPR repeat-containing protein (PPR_ARATH, NP_174349.1). (C) The TM parts of the human and sponge transporters were likewise aligned with the related segments of the sequences from human, the AE SLC4A3 (AESLC4A3_HOMO, AAN34939.1), the Na+-driven AE 1 CG4675-PB from D. melanogaster (NAEr_DROME, NP_723264.1), the probable electrogenic NBC co-transporter protein from C. elegans (NBCr_CAEEL, NP_492258.1), the yeast transporter (Ynl275wp from S. cerevisiae TPYl275_YEAST, NP_014124.1) and the plant (A. thaliana) probable AE protein (AEr_ARATH, G84911). The trees were computed and rooted with the plant sequences as the outgroup. The scale bar indicates an evolutionary distance of 0.1 aa substitution per position in the sequence.
Sequence comparisons revealed that the sponge NBC-related sequence shares highest similarity with the human family 4 NaHCO3 co-transporter (accession number NP_003606) having an ‘expect value’ (E; Blast-NCBI [39]) of e−155. The overall similarity value between the two sequences is high; 46% of the aas are similar and 28% of the aas are identical. The two AE consensus patterns [40] are found within the S. domuncula protein between aa665 and aa673 and between aa813 and aa822 respectively (Figure 3A). All conserved areas of aa identities or similarities found in other metazoan BTS members [41] are also present in the S. domuncula sequence (Figure 3A).
The AE transporters can be divided into two distinct functional domains, namely the N-terminal domain and the C-terminal TM domain [35]. The deduced sponge protein NBCSA_SUBDO shows in its both segments significant sequence similarity to human family 4 NBC (NP_003606). In the S. domuncula sequence, the N-terminal cytoplasmic domain ends at aa689. Subsequently the TM region proceeds which consists of ten putative TM segments, as concluded from the hydrophobicity plot (RAOARGOS algorithm [38]), with TM helix 1 ranging from aa690 to aa712, helix 2 from aa725 to aa760, helix 3 from aa778 to aa799, helix 4 from aa809 to aa826, helix 5 from aa900 to aa918, helix 6 from aa938 to aa954, helix 7 from aa983 to aa1017, helix 8 from aa1027 to aa1054, helix 9a from aa1088 to aa1110, helix 9b from aa1113 to aa1133, helix 10a from aa1162 to aa1177, and helix 10b from aa1182 to aa1205 (Figure 3A). Helices 9 and 10 contain two hydrophobic stretches. Whereas in higher metazoans the HCO3− transporters have a large extracellular loop between helix 5 and helix 6, this region is short in the deduced sponge sequence; in contrast, NBCSA_SUBDO has an extensive intracellular loop between helix 4 and helix 5 (Figure 3A). Within the TM region, NBCSA_SUBDO has four putative DIDS motifs; they are found at aa763–aa766 (disrupted [42]), aa826–aa828 (disrupted [42]), aa1027–aa1036 (intact [41]) and aa1176–aa1180 (intact [41]).
Sponge NBC-related sequence: phylogenetic analysis
Up until now, NBC transporters are known only from Metazoa. In our phylogenetic analysis the putative N-terminal cytoplasmic domain and the TM regions were analysed separately. The N-terminal cytoplasmic domain of NBCSA_SUBDO shows the highest sequence similarity to the human NBC co-transporter, member 7 (NBC4-2_HOMO, NP_003606); between these two molecules there exists 46% similarity and 28% identity. Less related are the two transporters from the invertebrates: an Na+-driven AE (NAE_DROME) from the fruitfly Drosophila melanogaster, and an NBC (NBCl_CAEEL) from the nematode worm Caenorhabditis elegans. Only distantly related are the most similar proteins: the protein Bre5p (BRE5p_YEAST) from baker's yeast (Saccharomyces cerevisiae), and the plant pentatricopeptide (PPR) repeat-containing protein, PPR_ARATH, from thale cress (Arabidopsis thaliana) (Figure 3B).
A similar relationship exists for the TM segment of the sponge NBC molecule. Here too the highest homology exists with the metazoan polypeptides, especially to the human NBC (NP_003606; NBC4-2_HOMO) and the SLC4A3 AE (AAN34939.1; AESLC4A3_HOMO) and likewise to the Na+-driven AE from D. melanogaster (NAEr_DROME) and the HCO3− co-transporter protein from C. elegans (NBCr_CAEEL); the similarity score of approx. 45% similarity and 28% identity is seen. The sequence relationship between the yeast and plant transporter/exchanger proteins is low. The rooted tree shows the significant grouping between the metazoan and non-metazoan molecules (Figure 3C).
Localization of SDNBCSA transcripts: Northern-blot analysis
Northern-blot experiments were performed to demonstrate that the full-length clone of the sponge NBC transporter had been isolated and also to show that the expression of the sponge gene is inducible by exposure to silicic acid. RNA was extracted from tissue that had been exposed to ambient (2 μM) or 60 μM silicic acid for up to 5 days. After size separation the blots were probed with parts of the cDNAs SDNBCSA (NBC transporter) and SDTUB (β-tubulin).
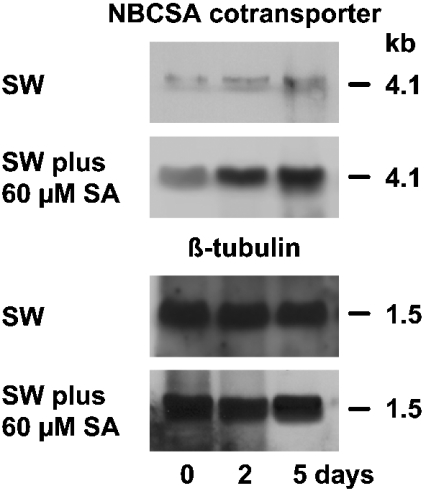
The size of the SDNBCSA signal is 4.1 kb (Figure 4), matching the length of the isolated cDNA of SDNBCSA. In the absence of additional silicic acid in the natural SW, the level of the transcript was low and did not change during the 5-day incubation period. However, if the animals were kept for 5 days in SW supplemented with 60 μM silicic acid, a strong increase in the steady-state level of the SDNBCSA transcripts was seen (Figure 4). The level of tubulin RNA did not change under both incubation conditions.
Figure 4. Steady-state level of SDNBCSA expression in tissue from S. domuncula.
The specimens were kept either in the absence of additional silicic acid in SW or were maintained in SW supplemented with 60 μM silicic acid (SA) for 2 or 5 days. Then RNA was isolated, electrophoresed and transferred. Finally the membranes were probed with the labelled cDNAs, SDNBCSA (NBCSA transporter) and SDTUB (β-tubulin). The tubulin probe was used to demonstrate that the same amounts of RNA were loaded on to the gels.
Localization of SDNBCSA transcripts: in situ analysis
Two approaches were taken to demonstrate the relationship between the spicule formation and the expression of the NBC-related sequence.
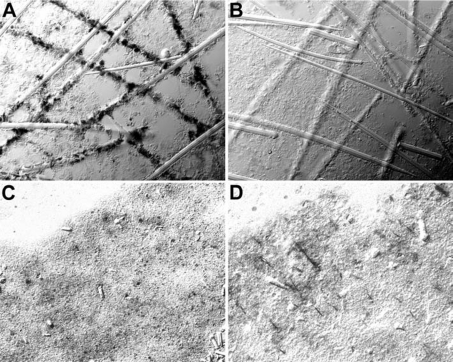
First, spicules obtained after limited dissociation of S. domuncula tissue which still remained partially associated with the adjacent cells were subjected to in situ hybridization. After treatment with the antisense SDNBCSA probe, almost all cells linked to the spicules became brightly stained, indicating that they strongly express the gene encoding the NBC-related sequence (Figure 5A). In controls, using the sense probe, no staining is seen (Figure 5B).
Figure 5. In-situ-hybridization studies to localize the cells expressing the gene encoding the NBC-related sequence.
(A) Spicules were isolated from sponge tissue by limited EDTA treatment. Applying the in-situ-hybridization technique and using the labelled antisense SDNBCSA as a probe, those cells which are closely associated with the spicules are positively stained (magnification ×160). (B) Control hybridization with the sense SDNBCSA (×160). (C) Sections through a primmorph cultivated in the absence of silicic acid or (D) kept for 5 days in medium, supplemented with 60 μM silicic acid (×40). After incubation with silicic acid (D), cell clusters arranged in an rod-like organization pattern which was stained with the antisense SDNBCSA.
To confirm this result, primmorphs were analysed. These three-dimensional cell cultures are formed from dissociated cells. If they were analysed 5 days after formation, only very few cells showed the signal after in situ hybridization with antisense SDNBCSA (Figure 5C). However, if they were cultivated for 5 days in SW supplemented with 60 μM silicic acid, such cell clusters, arranged in an rod-like pattern (Figure 5D), became positively stained with the antisense probe. From such cell arrangements, spicules are formed within the primmorphs [13,17,33].
DISCUSSION
CO2 is known to enter mammalian cells via the apical membrane of the renal tubules or their tight junctions [43], while silica is taken up as silicic acid (see [9,22,44]). The silica content in the SW is low, both in high-ionic-strength water, e.g. the Mediterranean [44], and in low-salinity marine environment, e.g. in the Baltic Sea (Kiel Bight [22]); the concentration amounts to ≤10 μM. Furthermore, experiments with the sponge Halichondria panicea suggested that silica transport is an energy-consuming process and the uptake rate depends on the ambient silica concentration [22]. Rates of 5 μmol of Si/h per g of ash-free dry weight have been measured [22]. Within the sponge body, silica is accumulated in the silicalemma. These unit-type membranes surround the immature spicules and are connected both with the axial filaments and the cell membranes of the sclerocytes. Since no silicon has been detected in archaeocytes or choanocytes (see [9]), it has been proposed that silicon/silicic acid enters the body through transient spaces between the epithelial cells [7]. However, on the basis of recent findings that sponges are surrounded by an epithelium which comprises, besides the cell–cell adhesion molecules, also specialized junction molecules [45], it appears to be more likely that silica uptake takes place in a highly controlled manner.
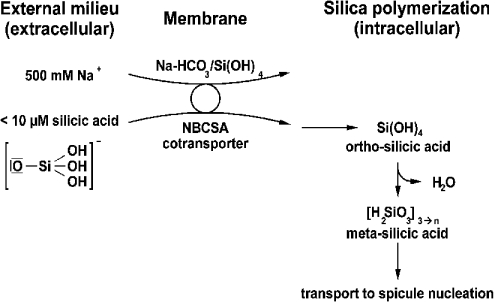
Sponges live in an external milieu of high salinity (about 0.5 M [46]). It is conceivable that silicic acid is taken up by a transporter mechanism in sponges; an NBC co-transporter has been considered a prime candidate. The cDNA of the putative transporter was isolated, which showed a considerable sequence similarity to that of the NBC co-transporter. On the basis of indirect cell-biological data suggesting the involvement of this transporter in silicic acid uptake, we termed the deduced protein ‘NBCSA-related molecule’. In the marine environment silicon exists to a small extent as [SiO(OH)3]−, but primarily in form of un-ionized silicic acid [Si(OH)4] [20]. In line with the uptake mechanism proposed for diatoms [47], we postulate at present that silica is also taken up by sponges in the ionized form (Scheme 1).
Scheme 1. Model for silicic acid transport in sponge cells.
It is proposed that the NBCSA-related transporter facilitates the uptake of silicic acid across the plasma membrane. Free silicic acid in the cytoplasm undergoes polymerization, driven by the high osmolarity.
Depending on whether the transporter is an electroneutral or an electrogenic co-transporter, the ratio between Na+ and HCO3− is either 1:2 [48] or 1:3 [43] respectively. Hence stoichiometrically more HCO3− than Na+ is co-transported by this transporter. Such transport would also be favourable for sponges, since the outside Na+ concentration is much higher than the silicic acid concentration. Again, in line with the experiments published on diatoms [19], we postulate that in sponges, too, silicic acid transport is coupled with that of sodium. For the diatom Cylindrotheca fusiformis, a silicic acid transporter was cloned, expressed and found, as expected from the large evolutionary distance between the two taxa, diatoms and sponge, to display no sequence similarity to any of the known transporters listed in the databases [18,21]. For metazoans a silica transporter has not yet been described. In our strategy to identify the putative transporter from a sponge, the cDNA library was screened with degenerate primers directed against the conserved region within NBC transporters from metazoans. One clone was identified that contains, in the deduced protein, like in diatoms [18], ten TM segments. Phylogenetic analyses convincingly showed that the sponge transporter identified shows, both in the N-terminal cytoplasmic domain and in the TM region, a high sequence similarity to other NBC transporters, which are restricted to Metazoa. With this sequence at hand we set out to elucidate whether this NBC transporter molecule is also involved in silicic acid uptake. First, it was found that the deduced protein has, besides the characteristic signature sequences for NBC transporters, several DIDS-binding motifs. Therefore incubation studies were performed with cells in the presence or absence of DIDS. The results revealed that this stilbene inhibitor [35] blocks the PDMPO-mediated fluorescence change caused by silicic acid almost completely.
Next we attempted to determine whether the expression of the gene encoding the NBCSA-related transporter depends on the external supply of silicic acid in the surrounding medium. Increasing the silicic acid concentration to 60 μM resulted in a strong up-regulation of the steady-state expression rate, indicating that the transporter can be grouped with the silicon-responsive genes. Earlier studies with other genes, e.g. collagen or silicatein genes, which code for proteins involved in spicule formation, proved that they are likewise strongly up-regulated in response to silicic acid.
The final strong support for our conclusion that the NBCSA-related transporter is indeed involved in the transport of silicic acid came from in-situ-hybridization studies. This technique, only recently established for sponges [37], had already been shown to be suitable for the elucidation of morphogenetic and developmental processes in these animals (reviewed in [49]). The results show that the expression of the NBCSA-related-transporter gene is primarily seen in those cells in adult animals which are adjacent to spicules. In a further series of experiments we studied whether this gene is also induced in defined areas in primmorphs. Recent histological studies showed that initiation of spicule formation occurs as soon as the gastrulation phase and, during this period, especially in the periphery [50]. Moreover, it was demonstrated that the cells are arranged around an axial filament forming spindle-like cell arrangements. Such cell clusters, which contain increased levels of transcripts for the NBCSA-related transporter, appeared in primmorphs after incubating them in increased concentrations of silicic acid. This result is taken as a strong indication that the transporter identified here is indeed involved in cellular silicic acid accumulation.
To conclude, sponge cells respond to extracellular silicic acid with an increase in fluorescence of PDMPO used as a tracer compound. This shift is temperature-dependent and can be blocked by DIDS. A cDNA for a molecule was cloned which coded for a putative NBCSA-related transporter. Inhibition experiments, gene expression, as well as in-situ-hybridization studies, were performed, and these led us to suggest that this transporter is involved in the TM transport of silicic acid.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Bundesministerium für Bildung und Forschung (project: Center of Excellence BIOTECmarin) and the European Commission.
References
- 1.Kruse M., Müller I. M., Müller W. E. G. Early evolution of metazoan serine/threonine- and tyrosine kinases: identification of selected kinases in marine sponges. Mol. Biol. Evol. 1997;14:1326–1334. doi: 10.1093/oxfordjournals.molbev.a025742. [DOI] [PubMed] [Google Scholar]
- 2.Kooistra W. H. C. F., De Stefano M., Mann D. G., Medlin L. K. The phylogeny of the diatoms. Progress Mol. Subcell. Biol. 2003;33:59–97. doi: 10.1007/978-3-642-55486-5_3. [DOI] [PubMed] [Google Scholar]
- 3.Brasier M., Green O., Shields G. Ediacarian sponge spicule clusters from southwest Mongolia and the origins of the Cambrian fauna. Geology. 1997;25:303–306. [Google Scholar]
- 4.Uriz M. J., Turon X., Becerro M. A., Agell G. Siliceous spicules and skeleton frameworks in sponges: origin, diversity, ultrastructural patterns, and biological functions. Microsc. Res. Tech. 2003;62:279–299. doi: 10.1002/jemt.10395. [DOI] [PubMed] [Google Scholar]
- 5.Simpson T. L. New York: Springer-Verlag; 1984. The Cell Biology of Sponges. [Google Scholar]
- 6.Shore R. E. Axial filament of siliceous sponge spicules, its organic components and synthesis. Biol. Bull. 1972;143:125–136. doi: 10.2307/1540191. [DOI] [PubMed] [Google Scholar]
- 7.Uriz M. J., Turon X., Becerro M. A. Silica deposition in Demospongiae: spiculogenesis in Crambe crambe. Cell Tissue Res. 2000;301:299–309. doi: 10.1007/s004410000234. [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson C. R., Garrone R. Ultrastructure of siliceous spicules and microsclerocytes in the marine sponge Neofibularia irata n. sp. J. Morph. 1980;166:51–64. doi: 10.1002/jmor.1051660105. [DOI] [PubMed] [Google Scholar]
- 9.Uriz M. J., Turon X., Beccero M. A. Silica deposition in demosponges. Prog. Mol. Subcell. Biol. 2003;33:163–193. doi: 10.1007/978-3-642-55486-5_7. [DOI] [PubMed] [Google Scholar]
- 10.Weaver J., Morse D. E. Molecular biology of demosponge axial filaments and their roles in biosilification. Microsc. Res. Tech. 2003;62:356–367. doi: 10.1002/jemt.10401. [DOI] [PubMed] [Google Scholar]
- 11.Shimizu K., Cha J., Stucky G. D., Morse D. E. Silicatein α: cathepsin L-like protein in sponge biosilica. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6234–6238. doi: 10.1073/pnas.95.11.6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cha J. N., Shimizu K., Zhou Y., Christianssen S. C., Chmelka B. F., Stucky G. D., Morse D. E. Silicatein filaments and subunits from a marine sponge direct the polymerization of silica and silicones in vitro. Proc. Natl. Acad. Sci. U.S.A. 1999;96:361–365. doi: 10.1073/pnas.96.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krasko A., Batel R., Schröder H. C., Müller I. M., Müller W. E. G. Expression of silicatein and collagen genes in the marine sponge Suberites domuncula is controlled by silicate and myotrophin. Eur. J. Biochem. 2000;267:4878–4887. doi: 10.1046/j.1432-1327.2000.01547.x. [DOI] [PubMed] [Google Scholar]
- 14.Schröder H. C., Perović-Ottstadt S., Wiens M., Batel R., Müller I. M., Müller W. E. G. Differentiation capacity of the epithelial cells in the sponge Suberites domuncula. Cell Tiss. Res. 2004;316:271–280. doi: 10.1007/s00441-004-0869-7. [DOI] [PubMed] [Google Scholar]
- 15.Sandford F. Physical and chemical analysis of siliceous skeletons in six sponges of two groups (Demospongiae and Hexactinellida) Microsc. Res. Tech. 2003;62:336–355. doi: 10.1002/jemt.10400. [DOI] [PubMed] [Google Scholar]
- 16.Arndt W. Porifera. In: Junk W., editor. Tabulae Biologicae. Berlin: W. Junk; 1930. pp. 39–120. [Google Scholar]
- 17.Schröder H. C., Krasko A., Le Pennec G., Adell T., Hassanein H., Müller I. M., Müller W. E. G. Silicase, an enzyme which degrades biogenous amorphous silica: contribution to the metabolism of silica deposition in the demosponge Suberites domuncula. Prog. Mol. Subcell. Biol. 2003;33:249–268. doi: 10.1007/978-3-642-55486-5_10. [DOI] [PubMed] [Google Scholar]
- 18.Hildebrandt M., Wetherbee R. Components and control of silicification in diatoms. Prog. Mol. Subcell. Biol. 2003;33:11–57. doi: 10.1007/978-3-642-55486-5_2. [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharyya P., Vulcani B. E. Sodium-dependent silicate transport in the apochlorotic marine diatom Nitzschia alba. Proc. Natl. Acad. Sci. U.S.A. 1980;77:6386–6390. doi: 10.1073/pnas.77.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Amo Y., Brzezinski M. A. The chemical form of dissolved Si taken up by marine diatoms. J. Phycol. 1999;35:1162–1170. [Google Scholar]
- 21.Hildebrandt M., Dahlin K., Volcani B. E. Characterization of a silicon transporter gene family in Cylindrotheca fusiformis: sequences, expression analysis, and indication of homologs in other diatoms. Mol. Gen. Genet. 1997;260:480–486. doi: 10.1007/s004380050920. [DOI] [PubMed] [Google Scholar]
- 22.Fröhlich H., Bartel D. Silica uptake of the marine sponge Halichondria panicea in Kiel Bight. Mar. Biol. 1997;128:115–125. [Google Scholar]
- 23.Maldonado M., Carmona M. C., Uriz M. J., Cruzado A. Decline in Mesozoic reef-building sponges explained by silicon limitations. Nature (London) 1999;401:785–788. [Google Scholar]
- 24.Jewell M. E. An ecological study of the fresh-water sponge of northern Wisconsin. Ecol. Monogr. 1935;5:461–504. [Google Scholar]
- 25.Weissenfels N. Stuttgart: Gustav Fischer Verlag; 1989. Biologie und Mikroskopische Anatomie der Süsswasserschwämme (Spongillidae) [Google Scholar]
- 26.Romero M. F., Henry D., Nelson S., Harte P. J., Dillon A. K., Sciortino C. M. Cloning and characterization of a Na+-driven anion exchanger (NDAE1) J. Biol. Chem. 2000;275:24552–24559. doi: 10.1074/jbc.M003476200. [DOI] [PubMed] [Google Scholar]
- 27.Hogan E. M., Cohen M. A., Boron W. F. K+- and HCO3−-dependent acid–base transport in squid gland axons. II. Base efflux. J. Gen. Physiol. 1995;106:821–844. doi: 10.1085/jgp.106.5.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas R. C. The role of bicarbonate, chloride and sodium ions in the regulation of intracellular pH in snail neurones. J. Physiol. (London) 1977;273:317–338. doi: 10.1113/jphysiol.1977.sp012096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu K., Del Amo Y., Brzezinski M. A., Stucky G. D., Morse D. E. A novel fluorescent tracer for silicification studies. Chem. Biol. 2001;8:1051–1060. doi: 10.1016/s1074-5521(01)00072-2. [DOI] [PubMed] [Google Scholar]
- 30.Wimmer W., Perovic S., Kruse M., Krasko A., Batel R., Müller W. E. G. Origin of the integrin-mediated signal transduction: functional studies with cell cultures from the sponge Suberites domuncula. Eur. J. Biochem. 1999;178:156–165. doi: 10.1046/j.1432-1327.1999.00146.x. [DOI] [PubMed] [Google Scholar]
- 31.Rottmann M., Schröder H. C., Gramzow M., Renneisen K., Kurelec B., Dorn A., Friese U., Müller W. E. G. Specific phosphorylation of proteins in pore complex–laminae from the sponge Geodia cydonium by the homologous aggregation factor and phorbol ester. Role of protein kinase C in the phosphorylation of DNA topoisomerase II. EMBO J. 1987;6:3939–3944. doi: 10.1002/j.1460-2075.1987.tb02735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller W. E. G., Wiens M., Batel R., Steffen R., Borojevic R., Custodio M. R. Establishment of a primary cell culture from a sponge: primmorphs from Suberites domuncula. Marine Ecol. Progr. Ser. 1999;178:205–219. [Google Scholar]
- 33.Krasko A., Schröder H. C., Batel R., Grebenjuk V. A., Steffen R., Müller I. M., Müller W. E. G. Iron induces proliferation and morphogenesis in primmorphs from the marine sponge Suberites domuncula. DNA Cell Biol. 2002;21:67–80. doi: 10.1089/10445490252810320. [DOI] [PubMed] [Google Scholar]
- 34.Diwu Z., Chen C. S., Zhang C., Klaubert D. H., Haugland R. P. A novel acidotropic pH indicator and its potential application in labelling acidic organelles of live cells. Chem. Biol. 1999;6:411–418. doi: 10.1016/s1074-5521(99)80059-3. [DOI] [PubMed] [Google Scholar]
- 35.Kopito R. R. Molecular biology of the anion exchanger gene family. Int. Rev. Cytol. 1990;123:177–199. doi: 10.1016/s0074-7696(08)60674-9. [DOI] [PubMed] [Google Scholar]
- 36.Polak J. M., McGee J. D. situ Hybridization. Oxford: Oxford University Press; 1998. [Google Scholar]
- 37.Perović S., Schröder H. C., Sudek S., Grebenjuk V. A., Batel R., Štifanić M., Müller I. M., Müller W. E. G. Expression of one sponge Iroquois homeobox gene in primmorphs from Suberites domuncula during canal formation. Evol. Dev. 2003;5:240–250. doi: 10.1046/j.1525-142x.2003.03023.x. [DOI] [PubMed] [Google Scholar]
- 38.PC/GENE. Mountain View, CA: IntelliGenetics Inc.; 1995. Data Banks CD-ROM; release 14.0. [Google Scholar]
- 39.Coligan J. E., Dunn B. M., Ploegh H. L., Speicher D. W., Wingfield P. T. Chichester: John Wiley and Sons; 2000. Current Protocols in Protein Science. [Google Scholar]
- 40.Reithmeier R. A. F. The erythrocyte anion transporter (band 3) Curr. Biol. 1993;3:513–523. [Google Scholar]
- 41.Romero M. F., Boron W. F. Electrogenic Na+/HCO3− cotransporters: cloning and physiology. Annu. Rev. Physiol. 1999;61:699–723. doi: 10.1146/annurev.physiol.61.1.699. [DOI] [PubMed] [Google Scholar]
- 42.Virkki L. V., Choi I., Davis B. A., Boron W. F. Cloning of a Na+-driven Cl/HCO3 exchanger from squid giant fiber lobe. Am. J. Physiol. Cell Physiol. 2003;285:C771–C780. doi: 10.1152/ajpcell.00439.2002. [DOI] [PubMed] [Google Scholar]
- 43.Boron W. F., Fong P., Hediger M. A., Boulpaep E. L., Romero M. F. The electogenic Na/HCO3 cotransporter. Wiener Klin. Wochenschr. 1997;108:445–456. [PubMed] [Google Scholar]
- 44.Maldonado M., Uriz M. J. Sexual propagation by sponge fragments. Nature (London) 1999;398:476. [Google Scholar]
- 45.Adell T., Nefkens I., Müller W. E. G. Polarity factor “Frizzled” in the demosponge Suberites domuncula: identification, expression and localization of the receptor in the epithelium/pinacoderm. FEBS Lett. 2003;554:363–368. doi: 10.1016/s0014-5793(03)01190-6. [DOI] [PubMed] [Google Scholar]
- 46.Kennish M. J. Boca Ratón, FL: CRC Press; 1994. Practical Handbook of Marine Science. [Google Scholar]
- 47.Riedel G. F., Nelson D. M. Silicon uptake by algae with no known Si requirement. II. Strong pH dependence of uptake kinetics parameters in Phaeodactylum tricornutum (Bacillariophyceae) J. Phycol. 1985;21:168–171. [Google Scholar]
- 48.Camilion de Hurtado M. C., Perez N. G., Cingolani H. E. An electrogenic sodium–bicarbonate cotransport in the regulation of myocardial intracellular pH. J. Mol. Cell Cardiol. 1995;27:231–242. [PubMed] [Google Scholar]
- 49.Müller W. E. G., Wiens M., Adell T., Gamulin V., Schröder H. C., Müller I. M. The bauplan of the Urmetazoa: the basis of the genetic complexity of Metazoa using the siliceous sponges [Porifera] as living fossils. Int. Rev. Cytol. 2004 doi: 10.1016/S0074-7696(04)35002-3. in the press. [DOI] [PubMed] [Google Scholar]
- 50.Leys S. P. Comparative study of spiculogenesis in demosponge and hexactinellid larvae. Microsc. Res. Tech. 2003;62:300–311. doi: 10.1002/jemt.10397. [DOI] [PubMed] [Google Scholar]