Abstract
With the goal of constructing a genetic alphabet consisting of a set of three base pairs, the fidelity of replication of the three base pairs TH (5-methyl-2-pyrimidinone)/HS (6-thiopurine; thiohypoxanthine), C/H (hypoxanthine) and T/A was evaluated using T7 DNA polymerase, a polymerase with a strong 3′→5′ exonuclease activity. An evaluation of the suitability of a new base pair for replication should include both the contribution of the fidelity of a polymerase activity and the contribution of proofreading by a 3′→5′ exonuclease activity. Using a steady-state kinetics method that included the contribution of the 3′→5′ exonuclease activity, the fidelity of replication was determined. The method determined the ratio of the apparent rate constant for the addition of a deoxynucleotide to the primer across from a template base by the polymerase activity and the rate constant for removal of the added deoxynucleotide from the primer by the 3′→5′ exonuclease activity. This ratio was designated the eni (efficiency of net incorporation). The eni of the base pair C/H was equal to or greater than the eni of T/A. The eni of the base pair TH/HS was 0.1 times that of A/T for TH in the template and 0.01 times that of A/T for HS in the template. The ratio of the eni of a mismatched deoxynucleotide to the eni of a matched deoxynucleotide was a measure of the error frequency. The error frequencies were as follows: thymine or TH opposite a template hypoxanthine, 2×10−6; HS opposite a template cytosine, <3×10−4. The remaining 24 mismatched combinations of bases gave no detectable net incorporation. Two mismatches, hypoxanthine opposite a template thymine or a template TH, showed trace incorporation in the presence of a standard dNTP complementary to the next template base. T7 DNA polymerase extended the primer beyond each of the matched base pairs of the set. The level of fidelity of replication of the three base pairs with T7 DNA polymerase suggests that they are adequate for a three-base-pair alphabet for DNA replication.
Keywords: analogue, code, fidelity, incorporation, replication, 6-thiopurine
Abbreviations: H, hypoxanthine; HS, 6-thiopurine (thiohypoxanthine); dI, hypoxanthine deoxynucleoside (deoxynucleoside of inosine); dIS, 6-thiopurine deoxynucleoside; TH, 5-methyl-2-pyrimidinone; Kf, Klenow fragement of Eschericha coli DNA polymerase I; T7p, T7 DNA polymerase; eni, efficiency of net incorporation
INTRODUCTION
The replication of a number of non-standard base pairs with DNA polymerases has been investigated for their fidelity of replication [1–10], with the goals of elucidating structural requirements for incorporation and for good fidelity of replication, and the development of a genetic alphabet consisting of three base pairs. The discovery that difluorotoluene deoxynucleotide is incorporated opposite a template thymine [9] and that 4-methylbenzimidazole deoxynucleotide is incorporated opposite a template thymine [10] demonstrated that hydrogen bonding between bases is not necessary for incorporation. X-ray diffraction determinations of the structures of several DNA polymerases, including T7p (T7 DNA polymerase) [11], with template/primer and dNTP (reviewed in [12–14]) showed that the size and form of a substrate-inducible pocket are essential constraints on the incorporation of a deoxynucleotide opposite a template base. In the in vivo environment, there is the additional requirement that the base pair must have sufficient stability so that the deoxynucleotide incorporated into the growing strand will not be removed rapidly by a 3′→5′ exonuclease activity. The rapid removal of a deoxynucleotide from the terminal base pairs difluorotoluene/adenine and 4-methylbenzimidazole/thymine by the 3′→5′ exonuclease of Kf (the large Klenow fragment of DNA polymerase I) [15] demonstrated that incorporation by a polymerase activity is not sufficient to produce replication when a 3′→5′ exonuclease activity is present.
A previous investigation [6] of the fidelity of replication of the two standard base pairs and 6-thioguanine/TH (5-methyl-2-pyrimidinone) with Kf demonstrated very significant mismatched incorporation of the base pairs 6-thioguanine/cytosine and guanine/TH. Two suggestions to reduce the frequency of mismatch incorporations were to substitute HS (6-thiopurine) for 6-thioguanine, and to substitute 5-methyl-2-thiopyrimidinone for TH. Each substitution would reduce the number of potential hydrogen bonds between the mismatched bases to one. It was also realized that Kf might not be an appropriate model for in vivo replication. The enzyme has neither strong processivity nor a strong 3′→5′ exonuclease activity for proofreading. A more appropriate model for in vivo replication is unmodified T7p, a polymerase that has strong processivity [16] and a very active exonuclease [17]. A previous study of fidelity with T7p [18] showed an increase in mismatch incorporations mediated by the strong processivity in the absence of the exonuclease activity, and the importance of the 3′→5′ exonuclease activity for correcting mismatch incorporations.
The availability of a new base pair with good fidelity of replication would allow several new investigations. Several investigators [19,20] have analysed the requirements for an extension of the genetic code so that non-standard amino acids could be incorporated into protein. A significant effort is required to design appropriate tRNAs and their cognate synthetases. There are applications for a new base pair that would not require further development. Several examples are as follows. (1) The incorporation of sequences of a new base pair into the genome of a pathogenic organism would result in an organism that could not easily replicate its genome without a source of the new deoxynucleosides. (2) A sequence of the new base pairs placed in a promoter region of the genome would allow investigation of the importance of groups on the bases for determining specific protein interactions. (3) The incorporation of a sequence of a new base pair in the DNA of a chromosome would allow an investigation of the specificity of nucleosome formation. If one of the new bases contained a sulphur group, the degree of protection of this group could be determined during nucleosome reorganization associated with replication, recombination and transcription.
In the present study, a steady-state kinetic method for evaluating the fidelity of replication was developed. The method determines the ratio of the rate constants for the addition of a deoxynucleotide to the primer across from a template base by the polymerase activity and the removal of the added deoxynucleotide from the primer by the 3′→5′ exonuclease activity. Using this kinetic method, the fidelity of replication of the set of three base pairs T/A, C/H (hypoxanthine) and TH/HS (Figure 1) was evaluated with T7p.
Figure 1. Schematic representation of the base pairs.
(A) TH (left base) and HS (right base); (B) cytosine (left base) and hypoxanthine (right base); (C) thymine (left base) and adenine (right base).
MATERIALS AND METHODS
Materials
Enzymes were obtained from New England Biolabs [T4 ligase, large Klenow fragment (exo−), unmodified T7p] and Sigma (bacterial alkaline phosphatase, snake venon phosphodiesterase). Chemicals were obtained from Epicenter Technologies (all standard deoxynucleoside triphosphates), Aldrich Chemical Co. (all anhydrous solvents and reagents for synthesis) and ICN Radiochemicals ([γ-32P]ATP). HPLC columns were obtained from Synchrom, Inc. Oligomers with standard bases and hypoxanthine were obtained from Integrated DNA Technologies, Inc. G-25 Sephadex columns (NAP™5) were obtained from Amersham Pharmacia.
HPLC
All studies used a Milton Roy fixed wavelength Monitor D at either 254 or 326 nm.
Electrophoresis
Preparative and analytical electrophoresis used 20–25% (w/v) acrylamide with 7 M urea in buffer containing 50 mM Tris/borate and 1 mM EDTA, pH 8.3. When oligomers containing HS were purified by electrophoresis, 1 mM glutathione was present in the cathode buffer, and pre-sample electrophoresis was for 30 min.
Autoradiography
The relative amounts of 32P-labelled oligomers on a gel were determined by densitometer measurements of exposed Kodak X-Omat film. The densities were determined by volume integration of the whole image using a Molecular Dynamics Personnel Densitometer 355A and software provided (now owned by Amersham). Comparing two or more different times of exposure confirmed the linear response of the film.
Deoxynucleotide synthesis
2′-Deoxyribosyl-5-methyl-2-pyrimidinone was from a previous synthesis [21]. 2′-Deoxyribosyl-6-thiopurine was synthesized as described by Xu et al. [22]. The compound was purified further by preparative layer chromatography.
Synthesis and purification of deoxyribonucleoside 5′-triphosphates
All of the non-standard deoxynucleoside 5′-triphosphates were synthesized using the procedure of Maybaum et al. [23]. Purification was carried out using HPLC with a weak anion-exchange column and a gradient of ammonium bicarbonate [6].
Oligodeoxynucleotide synthesis
The synthesis, purification, and nucleoside analysis of oligomers containing TH were as described previously [6]. To avoid any conversion of HS by chemical reagents, the oligomer containing HS was synthesized by an enzymic method [6]. The oligomers were purified by electrophoresis and the deoxynucleoside analysis used the method of Rappaport [6]. The sequences of all oligomers are listed in Table 1.
Table 1. Template and primer sequences.
T-X refers to a template label, and P-X refers to a primer label.
| Template/primer | Sequence |
|---|---|
| T-2 | TCGACGGACCCGTC |
| T-4 | GACACGGACCCGTC |
| T-6 | GATACGGACCCGTC |
| T-HS | GACGGGTCCGTHSAAAGTAGCGCTA |
| T-TH | TTTHACGGACCCGTC |
| T-H | GAHACGGACCCGTC |
| P-3 | GACGGGTCCG |
| P-4 | GACGGGTCCGT |
| P-5 | TAGCGCTACTTT |
General DNA polymerization reaction
The final concentrations for the T7p-catalysed reaction were 60 mM Tris/HCl, pH 7.6, 5 mM dithiothreitol, 10 mM MgCl2, 1 mg/ml BSA, 2–8 nM template, 2–8 nM primer, 0.4 unit/10 μl T7p, and the appropriate concentrations of the dNTPs. Reactions were started by the addition of polymerase, and incubated at 18.5 °C. Portions (1 μl) were removed at appropriate times and added to 8 μl of 90% (v/v) formamide containing 1 mM EDTA and 0.06% xylene cyanol dye for tracking during electrophoresis.
RESULTS
Scheme 1 shows the minimum number of kinetic steps assumed for T7p. The standard experiment was the addition of a saturating amount of T7p to annealed template and primer, and a specific dNTP. A temperature of 18.5 °C was chosen to lower the reaction rates to values that allowed convenient sampling of the reactions in time and to ensure the stability of the template/primer molecule. Using 32P-labelled primer, aliquots of the solution were taken at various times, and the amounts of primer with initial length n and extended length n+1 were determined from a PAGE separation.
Scheme 1. Kinetic steps for action of T7p.
For mathematical analysis, see the Appendix.
Mathematical description and experimental verification
The mathematical analysis of Scheme 1 is given in the Appendix. With the assumptions of saturating amounts of T7p for the template/primer molecules and the steady-state condition for the intermediate [polymerase(template/primer·dNTP); Dn·dNTP] and the product [polymerase (template/extended primer); Dn+1], the ratio of the concentration of the primer with length n+1 ([Dn+1]) to the concentration of the primer of length n ([Dn]+[Dn·dNTP]) is:
 |
(1) |
k2 is the rate constant for the intermediate, polymerase(template/primer·dNTP), adding the deoxynucleotide to the primer; k3 is the rate of removal of the added deoxynucleotide from the primer; and Km is the Michaelis constant for the dNTP. Scheme 1 only shows the decay of Dn to Dn−1. The detailed kinetics for the formation and degradation of primers with lengths less than n do not affect the steady-state analysis of primers with length n and n+1, because of two conditions: (1) the concentration of polymerase saturated the template/primer molecules; and (2) a primer of length n − 1 (Dn−1) cannot return to a primer of length n (Dn) due to the absence of the correct dNTP.
Fersht et al. [24] suggested that the efficiency of incorporation of a deoxynucleotide can be measured by Vmax/Km when employing the standard steady-state method. Vmax/Km is the rate constant for the incorporation of a deoxynucleotide when the concentration of the dNTP is much less than its Km. Boosalis et al. [25] used the ratio of the Vmax/Km of a mismatched deoxynucleotide to the Vmax/Km of the matched deoxynucleotide as a measure of the error frequency. When both polymerase and exonuclease activities are present, an appropriate measure should take into account both activities. An appropriate measure is the ratio of the apparent second-order rate constant for the reaction of the polymerase(template/primer), Dn, and a dNTP to give polymerase(template/extended primer), Dn+1, and the first-order rate constant for the removal of the added deoxynucleotide from the extended primer. The expression k2/Km is the second-order rate constant under steady-state conditions (eqn A6 and interpretation in the Appendix). The first-order rate constant for removal of the deoxynucleotide is k3. The expression k2/(Km×k3) is defined as the eni (efficiency of net incorporation). The experimental justifications for the assumption of a steady state and for the fact that a saturating concentration of the polymerase was used are given in the next paragraph.
An example of the experimental data is shown in Figure 2. The experiment measured the incorporation of hypoxanthine deoxynucleotide opposite the template base cytosine (primer P-4, template T-4; see Table 1). The X-ray film recorded the decay of 32P as phosphate attached to the 5′ end of the initial primer (11 mer), the extended primer (12 mer), and the first degraded length (10 mer), after PAGE separation of the oligomers. Because the PAGE separation had 7 M urea in the gel, the amount of 11 mer is the sum of Dn+Dn·dNTP. It was not necessary to take into account the fact that the primer in Dn was degraded continuously during the experiment. The reasons for this were: (1) only the ratio of the concentration of extended primer to the concentration of initial primer was used in the analysis; and (2) Dn−1 could not return to Dn because the correct dNTP was not present. The decreases in density in Figure 2 that occurred from 10 to 60 min were due to the increasing amounts of primer with lengths of less than n − 1 produced by the exonuclease activity. Each panel is labelled with the time of the sample (10, 40 or 60 min), and the concentration of the dNTP (μM) is indicated above each lane. Primer lengths of <10 are not shown in Figure 2. Table 2(B) records the data from the X-ray film shown in Figure 2. In addition, typical data are shown for the net incorporation of guanine deoxynucleotide opposite the template base cytosine (Table 2A) and the net incorporation of HS deoxynucleotide opposite the template base TH (Table 2C). The assumption for the derivation of eqn (1) that the steady-state condition holds for the intermediate Dn·dNTP and the product Dn+1 was verified by the constancy of the ratio of the amount of the extended primer (12 mer) to that of the initial primer (11 mer) from 10 to 60 min, even though there was continuous and significant degradation of the initial primer to shorter lengths with increasing time. The assumption that a saturating amount of T7p was present was demonstrated by the fact that no change occurred in the ratios with increasing amounts of T7p above 40 units/ml.
Figure 2. Products of polymerase and exonuclease activities.
Shown are X-ray film recordings of 32P decay, as 5′ phosphate, from PAGE separations of the initial primer (11 mer), the extended primer (12 mer), and a primer degraded by one nucleotide (10 mer). The incorporation is hypoxanthine deoxynucleotide opposite the template base cytosine (T-4/P-4; see Table 1) for reaction times of 0 min (A), 10 min (B), 40 min (C) and 60 min (D). The concentration of hypoxanthine dNTP in each reaction is shown above the lanes. The zero-time sample is overexposed to indicate no 12 mer. The contrast has been changed from that of the original X-ray films for publication.
Table 2. Ratio of amount of extended primer (12 mer) to initial primer (11 mer) as a function of time and dNTP concentration in reactions catalysed by T7p.
Data are from the density analysis of the PAGE separation of 32P (as 5′-phosphate)-labelled extended primer (12 mer) and initial primer (11 mer), using X-ray film. The three sets of data are: (A) incorporation of dG into primer opposite a template cytosine (T-4/P-4); (B) incorporation of hypoxanthine deoxynucleotide (dI) into primer opposite a template cytosine (T-4/P-4); (C) incorporation of HS deoxynucleotide (dIS) into a primer opposite a template, TH (T-TH/P-3). The ratio of the amount of extended primer to initial primer is given as a function of time and the concentrations of the dNTPs used in reactions catalysed by T7p.
| (A) dG/C | ||||
|---|---|---|---|---|
| Ratio (extended/initial) | ||||
| Concentration (μM) | 20 min | 40 min | 60 min | Average |
| 25 | 72 | 59 | 69 | 67 |
| 12.5 | 62 | 59 | 58 | 59 |
| 6.3 | 30 | 29 | 43 | 34 |
| 3.2 | 17 | 22 | 16 | 18 |
| (B) dI/C | ||||
| Ratio (extended/initial) | ||||
| Concentration | 10 min | 40 min | 60 min | Average |
| 125 | 13 | 8.4 | 7.3 | 9.5 |
| 62.5 | 8.0 | 7.5 | 8.3 | 7.9 |
| 31.5 | 9.0 | 6.1 | 6.9 | 7.1 |
| 15.7 | 3.2 | 5.3 | 5.3 | 4.6 |
| 7.8 | 2.3 | 2.5 | 2.2 | 2.3 |
| (C) dIS/TH | ||||
| Ratio (extended/initial) | ||||
| Concentration | 20 min | 40 min | 60 min | Average |
| 100 | 1.4 | 1.6 | 1.2 | 1.4 |
| 50 | 1.2 | 1.3 | 1.2 | 1.2 |
| 25 | 0.8 | 1.0 | 1.0 | 0.9 |
| 12.5 | 0.6 | 0.7 | 0.6 | 0.6 |
Sensitivity of assay
The sensitivity of the assay in terms of the eni, k2/(k3×Km), was estimated to be 2×10−5 μM−1. The calculation used a ratio of [Dn+1]/([Dn]+[Dn·dNTP]) of 1:200 from the experimentally determined trace amount that could be detected. A concentration of 500 μM was used for the dNTP, the maximum concentration employed to detect incorporation. It was assumed that trace incorporation was detected when the rate of product formation was half of the maximum rate, i.e. Km=[dNTP] in eqn (1).
Choice of deoxynucleotides
Preliminary experiments indicated that significant net incorporation of guanine deoxynucleotide opposite template TH occurred. Since model building indicated that guanine had the potential to form two hydrogen bonds with TH, guanine was replaced with hypoxanthine, which has the potential to form one hydrogen bond with TH but retains the potential for two hydrogen bonds with cytosine.
Kinetic parameters
The kinetic parameters k2/k3 and Km were determined from graphs of k3/v against 1/[dNTP], illustrated in Figure 3. The intercepts of the line with the k3/v axis and the 1/[dNTP] axis gave k3/k2 and −1/Km respectively. The values of v/k3 were computed from the experimentally determined amounts of Dn+1 and Dn as given by eqn (1). All combinations of the six bases were tested except the pairs C/A, C/T, C/C, T/T and A/A, a total of 31 combinations. Table 3 lists the values of k2/k3, Km and k2/(k3×Km) for those combinations of the six bases that gave detectable incorporation with T7p. dG/C, dG/T and dT/G are included for reference. For the base-pair combinations that did not give detectable net incorporation, the maximum concentration of the dNTP employed was 500 μM.
Figure 3. Plots of kinetics data from Table 2.
k3/v is plotted against the reciprocal concentration of dNTP. k3 is the rate of removal of the 3′ deoxynucleotide from the extended primer by the exonuclease, and v is the rate of addition of deoxynucleotide to the primer per unit enzyme concentration. (A) Incorporation of guanine deoxynucleotide opposite a template cytosine; (B) incorporation of hypoxanthine deoxynucleotide opposite a template cytosine; (C) incorporation of HS deoxynucleotide opposite a template TH.
Table 3. Summary of kinetic parameters for deoxynucleotides with net incorporation by T7p.
The kinetic parameters for the incorporation of a deoxynucleotide (dN) opposite a template base (B) with T7p are given. Template/primer sequences are listed in Table 1. k2 is the rate of incorporation of the deoxynucleotide by the polymerase, k3 is the rate of removal of the incorporated deoxynucleotide by the 3′→5′ exonuclease, and Km is the Michaelis constant for the dNTP. In an experiment separate from those determining the kinetic parameters, in addition to the designated deoxynucleotide a standard dNTP complementary to the template base following the designated template base was present in saturating amounts. The incorporation of the standard deoxynucleotide on to the extended primer under steady-state conditions is shown by: (+), good extension; (±), moderate extension; (−), no extension. All combinations of the bases adenine (A), thymine (T), hypoxanthine (H), cytosine (C), HS and TH were examined, except for A/C, C/T, C/C, T/T and A/A. The combinations dG/C, dG/T and dT/G are given for reference. The standard error was 0.4 or less for each parameter. n is the number of independent experiments. Only combinations with detectable incorporation are shown.
| dN/B | k2/k3 | Km (μM) | k2/(k3×Km) (μM−1) | Extension | Template/primer | n |
|---|---|---|---|---|---|---|
| dG/C | 1×102 | 1.3×10 | 7.7 | (+) | T-4/P-4 | 3 |
| dT/G | trace | – | ∼2×10−5 | trace | T-7/P-4 | 2 |
| dG/T | – | – | – | trace | T-6/P-4 | 2 |
| dA/T | 1×10 | 1.6×10 | 6×10−1 | (+) | T-6/P-4 | 2 |
| dT/A | 2×10 | 3.3×10 | 6.1×10−1 | (+) | T-2/P-3 | 2 |
| dIS/TH | 1.7 | 2.0×10 | 8.5×10−2 | (+) | T-TH/P-3 | 2 |
| dTH/HS | 8×10−2 | 1.1×10 | 7.3×10 −3 | (±) | T-HS/P-5 | 2 |
| dI/C | 9 | 1.8×10 | 5.0×10−1 | (+) | T-4/P-4 | 2 |
| dC/H | 4.5×10 | 5.0 | 9.0 | (+) | T-HP-4 | 2 |
| dT/H | trace | – | ∼2×10−5 | trace | T-H/P-4 | 2 |
| dI/T | – | – | – | trace | T-6/P-4 | 2 |
| dTH/H | trace | – | ∼2×10−5 | trace | T-H/P-4 | 2 |
| dI/TH | – | – | – | trace | T-TH/P-3 | 2 |
| dIS/C | see text | see text | <2×10−4 | trace | T-4/P-4 | 2 |
Potential sources of error
There are three major sources of possible error in determining the kinetic parameters: (1) if the concentration of the dNTP changed significantly during the reaction; (2) if the dNTP acted as an inhibitor of the exonuclease; and (3) if there were molecules of the primer or template/primer that could not be extended. Both possibilities 1 and 2 would lead to plots of k3/v against 1/[dNTP] that were not linear. If the concentration of the dNTP changed significantly during the reaction, the lowest initial concentration would show the greatest deviation from linearity. If the dNTP was an inhibitor of the exonuclease, the greatest effect would occur at the highest concentration. No deviations from linearity at the lowest concentration were observed for any case with detectable net incorporation, as illustrated in Figure 3. In one case, with HS deoxynucleotide opposite a template cytosine, inhibition was observed at a high concentration of HS dNTP. The third possible source of error, i.e. that some primer molecules could not be extended, would distort the measured ratio of [Dn+1]/([Dn]+[Dn·dNTP]) from the actual steady-state value. The distortion could be particularly significant at high concentrations of dNTP, where most of the extendable primer molecules would have a length of n+1. Primer molecules, as both primer and template/primer, were degraded rapidly to oligomers of lengths of two or three deoxynucleotides when no dNTP was present. More than 98% of the initial primer molecules were degraded within 10 min, and no primer molecules of length n were detected at 40 min. This result showed that all detectable primer molecules, regardless of whether or not they could be extended, were degraded by the exonuclease activity in 40 min. The sample data of Table 2 do not show a change in the measured ratio [Dn+1]/([Dn]+[Dn·dNTP]) from 10 to 60 min for any concentration of dNTP employed. The same was true for all incorporations that could be detected. If primer molecules that could not be extended made a significant contribution to the measured [Dn]+[Dn·dNTP] value during the time interval from 10 to 60 min, then the measured ratio [Dn+1]/([Dn]+[Dn·dNTP]) should have increased from 10 to 60 min as the non-extendable primer molecules were degraded.
Estimation of k2/(k3×Km) for trace incorporation
The sensitivity of detection with X-ray film of the extended primer, Dn+1, was 1/200 of the density of the initial primer, Dn. Assuming that trace net incorporation occurred when the velocity of production of the extended primer was half the maximum velocity ([dNTP]=Km), k2/k3 was 10−2 (eqn 1). Since trace incorporation occurred at a concentration of 500 μM dNTP, the estimate of k2/(k3×Km) was 2×10−5 μM−1.
Comparison of kinetic parameters and error frequencies
The values of k2/k3 for dIS (6-thiopurine deoxynucleoside)/TH and dTH/HS were much smaller than the values for dA/T, dT/A, dI (hypoxanthine deoxynucleoside)/C and dC/H. The values of Km for dIS/TH and dTH/HS were within the range of Kms for dA/T, dT/A, dI/C and dC/H. It is the low values of k2/k3 for dIS/TH and dTH/HS that place the eni [k2/(k3×Km)] of dIS/TH and dTH/HS much lower than that of the other two base pairs. The difference in k2/k3 between dIS/TH and dTH/HS, i.e. a factor of 20, is much greater than the difference between the values of k2/k3 for dI/C and dC/H, or dT/A and dA/T. An experiment separate from those determining the kinetic parameters was used to determine if extension of the primer beyond each of the two combinations of base pairs, for example dTH/HS and dIS/TH, could occur. In these experiments a standard dNTP at a saturating concentration was present in addition to the designated dNTP. The standard dNTP was complementary to the template base following the designated template base. A qualitative determination of the amount of incorporation of the standard deoxynucleotide opposite its complementary template base was made, with categories of good extension (+), moderate extension (±) and no extension (−) (Table 3). There was significant extension of the primer beyond each of the two combinations of the three base pairs, i.e. dT/A and dA/T, dI/C and dC/H, and dTH/HS and dIS/TH.
Trace net incorporation of the mismatch thymine deoxynucleotide or TH deoxynucleotide occurred opposite hypoxanthine. The eni of thymine deoxynucleotide or TH deoxynucleotide opposite hypoxanthine was 2×10−5 μM−1, and the eni of cytosine deoxynucleotide opposite hypoxanthine was 9 μM−1 (Table 3). The error frequency (the ratio of the mismatched to the matched eni) was one thymine deoxynucleotide or TH deoxynucleotide to 5×105 cytosine deoxynucleotides incorporated opposite hypoxanthine.
Only an estimate of the maximum eni of HS deoxynucleotide opposite cytosine could be calculated, because HS dNTP inhibited the exonuclease activity at high concentrations. The inhibition was manifested by a much lower rate of degradation of the primer into shorter lengths than was observed with a saturating concentration of guanine dNTP opposite a template cytosine. The calculation is based on the following results: (1) no detectable incorporation occurred up to 60 μM HS dNTP; and (2) at a concentration of 125 μM HS dNTP, net incorporation was observed, and inhibition of exonuclease activity occurred. To estimate the maximum eni, two conditions were assumed. (1) At 60 μM HS dNTP, a trace amount of 12 mer was present. The ratio of 12 mer/11 mer was then 1:200, the trace amount that was detectable. (2) For trace incorporation to have occurred, the rate of product formation was half of the maximum rate. Using eqn (1), the value of k2/(k3×Km), i.e. the eni, was less than 1.7×10−4 μM−1. The eni of hypoxanthine deoxynucleotide opposite cytosine was 5×10−1 μM−1. The error frequency was thus one HS deoxynucleotide to 3×103 hypoxanthine deoxynucleotides incorporated opposite cytosine. There was no net incorporation of cytosine deoxynucleotide opposite a template HS at 500 μM.
In the three cases of trace incorporation, i.e. dT/G, dT/H and dTH/H, trace extension of the primer beyond the base pairs was detected. Although no net incorporation of HS deoxynucleotide was detected opposite a template cytosine at a concentration of 60 μM, a trace amount of extension (Dn+2) occurred when a saturating amount of thymine dNTP was used opposite the next base of the template, adenine. No net incorporation of guanine or hypoxanthine deoxynucleotide was detected opposite a template thymine, but a trace amount of extension (Dn+2) was found in both cases. No other combination of bases showed extension to Dn+2 with saturating concentrations of a standard dNTP complementary to the next base in the template.
DISCUSSION
The goal of this investigation was to construct a set of three base pairs with the minimum change in the standard two base pairs that would have good fidelity of replication with a DNA polymerase. For an investigation of the fidelity of replication of new bases to have relevance for in vivo replication, both the accuracy of a polymerase activity and the proofreading of a 3′→5′ exonuclease activity should be evaluated together. T7p was chosen for the in vitro investigation because of its high polymerase and 3′→5′ exonuclease activities [17].
There are two properties that a base pair should possess in order to show significant net incorporation with a DNA polymerase that has strong 3′→5′ exonuclease activity: (1) a significant rate of incorporation of each deoxynucleotide occurs opposite its complementary template base; and (2) the base pair has sufficient stability that the rate of removal of the added deoxynucleotide from the primer by the 3′→5′ exonuclease activity is low. The results of Morales and Kool [26] showed that the base pair 2,4-difluorotoluene/adenine, with no hydrogen bonds between the bases, had a large difference in the efficiency of incorporation between Kf and T7p. Although the base pairs 2,4-difluorotoluene/adenine and 4-methylbenzimidazole/thymine have significant rates of incorporation with Kf, they do not always show stability towards a 3′→5′ exonuclease activity [15]. The latter result suggested that an essential property to incorporate into the design of a new base pair is stability towards a 3′→5′ exonuclease activity. Although base stacking interactions are important for stability, the specificity of different base-pair stabilities is determined by steric effects and hydrogen bonding [27].
A previous investigation [6] of the fidelity of replication of the two standard base pairs and 6-thioguanine/TH with Kf demonstrated very significant mismatched incorporation of the base pairs 6-thioguanine/cytosine and guanine/TH. Two suggestions to reduce the frequency of mismatch incorporations were to substitute HS for 6-thioguanine and to substitute 5-methyl-2-thiopyrimidinone for TH. Both of these suggestions result in one potential hydrogen bond between the mismatched pairs.
Two physical processes have been proposed to distinguish between matched and mismatched base pairs at the 3′ end of the primer when the primer is positioned at the polymerase site. Since only single-stranded DNA is bound at the exonuclease site, and the distance between the exonuclease site andthe polymerase site is 30 Å (3 nm) [12–14], both processes are assumed to allow at least four base pairs at the 3′ end of the primer to unwind from the template. The 3′ end of the primer then moves to the exonuclease site by Brownian motion, where the deoxynucleotide is removed. The physical processes are as follows: (1) the interactions of the template and primer bases at the 3′ end of the primer determine the probability of the ends fraying [28,29]; and (2) the interactions of amino acid side chains of the polymerase with the bases are different for matched and mismatched bases [30]. With T7p, two important interactions are hydrogen bonds between Arg-429 and Gln-615 with the N-3 of the purine ring and the O-2 of thymine or cytosine [11,14]. Because of the strong experimental evidence for both processes, their joint requirements for stability essentially determined the choice of what base change to make. The substitution of 5-methyl-2-thiopyrimidinone for TH in a base pair with a purine would remove the stabilizing interaction of a hydrogen bond between the polymerase and the O-2 atom of the pyrimidine. In addition, the high tendency of the sulphur atom to polarize could increase base stacking, and possibly cause a mismatch of a 5-methyl-2-thiopyrimidinone deoxynucleotide with a template adenine to be too stable. The other possibility to decrease the stability of the base pair guanine/TH was to change guanine. The choice was to substitute hypoxanthine for guanine. The two potential hydrogen bonds of guanine deoxynucleotide opposite a template TH were reduced in number to one potential hydrogen bond between hypoxanthine deoxynucleotide and the template TH. However, hypoxanthine retained the potential to form two hydrogen bonds with cytosine. The experimental results showed no detectable net incorporation of hypoxanthine deoxynucleotide opposite a template TH, and only trace incorporation with the opposite configuration (Table 3). The error frequency of the incorporation of TH compared with the incorporation of cytosine opposite hypoxanthine was 2×10−6. The stability of the hypoxanthine/cytosine base pair was maintained, and very good net incorporation occurred (Table 3). To reduce the stability of the base pair 6-thioguanine/cytosine, HS was substituted for 6-thioguanine. The use of HS instead of 6-thioguanine lowered the number of potential hydrogen bonds with the mismatched base cytosine from two to one. No net incorporation of cytosine deoxynucleotide opposite a template HS was found, and the eni of HS deoxynucleotide opposite a template cytosine was estimated to be less than 3×10−4 μM−1.
The base pairs HS/TH and H/TH each have the potential to form one hydrogen bond. However, H/TH showed only trace net incorporation, while HS/TH showed significant net incorporation in both configurations, with dIS/TH having a value of k2/k3 that was only a factor of 5 less than that of dI/C. There are two significant differences between sulphur and oxygen in the context of incorporation and the stability of a base pair towards 3′→5′ exonuclease activity. (1) Gas-phase spectroscopy [31] measurements show that the force of interaction of hydrogen with sulphur via a hydrogen bond is half that of hydrogen with oxygen, and that the angle with the maximum interaction, measured from the carbon–sulphur (oxygen) bond, is 90° with sulphur compared with 45° with oxygen. (2) X-ray structure determinations of crystals of thiopurines and thiopyrimidines are interpreted to indicate strong stacking interactions due to the high polarizability of the sulphur atom [32,33]. Both base pairs, H/TH and HS/TH, have N-3 and O-2 atoms that can accept hydrogen bonds from the polymerase. The polarizability of the sulphur on HS has the potential to cause a significant stacking interaction with the adjacent base. This stacking interaction would contribute most significantly when HS is at the 3′ end of the primer, because this is the deoxynucleotide excised if the 3′ end of the primer reaches the exonuclease site. The two configurations of the base pair, dIS/TH and dTH/HS, showed a 20-fold difference in the value of k2/k3, with the value for dIS/TH being greater. This large difference can be understood on the basis of the large stacking interaction of HS with the previous base at the 3′ end of the primer. The stacking interaction would lower the probability of the primer unwinding from the template and then moving to the exonuclease site. Kool [34] has suggested that water molecules bound to amino and oxygen groups of the bases through hydrogen bonds may contribute to discrimination between matched and mismatched incorporation by steric interference. If hypoxanthine had a water molecule bound to its O-6 atom, then there is no appropriate group on TH to displace the water molecule. On the basis of the gas-phase measurement, it is possible that the interaction energy of a hydrogen bond between S-6 on HS and water could be half the interaction energy between O-6 on hypoxanthine and water. Assuming the interaction energy of the hydrogen bond between O-6 of hypoxanthine and a water molecule to be 21 kJ/mol (5 kcal/mol), the probability of HS having a bound water molecule compared with hypoxanthine having a bound water molecule is 1.8×10−2 at 300 K. If O-6 and S-6 interact with two water molecules, and both must be removed for correct base pairing to occur, then the ratio of the probabilities becomes 3.2×10−4. From this model, when the base HS approaches the template base TH, it carries a water molecule less often than the base hypoxanthine.
The frequency of incorporation of the mismatched base pair G/T compared with the matched base pair G/C for a large number of DNA polymerases with and without 3′→5′ exonuclease activity ranges from 10−3 to 10−5. The frequency of the net incorporation of dT/G compared with dC/G with T7p was estimated to be 1.6×10−5 (Table 3). The frequency for dT/H was the same. The opposite configurations, i.e. dG/T and dI/T, were detectable when a saturating amount of a standard dNTP was present for the next primer position opposite a complementary template base. Given the usually assumed geometry for the base pair T/G [35], i.e. a hydrogen bond between O-2 of thymine and H-1 of guanine, and a hydrogen bond between H-3 of thymine and O-6 of guanine, it would be expected that dT/G and dT/H would have the same error frequency, because the 2-amino group of guanine in the template does not interact with the thymine, and there is probably no bridging water between the 2-amino group of guanine and O-2 of thymine in the catalytic site of the polymerase. In the opposite configurations, i.e with guanine and hypoxanthine as entering deoxynucleotides, the 2-amino group does not interact with the thymine or the polymerase.
T7p is a processive polymerase [16]. Huber et al. [36] showed that the degree of processivity increased with increasing length of the template/primer molecule. Patel et al. [37] stated that the strength of binding of a templete/primer molecule to T7p decreased when the length was less than a 32/25 mer. The present investigation used template/primer molecules of 14/10 and 24/12 mer, and the degree of processivity was not determined. The level of processivity should not have affected the measurement of the rate of addition of a deoxynucleotide to the primer, or the unwinding of the 3′ end of the primer from the template and its movement to the exonuclease site. This is because the template/primer molecules always had T7p bound, there was a saturating concentration of T7p, and only a single deoxynucleotide extension occurred.
Although error frequency is dependent on deoxynucleotide sequence [38] and the sequence context was not examined in the present set of experiments, analysis of the results suggests that the set of three base pairs adenine/thymine, hypoxanthine/cytosine and HS/TH may have adequate fidelity for use in replication and as a basis for a genetic alphabet comprising three base pairs.
Acknowledgments
I thank the former provost of Temple University, Barbara Brownstein, for intellectual interest and administrative support; David George, Richard Waldon and Ryan Yip for initial experiments before starting their own projects; Bill Brinigar for many useful technical discussions; and Karen Palter, Richard C. Waring and Greg Smutzer for important comments about the manuscript. The experimental work was supported by a grant from Temple University and a private foundation.
APPENDIX
The minimum number of kinetic steps assumed for T7p are shown at the beginning of the Results section in Scheme 1. The Dns are the template/primers with primers of length n. There are four essential assumptions, as follows. (1) Every template/primer molecule has a bound polymerase molecule. (2) The intermediate (Dn·dNTP) and product (Dn+1) are essentially in the steady state, i.e. the k4 rate constant is sufficiently small compared with k1 that the cycle is maintained near a steady-state level. (3) The concentration of the dNTP does not change significantly during the reaction. (4) Dn−1 cannot return to Dn because of the absence of the appropriate dNTP. The rate equations for Scheme 1 are:
 |
(A1) |
 |
(A2) |
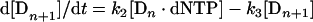 |
(A3) |
Eqns (A2) and (A3) with the steady-state condition give:
 |
(A4) |
where Km=(k−1+k2)/k1
 |
(A5) |
Substituting for [Dn·dNTP] in eqn (A5) with the left side of eqn (A4) gives:
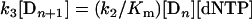 |
(A6) |
Since at steady state the concentration of Dn+1 is not changing, the velocity of loss of Dn+1 is equal to the velocity of production of Dn+1. Eqn (A6) states that the velocity of loss of Dn+1 (left side) is equal to the velocity of production (right side). Thus k2/Km on the right side can be interpreted as the second-order rate constant at steady state for polymerase(template/primer), Dn, and dNTP going to polymerase(template/extended primer), Dn+1. Using eqn (A6) for Dn+1 in terms of [Dn][dNTP] and eqn (A4) for [Dn·dNTP] in terms of [Dn][dNTP] gives:
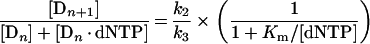 |
(A7) |
Setting v=k2/(1+Km/[dNTP]) gives:
 |
(A8) |
i.e. eqn (1) in the main text. v may be interpreted as the rate of product formation per unit of enzyme concentration.
For completeness, the equation that describes the fraction of primer present as the sum Dn+Dn·dNTP+Dn+1, as a function of time after the steady-state condition occurs, is given below (eqn A9). The equation is derived from eqn (A1), the conservation in time of the sum of primers of all lengths (1 to n+1), and the steady-state conditions expressed by eqns (A4) and (A5). For two times, t1 and t2, where t2 > t1, the fraction of primer with lengths n and n+1 (f) is given by:
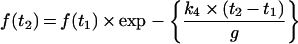 |
(A9) |
where g=1+([dNTP]/Km)×[1+(k2/k3)].
References
- 1.Tac E. L., Wu Y., Xia G., Schultz P. G., Romesburg F. E. Efforts toward expansion of the genetic alphabet: replication of DNA with three base pairs. J. Am. Chem. Soc. 2000;123:7439–7440. doi: 10.1021/ja010731e. [DOI] [PubMed] [Google Scholar]
- 2.Bergen M., Wu Y., Ogawa A. K., McMinn D. L., Schultz P. G., Romesburg F. E. Universal bases for hybrization, replication, and chain termination. Nucleic Acids Res. 2000;28:2911–2914. doi: 10.1093/nar/28.15.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo M. J., Hildbrand S., Leumann C. L., McLaughlin L. W., Waring M. Inhibition of DNA polymerase reactions by pyrimidine nucleotide analogues lacking the 2-keto group. Nucleic Acids Res. 1998;26:1863–1869. doi: 10.1093/nar/26.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lutz M. J., Held H. A., Hottiger M., Hubscher U., Benner S. A. Differential discrimination of DNA polymerases for variants of the non-standard nucleobase pair between xanthosine and 2,4-diaminopyrimidine, two components of an expanded genetic alphabet. Nucleic Acids Res. 1996;24:1308–1313. doi: 10.1093/nar/24.7.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horlacher J., Hottiger M., Podust V. M., Hubscher U., Benner S. A. Recognition by viral and cellular DNA polymerases of nucleosides bearing bases with non-standard hydrogen bonding patterns. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6329–6334. doi: 10.1073/pnas.92.14.6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rappaport H. P. Replication of the base pair 6-thioguanine/5-methyl-2-pyrimidinone with the large Klenow fragement of Escherichia coli DNA polymerase I. Biochemistry. 1993;32:3047–3057. doi: 10.1021/bi00063a016. [DOI] [PubMed] [Google Scholar]
- 7.Piccirilli J. A., Krauch T., Moroney S. E., Benner S. A. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature (London) 1990;343:33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- 8.Switzer C. Y., Moroney S. E., Benner S. A. Enzymatic incorporation of a new base pair into DNA and RNA. J. Am. Chem. Soc. 1989;111:8322–8327. [Google Scholar]
- 9.Moran S., Ren R. X., Kool E. T. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morales J. C., Guckian K. M., Sheils C. J., Kool E. T. Structure and base pairing properties of a replication-competent nonpolar isostere for deoxyadenosine. J. Org. Chem. 1998;63:9652–9656. doi: 10.1021/jo9805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublie S., Tabor S., Long A. M., Richardson C. C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature (London) 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 12.Kunkel T. A., Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2001;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 13.Patel P. H., Suzuki M., Adman E., Shinkai A., Loeb L. A. Prokaryotic DNA polymerase I: evolution, structure, and base flipping mechanism for nucleotide selection. J. Mol. Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 14.Doublie S., Swaya M. R., Ellenberger T. An open and closed case for all polymerases. Structure. 1999;7:1731–1735. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 15.Morales J. C., Kool E. T. Importance of terminal base pair hydrogen-bonding in 3′-end proofreading by the Klenow fragment of DNA polymerase I. Biochemistry. 2000;39:2626–2636. doi: 10.1021/bi992173a. [DOI] [PubMed] [Google Scholar]
- 16.Tabor S., Huber H. E., Richardson C. C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of gene 5 protein of bacteriophage T7. J. Biol. Chem. 1987;262:16212–16223. [PubMed] [Google Scholar]
- 17.Hori K., Mark D. F., Richardson C. C. Deoxyribonucleic acid polymerase of bacteriophage T7. Characterization of the exonuclease activities of the gene 5 protein and the reconstituted polymerase. J. Biol. Chem. 1979;254:11598–11604. [PubMed] [Google Scholar]
- 18.Kunkel T. A., Patel S. S., Johnson K. A. Error-prone replication of repeated DNA sequences by T7 DNA polymerase in the absence of its processivity subunit. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6830–6834. doi: 10.1073/pnas.91.15.6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu D. R., Schultz P. G. Progress towards the evolution of an organism with an expanded genetic code. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4780–4785. doi: 10.1073/pnas.96.9.4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowal A. K., Köhrer C., RajBhandary U. L. Twenty-first aminoacyl-tRNA synthetase-suppressor tRNA pairs for possible use in site-specific incorporation of amino acid analogues into proteins in eukaryotes and in eubacteria. Proc. Natl. Acad. Sci. U.S.A. 2000;98:2268–2273. doi: 10.1073/pnas.031488298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rappaport H. P. The 6-thioguanine/5-methyl-2-pyrimidinone base pair. Nucleic Acids Res. 1988;16:7253–7267. doi: 10.1093/nar/16.15.7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y.-Z., Zheny Q., Swann R. F. Synthesis and duplex stability of oligodeoxynucleotides containing 6-mercaptopurine. Tetrahedron Lett. 1992;33:5837–5840. [Google Scholar]
- 23.Maybaum J., Bainnson A. N., Roethal W. M., Ajmera S., Iwaniee L. M., TerBush D. R., Kroll J. Effects of incorporation of 6-thioguanine into SV40 DNA. Mol. Pharmacol. 1987;32:606–614. [PubMed] [Google Scholar]
- 24.Fersht A. R., Knill-Jones J. W., Tsui W. C. Kinetic bases of spontaneous mutation: misinsertion frequencies, proofreading specificities and cost of proofreading by DNA polymerase of Escherichia coli. J. Mol. Biol. 1982;156:37–48. doi: 10.1016/0022-2836(82)90457-0. [DOI] [PubMed] [Google Scholar]
- 25.Boosalis M. S., Petruska J., Goodman M. F. DNA polymerase insertion fidelity. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 26.Morales J. C., Kool E. T. Varied molecular interactions in the active sites of several DNA polymerases: non polar nucleoside isosteres as probes. J. Am. Chem. Soc. 2000;122:1001–1007. doi: 10.1021/ja993464+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kool E. T. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Brenowitz S., Kwack S., Goodman M. F., O'Donnell M., Echols H. Specificity and enzymatic mechanism of the editing exonuclease of Eschericha coli DNA polymerase III. J. Biol. Chem. 1991;266:7888–7892. [PubMed] [Google Scholar]
- 29.Goodman M. F. Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minnick D. T., Liu L., Grindley N. D. F., Kunkel T. A., Joyce C. M. Discrimination against purine-pyrimidine mispairs in the polymerase active site of DNA polymerase I. A structural explanation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1194–1199. doi: 10.1073/pnas.032457899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leon A. C., Miller D. J. Directional character, strength, and nature of the hydrogen bond in gas phase dimers. Acc. Chem. Res. 1987;20:39–46. [Google Scholar]
- 32.Saenger W., Suck D. The relationship between hydrogen bonding and base stacking in crystalline 4-thiouridine derivatives. Eur. J. Biochem. 1973;32:473–478. doi: 10.1111/j.1432-1033.1973.tb02630.x. [DOI] [PubMed] [Google Scholar]
- 33.Thewalt U., Bugg C. E. Effects of sulfur substituents on base stacking and hydrogen bonding. The crystal structure of 6-thioguanosine monohydrate. J. Am. Chem. Soc. 1972;94:8892–8898. [Google Scholar]
- 34.Kool E. T. A mechanism for shape selection during DNA synthesis. Biopolymers. 1998;48:3–17. doi: 10.1002/(SICI)1097-0282(1998)48:1<3::AID-BIP2>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Hunter W. N., Brown T., Kneale G., Anand N. N., Rabinowitz D., Kennard O. The structure of guanosine-thymidine mismatches in B-DNA at 2.5-Å resolution. J. Biol. Chem. 1987;262:9962–9966. doi: 10.2210/pdb113d/pdb. [DOI] [PubMed] [Google Scholar]
- 36.Huber H. E., Tabor S., Richardson C. C. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J. Biol. Chem. 1987;262:16224–16232. [PubMed] [Google Scholar]
- 37.Patel S. S., Wong I., Johnson K. Kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 38.Mendelman L. V., Boosalis M. S., Petruska J., Goodman M. F. Nearest neighbor influence on DNA polymerase insertion fidelity. J. Biol. Chem. 1989;270:4759–4774. [PubMed] [Google Scholar]






