Abstract
Cardiac hypertrophy and remodelling in chagasic disease might be associated with mitochondrial dysfunction. In the present study, we characterized the cardiac metabolic responses to Trypanosoma cruzi infection and progressive disease severity using a custom-designed mitoarray (mitochondrial function-related gene array). Mitoarrays consisting of known, well-characterized mitochondrial function-related cDNAs were hybridized with 32P-labelled cDNA probes generated from the myocardium of mice during immediate early, acute and chronic phases of infection and disease development. The mitoarray successfully identified novel aspects of the T. cruzi-induced alterations in the expression of the genes related to mitochondrial function and biogenesis that were further confirmed by real-time reverse transcriptase–PCRs. Of note is the up-regulation of transcripts essential for fatty acid metabolism associated with repression of the mRNAs for pyruvate dehydrogenase complex in infected hearts. We observed no statistically significant changes in mRNAs for the enzymes of tricarboxylic acid cycle. These results suggest that fatty acid metabolism compensates the pyruvate dehydrogenase complex deficiencies for the supply of acetyl-CoA for a tricarboxylic acid cycle, and chagasic hearts may not be limited in reduced energy (NADH and FADH2). The observation of a decrease in mRNA level for several subunits of the respiratory chain complexes by mitoarray as well as global genome analysis suggests a limitation in mitochondrial oxidative phosphorylation-mediated ATP-generation capacity as the probable basis for cardiac homoeostasis in chagasic disease.
Keywords: chagasic cardiomyopathy, custom-designed microarray, gene expression, mitochondria, Trypanosoma cruzi
Abbreviations: AMPK, AMP-activated protein kinase; BZRP, peripheral benzodiazepine receptor; CCM, chronic chagasic cardiomyopathy; CS, citrate synthase; dpi, days post-infection; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MPTP, mitochondrial membrane transition pore; PDC, pyruvate dehydrogenase complex; RT, reverse transcriptase; SDH, succinate-ubiquinone oxidoreductase; VDAC, voltage-dependent anion channel
INTRODUCTION
Trypanosoma cruzi is estimated to infect approx. 20 million people in Latin America, and an additional 100 million people are at risk of infection in endemic countries [1,2]. The ultimate consequence of T. cruzi infection is the development of CCM (chronic chagasic cardiomyopathy), considered to be the major cause of congestive heart failure and sudden death among young adults [3]. Despite decades of research, limited information is available on the cellular and molecular mechanisms by which cardiovascular structure and functions are adversely affected in T. cruzi-infected patients and experimental models. In recent studies, we [4] and others [5] have profiled the changes in host-gene expression associated with progressive cardiomyopathy in experimental models of T. cruzi infection. Many of the differentially expressed genes identified in these studies were found to encode proteins associated with cardiac hypertrophic and remodelling processes and might have implications in decreased cardiac performance with disease severity. The striking feature of these studies is, however, the observation of changes in the expression of a variety of genes encoding components of the respiratory complexes in infected murine hearts [4], thus providing the first indication of mitochondrial defects in CCM.
The heart, a highly oxidative tissue, is essentially dependent on mitochondria that provide ∼90% of the cellular energy required for contractile and other metabolic activities of the myocardium [6,7]. The β-oxidation of fatty acids, tricarboxylic acid cycle and respiratory chain-mediated oxidative phosphorylation, the three important linked metabolic pathways of energy, are all carried out in mitochondria [7–10]. The fatty acid β-oxidation and tricarboxylic acid cycle are highly efficient processes for the generation of reduced high-energy electron carriers NADH and/or FADH2. The NADH and FADH2 are oxidized by the respiratory chain and the electron energy utilized in oxidative phosphorylation and ATP synthesis. Consequently, a defect in any of the constituents of the fatty acid β-oxidation pathway, tricarboxylic acid cycle and/or the respiratory chain that are involved in normal mitochondrial function may compromise the oxidative phosphorylation capacity and ATP production [11–13] and potentially impair cardiac performance in CCM patients.
We report here our ongoing genomic analysis to specify mitochondrial dysfunction in the myocardium exposed to various stimuli. Our first goal was to develop a custom mitoarray (mitochondrial function-related gene array) prototype that can be used to identify the molecular and physiological changes in cardiac mitochondria in response to various toxic and pathogenic stimuli. All of the genes printed on the arrays are related to mitochondrial function and are representative of various metabolic pathways. We demonstrate the usefulness of these arrays in time-course studies and diagnosis of mitochondrial dysfunction with progressive development of T. cruzi-induced chagasic cardiomyopathy. Mitoarray results were confirmed by traditional and/or real-time RT (reverse transcriptase)–PCR approaches. Our results implicate a specific pattern of mitochondrial metabolic alterations with cardiac homoeostasis in CCM and raise the possibility that treatments capable of reducing cardiac pathology in chagasic patients may exert their effects through normalizing at least some of the changes associated with mitochondrial function.
MATERIALS AND METHODS
Mice and parasites
C3H/HeN male mice (6–8-week old) were purchased from Harlan (Indianapolis, IN, U.S.A.). T. cruzi (Sylvio X10/4 strain) and C2C12 cells (murine skeletal-muscle cell line) were purchased from A.T.C.C. T. cruzi trypomastigotes were maintained and propagated by the continuous in vitro passage of parasites in monolayers of C2C12 cells [14]. Mice were infected by intraperitoneal injection of 50 000 culture-derived trypomastigotes. Animal experiments were performed according to the National Institutes of Health Guide for Care and Use of Experimental Animals and approved by the UTMB Animal Care and Use Committee.
Plasmid library construction and mitoarray printing
To produce a prototype of the custom-made cDNA mitoarray, a total of 95 genes were selected (Table 1). Genes, encoding GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and β-actin, and media blanks were incorporated as positive and negative controls respectively. All genes were PCR-amplified using murine cDNA as a template and gene-specific oligonucleotides as forward and reverse primers. All primers were designed with the use of Primer 3 v 0.2 software and purchased from Sigma-Genosys. PCRs were performed using a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, U.S.A.) in total volume of 25 μl, containing 2.5 units Taq polymerase (Invitrogen, New York, NY, U.S.A.), 50 ng of cDNA template and 200 nM gene-specific primer pairs with the following cycling conditions: a 95 °C predenaturation/activation step for 5 min, followed by 30 cycles at 94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and 72 °C final extension for 10 min. Individual amplicons were electrophoresed on 1.5% agarose gels, stained with 0.5 μg/ml ethidium bromide, and visualized under long-wave UV light using a MultiImage Light Cabinet (Alpha Innotech, San Leandro, CA, U.S.A.). All PCR amplicons (average size, 600 bp) were TA-cloned in PCR 2.1-TOPO vector (Invitrogen). Recombinant plasmids were transformed in One Shot Chemically Competent Escherichia coli (Invitrogen), and glycerol stocks were prepared in 96-well plates and stored at − 80 °C. The sequence of the successfully cloned amplicons was confirmed using an ABI Prism 310 Genetic Analyzer (Applied Biosystems) in the UTMB Recombinant DNA Core Facility.
Table 1. Genes included on the custom-designed mitoarrays.
LFAIM, lipid, fatty acid and isoprenoid metabolism; VCPM, vitamin, cofactor and prosthetic-group metabolism; ETC, electron-transport chain.
| Orientation grid | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | A9 | A10 | A11 | A12 |
| B1 | B2 | B3 | B4 | B5 | B6 | B7 | B8 | B9 | B10 | B11 | B12 |
| C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | C11 | C12 |
| D1 | D2 | D3 | D4 | D5 | D6 | D7 | D8 | D9 | D10 | D11 | D12 |
| E1 | E2 | E3 | E4 | E5 | E6 | E7 | E8 | E9 | E10 | E11 | E12 |
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | F11 | F12 |
| G1 | G2 | G3 | G4 | G5 | G6 | G7 | G8 | G9 | G10 | G11 | G12 |
| H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | H10 | H11 | H12 |
| Spot | Gene name | Product | NCBI acc. no. | Functional classification | |||||||
| A1 | ACADL | Acyl-CoA dehydrogenase, long chain | U21489 | LFAIM | |||||||
| A2 | ACADM | Acyl-CoA dehydrogenase, medium chain | U07159 | LFAIM | |||||||
| A3 | ACADS | Acyl-CoA dehydrogenase, short chain | L11163 | LFAIM | |||||||
| A4 | AIF | Apoptosis-inducing factor | AF100927 | Cell defence, cell death and aging | |||||||
| A5 | ALAS2 | 5-Aminolaevulinate synthase | M63244 | VCPM | |||||||
| A6 | ALDH2 | Aldehyde dehydrogenase 2 | U07235 | C-compound and carbohydrate metabolism | |||||||
| A7 | ANT1 | ADP-ATP carrier protein T1, heart | U27315 | Nuclear metabolism and transport | |||||||
| A8 | ANT2 | ADP-ATP carrier protein T2, fibroblast | U27316 | Nuclear metabolism and transport | |||||||
| A9 | AOP1/2 | MER5 protein | M28723 | Cell defence, cell death and aging | |||||||
| A10 | ATP5A1 | H+-transporting ATP synthase α | L01062 | Mitochondrial transport, transport ATPase | |||||||
| A11 | ATP5G1 | ATP synthase lipid-binding protein P1 | L19737 | Mitochondrial transport, transport ATPase | |||||||
| A12 | ATP7B | Copper transporting P-type ATPase | U38477 | Metal ion transport, transport ATPase | |||||||
| B1 | BAX | Apoptosis regulatory protein membrane α | L22472 | Cell defence, cell death and aging | |||||||
| B2 | BCKDHA | α-Oxoacid dehydrogenase E1α | L47335 | Amino acid metabolism | |||||||
| B3 | BCKDHB | 3-Methyl-2-oxobutanoate dehydrogenase | L16992 | Amino acid metabolism | |||||||
| B4 | BCL2 | Transforming protein bcl 2-β | M16506 | Cell defence, cell death and aging | |||||||
| B5 | BZRP | Peripheral-type benzodiazepine receptor 1 | D21207 | LFAIM | |||||||
| B6 | CAR5 | Carbonate dehydratase, hepatic | X51971 | C-compound and carbohydrate metabolism | |||||||
| B7 | CKMT1 | Creatine kinase (mitochondrial) | Z13968 | Energy (phosphate) metabolism | |||||||
| B8 | COX4 | Cytochrome c oxidase, chain IV | M37829 | Energy metabolism, respiration, ETC | |||||||
| B9 | COX7A2 | Cytochrome c oxidase, chain VIIa | X58486 | Energy metabolism, respiration, ETC | |||||||
| B10 | COX8A | Cytochrome c oxidase, chain VIII | U37721 | Energy metabolism, respiration, ETC | |||||||
| B11 | CPO | Coproporphyrinogen oxidase | D16333 | VCPM, plasma membrane biogenesis | |||||||
| B12 | CPT2 | Carnitine-O-palmitoyltransferase II, mitochondrial | U01163 | LFAIM, mitochondrial lipid transport | |||||||
| C1 | CRAT | Carnitine-O-acetyltransferase | X85983 | Mitochondrial lipid transport | |||||||
| C2 | CYCS | Cytochrome c, somatic | M20625 | Energy metabolism, respiration | |||||||
| C3 | DBT | Dihydrolipoamide transacylase | L42996 | C-compound and carbohydrate metabolism | |||||||
| C4 | DCI | 3,2-trans-Enoyl-CoA isomerase, mitochondrial | Z14049 | LFAIM | |||||||
| C5 | DLD | Dihydrolipoamide dehydrogenase E3 | U73445 | Amino acid and carbohydrate metabolism, tricarboxylic acid cycle | |||||||
| C6 | FDX1 | Adrenodoxin precursor | D43689 | LFAIM | |||||||
| C7 | FDXR | Ferredoxin–NADP+ reductase | D49920 | LFAIM | |||||||
| C8 | Blank | ||||||||||
| C9 | FPGS | Tetrahydrofolylpolyglutamate synthase | U32197 | VCPM | |||||||
| C10 | FRDA | Friedreich ataxia (cellulase) | U95736 | Mitochondria transport | |||||||
| C11 | GCDH | Glutaryl-CoA dehydrogenase | U18992 | Amino acid metabolism | |||||||
| C12 | GLUD | Glutamate dehydrogenase (NADP+) | X57024 | Amino acid metabolism, N-S metabolism | |||||||
| D1 | GOT2 | Glutamate oxaloacetate transaminase 2 | J02622 | Nitrogen sulphur metabolism | |||||||
| D2 | HADH | 3-Hydroxyacyl-CoA dehydrogenase, short chain | D29639 | LFAIM | |||||||
| D3 | HCCS | Cytochrome c type haem lyase | U36788 | Energy metabolism, respiration | |||||||
| D4 | HK1 | Hexokinase I | J05277 | Carbohydrate metabolism, glycolysis | |||||||
| D5 | HMGCL | Hydroxymethylglutaryl-CoA lyase | S65036 | Amino acid metabolism | |||||||
| D6 | HMGCS2 | Hydroxymethylglutaryl-CoA synthase, mit. | U12790 | Amino acid metabolism | |||||||
| D7 | HSC70T | Heat-shock protein cognate 70, testis | D85732 | NA transport, cell defence, cell-wall biogenesis | |||||||
| D8 | HSD3B1 | 3-β-Hydroxy-5-ene steroid dehydrogenase type I | M58567 | LFAIM | |||||||
| D9 | HSP60 | Heat-shock protein 60 (groEL) precursor | X55023 | Protein folding and transport, cell defence/death | |||||||
| D10 | HSP70-1 | Heat-shock protein, 70K (hsp68) | M35021 | Protein folding and transport, cell-wall biogenesis | |||||||
| D11 | HSP74 | Heat-shock protein, 70 precursor | D17556 | Protein folding and transport, transcription, processing | |||||||
| D12 | HSPE1 | Chaperonin-10 | U09659 | Protein transport and trafficking | |||||||
| E1 | IDH2 | Isocitrate dehydrogenase (NADP+), mitochondrial | U51167 | C-compound and carbohydrate metabolism, tricarboxylic acid cycle | |||||||
| E2 | TIM44 | Mitochondrial inner-membrane import protein | U69898 | Mitochondrial protein transport and trafficking | |||||||
| E3 | MDH1 | Malate dehydrogenase, mitochondrial | M16229 | C-compound and carbohydrate metabolism, tricarboxylic acid cycle | |||||||
| E4 | CBR2 | Carbonyl reductase (NADPH) | D26123 | Unclassified | |||||||
| E5 | COX6A1 | Cytochrome c oxidase chain VIa, heart | U08439 | Energy metabolism, respiration, ETC | |||||||
| E6 | MTHFD | Methylenetetrahydrofolate DH | J04627 | Unclassified | |||||||
| E7 | MUT | Methylmalonyl-CoA mutase α | X51941 | Amino acid metabolism | |||||||
| E8 | NNT | NADP+ transhydrogenase (β-specific) | Z49204 | Unclassified | |||||||
| E9 | OAT | Ornithine–oxoacid transaminase | X64837 | Amino acid metabolism | |||||||
| E10 | OIAS1 | 2′-5′-Oligoadenylate synthetase 1 | M33863 | Unclassified | |||||||
| E11 | OTC | Ornithine carbamoyltransferase | M17030 | Amino acid metabolism, cell-wall biogenesis | |||||||
| E12 | PCX | Pyruvate carboxylase | L09192 | Carbohydrate metabolism, glycolysis, LFAIM | |||||||
| F1 | PDHA1 | Pyruvate dehydrogenase (lipoamide) | M76727 | Carbohydrate metabolism, glycolysis | |||||||
| F2 | SDH1 | Succinate dehydrogenase, subunit b | BC013509 | Energy metabolism, respiration, ETC | |||||||
| F3 | POLG | DNA polymerase, γ | U53584 | DNA synthesis and replication | |||||||
| F4 | SDH2 | Succinate dehydrogenase, subunit a | XM_127445 | Energy metabolism, respiration, ETC | |||||||
| F5 | SDHC | Succinate dehydrogenase, subunit CII-3 | NM_025321 | Energy metabolism, respiration, ETC | |||||||
| F6 | SCP2 | Sterol carrier protein X | M91458 | Lipid transporter | |||||||
| F7 | SOD2 | Superoxide dismutase (Mn2+) | X04972 | Cell death and cell defence | |||||||
| F8 | STAR | Steroidogenic acute regulatory protein | L36062 | LFAIM | |||||||
| F9 | TFAM | Mitochondrial transcription factor A | U57939 | Mitochondrial DNA replication, transport | |||||||
| F10 | TST | Thiosulphate–sulphur transferase | U35741 | Nitrogen sulphur metabolism | |||||||
| F11 | UNG | Uracil DNA glycosylase | P97931 | Recombination and DNA repair | |||||||
| F12 | VDAC1 | VDAC1 | U30840 | Mitochondrial transport | |||||||
| G1 | VDAC2 | VDAC2 | U30838 | Mitochondrial transport | |||||||
| G2 | VDAC3 | VDAC3 | U30839 | Mitochondrial transport | |||||||
| G3 | YWHAZ | 14-3-3 protein ζ/δ | U79231 | Mitochondrial import, unclassified | |||||||
| G4 | WS-3 | WS-3 protein, heart muscle isoform | AF124788 | Mitochondrial biogenesis, cell defence/death | |||||||
| G5 | SKD3 | Putative ATPase, HSP104 family member | U09874 | Mitochondrial biogenesis, cell defence/death | |||||||
| G6 | Myosin 1 | L00923 | Cell structure, cytoskeletal | ||||||||
| G7 | GAPDH | GAPDH | M32599 | LFAIM | |||||||
| G8 | HSD3B5 | 3-Oxosteroid reductase | L41519 | ||||||||
| G9 | APE 1 | Apurinic/apyrimidinic endonuclease 1 | P28352 | DNA repair | |||||||
| G10 | OGDH | 2-Oxoglutarate dehydrogenase E1 | U02971 | C-compound and carbohydrate metabolism, tricarboxylic acid cycle | |||||||
| G11 | ACADV | Acyl-CoA dehydrogenase, very long chain | U41497 | LFAIM | |||||||
| G12 | SLC1A1 | Excitatory amino acid transporter 3 | D43797 | Amino acid transporter | |||||||
| H1 | HPRT | Hypoxanthine phosphoribosyltransferase | J00423 | DNA recombination | |||||||
| H2 | PLA2 | Phospholipase A2 | D78647 | Carrier protein, calcium metabolism | |||||||
| H3 | CAB45 | Calcium-binding protein Cab45 | U45977 | Cellular transport and trafficking | |||||||
| H4 | NRF1 | Nuclear respiratory factor 1 | NM_010938 | Cell defence, cell death and aging | |||||||
| H5 | COX5B | Cytochrome c oxidase, chain Vb | X53157 | Energy metabolism, respiration, ETC | |||||||
| H6 | COX 6A2 | Cytochrome c oxidase, chain VIa liver | L06465 | Energy metabolism, respiration, ETC | |||||||
| H7 | ATP5K | ATP synthase H+ transporting chain e | S52977 | Energy metabolism, respiration, ETC | |||||||
| H8 | β-ACTIN | β-Actin | X03672 | Cell structure, cytoskeletal | |||||||
| H9 | ODC | Ornithine decarboxylase | M10624 | Polyamines biosynthesis | |||||||
| H10 | TOM40 | Mitochondrial outer-membrane protein | AF043249 | Mitochondrial transport | |||||||
| H11 | GPAM | Glycerol-3-phosphate acyltransferase | NM_008149 | Glycerol and fatty acid metabolism | |||||||
| H12 | SDHD | Succinate dehydrogenase, small subunit | XM_134803 | Energy metabolism, respiration, ETC | |||||||
For mitoarray printing, 96-well culture boxes were inoculated from the glycerol stocks using robotics, incubated at 37 °C overnight, and plasmids were purified using a NucleoSpin Robot 96 Extract kit (Macherey-Nagel, Düren, Germany). The purified plasmid DNAs were estimated using a SPECTRAmax PLUS 384 (Molecular Devices, Sunnyvale, CA, U.S.A.) and quantitatively normalized to 40 ng/μl average concentration using SOFTmax PRO 4.0 software and robotics. Purified and normalized plasmids were denatured in 0.2 M NaOH at 37 °C for 30 min, and were spotted, in duplicate, on Zetaprobe GT membranes (Bio-Rad, Hercules, CA, U.S.A.) using a BioMek 2000 Laboratory Work-station (Beckman, Fullerton, CA, U.S.A.). The resulting spotted membranes were cross-linked in a UV Stratalinker 1800 (Stratagene, La Jolla, CA, U.S.A.) and stored at − 20 °C until use. Other researchers interested in using the mitoarrays are encouraged to contact the authors (N. G. or J. P.).
Probe synthesis and mitoarray hybridization
Tissue samples were harvested from normal and infected mice at various time points post-infection, flash-frozen in liquid nitrogen and stored at − 80 °C. The frozen tissues were individually transferred into guanidine–phenol solution, processed with a tissue homogenizer, and the total RNA was isolated as described in [15] with slight modifications [4]. The DNA that might be contaminating the RNA preparation was removed using an RNA purification kit (Ambion, Austin, TX, U.S.A.). Total RNA was analysed for quality and quantity using SPECTRAmax PLUS 384 and for integrity using an Agilent 2100 Bioanalyzer. The 32P-labelled cDNA probe was then generated by reverse transcription of 2 μg of total RNA using a gene-specific primer mix and Superscript II reverse transcriptase (Invitrogen) in the presence of 5 μl of [α-32P]dATP (3000 μCi/mmol, 10 μCi/μl; Amersham Biosciences, Piscataway, NJ, U.S.A.). The labelled cDNA probe was purified from unincorporated 32P-labelled nucleotides and small (< 0.1 kb) cDNA fragments using nucleospin extraction chromatography (ClonTech Laboratories, Palo Alto, CA, U.S.A.), and the specific activity of the labelled probe was determined on an LS 6000 SC liquid-scintillation counter (Beckman).
Individual RNA preparations from 4–6 murine hearts/group were labelled and used separately for hybridization. The hybridization and wash procedures were performed essentially as described in [4]. Membranes were exposed to a phosphorimaging screen for 2–5 days, and the images were captured on a Storm 860 phosphorimager (Amersham Biosciences). The results from different arrays were imported in Microsoft Excel spreadsheets, normalized using a set of highly and steadily expressed genes and averaged among replicate arrays. The differential expression of a gene in infected murine hearts relative to controls was evaluated by calculating the corresponding P value using the native Excel t test option.
Real-time RT–PCR
To validate the mitoarray results, we examined the differential expression of selected genes by real-time PCR. First-strand cDNA was synthesized from DNase-treated total RNA (2 μg) using 2.5 units of Superscript II reverse transcriptase and 1 μM poly(dT)18 oligonucleotide at 42 °C for 50 min in 20 μl reaction volume. Real-time PCRs (25 μl volume) were performed in an iCycler Thermal Cycler (Bio-Rad) using 2 μl of the 10-fold diluted cDNA and 5 μl of 1 μM gene-specific primer pair (listed in Table 2) with SYBR-Green Supermix (Bio-Rad). The PCR Base Line Subtracted Curve Fit mode was applied for determining the threshold cycle, Ct, using iCycler iQ Real-Time Detection System Software, Version 3.0a (Bio-Rad). For each target gene, the Ct values were normalized to GAPDH expression and represent the means for triplicate samples in two independent experiments. The relative expression level of each target gene in infected mice was calculated as fold change=2−ΔCt, where ΔCt represents the Ct (infected sample)−Ct (control). To visualize better the mRNA levels in all groups, bar graphs were generated by plotting the 1/2Ct values on the y-axis.
Table 2. Oligonucleotides used in the present study for the confirmation of mitoarray data by traditional and real-time RT–PCRs.
| Gene name | Product | Forward primer (5′→3′) | Reverse primer (5′→3′) | Size (bp) |
|---|---|---|---|---|
| ALAS2 | 5-Aminolaevulinate synthase | ggagctggctgaactacacc | ccttgagtaggcgcacagat | 596 |
| BZRP | Peripheral benzodiazone receptor | ggcttcatgggagcctactt | gccagagttatcacgccatac | 426 |
| DBT | Dihydrolipomide transacylase E2 | cctccctcaccaaaatctga | cttccggtggcaatatcact | 601 |
| DLD | Dihydrolipomide dehydrogenase E3 | gctgcttctggtggtaaagc | gcctctgataaggtcggatg | 596 |
| IDH2 | Isocitrate dehydrogenase | aaggtggagaagccggtagt | cggccatttcttctggatag | 603 |
| MDH | Malate dehydrogenase | aagccatggtttgcatcatt | caaagtcctcgcctttcttg | 600 |
| PDHA1 | Pyruvate dehydrogenase E1 | aatgcacatgtacgccaaga | gcagcatcctcgatttcttt | 594 |
| SDH2 | Succinate dehydrogenase | gccaggaacactccaaaaac | atcagccacacagcaacacc | 517 |
| SOD2 | Superoxide dismutase Mn2+ | acaagcacagcctcccagac | cttgcaagctgtgtatctttcag | 593 |
| VDAC2 | VDAC2 | aacaaaggatttggctttgg | cgagtgcagttggtacctga | 599 |
| GAPDH | GAPDH | tggcaaagtggagattgttg | ttcagctctgggatgacctt | 600 |
| ATP5K | ATP synthase, subunit e | cggttcaggtctctccactc | tgctgtcatcttgagcttcc | 195 |
In some experiments, traditional RT–PCR was employed to determine the mRNA level for differentially expressed genes. For this, 2 μl of the 10-fold diluted cDNA samples were amplified by PCR for 25–36 cycles. Individual amplicons (8 μl) were electrophoresed on 1.2% agarose gels, stained with ethidium bromide and visualized in long-wave UV light. Densitometric analysis was performed on a FluroChem Imaging System (Alpha Innotech).
Spectrophotometric analysis of enzyme activities
Mitochondria were isolated from normal and infected murine hearts as described in [4]. Proteins were determined using the Bradford method [16] with BSA as a standard. The activity of PDC (pyruvate dehydrogenase complex) was measured following the reduction of NAD+ at 340 nm. Mitochondria (5–10 μg of protein) were sonicated (30 s, setting of 3.0, 100% pulse rate, VWR Scientific) in 950 μl reaction buffer containing 35 mM KH2PO4, 5.0 mM MgCl2, 2.0 mM NaCN, and 0.5 mM EDTA at pH 7.25 [17]. The enzymic reaction was started with 0.5 mM NAD+, 200 μM thymine pyrophosphate, 40 μM CoASH and 4.0 mM sodium pyruvate, and a decrease in absorbance measured on an UltraSpec 2000 Spectrophotometer (Amersham Biosciences, Piscataway, NJ, U.S.A.) for 5 min. The assay was repeated in the presence of 2.5 μM rotenone to prevent NADH consumption by complex I. The enzymic activities of SDH (succinate-ubiquinone oxidoreductase) and CS (citrate synthase) were measured as described in [18–20].
RESULTS
We designed the customized mitoarray prototype to analyse the time course of the T. cruzi infection-induced mitochondrial profile in murine hearts. All of the genes printed on the arrays are related to mitochondrial function and are representative of various metabolic pathways. For example, several cDNAs encoding components of fatty acid metabolism and transport (13 genes), tricarboxylic acid cycle (11 genes) and respiratory complexes (14 genes) are printed on the mitoarrays (Table 1). The genes in the selected subset include those that showed up- or down-regulation in relation to T. cruzi infection, as determined by large-scale profiling of gene expression using mouse global microarray systems [4] (N. Garg and V. Bhatia, unpublished work). The cDNA fragments for each gene are spotted in duplicate, yielding a pair of data sets from one hybridization.
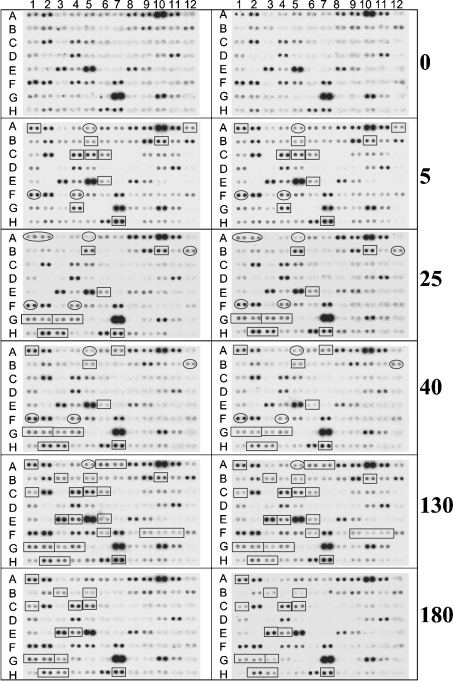
As a model system, we chose C3H/HeN mice infected with the SylvioX10/4 strain of T. cruzi. This mouse–parasite combination mimics the symptoms of chagasic disease and has been extensively characterized in our laboratory as one of the standard models of human CCM [4]. The T. cruzi-infected mice were killed during the immediate early (3–5 days post-infection, dpi), acute (25–40 dpi) and chronic (130–180 dpi) phases of infection and disease development, and the total RNA isolated from the heart was used for the mitoarray hybridizations. The expression pattern of the 95 genes printed on mitoarrays was captured by phosphorimaging analysis (Figure 1), quantified by densitometric analysis and the differential expression calculated relative to controls (Table 3). The reliability of the hybridization was demonstrated by similar expression patterns of the pairs of spots and the tight correlation coefficients of the expression pattern obtained from triplicate samples per time point post-infection.
Figure 1. Mitoarray image of the gene expression profile in T. cruzi-infected murine hearts.
C3H/HeN mice were infected by intraperitoneal injection with culture-derived trypomastigotes of T. cruzi. Heart ventricle sections were obtained at various time intervals post-infection, and total RNA was isolated and then used for prototype customized mitoarray analysis as described in the Materials and methods section. Replicate phosphorimages of the mitoarrays hybridized with P32-labelled cDNA probes generated from total RNA of the murine hearts killed at 0, 5, 25, 40, 130 and 180 dpi are shown. Selected genes whose expression was increased or decreased in infected murine hearts relative to normal controls are enclosed by a rectangle or oval structure respectively.
Table 3. Altered expression pattern of the selected mitochondrial genes in the myocardium of T. cruzi-infected mice relative to normal controls.
↑and ↓arrows indicate an increase and decrease in expression respectively, relative to normal controls.
| Dpi | |||||||
|---|---|---|---|---|---|---|---|
| Spot position | Gene name | Description | 5 | 25 | 40 | 130 | 180 |
| A1 | ACADL | Acyl-CoA dehydrogenase, long chain | ↑ | ↓ | ↑ | ↑ | – |
| A5 | ALAS2 | 5-Aminolevulinate synthase | ↓ | ↓ | ↓ | ↓ | – |
| A12 | ATP7B | Copper transporting P-type ATPase | ↑ | – | – | – | – |
| B5 | BZRP | Peripheral benzodiazepine receptor 1 | ↑ | ↑ | ↑ | ↑ | ↑ |
| B10 | COX8A | Cytochrome c oxidase, chain VIII | ↑ | ↑ | – | ↑ | – |
| B12 | CPT2 | Carnitine-o-palmitoyltransferase II | – | ↓ | ↓ | – | – |
| C1 | CRAT | Carnitine-o-acetyltransferase | – | – | ↑ | ↑ | ↑ |
| C3 | DBT | Dihydrolipoamide transacylase | ↓ | ↓ | ↓ | ↓ | – |
| C4 | DCI | 3,2-trans-Enoyl CoA isomerase | ↑ | – | – | ↑ | ↑ |
| C5 | DLD | Dihydrolipoamide dehydrogenase E3 | ↑ | ↑ | – | ↑ | ↑ |
| E1 | IDH2 | Isocitrate dehydrogenase | ↓ | – | – | – | – |
| E3 | MDH | Malate dehydrogenase | – | – | – | ↑ | ↑ |
| E4 | CBR2 | Carbonyl reductase (NADP+) | – | – | – | ↑ | ↑ |
| E6 | MTHFD | Methylenetetrahydrofolate DH | ↑ | ↑ | ↑ | ↑ | – |
| F1 | PDHA1 | Pyruvate dehydrogenase | ↓ | ↓ | ↓ | – | – |
| F4 | SDH2 | Succinate dehydrogenase, subunit a | ↓ | ↓ | ↓ | – | – |
| F7 | SOD2 | Superoxide dismutase 2 | ↑ | ↓ | ↓ | – | – |
| G1 | VDAC2 | VDAC2 | – | ↑ | ↑ | ↑ | ↑ |
| G2 | VDAC3 | VDAC3 | – | ↑ | ↑ | ↑ | ↑ |
| G3 | YWHAZ | 14-3-3 protein ζ/δ | – | ↑ | ↑ | ↑ | ↑ |
| G4 | WS-3 | ↑ | ↑ | ↑ | ↑ | – | |
| H2 | PLA2 | Phospholipase A2 | – | ↑ | ↑ | ↑ | ↑ |
| H3 | CAB45 | Calcium-binding protein Cab45a | – | ↑ | ↑ | ↑ | ↑ |
| H7 | ATP5K | F1Fo-ATPas synthase, subunit e | ↑ | ↑ | ↑ | ↑ | ↑ |
We adopted an arbitrary threshold cutoff, used by others, to consider genes that were changed by > 40% compared with controls, i.e. a ratio of 1.4 or 0.7 over controls. This criterion clearly highlighted the genes that were differentially expressed during the immediate early phase of parasite invasion and replication (early responsive genes) and whose expression was altered with progressive severity of chronic disease (late-responsive genes) (Figure 1, Table 3). We did not observe an increase or decrease in the level of gene expression greater than 4-fold over controls at either time point post-infection. The overall expression profile suggested an up-regulation of the mRNAs essential for fatty acid metabolism, alterations in the transcripts level for respiratory complexes and repression of the mRNAs encoding subunits of the PDC. We observed no substantial changes in transcripts for the enzymes of tricarboxylic acid cycle.
To confirm the T. cruzi-responsive gene expression profile obtained by custom mitoarray analysis, we selected several genes from those marked in Figure 1 (listed in Table 3) and performed traditional or real-time RT–PCR analysis. These included ALAS2, DBT, PDHA1 and SDH2, noted to be repressed early in response to infection (3–5 dpi) and parasite replication (25–40 dpi) in murine hearts, in mitoarray studies; IDH2 repressed during the immediate early (3–5 dpi) phase; ATP5K, BZRP, DLD and VDAC2, consistently induced during infection and disease development; MDH, primarily up-regulated during the chronic phase; and GAPDH that exhibited no change in expression in infected and normal mice.
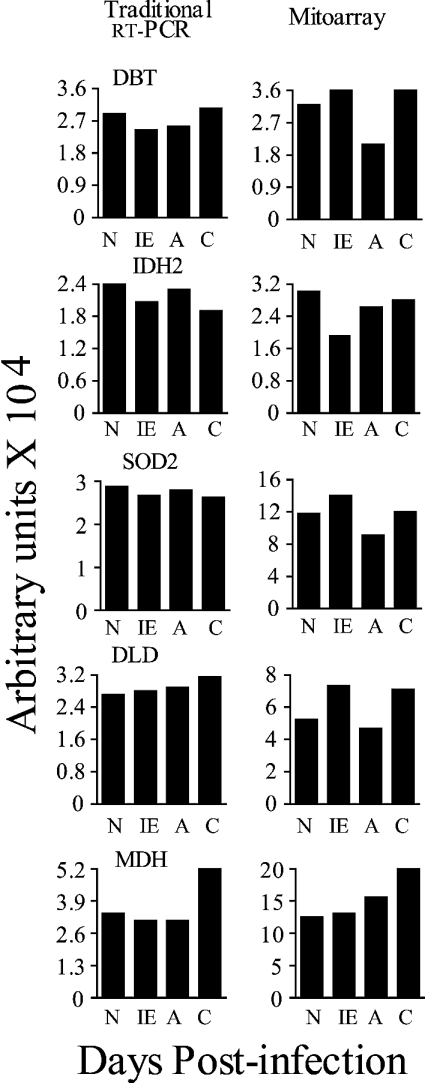
We first determined the expression profile of five genes by traditional RT–PCR. Similar to mitoarray findings, RT–PCR analysis demonstrated an ∼2-fold increase in MDH transcript in chronically infected murine hearts compared with normal controls (Figure 2). However, the differential expression profile of DBT, DLD, IDH2 and SOD2 was not as pronounced in traditional RT–PCR analysis as was observed by mitoarray studies (Figure 2). RT–PCR is semi-quantitative and presents the endpoint after exponential amplification of cDNA for a fixed number of cycles. The optimization of the cycle number for each gene is tedious and less accurate in traditional RT–PCR. Also, the densitometric analysis of the ethidium bromide-stained gels is not very sensitive. Therefore we decided to confirm our mitoarray results by real-time PCR.
Figure 2. Traditional RT–PCR analysis of the selected genes in heart tissue samples from T. cruzi-infected mice.
Total RNA was isolated from the heart tissue of normal (N) mice and mice killed during the immediate early (IE, 3–5 dpi), acute (A, 25–40 dpi) and chronic (C, 130–180 dpi) phases of disease progression. First-strand cDNA was synthesized from 2 μg of the total RNA samples in a 20 μl reaction mixture. Subsequently, PCR was performed for 28–32 cycles using 2 μl of the 10-fold diluted cDNA as template and gene-specific primers. The ethidium bromide-stained gel images were quantified on a FluorChem 8800 Image Analyzing System. The quantitative results represent the mean values obtained from three independent experiments. The S.D. for all the data points was < 12%. All gene names are defined in Table 1. Results from traditional RT–PCR are shown in the left panels. The densitometric quantification of the mitoarray results is shown in the right panels.
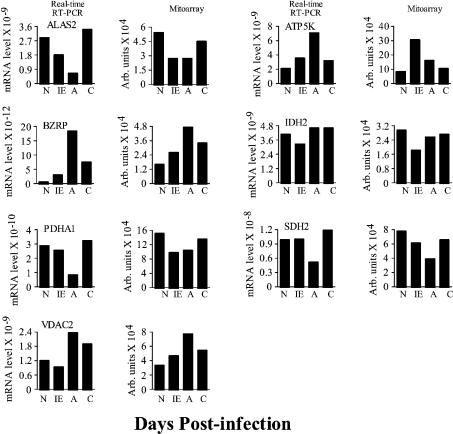
As shown in Figure 3, ALAS2, IDH2, PDHA1 and SDH2 in the early responsive genes group showed a similar expression pattern in the heart tissue of infected mice by real-time PCR, as was observed in mitoarray analysis (Figure 3). The expression of ALAS2, IDH2 and PDHA1 transcripts was rapidly and significantly decreased in response to T. cruzi infection (showing 4-, 3- and 2.5-fold decrease respectively at 5 dpi) in infected murine hearts relative to controls and remained repressed throughout the acute phase of parasite replication and immune activation. In comparison, the repression of DBT mRNA (∼60% decrease) was observed in acutely infected murine myocardium only (Figure 2, real-time PCR results not shown). For ATP5K, BZRP and VDAC2, in agreement with the mitoarray results, a substantial increase in mRNA abundance was noticed in infected mice during the entire course of infection and disease progression. The densitometric analysis of the mitoarray results suggested a 3.5-, 4- and 2.8-fold increase in ATP5K, BZRP and VDAC2 transcripts respectively in infected mice when compared with that detected in normal controls. In comparison, real-time PCR analysis indicated a 3.5-, > 30- and 2-fold increase in ATP5K, BZRP and VDAC2 transcripts respectively in infected mice relative to controls (Figure 3). Overall, real-time PCR has provided independent confirmation of mitoarray results for the induction and repression of several of the mitochondrial function-related genes in response to T. cruzi infection and disease development.
Figure 3. Confirmation of mitoarray expression results by real-time PCR analysis.
Total RNA was isolated from the heart tissue of normal and infected mice and first-strand cDNA synthesized (as in Figure 2). Subsequently, real-time PCR was performed using 2 μl of the 10-fold diluted cDNA as template and gene-specific primers. The results were normalized to GAPDH and represent mean values obtained from two independent experiments using heart tissue of at least three mice/experiment. The S.D. for all the data points was < 12%. All gene names are defined in Table 1. The bar graphs on the left show the relative mRNA level for target genes determined by real-time RT–PCR. 1/2Ct values were plotted on the y-axis. Densitometric quantifications of the mitoarray results are shown in the right panels.
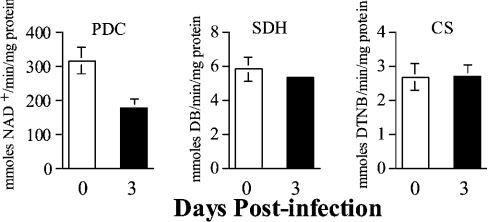
Of note is the expression profile of the genes involved in pyruvate metabolism. Among the five genes related to carbohydrate metabolism (DBT, DLD, IDH2, MDH and PDHA1) that were differentially expressed in infected murine hearts relative to controls (Figure 1, Table 3), three (DBT, DLD, and PDHA1) encoded the components of PDC (Figures 1–3). Whereas the transcripts for the PDHA1 and DBT subunits of the PDC were repressed, transcript for the DLD subunit of the PDC complex appeared to be increased in infected murine hearts (Figures 1–3, Table 3). The decrease in mRNA for PDHA1 was detectable as early as 5 dpi, followed by repression of DBT (Table 3), suggesting early defects in PDC. To confirm if such is the case, we estimated the enzymic activity of the PDC in mitochondria isolated from the cardiac tissue of normal and infected mice by spectrophotometric assays. The cardiac mitochondria isolated from infected mice showed an approx. 40% decrease in PDC activity as early as 3 dpi (Figure 4) that remained consistently low in infected murine hearts during the course of disease progression (results not shown) when compared with controls. The decrease in the enzymic activity of the PDC in infected mice provides further evidence for the accuracy of the mitoarray results. The nuclear-encoded SDH and CS activities are commonly used as controls for verifying the efficiency of mitochondria isolation in different extractions. We detected similar levels of SDH activity in all cardiac mitochondria samples extracted from infected and normal tissues (Figure 4, results not shown) suggesting that the efficiency of mitochondria isolation was relatively similar in all experiments.
Figure 4. Enzymic activity of PDC is reduced in the myocardium of T. cruzi-infected mice.
C3H/HeN mice were infected by intraperitoneal injection with T. cruzi. Enzymic activities of PDC, SDH and CS were measured in cardiac mitochondria isolated from normal mice and T. cruzi-infected mice killed at various time points post-infection. Specific activity [mmol·min−1·(mg of protein)−1] was calculated as means±S.D. obtained from duplicates estimated in three independent experiments.
DISCUSSION
We report, in the present study, on our ongoing cDNA microarray analyses to identify the mitochondrial dysfunction related to various toxic and pathogenic stimuli. Our primary focus was to develop a custom mitoarray prototype that can be used to specify the alterations in mitochondrial pathways involved in ATP production, namely β-oxidation of fatty acids, tricarboxylic acid cycle and respiratory chain-mediated oxidative phosphorylation. The usefulness of these arrays in the time-course studies and diagnosis of mitochondrial dysfunction with progressive development of T. cruzi-induced chagasic cardiomyopathy in a murine model was then explored. Our results clearly show the changes in the expression level of transcripts related to the above-mentioned mitochondrial metabolic pathways in response to T. cruzi infection and disease progression in murine myocardium. The overall changes suggest that the up-regulation of the fatty acid β-oxidation might compensate for the deficiencies of glycolysis-linked pyruvate oxidation in the infected myocardium, and provide a substrate for the maximal efficiency of the tricarboxylic acid cycle in cardiac mitochondria of infected mice. However, the repression of respiratory chain components implies that the loss in mitochondrial oxidative phosphorylation-mediated ATP generation capacity would be the probable basis for energy deficiency in CCM.
The prime source of energy for heart muscle is fatty acid β-oxidation, with the rest provided by glycolysis-linked pyruvate oxidation and tricarboxylic acid cycle [9,10]. Our mitoarray analysis shows that the expression of genes involved in β-fatty acid oxidation is increased in T. cruzi-infected murine hearts. For example, the level of transcripts for long-chain acyl-CoA dehydrogenase (the rate-limiting enzyme that catalyses the first dehydrogenation of fatty acyl-CoA yielding a trans-Δ2-enoyl-CoA), trans-enoyl-CoA isomerase or dodecenoyl-CoA δ-isomerase (catalyses the hydration step to form β-hydroxy-acyl-CoA) and carnitine-o-acetyltransferase (transports acetyl-CoA and short-chain fatty acids to mitochondria) were enhanced in infected murine hearts relative to controls. Our mitoarray findings concur well with our previous global array studies [4], in which we found in the myocardium of infected mice, relative to controls, an increased mRNA level for long-chain acyl-CoA dehydrogenase, medium-chain acyl-CoA dehydrogenase, dodecenoyl-CoA δ-isomerase and 3-hydroxyacyl-CoA dehydrogenase (catalyses the third step of dehydrogenation to form β-ketoacyl CoA). A few other studies [5,21] have also demonstrated alterations in the mRNA and protein levels of enzymes involved in β-oxidation of fatty acids in hypertrophic and CCM hearts.
Of the 11 genes related to carbohydrate metabolism that were screened on the mitoarrays, we found differential expression of all three subunits of the PDC. Subsequently, a decrease in the PDC activity (Figure 4) was also noted in the myocardium of infected mice. PDC associated with the inner mitochondrial membrane is a key enzyme in oxygen-dependent metabolic pathways by shifting intermediates of glucose metabolism into the tricarboxylic acid cycle and to subsequent oxidation in the respiratory chain. PDC is sensitive to inflammatory cytokines [TNFα (tumour necrosis factor α) and interleukin 1] [22]. In the myocardium of infected mice, inflammatory response marked by TNF expression [23,24] and PDC repression (Figure 4) are observed during the immediate early phase of T. cruzi infection. We surmise that primary inhibition of PDC in the myocardium of infected mice might be mediated by inflammatory response. Our hypothesis is supported by the observations that PDC activity is inhibited in neonatal cardiomyocytes within a few hours of TNFα and interleukin 1 exposure [25].
The oxidative phosphorylation pathway comprising five large enzyme complexes couples the NADH and FADH2 oxidation with phosphorylation and provides > 90% of cellular ATP in the myocardium [26]. All complexes are located in the inner mitochondrial membrane and designated as CI (NADH-ubiquinone oxidoreductase), CII (succinate-ubiquinone oxidoreductase), CIII (ubiquinol-cytochrome c oxidoreductase), CIV (cytochrome c oxidase) and CV (F1Fo-ATPase synthase). Our mitoarray study showed differential expression of transcripts for two subunits of CII, two subunits of CIV and for the ATP5K subunit of CV. These changes, although relatively modest, were reproducible in multiple experiments and statistically significant, and consistent with the published global array studies [4]. Recent molecular and biochemical studies documenting the repression of mitochondria-encoded subunits of the respiratory complexes and a decrease in the activities of the respiratory chain complexes in the myocardium of T. cruzi-infected mice with progressive disease severity [18] provide additional support to the mitoarray observations.
Taken together, the mitoarray analysis of the cardiac mitochondria from mice shows an adaptation by the heart metabolism in response to T. cruzi infection and CCM development. This includes up-regulation of fatty acid β-oxidation in an early responsive phase associated with down-regulation of PDC. Both fatty acid oxidation and PDC provide acetyl CoA for the tricarboxylic acid cycle. Given that no significant change was detected in the transcripts for tricarboxylic acid cycle enzymes in our model system, we surmise that a decrease in PDC may not limit the substrate availability for the tricarboxylic acid cycle. Furthermore, the efficiency of the tricarboxylic acid cycle to generate NADH and FADH2 for the respiratory chain may also remain unaffected. However, the down-regulation of respiratory chain components is expected to be accompanied by a decrease in the ATP generation capacity. In support of this hypothesis, we have recently found that mitochondrial ATP production rate is decreased by > 40% in chagasic hearts relative to controls [18]. A decrease in ATP production coupled with increase in AMP level affects the cellular energy charge and stimulates the AMPK (AMP-activated protein kinase) (see [27] and references therein). When induced, AMPKs shut off the energy-consuming processes and switch on the catabolic processes (e.g. fatty acid oxidation) that generate ATP [27]. It is probable that activation of fatty acid β-oxidation in chagasic hearts might be an AMPK-mediated adaptive response to maintain the availability of ATP for cardiac function.
Among the other genes screened on mitoarrays, we observed up-regulation of the transcripts for peripheral benzodiazepine receptor BZRP and VDACs (voltage-dependent anion channels VDAC2, VDAC3) in the myocardium of infected mice throughout the infection and disease phases, also documented in a previous study [4]. In mitochondria, BZRP associates with VDACs, CK (creatine kinase) and ATP/ADP transport protein to form the MPTP (mitochondrial membrane transition pore) [28,29]. The MPTP plays an important role in maintaining the metabolic co-ordination between the cytosol, the mitochondrial intermembrane space and the matrix [30]. The MPTP-mediated regulation of mitochondrial membrane potential facilitates oxidative phosphorylation, ATP synthesis and ATP/ADP transfer from the matrix (reviewed in [29]). BZRP in MPTP forms a channel for the transport of cholesterol for steroid biosynthesis [31]. A dysregulation in MPTP at molecular or functional level can result in an abnormal flux of anionic metabolites, leading to impaired mitochondrial membrane permeability and mitochondrial dysfunction, followed by apoptotic cell death [32,33]. Accordingly, aberrations in VDAC expressions are linked to disturbed oxidative phosphorylation and impaired rates of pyruvate oxidation and ATP production in mitochondriopathy patients [34]. Enhanced expression of BZRP, observed in a variety of neurodegenerative disorders and cardiac ischaemia, is in general related to its role in regulating steroidogenesis and mitochondrial oxidative processes [29]. The role of VDACs and BZRP dysregulation in mitochondrial metabolism in T. cruzi-infected mice remains to be elucidated in future studies.
The only discrepancy between mitoarray analysis and the published results [4] was the expression level of MDH transcript in T. cruzi-infected mice. The MDH transcript evidenced to be decreased in chronic chagasic hearts in a previous report [4] was found to be increased in the infected myocardium in the present study (Figure 2). These discrepancies indicate the importance of validation of the global expression results with methods such as traditional RT–PCR, real-time RT–PCR or Northern blotting. We have used real-time PCR to confirm the mitoarray analysis of expression levels of several of the mitochondrial function-related genes during the course of infection and CCM development. Additionally, biochemical assays were employed to confirm further the deficiency of PDC during CCM progression.
In summary, the development of mitoarray has facilitated the molecular printing of the changes in mitochondrial metabolic activities in response to various toxic and pathogenic stimuli. Our results suggest a specific pattern of mitochondrial metabolic alterations with cardiac homoeostasis in CCM and increase the possibility that treatments capable of reducing the cardiac pathology in chagasic patients may exert their effects by normalizing at least some of the changes associated with mitochondrial function.
Acknowledgments
This work was partially supported by grants from the American Heart Association (0160074Y) and National Institutes of Health (AI053098-01) to N. G. and grants from National Institutes of Health (AG10514) and The University of Texas Medical Branch Sealy Center on Aging to J. P. This research was conducted in part by the Clayton Foundation for Research.
References
- 1.World Health Organization. Chagas disease: tropical diseases progress in research, 1997–1998. WHO Techn. Rep. Series. 1999;1:1.
- 2.Moncayo A. Chagas disease: epidemiology and prospects for interruption of transmission in the Americas. World Health Stat. Q. 1992;45:276–279. [PubMed] [Google Scholar]
- 3.Higuchi M. L., Benvenuti L. A., Martins Reis M., Metzger M. Pathophysiology of the heart in Chagas’ disease: current status and new developments. Cardiovasc. Res. 2003;60:96–107. doi: 10.1016/s0008-6363(03)00361-4. [DOI] [PubMed] [Google Scholar]
- 4.Garg N., Popov V. L., Papaconstantinou J. Profiling gene transcription reveals a deficiency of mitochondrial oxidative phosphorylation in Trypanosoma cruzi-infected murine hearts: implications in chagasic myocarditis development. Biochim. Biophys. Acta. 2003;1638:106–120. doi: 10.1016/s0925-4439(03)00060-7. [DOI] [PubMed] [Google Scholar]
- 5.Mukherjee S., Belbin T. J., Spray D. C., Lacobas D. A., Weiss L. M., Kitsis R. N., et al. Microarray analysis of changes in gene expression in a murine model of chronic chagasic cardiomyopathy. Parasitol. Res. 2003;91:187–196. doi: 10.1007/s00436-003-0937-z. [DOI] [PubMed] [Google Scholar]
- 6.Starling R. C., Hammer D. F., Altschuld R. A. Human myocardial ATP content and in vivo contractile function. Mol. Cell. Biochem. 1998;180:171–177. [PubMed] [Google Scholar]
- 7.Carvajal K., Moreno-Sánchez R. Heart metabolic disturbances in cardiovascular diseases. Arch. Med. Res. 2003;3:89–99. doi: 10.1016/S0188-4409(03)00004-3. [DOI] [PubMed] [Google Scholar]
- 8.Braunwald E. Control of myocardial oxygen consumption: physiologic and clinical considerations. Am. J. Cardiol. 1971;27:416–432. doi: 10.1016/0002-9149(71)90439-5. [DOI] [PubMed] [Google Scholar]
- 9.Neely J. R., Rovetto M. J., Oram J. F. Myocardial utilization of carbohydrate and lipids. Prog. Cardiovasc. Dis. 1972;15:289–329. doi: 10.1016/0033-0620(72)90029-1. [DOI] [PubMed] [Google Scholar]
- 10.Zierler K. L. Fatty acids as substrates for heart and skeletal muscle. Circ. Res. 1976;38:459–463. doi: 10.1161/01.res.38.6.459. [DOI] [PubMed] [Google Scholar]
- 11.Rodrigues B., McNeill J. H. The diabetic heart: metabolic causes for the development of a cardiomyopathy. Cardiovasc. Res. 1992;26:913–922. doi: 10.1093/cvr/26.10.913. [DOI] [PubMed] [Google Scholar]
- 12.Lopaschuk G. D., Wambolt R. B., Barr R. L. An imbalance between glycolysis and glucose oxidation is a possible explanation for the detrimental effects of high levels of fatty acids during aerobic reperfusion of ischemic hearts. J. Pharmacol. Exp. Ther. 1993;264:135–144. [PubMed] [Google Scholar]
- 13.Ferrari R., Pepi P., Ferrari F., Nesta F., Benigno M., Visioli O. Metabolic derangement in ischemic heart disease and its therapeutic control. Am. J. Cardiol. 1998;82:2K–13K. doi: 10.1016/s0002-9149(98)00531-1. [DOI] [PubMed] [Google Scholar]
- 14.Plata F., Garcia Pons F., Eisen H. Antigenic polymorphism of Trypanosoma cruzi: clonal analysis of trypomastigote surface antigens. Eur. J. Immunol. 1984;14:392–399. doi: 10.1002/eji.1830140503. [DOI] [PubMed] [Google Scholar]
- 15.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 16.Bradford M. A. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein–DNA binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Lib M., Rodriguez-Mari A., Marusich M. F., Capaldi R. A. Immunocapture and microplate-based activity measurement of mammalian pyruvate dehydrogenase complex. Anal. Biochem. 2003;314:121–127. doi: 10.1016/s0003-2697(02)00645-0. [DOI] [PubMed] [Google Scholar]
- 18.Vyatkina G., Bhatia V., Gerstner A., Papaconstantinou J., Garg N. Impaired mitochondrial respiratory chain and bioenergetics during chagasic cardiomyopathy development. Biochim. Biophys. Acta. 2004 doi: 10.1016/j.bbadis.2004.03.005. in the press. [DOI] [PubMed] [Google Scholar]
- 19.Trounce I. A., Kim Y. L., Jun A. S., Wallace D. C. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 20.Jarreta D., Orus J., Barrientos A., Miro O., Roig E., Heras M., Moraes C. T., Cardellach F., Casademont J. Mitochondrial function in heart muscle from patients with idiopathic dilated cardiomyopathy. Cardiovasc. Res. 2000;45:860–865. doi: 10.1016/s0008-6363(99)00388-0. [DOI] [PubMed] [Google Scholar]
- 21.Calvani M., Reda E., Arrigoni-Martelli E. Regulation by carnitine of myocardial fatty acid and carbohydrate metabolism under normal and pathological conditions. Basic Res. Cardiol. 2000;95:75–83. doi: 10.1007/s003950050167. [DOI] [PubMed] [Google Scholar]
- 22.Tredget E. E., Yu Y. M., Zhong S., Burini R., Okusawa S., Gelfand J. A., et al. Role of interleukin 1 and tumor necrosis factor on energy metabolism in rabbits. Am. J. Physiol. 1988;255:E760–E768. doi: 10.1152/ajpendo.1988.255.6.E760. [DOI] [PubMed] [Google Scholar]
- 23.Chandrasekar B., Melby P. C., Troyer D. A., Colston J. T., Freeman G. L. Temporal expression of pro-inflammatory cytokines and inducible nitric oxide synthase in experimental acute chagasic cardiomyopathy. Am. J. Pathol. 1998;152:925–934. [PMC free article] [PubMed] [Google Scholar]
- 24.Talvani A., Ribeiro C. S., Aliberti J. C., Michailowsky V., Santos P. V., Murta S. M., Romanha A. J., Almeida I. C., Farber J., Lannes-Vieira J., et al. Kinetics of cytokine gene expression in experimental chagasic cardiomyopathy: tissue parasitism and endogenous IFN-γ as important determinants of chemokine mRNA expression during infection with Trypanosoma cruzi. Microbes Infect. 2000;2:851–866. doi: 10.1016/s1286-4579(00)00388-9. [DOI] [PubMed] [Google Scholar]
- 25.Zell R., Geck P., Werdan K., Boekstegers P. TNF-α and IL-1 α inhibit both pyruvate dehydrogenase activity and mitochondrial function in cardiomyocytes: evidence for primary impairment of mitochondrial function. Mol. Cell. Biochem. 1997;177:61–67. doi: 10.1023/a:1006896832582. [DOI] [PubMed] [Google Scholar]
- 26.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 27.Hardie D. G., Hawley S. A. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays. 2001;23:1112–1119. doi: 10.1002/bies.10009. [DOI] [PubMed] [Google Scholar]
- 28.McEnery M. W., Snowman A. M., Trifiletti R. R., Snyder S. H. Isolation of the mitochondrial benzodiazepine receptor: association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc. Natl. Acad. Sci. U.S.A. 1992;89:3170–3174. doi: 10.1073/pnas.89.8.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casellas P., Galiegue S., Basile A. S. Peripheral benzodiazepine receptors and mitochondrial function. Neurochem. Int. 2002;40:475–486. doi: 10.1016/s0197-0186(01)00118-8. [DOI] [PubMed] [Google Scholar]
- 30.Halestrap A. P. The mitochondrial permeability transition: its molecular mechanism and role in reperfusion injury. Biochem. Soc. Symp. 1999;66:181–203. doi: 10.1042/bss0660181. [DOI] [PubMed] [Google Scholar]
- 31.Papadopoulo V. Peripheral benzodiazepine receptor: structure and function in health and disease. Ann. Pharmacol. Fr. 2003;61:30–50. [PubMed] [Google Scholar]
- 32.Colombini M. VDAC: the channel at the interface between mitochondria and the cytosol. Mol. Cell. Biochem. 2004;256/257:107–115. doi: 10.1023/b:mcbi.0000009862.17396.8d. [DOI] [PubMed] [Google Scholar]
- 33.Granville D. J., Gottlieb R. A. Mitochondria: regulators of cell death and survival. Sci. World J. 2002;2:1569–1578. doi: 10.1100/tsw.2002.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huizing M., Ruitenbeek W., Thinnes F. P., DePinto V., Wendel U., Trijbels F. J., Smit L. M., ter Laak H. J., van den Heuvel L. P. Deficiency of the voltage-dependent anion channel: a novel cause of mitochondriopathy. Pediatr. Res. 1996;39:760–765. doi: 10.1203/00006450-199605000-00003. [DOI] [PubMed] [Google Scholar]