Abstract
Breast cancer is the most frequent malignancy in women, constituting 15.2% of all new cancers diagnosed in the United States. Distant breast cancer metastasis accounts for the majority of breast cancer-related deaths; brain metastasis is the third most common site for metastatic breast cancer but is associated with worst prognosis of approximately eight months of survival. Current treatment options for breast cancer brain metastasis (BCBM) are limited and ineffective. To help identify new and effective therapies for BCBM, it is important to investigate the mechanisms by which breast cancer cells metastasize to the brain and thrive in the brain microenvironment. To this end, studies have reported that primary breast tumor cells can prime brain microenvironmental cells, including, astrocytes and microglia, to promote the formation of BCBM through the release of extracellular vesicle-microRNAs (miRNAs). Breast tumor-derived miRNAs can also promote breast cancer cell invasion through the blood-brain barrier by disrupting the integrity of the brain microvascular endothelial cells. In this review, we summarize current literature on breast cancer-derived BCBM-promoting miRNAs, cover their roles in the complex steps of BCBM particularly their interactions with microenvironmental cells within the brain metastatic niche, and finally discuss their therapeutic applications in the management of BCBM.
Keywords: breast cancer, microRNA, brain microenvironment, blood-brain barrier, brain metastasis
1. Introduction
According to the latest statistics by the American Cancer Society, the estimated number of new breast cancer cases in 2024 is 310,720, accounting for 32% of all cancers diagnosed in women (1). Breast cancer patients have the second highest rate of brain metastasis behind lung cancer (2). Furthermore, subtype analysis of breast cancer brain metastasis (BCBM) patients showed that HER2-enriched breast cancer and triple-negative breast cancer (TNBC) subtypes have a higher potential to develop brain metastasis, underscoring the particular importance of studying BCBM in these two subtypes of breast cancer (3).
In 1993, Ambros and his team made the groundbreaking discovery of a microRNA (miRNA) in Caenorhabditis elegans (4). The miRNA, transcribed from the lin-4 gene, exhibits complementarity to and consequently regulates the expression of the lin-14 protein (4). miRNAs are part of the small non-coding RNA family that are endogenous, single-stranded RNAs with an average length of 20–22 nucleotides, known to regulate gene expression in many physiological processes (5, 6). Studies have shown that miRNAs play a pleiotropic role acting as tumor suppressors and/or oncogenes in different cancers (7, 8). Numerous studies have now implicated miRNAs in every step of brain metastasis beginning from epithelial-mesenchymal transition to colonization in the brain parenchyma (9–29).
Brain organotropism in breast cancer is influenced by several factors: breast cancer subtype, molecular features of circulating tumor cells, extracellular vesicle-derived miRNA expression profile, tumor microenvironment, and the ability of breast cancer cells to penetrate the blood-brain barrier (BBB) (30). BBB is a specialized neurovascular unit adjoining blood capillaries with brain parenchyma, comprising of brain microvascular endothelial cells (BMECs), astrocytic end-feet, and pericytes (31). BMECs line the luminal and abluminal membranes and are held together by tight and adherens junctions (32). They tightly regulate the transport of cells and molecules from blood to the brain parenchyma. BMECs lack leukocyte adhesion molecules and have a higher concentration of mitochondria, which limits the influx of immune cells from blood into central nervous system (CNS) and indicates the prevalence of high-energy requiring role of BMECs respectively (33). In response to pathological changes in the CNS, astrocytes undergo molecular, functional, and morphological transformation and are termed as “reactive astrocytes”. Studies have shown the reactive astrocytes stimulate BMECs through secretion of SERPINA3 by activating NF-κB/STAT3 signaling axis (34). Pericytes play a few roles in the maintenance of BBB integrity including regulating microvascular stability, angioarchitecture, and clearance of foreign proteins and tissue debris (35).
The interactions between tumor cells and brain microenvironmental cells, primarily astrocytes and microglia, facilitate various stages of metastasis. Reactive astrocytes through the secretion of inflammatory chemokines such as interferon-α (IFNα) and Ciliary Neurotrophic Factor promote tumor growth by activating transcriptional and cell survival pathways (36–38). Microglial cells, known as the resident macrophages of the CNS, are often polarized from M1 to M2 microglia to secrete immunosuppressive chemokines (39). These modulations in the environment play a key role in promoting the growth of brain metastasis. Through regulating the brain-metastatic microenvironmental cells and their interaction with breast cancer cells, miRNAs can influence breast cancer metastasis to the brain and progression within the brain.
2. MicroRNA biogenesis and mechanism of action
The biogenesis of miRNAs is classified into canonical and non-canonical pathways. The canonical pathway begins with the transcription of a hairpin-containing primary miRNA (pri-miRNA) in the nucleus (8). The pri-miRNA transcript is then cleaved by the microprocessor complex containing DiGeorge Syndrome Critical Region 8 (DGCR8) and Drosha to form the precursor miRNA (pre-miRNA). DGCR8 is an RNA-binding protein that recognizes and binds N6-methyladenosine GGAC motif in the pri-miRNA, and the RNase III enzyme, Drosha, recognizes and cleaves the base of the hairpin structure (40, 41). The pre-miRNA is transported from the nucleus to the cytoplasm through an exportin5/RanGTP complex where RNase III endonuclease Dicer cleaves the hairpin loop structure and leads to the formation of mature double-stranded miRNA (40, 42, 43).
There are two non-canonical pathways: Drosha/DGCR8-independent and Dicer-independent pathways. In the former pathway, (mirtrons) RNAs are exported to the cytoplasm through exportin 1 without undergoing Drosha processing; and in most cases, the 3p strand is loaded onto the AGO protein due to the presence of a 7-methylguanosine cap at the 5’ end (44). In the Dicer independent pathway, shRNA transcripts are processed by Drosha/DGCR8 complex and exported to the cytoplasm by exportin5/RanGTP where they are loaded onto AGO2 and processed (45, 46).
miRNAs binding to a specific seed sequence at either the 3’ or 5’ untranslated region of the target mRNA can lead to mRNA degradation or translational repression, leading to gene silencing (47–50). miRNAs can also bind at the promoter region of target mRNAs leading to transcriptional activation (51). miRNAs can regulate multiple biological pathways such as cell proliferation, cell death, immune evasion, invasion, metastasis, and angiogenesis. miRNAs are classified as tumor suppressors or oncogenes depending on their target gene and cell type (8).
3. Development of brain metastases
For the initiation of metastasis to occur, cancer cells undergo epithelial-mesenchymal transition (EMT) demonstrated by an increase in self-renewing stem cells, anoikis resistance, and dissemination (52–56). TWIST1, SNAIL1, and SLUG are some of the most heavily studied transcription factors in the context of breast cancer metastasis (57). Under regulation of pathways like the Notch signaling pathway, these transformed cells can penetrate the vascular endothelium where endothelial cells promote membrane remodeling and cancer cells enter blood vasculature (58–61). An important part of metastasis is the development of a tumor-supportive environment in distant target organs. Cancer cells prime a secondary site by secreting tumor-promoting extracellular vesicles and inflammatory chemokines, forming the premetastatic niche (62, 63). Extravasation into the brain requires tampering with the BBB permeability. The cross-talk between cancer cells and BMEC is stimulated by the expression of cellular adhesion molecules (E-selectin, VCAM-1) on cancer cells and degradation of the BBB by matrix metalloproteinases (64–66). Extravasation is followed by mesenchymal-epithelial transition (MET) or partial MET lending a higher aggressive phenotype to the cancer cell (67–69). Reactive astrocytes play a dichotomous role by initially inhibiting brain metastases and switching to a pro-metastatic role in later stages (37, 62, 70–72). Glial cells such as tumor-associated microglia/macrophages lend a supportive hand to cancer cells by stimulation of TGF-β1 signaling pathway (73).
4. miRNAs implicated in the cross-talk between breast cancer cells and brain cells
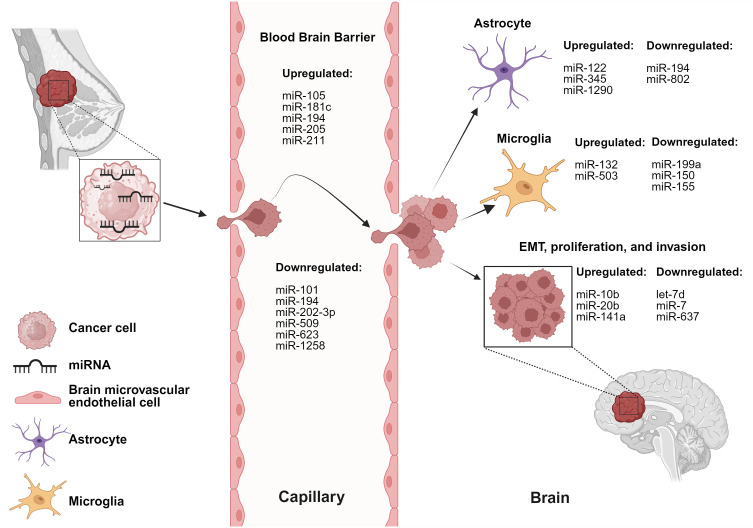
The cross-talk between breast cancer cells and astrocytes/microglia at any stage of brain metastasis leads to microenvironmental modulation that subsequently facilitates the progression of brain metastasis. miRNAs involved in these interactions are listed in Table 1 and depicted in Figure 1 .
Table 1.
miRNAs mediate dysregulation of various cells and stages of BCBM.
| miRNA | Activity | Validated Target(s) | In-vitro Models | Additional Model(s) | Reference |
|---|---|---|---|---|---|
| let-7d | Promote brain metastatic colonization | Pdgfb/PGDFA | 4T1, D2A1, MDA-MB-231, MDA-MB-231-BrM2 | Mouse | (74) |
| miR-7 | Suppress brain metastases formation by inhibiting proliferation, invasion, and transmigration of CSCs | KLF4 | MDA-MB-231, 231BoM-1833, 231BrM-2a, CN34, CN34-BoM2d, CN34-BrM2c, MCF7, MCF7-BoM2d | Mouse | (75) |
| miR-10b | Promote invasion of BC cells | No specific target | MDA-MB-231, MDA-MB-468 | Patient samples | (76) |
| miR-20b | Promote colony formation and invasion of BC cells | No specific target | MCF-7, MDA-MB-231, MDA-MB-231-brain and bone derivatives | Patient samples | (77) |
| miR-101-3p | Promote extravasation and trans-endothelial migration of BC cells | COX2 | MCF-7, MDA-MB-231, MDA-MB-231-BrM2, MDA-MB-231-TGL | N/A | (78) |
| miR-105 | Promote tumor cell invasion and disrupt vascular endothelium | ZO-1 | MDA-MB-231, MCF-10A, MCFDCIS, MDA-231-HM, Primary HMVECs | Patient samples | (79) |
| miR-122 | Inhibit glucose uptake by astrocytes | PKM | MCF10A, MDA-MB-231 | Mouse | (80) |
| miR-132-3p, miR-199a, miR-150 and miR-155 | Promote tumor angiogenesis and colonization of BC cells | MET | Patient samples | (81) | |
| miR-181c | Downregulate PDPK1 promoting BBB destruction | PDPK1 | MDA-MB-231-luc-D3H1, MDA-MB-231-luc-D3H2LN, BMD2a, BMD2b | Mouse | (82) |
| miR-194 and miR-802 | Upregulate MEF2C highly expressed in peritumoral astrocytes promoting cross-talk | MEF2C | 4T1 | Mouse, patient samples | (83) |
| miR-194, miR-181a-1-3p, miR-205, | Mediate cross-talk between BC cells and BMECs | No specific target | b.End5, 4T1 | Mouse | (84) |
| miR-200 family (miR-141) | Induce MET and brain metastatic colonization | E-cadherin | SUM149, SUM159, MDA-IBC3, MDA-231, MCF7 | Mouse, patient samples | (18) |
| miR-202-3p | Induce MMP-1 and promote extravasation of BC cells | MMP-1 | MCF-7, MDA-MB-231-TGL, MDA-MB-231-BrM2, hCMEC/D3 | Mouse | (85) |
| miR-211 | Promote early colonization and BBB adherence through regulation of SOX11/NGN2 axis | SOX11/NGN2 | MDA-MB-231, HCC1806, LM2-4175, BrM-831 | Mouse | (86) |
| miR-345 | Induce cancer progression through cross-talk and niche formation | KISS1, E-cadherin | CN34TGL, MDA-MB-231, CN34Br, MDA231Br | Mouse | (87) |
| miR-503 | Induce M1 to M2 polarization of microglia | STAT3/NF-κB pathways | MCF7, ZR75-1, SKBR3, MDA-MB-231, SKBrM3, MDA-MB-231BrM2a, SIM-A9 | Mouse, patient samples | (26) |
| miR-509 | Suppress RhoC and TNFα mediated transmigration and invasion of BC cells | RhoC, TNF-α | MDA-MB-231, MDA-MB231BrM-2a, CN34, CN34-BrM2c, MCF7, MDA-MB-231-HM, 293TN, mBrEC | Mouse, patient samples | (88) |
| miR-623 | Promote extravasation and trans-endothelial migration of BC cells | MMP-1 | MDA-MB-231-BrM2, MDA-MB-231-TGL, hCMEC/D3 | N/A | (89) |
| miR-637 | Promote autophagy and invasion of TNBC cells | STAT3 | MCF10A, BT549, MDA-MB-361, MDA-MB-453, MDA-MB-, MDA-MB-468, SUM-159, HCC-1806 | Mouse, patient samples | (90) |
| miR-1258 | Inhibit Heparanase to promote invasion | HPSE | MDA-MB-231BR1, MDA-MB-231BR3, SUM-225, SUM-149, HMEC, MCF-10A | Mouse, patient samples | (25) |
| miR-1290 | Activate astrocytes in the brain metastatic microenvironment via the FOXA2→CNTF axis | FOXA2 | MDA-MB-231, MDA-231-BRM, SKBR3, CN34, SKBRM-tGLI1, immortalized human astrocytes | Mouse | (36) |
Figure 1.
miRNAs are involved in dysregulation of various cells and stages of BCBM. BC-derived miRNAs alter functionality of brain and BBB microenvironmental cells. Created with BioRender.com.
miR-122 is upregulated in the conditioned media of breast cancer cells (80). Uptake of breast cancer-derived miR-122 by astrocytes led to a reprogramming of glucose uptake, notably through the downregulation of PKM1/2 and GLUT1, resulting in decreased uptake of 2-NBDG (a fluorescent glucose analogue). The authors reported reduced glucose uptake by tumor cells led to inhibition of tumor cell proliferation in primary tumors, while simultaneously supporting metastatic tumor cell colonization in the pre-metastatic niche (80).
miR-194 and miR-802 were downregulated in plasma samples collected from brain metastasis model of a 4T1-injected mice (83). MEF2C was validated as a target gene using an in vivo model through immunofluorescence staining. MEF2C was found to be highly expressed in established metastatic cells in the brain parenchyma and peritumoral astrocytes. These findings were further validated in human TNBC brain metastases samples (83). Moreover, miR-802 inhibits FoxM1 decreasing the proliferation of breast cancer cells and miR-194 inhibits proliferation and migration of breast cancer cells, supporting the tumor-suppressive roles of these miRNAs (91, 92).
Breast cancer-derived miR-345 expression is upregulated due to increased astrocytic secretion of CCL2 and CXCL12 (87). miR-345 downregulates KISS1 which in turn leads to localization of breast cancer cells in the brain microenvironment (87). Another study reports that miR-345-mediated KISS1 transcriptional inhibition plays a role in promoting autophagy and invasiveness of breast cancer cells (87, 93).
miR-1290 is upregulated in the sera of breast cancer patients (36). miR-1290 activates astrocytes and promotes cancer stemness factors, subsequently aiding the progression of BCBM. miR-1290 binds to FOXA2 and induces transcriptional repression, leading to upregulation of CNTF expression in astrocytes. Astrocytes activated by miR-1290 promoted intracranial growth of co-implanted breast cancer cells in the brain in vivo (36).
miR-199a, miR-150, and miR-155 are downregulated, whereas miR-132-3p is upregulated in tissue obtained from BCBM patients (81). The authors report an increase in miR-132-3p correlated with improved brain metastasis-free survival (BMFS) and overall survival (OS) whereas miR-199a, miR-150 and miR-155 correlated with poorer BMFS and OS. MET was identified as a target oncogene and reported to be overexpressed in microglial cells responsible for tumor angiogenesis and colonization of breast cancer cells (81). miR-503 is upregulated in the sera of BCBM patients and is reported to promote the M1 to M2 polarization of microglia, demonstrated by increased phosphorylation of STAT3 along with decreased phosphorylation of NF-κB (26). Taken together, these findings suggest an important role for miRNAs in regulating the brain microenvironmental cells.
5. miRNAs implicated in the cross-talk between breast cancer cells and BBB cells
A pivotal aspect of brain metastasis involves the disruption of the BBB integrity. Thus, studying the interactions between breast cancer cells and BBB cells is of utmost importance. miRNAs involved in mediating cross-talk between breast cancer cells and BBB cells are listed in Table 1 .
miR-101-3p is decreased in BCBM leading to increased expression of COX-2 and stimulation of COX-2/MMP-1 signaling pathway, promoting trans-endothelial migration of breast cancer cells and extravasation across the BBB (78). miR-105 downregulates ZO-1 expression in endothelial cells hindering the integrity of endothelial and epithelial tight junctions leading to trans-endothelial invasion of breast cancer cells. Disruption of the endothelial barriers and increased vascular permeability promoted distant metastases formation in lung and brain (79). miR-181c is increased in sera of brain metastasis patients compared to non-brain metastasis patients. The study reports that miR-181c regulates the expression of PDPK1 in BMECs, where PDPK1 plays a role in localization of N-cadherin and actin filaments in BMECs. Delocalization of actin in the BMECs plays a role in BBB dysregulation (82).
miR-202-3p is decreased in brain-tropic breast cancer cell lines. The authors report restoration of miR-202-3p leading to MMP-1 suppression results in inhibited extravasation of BCBM cells (85). MMP-1 has previously been reported to promote trans-endothelial migration of breast cancer cells by degrading endothelial junctions in BMECs and permeabilizing the endothelial barrier. One study reported the upregulation of miR-205 and miR-181a-1-3p along with downregulation of miR-194 in co-culture models. The authors report overexpression of miR-181a-1-3p results from the interaction between breast cancer cells and BMECs, and that BMECs contribute to the downregulation of miR-194, while breast cancer cells upregulate the expression of miR-205 (84).
miR-211 is upregulated in the brain-tropic TNBC cells and human breast cancer tissue from TNBC and non-TNBC patients. miR-211 overexpressing breast cancer cells promote brain metastases in vivo; inhibition of miR-211 with anti-miR-211 treatment suppresses brain metastases in vivo. Increased expression of miR-211 promoted migration and invasion of TNBC cells and enhanced adherence of cancer cells to the BBB through downregulation of SOX11/NGN2 axis (86).
miR-509 is downregulated in brain metastases compared to primary breast tumors and targets RhoC, a critical mediator of metastasis and invasion. Additionally, miR-509 suppressed the trans-endothelial migration of breast cancer cells and contributed to the suppression of MMP9 via modulation of RhoC (94). miR-509 also indirectly represses TNFα leading to decreased BBB permeability (88). miR-623 is downregulated in brain metastatic lesions in comparison to primary breast tumors. MMP-1 is known to play a significant role in promoting extravasation of TNBC cells into the brain endothelium and is suppressed by miR-623. This study reports the restoration of miR-623 inhibits trans-endothelial migration of brain-tropic TNBC cells, thereby suppressing BCBM (89). Downregulation of miR-1258 was associated with an increase in HPSE levels in BCBM cell lines, paired primary breast tissue, and BCBM tissue. miR-1258 expression leads to a decrease in HPSE levels and HPSE-related proteins: p-Akt, p-EGFR, MMP-9, COX2 consequently, leading to inhibition of brain metastasis by limiting breast cancer cell invasion (25). These studies are further proof of the crucial role of miRNAs in maintenance of BBB integrity.
6. miRNAs implicated in EMT, invasion, and colonization of breast cancer cells in BCBM
EMT, invasion, and colonization serve an essential role in metastasis of cancer cells. miRNAs involved in these processes are briefly described in Table 1 .
miR-7 is downregulated in mammospheres and brain-tropic breast cancer cell lines compared to parental cells (75). KLF4 is a miR-7 target gene and in vivo studies report miR-7 inhibits the expression of KLF4 downregulating the proliferation, invasion, and transmigration of brain-tropic cancer stem cells (CSCs). The miR-7 and KLF4 correlation was further validated in human samples from primary breast tumor and brain metastatic lesions, and the authors suggest interaction between CSCs and brain cells promotes formation of a pre-metastatic niche (75).
miR-10b is significantly upregulated in tumor samples of BCBM patients when compared to primary breast tumors without brain metastasis (76). It was reported that higher levels of miR-10b were correlated with increased invasiveness of breast cancer cells (76). miR-20b is increased in brain metastatic lesions of BCBM patients compared to breast cancer patients without brain metastasis (77). miR-20b is also upregulated in brain-tropic breast cancer cells compared to bone-tropic breast cancer cells highlighting brain-tropism of miR-20b. miR-20b overexpression resulted in increased colony formation and invasiveness of breast cancer cells (77).
let-7d is downregulated in brain metastatic breast cancer cells and regulates PGDFA expression (74). PGDFA inhibition leads to decreased brain metastases formation in mice models due to the loss of autocrine proliferation loop activity which promotes metastatic colonization. The authors also reported HIF1 activity is negatively regulated by let-7d (74). miR-141 expression is upregulated in sera from BCBM patients (18). Upregulation of miR-141 was correlated with increased E-cadherin expression, which suggests miR-141 plays a role in EMT. The role of miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, miR-429) in EMT has been well established in various metastatic models and the authors propose a possible role in breast cancer brain metastatic colonization (18). Moreover, circKIF4A sponges miR-637 to suppress its expression in brain metastatic lesions compared to primary breast tumors. miR-637 inhibits STAT3, therefore, miR-637 inhibition increases STAT3 protein levels and promotes the brain metastatic properties of TNBC cells through autophagic activation (90). These studies suggest that miRNAs play key roles in regulating multiple stages of BCBM; however, further investigations into these mechanisms are necessary.
7. Application of miRNAs in cancer therapeutics
The dismal overall survival of patients with BCBM is partly due to the lack of early biomarkers and targeted BBB-penetrant therapies (95–97), and miRNAs could possibly be used to address both deficiencies. The ubiquitous presence of miRNAs in peripheral blood, urine, and saliva makes it a highly valuable and non-invasive biomarker of disease burden, progression, treatment response, and resistance (98–100). Currently, there are several clinical trials assessing the potential application of miRNAs as biomarkers. Project CADENCE (NCT05633342) is a cohort study aimed at investigating miRNA expression along with other biomarkers and ultimately developing in-vitro diagnostic assays for screening nine highly prevalent cancers: breast, colorectal, lung, prostate, liver, pancreatic, gastric, ovarian and esophageal (101). Oncoliq US (NCT06439940) is actively recruiting for a prospective cohort study to identify early diagnosis markers for breast cancer utilizing liquid biopsies and miRNAs (102). Another prospective cohort study (NCT05417048) at Peking University is currently investigating the performance of a blood-based assay utilizing miRNAs to differentiate between benign and malignant breast disease (103). MiraKind is currently running a prospective cohort study (NCT02253251) to validate the role of mutations at miRNA binding sites in breast and ovarian cancer patients (104). A randomized diagnostic clinical trial (NCT04516330) is investigating the role of an 84-miRNA panel in predicting multicentricity in breast cancer (105). City of Hope Medical Center conducted a cohort study (NCT01231386) to perform miRNA profiling in patients undergoing treatment for locally advanced or inflammatory breast cancer (106).
miRNA profiling has also been used as a non-surgical tool to differentiate between medulloblastoma, glioblastoma, BCBM, and lung cancer brain metastasis (107). High expression of miR-200 family in the cerebrospinal fluid of brain metastasis patients helps differentiate between cases of brain metastasis and glioblastoma (108).
Therapeutic miRNA development is being explored given the significant roles that miRNAs play in dysregulation of multiple genes leading to tumor initiation, progression, and metastasis (95–97). MRG-106, an inhibitor of miR-155, was investigated for the treatment of cutaneous T-cell lymphoma, mycosis fungoides subtype. The Phase I study reported tolerability and reduction in the Composite Assessment of Index Lesion Severity score and modified Severity Weighted Assessment Tool used to measure skin lesions/disease (109). However, the study by Miragen Therapeutics was discontinued during Phase II (NCT03713320) due to business reasons. Miragen Therapeutics also completed a Phase I study (NCT03603431) with MRG-110, a miR-92a inhibitor, which was investigated in healthy volunteers and was reported to augment wound healing and angiogenesis (110). TransCode Therapeutics recently entered a Phase I/II dose-escalation study (NCT06260774) with TTX-MC138, a miR-10b inhibitor, which has previously been implicated in metastatic lesions arising from advanced solid tumors (111, 112). MRX34, miR-34a mimic, was investigated in patients with unresectable primary liver cancer, hematological malignancies and advanced solid tumors (NCT01829971). MiRNA therapeutics reported treatment with MRX34 demonstrated some clinical activity, however, treatment-associated severe adverse events led to termination of the study (113). CDR132L, a selective miR-132-3p inhibitor, is currently in Phase II clinical trial (NCT05350969) for patients with reduced Left Ventricular Ejection fraction post-myocardial infarction (114). In 2019, Regulus Therapeutics announced pre-clinical success of RGLS5579, an anti-miR-10b, in combination with temozolomide (TMZ) in glioblastoma animal models (115). Regulus Therapeutics reported the median survival rate of glioblastoma-bearing mice models treated with anti-miR-10b, anti-miR-10b in combination with TMZ, and TMZ alone increased by 18%, >120%, and 27% respectively. Combination of tumor suppressive miRNAs with conventional chemotherapy is another promising avenue. Tumor suppressive miR-770 inhibited doxorubicin resistance in TNBC and promoted sensitivity to trastuzumab in HER2-positive breast cancer (116, 117). Overexpression of miR-298 sensitizes doxorubicin-resistant breast cancer cells to treatment by targeting MDR1 (118). Other studies have also reported targeting of ABCG2 overexpression of miR-181a or miR-328 in breast cancer cells led to increased sensitivity to mitoxantrone (119, 120). miRNAs may be the key to improving targeted therapeutics for cancer patients; however, it is worth noting that many Phase I and II clinical trials have been halted in the past due to severe adverse effects (96, 121, 122). Nonetheless, these studies highlight the importance of further investigations into the role of miRNAs as biomarkers and for the advancement of miRNA incorporation into cancer therapeutics.
8. Discussion
There has been a rise in incidence of BCBM, due to significantly advanced and effective therapies that prolong patient survival. This extended survival period allows latent metastatic cells greater opportunity to penetrate the BBB and colonize in the brain parenchyma. Given the rise in frequency and limited treatment opportunities of brain metastasis, there is an urgent need for new predictive, diagnostic, and prognostic biomarkers to assess brain metastasis. miRNAs are small but mighty in the regulation of every step of brain metastasis starting from the cancer stemness genes, genes responsible for intravasation and extravasation into a foreign site, organ tropism, and colonization-related genes. The specificity of dysregulated miRNAs in brain metastasis from various primary tumors can be utilized to differentiate tumor types and identify the origin of primary tumors in unknown cases. Certainly, further research aimed at identifying novel miRNAs, elucidating their biological functions, and uncovering their target genes will significantly enhance our understanding of the role miRNAs play in metastases formation and progression. This will, in turn, lay the groundwork for the advancement of miRNA-related approaches for cancer prognosis, diagnosis, and treatment.
Author contributions
MK: Writing – original draft, Conceptualization. GW: Writing – review & editing. CZ: Writing – review & editing. MN: Writing – review & editing, Visualization. H-WL: Writing – review & editing, Supervision, Resources, Funding acquisition, Conceptualization.
Acknowledgments
The authors would like to thank Texas Medical Center Library at The University of Texas MD Anderson Cancer Center UTHealth Graduate School of Biomedical Sciences for open-access literature support.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We acknowledge funding support for this project from DoD grants W81XWH-19-1-0072 (H-WL), W81XWH-20-1-0044 (H-WL), and W81XWH-19-1-0753 (H-WL), NIH grant R01CA228137 (H-WL), as well as, MetaVivor Translational Research Grant (H-WL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA: A Cancer J Clin. (2024) 74:12–49. doi: 10.3322/caac.21820 [DOI] [PubMed] [Google Scholar]
- 2. Barnholtz-Sloan JS, Sloan AE, Davis FG, Vigneau FD, Lai P, Sawaya RE. Incidence proportions of brain metastases in patients diagnosed (1973 to 2001) in the Metropolitan Detroit Cancer Surveillance System. J Clin Oncol. (2004) 22:2865–72. doi: 10.1200/JCO.2004.12.149 [DOI] [PubMed] [Google Scholar]
- 3. Kuksis M, Gao Y, Tran W, Hoey C, Kiss A, Komorowski AS, et al. The incidence of brain metastases among patients with metastatic breast cancer: a systematic review and meta-analysis. Neuro-oncology. (2021) 23:894–904. doi: 10.1093/neuonc/noaa285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee RC, Feinbaum RL, Ambrost V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. (1993) 75:843–54. doi: 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- 5. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. (2008) 9:102–14. doi: 10.1038/nrg2290 [DOI] [PubMed] [Google Scholar]
- 6. Ambros V. The functions of animal microRNAs. Nature. (2004) 431(7006):350–5. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 7. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci United States America. (2002) 99:15524–9. doi: 10.1073/pnas.242606799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng Y, Croce CM. The role of microRNAs in human cancer. Signal Transduction Targeted Ther. (2016) 1:15004. doi: 10.1038/sigtrans.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee JW, Guan W, Han S, Hong DK, Kim LS, Kim H. MicroRNA-708-3p mediates metastasis and chemoresistance through inhibition of epithelial-to-mesenchymal transition in breast cancer. Cancer Science. (2018) 109:1404–13. doi: 10.1111/cas.13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gao Y, Ma H, Gao C, Lv Y, Chen XH, Xu R, et al. Tumor-promoting properties of miR-8084 in breast cancer through enhancing proliferation, suppressing apoptosis and inducing epithelial-mesenchymal transition. J Trans Med. (2018) 16:38. doi: 10.1186/s12967-018-1419-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mansoori B, Mohammadi A, Ghasabi M, Shirjang S, Dehghan R, Montazeri V, et al. miR-142-3p as tumor suppressor miRNA in the regulation of tumorigenicity, invasion and migration of human breast cancer by targeting Bach-1 expression. J Cell Physiol. (2019) 234:9816–25. doi: 10.1002/jcp.27670 [DOI] [PubMed] [Google Scholar]
- 12. Harquail J, Leblanc N, Ouellette RJ, Robichaud GA. miRNAs 484 and 210 regulate Pax-5 expression and function in breast cancer cells. Carcinogenesis. (2019) 40:1010–20. doi: 10.1093/carcin/bgy191 [DOI] [PubMed] [Google Scholar]
- 13. Augoff K, Das M, Bialkowska K, McCue B, Plow EF, Sossey-Alaoui K. MiRNAs 484 and 210 regulate Pax-5 expression and function in breast cancer cells. Mol Cancer Res. (2011) 9:1500–08. doi: 10.1158/1541-7786.MCR-11-0311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pakravan K, Babashah S, Sadeghizadeh M, Mowla SJ, Mossahebi-Mohammadi M, Ataei F, et al. MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes suppresses in vitro angiogenesis through modulating the mTOR/HIF-1α/VEGF signaling axis in breast cancer cells. Cell Oncol. (2017) 40:457–70. doi: 10.1007/s13402-017-0335-7 [DOI] [PubMed] [Google Scholar]
- 15. Flores-Pérez A, Marchat LA, Rodríguez-Cuevas S, Bautista-Piña V, Hidalgo-Miranda A, Ocampo EA, et al. Dual targeting of ANGPT1 and TGFBR2 genes by miR-204 controls angiogenesis in breast cancer. Sci Rep. (2016) 6:34504. doi: 10.1038/srep34504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Keklikoglou I, Koerner C, Schmidt C, Zhang JD, Heckmann D, Shavinskaya A, et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene. (2012) 31:4150–63. doi: 10.1038/onc.2011.571 [DOI] [PubMed] [Google Scholar]
- 17. Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, et al. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol. (2012) 32:633–51. doi: 10.1128/MCB.06212-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Debeb BG, Lacerda L, Anfossi S, Diagaradjane P, Chu K, Bambhroliya A, et al. miR-141-mediated regulation of brain metastasis from breast cancer. J Natl Cancer Institute. (2016) 108:djw026. doi: 10.1093/jnci/djw026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hong L, Yang J, Han Y, Lu Q, Cao J, Syed L. High expression of miR-210 predicts poor survival in patients with breast cancer: A meta-analysis. Gene. (2012) 507:135–8. doi: 10.1016/j.gene.2012.07.025 [DOI] [PubMed] [Google Scholar]
- 20. Doberstein K, Bretz NP, Schirmer U, Fiegl H, Blaheta R, Breunig C, et al. MiR-21-3p is a positive regulator of L1CAM in several human carcinomas. Cancer Letters. (2014) 354:455–66. doi: 10.1016/j.canlet.2014.08.020 [DOI] [PubMed] [Google Scholar]
- 21. Donatelli SS, Zhou JM, Gilvary DL, Eksioglu EA, Chen X, Cress WD, et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci. (2014) 111:4203–8. doi: 10.1073/pnas.1319269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bai Y, Zhang Y, Hua J, Yang X, Zhang X, Duan M, et al. Silencing microRNA-143 protects the integrity of the blood-brain barrier: Implications for methamphetamine abuse. Sci Rep. (2016) 6:35642. doi: 10.1038/srep35642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma Q, Dasgupta C, Li Y, Huang L, Zhang L. MicroRNA-210 suppresses junction proteins and disrupts blood-brain barrier integrity in neonatal rat hypoxic-ischemic brain injury. Int J Mol Sci. (2017) 18:1356. doi: 10.3390/ijms18071356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reijerkerk A, Alejandro Lopez-Ramirez M, van het Hof B, Drexhage JAR, Kamphuis WW, Kooij G, et al. MicroRNAs regulate human brain endothelial cell-barrier function in inflammation: Implications for multiple sclerosis. J Neurosci. (2013) 33:6857–63. doi: 10.1523/JNEUROSCI.3965-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L, Sullivan PS, Goodman JC, Gunaratne PH, Marchetti D. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res. (2011) 71:645–54. doi: 10.1158/0008-5472.CAN-10-1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo YY, et al. Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. (2018) 78:4316–30. doi: 10.1158/0008-5472.CAN-18-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. (2008) 22:894–907. doi: 10.1101/gad.1640608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paterson EL, Kolesnikoff N, Gregory PA, Bert AG, Khew-Goodall Y, Goodall GJ. The microRNA-200 family regulates epithelial to mesenchymal transition. Sci World J. (2008) 8:901–4. doi: 10.1100/tsw.2008.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. (2008) 10:593–601. doi: 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- 30. Chen W, Hoffmann AD, Liu H, Liu X. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis Oncol. (2018) 2:4. doi: 10.1038/s41698-018-0047-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Steeg PS. The blood–tumour barrier in cancer biology and therapy. Nat Rev Clin Oncol. (2021) 18:696–714. doi: 10.1038/s41571-021-00529-6 [DOI] [PubMed] [Google Scholar]
- 32. Daneman R, Prat A. The blood-brain barrier. Cold Spring Harbor Perspect Biol. (2015) 7:a020412–a. doi: 10.1101/cshperspect.a020412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu D, Chen Q, Chen X, Han F, Chen Z, Wang Y. The blood–brain barrier: structure, regulation, and drug delivery. Signal Transduction Targeted Ther. (2023) 8:217. doi: 10.1038/s41392-023-01481-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim H, Leng K, Park J, Sorets AG, Kim S, Shostak A, et al. Reactive astrocytes transduce inflammation in a blood-brain barrier model through a TNF-STAT3 signaling axis and secretion of alpha 1-antichymotrypsin. Nat Commun. (2022) 13:6581. doi: 10.1038/s41467-022-34412-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sweeney MD, Ayyadurai S, Zlokovic BV. Pericytes of the neurovascular unit: Key functions and signaling pathways. Nat Neurosci. (2016) 19:771–83. doi: 10.1038/nn.4288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sirkisoon SR, Wong GL, Aguayo NR, Doheny DL, Zhu D, Regua AT, et al. Breast cancer extracellular vesicles-derived miR-1290 activates astrocytes in the brain metastatic microenvironment via the FOXA2→CNTF axis to promote progression of brain metastases. Cancer letters. (2022) 540:215726–. doi: 10.1016/j.canlet.2022.215726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature. (2016) 533:493–8. doi: 10.1038/nature18268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim SJ, Kim JS, Park ES, Lee JS, Lin Q, Langley RR, et al. Astrocytes upregulate survival genes in tumor cells and induce protection from chemotherapy. Neoplasia. (2011) 13:286–98. doi: 10.1593/neo.11112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei J, Gabrusiewicz K, Heimberger A. The controversial role of microglia in Malignant gliomas. J Immunol Res. (2013) 2013:285246–58. doi: 10.1155/2013/285246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. (2004) 432:231–5. doi: 10.1038/nature03049 [DOI] [PubMed] [Google Scholar]
- 41. Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. (2015) 519:482–5. doi: 10.1038/nature14281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Sci (New York NY). (2009) 326:1275–9. doi: 10.1126/science.1178705 [DOI] [PubMed] [Google Scholar]
- 43. Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. (2004) 18:3016–27. doi: 10.1101/gad.1262504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, et al. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. (2013) 155:1568–80. doi: 10.1016/j.cell.2013.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cheloufi S, Dos Santos CO, Chong MMW. Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. (2010) 465:584–9. doi: 10.1038/nature09092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang S, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, et al. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci. (2010) 107:15163–8. doi: 10.1073/pnas.1006432107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. (2011) 12:99–110. doi: 10.1038/nrg2936 [DOI] [PubMed] [Google Scholar]
- 48. Ipsaro JJ, Joshua-Tor L. From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat Struct Mol Biol. (2015) 22:20–8. doi: 10.1038/nsmb.2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci. (2008) 105:14879–84. doi: 10.1073/pnas.0803230105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. (2005) 120:15–20. doi: 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 51. Dharap A, Pokrzywa C, Murali S, Pandi G, Vemuganti R. MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PloS One. (2013) 8:e79467. doi: 10.1371/journal.pone.0079467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Brabletz T. To differentiate or not-routes towards metastasis. Nat Rev Cancer. (2012) 12:425–36. doi: 10.1038/nrc3265 [DOI] [PubMed] [Google Scholar]
- 53. Rettig M, Trinidad K, Pezeshkpour G, Frost P, Sharma S, Moatamed F, et al. PAK1 Kinase promotes cell motility and invasiveness through CRK-II serine phosphorylation in non-small cell lung cancer cells. PloS One. (2012) 7:e42012. doi: 10.1371/journal.pone.0042012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Craene BD, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. (2013) 13:97–110. doi: 10.1038/nrc3447 [DOI] [PubMed] [Google Scholar]
- 55. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. (2008) 133:704–15. doi: 10.1016/j.cell.2008.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Letters. (2008) 272:177–85. doi: 10.1016/j.canlet.2008.05.029 [DOI] [PubMed] [Google Scholar]
- 57. Imani S, Hosseinifard H, Cheng J, Wei C, Fu J. Prognostic value of EMT-inducing transcription factors (EMT-TFs) in metastatic breast cancer: A systematic review and meta-analysis. Sci Rep. (2016) 6:28587. doi: 10.1038/srep28587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wong AD, Searson PC. Live-cell imaging of invasion and intravasation in an artificial microvessel platform. Cancer Res. (2014) 74:4937–45. doi: 10.1158/0008-5472.CAN-14-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bolós V, Mira E, Martínez-Poveda B, Luxán G, Cañamero M, Martínez-A C, et al. Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res. (2013) 15:R54. doi: 10.1186/bcr3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sonoshita M, Aoki M, Fuwa H, Aoki K, Hosogi H, Sakai Y, et al. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. (2011) 19:125–37. doi: 10.1016/j.ccr.2010.11.008 [DOI] [PubMed] [Google Scholar]
- 61. Khuon S, Liang L, Dettman RW, Sporn PHS, Wysolmerski RB, Chew TL. Myosin light chain kinase mediates transcellular intravasation of breast cancer cells through the underlying endothelial cells: a three-dimensional FRET study. J Cell Science. (2010) 123:431–40. doi: 10.1242/jcs.053793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. (2015) 527:100–4. doi: 10.1038/nature15376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu Y, Kosaka A, Ikeura M, Kohanbash G, Fellows-Mayle W, Snyder LA, et al. Premetastatic soil and prevention of breast cancer brain metastasis. Neuro-Oncology. (2013) 15:891–903. doi: 10.1093/neuonc/not031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Soto MS, Serres S, Anthony DC, Sibson NR. Functional role of endothelial adhesion molecules in the early stages of brain metastasis. Neuro-Oncology. (2014) 16:540–51. doi: 10.1093/neuonc/not222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu K, Fukuda K, Xing F, Zhang Y, Sharma S, Liu Y, et al. Roles of the cyclooxygenase 2 matrix metalloproteinase 1 pathway in brain metastasis of breast cancer. J Biol Chem. (2015) 290:9842–54. doi: 10.1074/jbc.M114.602185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rempe RG, Hartz AMS, Bauer B. Matrix metalloproteinases in the brain and blood-brain barrier: Versatile breakers and makers. J Cereb Blood Flow Metab. (2016) 36:1481–507. doi: 10.1177/0271678X16655551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gunasinghe NPAD, Wells A, Thompson EW, Hugo HJ. Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev. (2012) 31:469–78. doi: 10.1007/s10555-012-9377-5 [DOI] [PubMed] [Google Scholar]
- 68. Yoshida T, Ozawa Y, Kimura T, Sato Y, Kuznetsov G, Xu S, et al. Eribulin mesilate suppresses experimental metastasis of breast cancer cells by reversing phenotype from epithelial–mesenchymal transition (EMT) to mesenchymal–epithelial transition (MET) states. Br J Cancer. (2014) 110:1497–505. doi: 10.1038/bjc.2014.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chao Y, Wu Q, Acquafondata M, Dhir R, Wells A. Partial mesenchymal to epithelial reverting transition in breast and prostate cancer metastases. Cancer Microenviron. (2012) 5:19–28. doi: 10.1007/s12307-011-0085-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim SW, Choi HJ, Lee HJ, He J, Wu Q, Langley RR, et al. Role of the endothelin axis in astrocyte- and endothelial cell-mediated chemoprotection of cancer cells. Neuro-Oncology. (2014) 16:1585–98. doi: 10.1093/neuonc/nou128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XHF, Lee DJ, et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. (2014) 156:1002–16. doi: 10.1016/j.cell.2014.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Choy C, Ansari KI, Neman J, Hsu S, Duenas MJ, Li H, et al. Cooperation of neurotrophin receptor TrkB and Her2 in breast cancer cells facilitates brain metastases. Breast Cancer Res. (2017) 19:51. doi: 10.1186/s13058-017-0844-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ye X-Z, Xu S-L, Xin Y-H, Yu S-C, Ping Y-F, Chen L, et al. Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGF-β1 signaling pathway. J Immunol. (2012) 189:444–53. doi: 10.4049/jimmunol.1103248 [DOI] [PubMed] [Google Scholar]
- 74. Wyss CB, Duffey N, Peyvandi S, Barras D, Usatorre M, Coquoz O, et al. Gain of HIF1 activity and loss of miRNA let-7d promote breast cancer metastasis to the brain via the PDGF/PDGFR axis. Cancer Res. (2021) 81:594–605. doi: 10.1158/0008-5472.CAN-19-3560 [DOI] [PubMed] [Google Scholar]
- 75. Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. (2013) 73:1434–44. doi: 10.1158/0008-5472.CAN-12-2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ahmad A, Sethi S, Chen W, Ali-Fehmi R, Mittal S, Sarkar FH. Up-regulation of microRNA-10b is associated with the development of breast cancer brain metastasis. Am J Trans Res. (2014) 6:384–90. [PMC free article] [PubMed] [Google Scholar]
- 77. Ahmad A, Ginnebaugh KR, Sethi S, Chen W, Ali R, Mittal S, et al. miR-20b is up-regulated in brain metastases from primary breast cancers. Oncotarget. (2015) 6:12188–95. doi: 10.18632/oncotarget.v6i14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Harati R, Mohammad MG, Tlili A, El-Awady RA, Hamoudi R. Loss of miR-101-3p promotes transmigration of metastatic breast cancer cells through the brain endothelium by inducing COX-2/MMP1 signaling. Pharmaceuticals. (2020) 13:1–19. doi: 10.3390/ph13070144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer-Secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. (2014) 25:501–15. doi: 10.1016/j.ccr.2014.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, et al. Breast cancer-secreted miR-122 reprograms glucose metabolism in pre-metastatic niche to promote metastasis. Nat Cell Biol. (2015) 17:183–94. doi: 10.1038/ncb3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Giannoudis A, Clarke K, Zakaria R, Varešlija D, Farahani M, Rainbow L, et al. A novel panel of differentially-expressed microRNAs in breast cancer brain metastasis may predict patient survival. Sci Rep. (2019) 9:18518. doi: 10.1038/s41598-019-55084-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tominaga N, Kosaka N, Ono M, Katsuda T, Yoshioka Y, Tamura K, et al. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood–brain barrier. Nat Commun. (2015) 6:1–12. doi: 10.1038/ncomms7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sereno M, Haskó J, Molnár K, Medina SJ, Reisz Z, Malhó R, et al. Downregulation of circulating miR 802-5p and miR 194-5p and upregulation of brain MEF2C along breast cancer brain metastasization. Mol Oncol. (2020) 14:520–38. doi: 10.1002/1878-0261.12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Figueira I, Godinho-Pereira J, Galego S, Maia J, Haskó J, Molnár K, et al. MicroRNAs and extracellular vesicles as distinctive biomarkers of precocious and advanced stages of breast cancer brain metastases development. Int J Mol Sci. (2021) 22:5214. doi: 10.3390/ijms22105214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Harati R, Hafezi S, Mabondzo A, Tlili A. Silencing miR-202-3p increases MMP-1 and promotes a brain invasive phenotype in metastatic breast cancer cells. PloS One. (2020) 15:e0239292–e. doi: 10.1371/journal.pone.0239292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pan J-K, Lin C-H, Kuo Y-L, Ger L-P, Cheng H-C, Yao Y-C, et al. MiR-211 determines brain metastasis specificity through SOX11/NGN2 axis in triple-negative breast cancer. Oncogene. (2021) 40:1737–51. doi: 10.1038/s41388-021-01654-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kaverina N, Borovjagin AV, Kadagidze Z, Baryshnikov A, Baryshnikova M, Malin D, et al. Astrocytes promote progression of breast cancer metastases to the brain via a KISS1-mediated autophagy. Autophagy. (2017) 13:1905–23. doi: 10.1080/15548627.2017.1360466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Xing F, Sharma S, Liu Y, Mo YY, Wu K, Zhang YY, et al. miR-509 suppresses brain metastasis of breast cancer cells by modulating RhoC and TNF-α. Oncogene. (2015) 34:4890–900. doi: 10.1038/onc.2014.412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hammash D, Mahfood M, Khoder G, Ahmed M, Tlili A, Hamoudi R, et al. miR-623 targets Metalloproteinase-1 and attenuates extravasation of brain metastatic triple-negative breast cancer cells. Breast Cancer: Targets Ther. (2022) 14:187–98. doi: 10.2147/BCTT.S372083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wu S, Lu J, Zhu H, Wu F, Mo Y, Xie L, et al. A novel axis of circKIF4A-miR-637-STAT3 promotes brain metastasis in triple-negative breast cancer. Cancer Lett. (2024) 581:216508. doi: 10.1016/j.canlet.2023.216508 [DOI] [PubMed] [Google Scholar]
- 91. Le X-F, Almeida MI, Mao W, Spizzo R, Rossi S, Nicoloso MS, et al. Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PloS One. (2012) 7:e41170–e. doi: 10.1371/journal.pone.0041170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yuan F, Wang W. MicroRNA-802 suppresses breast cancer proliferation through downregulation of FoxM1. Mol Med Rep. (2015) 12:4647–51. doi: 10.3892/mmr.2015.3921 [DOI] [PubMed] [Google Scholar]
- 93. Ulasov I, Borovjagin A, Fares J, Yakushov S, Malin D, Timashev P, et al. MicroRNA 345 (miR345) regulates KISS1-E-cadherin functional interaction in breast cancer brain metastases. Cancer Letters. (2020) 481:24–31. doi: 10.1016/j.canlet.2020.03.025 [DOI] [PubMed] [Google Scholar]
- 94. Iiizumi M, Bandyopadhyay S, Pai SK, Watabe M, Hirota S, Hosobe S, et al. RhoC promotes metastasis via activation of the Pyk2 pathway in prostate cancer. Cancer Res. (2008) 68:7613–20. doi: 10.1158/0008-5472.CAN-07-6700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. (1998) 391:806–11. doi: 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 96. Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol Ther Nucleic Acids. (2017) 8:132–43. doi: 10.1016/j.omtn.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. (2012) 489:57–74. doi: 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. (2010) 56:1733–41. doi: 10.1373/clinchem.2010.147405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PloS One. (2012) 7:e30679. doi: 10.1371/journal.pone.0030679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sun XY, Lu J, Zhang L, Song HT, Zhao L, Fan HM, et al. Aberrant microRNA expression in peripheral plasma and mononuclear cells as specific blood-based biomarkers in schizophrenia patients. J Clin Neurosci. (2015) 22:570–4. doi: 10.1016/j.jocn.2014.08.018 [DOI] [PubMed] [Google Scholar]
- 101. Seyhan AA. Circulating microRNAs as Potential Biomarkers in Pancreatic Cancer—Advances and Challenges. International Journal of Molecular Sciences. 2023;24(17):13340. doi: 10.3390/ijms241713340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Simian M, Farré PL, Duca R, Graña K, Magi G, Moro J, et al. Plasma microRNA signature as a potential early detection biomarker in breast cancer [abstract]. Journal of Clinical Oncology. (2024) 42(16):e13780. doi: 10.1200/JCO.2024.42.16_suppl.e13780 [DOI] [Google Scholar]
- 103. Chen Y, Huang Y, Deng Y, Liu X, Ye J, Li Q, et al. Cancer Therapy Empowered by Extracellular Vesicle-Mediated Targeted Delivery. Biological and Pharmaceutical Bulletin. (2023) 46(10):1353–64. doi: 10.1248/bpb.b23-00378 [DOI] [PubMed] [Google Scholar]
- 104. Chung CH, Lee JW, Slebos RJ, Howard JD, Perez J, Kang H, et al. A 3'-UTR KRAS-variant is associated with cisplatin resistance in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Annals of oncology : official journal of the European Society for Medical Oncology. (2014) 25(11):2230–6. doi: 10.1093/annonc/mdu367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Akbulut H, Ersoy YE, Coskunpinar E, Gucin Z, Yildiz S, Malya FU, et al. The role of miRNAs as a predictor of multicentricity in breast cancer. Molecular Biology Reports. (2019) 46(2):1787–96. doi: 10.1007/s11033-019-04629-6 [DOI] [PubMed] [Google Scholar]
- 106. Wu X, Somlo G, Yu Y, Palomares MR, Li AX, Zhou W, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. Journal of Translational Medicine. (2012) 10:42. doi: 10.1186/1479-5876-10-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Drusco A, Bottoni A, Laganà A, Acunzo M, Fassan M, Cascione L, et al. A differentially expressed set of microRNAs in cerebro-spinal fluid (CSF) can diagnose CNS Malignancies. Oncotarget. (2015) 6:20829–39. doi: 10.18632/oncotarget.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro-Oncology. (2012) 14:689–700. doi: 10.1093/neuonc/nos074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Querfeld C, Foss FM, Pinter-Brown LC, Porcu P, William BM, Pacheco T, et al. Phase 1 Study of the Safety and Efficacy of MRG-106, a Synthetic Inhibitor of microRNA-155, in CTCL Patients. Blood. (2017) 130:820. doi: 10.1182/blood.V130.Suppl_1.820.820 [DOI] [Google Scholar]
- 110. Gallant-Behm CL, Piper J, Dickinson BA, Dalby CM, Pestano LA, Jackson AL. A synthetic microRNA-92a inhibitor (MRG-110) accelerates angiogenesis and wound healing in diabetic and nondiabetic wounds. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society. (2018) 26(4):311–23. doi: 10.1111/wrr.12660 [DOI] [PubMed] [Google Scholar]
- 111. Varkaris A, Caravan P, Robertson N, Ghosh S, Warren M, Medarova Z, et al. Abstract CT246: An open-label, single-center, phase 0, microdose study to demonstrate delivery of TTX-MC138-NODAGA-Cu64 to radiographically confirmed metastases in subjects with advanced solid tumors [abstract]. Cancer Research. (2023) 83:CT246. doi: 10.1158/1538-7445.AM2023-CT246 [DOI] [Google Scholar]
- 112. Medarova Z, Robertson N, Ghosh S, Duggan S, Varkaris A. Initial clinical experience with the first-in-class anti-metastasis therapeutic TTX-MC138 [abstract]. Journal of Clinical Oncology. (2024) 42(Number 16_supplement):e15072. doi: 10.1200/JCO.2024.42.16_suppl.e15072 [DOI] [Google Scholar]
- 113. Hong DS, Kang Y-K, Borad M, Sachdev J, Ejadi S, Lim HY, et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. British Journal of Cancer. (2020) 122(11):1630–7. doi: 10.1038/s41416-020-0802-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Bauersachs J, Solomon SD, Anker SD, Antorrena-Miranda I, Batkai S, Viereck J, et al. Efficacy and safety of CDR132L in patients with reduced left ventricular ejection fraction after myocardial infarction: Rationale and design of the HF-REVERT trial. European journal of heart failure. (2024) 26(3):674–82. doi: 10.1002/ejhf.3139 [DOI] [PubMed] [Google Scholar]
- 115. Volpini L, Monaco F, Santarelli L, Neuzil J, Tomasetti M. Advances in RNA cancer therapeutics: New insight into exosomes as miRNA delivery. Aspects Mol Med. (2023) 1:100005. doi: 10.1016/j.amolm.2023.100005 [DOI] [Google Scholar]
- 116. Li Y, Liang Y, Sang Y, Song X, Zhang H, Liu Y, et al. MiR-770 suppresses the chemo-resistance and metastasis of triple negative breast cancer via direct targeting of STMN1. Cell Death Dis. (2018) 9:14. doi: 10.1038/s41419-017-0030-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Noyan S, Gurdal H, Dedeoglu BG. Involvement of miR-770-5p in trastuzumab response in HER2 positive breast cancer cells. PloS One. (2019) 14:e0215894. doi: 10.1371/journal.pone.0215894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bao L, Hazari S, Mehra S, Kaushal D, Moroz K, Dash S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am J Pathology. (2012) 180:2490–503. doi: 10.1016/j.ajpath.2012.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Jiao X, Zhao L, Ma M, Bai X, He M, Yan Y, et al. MiR-181a enhances drug sensitivity in mitoxantone-resistant breast cancer cells by targeting breast cancer resistance protein (BCRP/ABCG2). Breast Cancer Res Treat. (2013) 139:717–30. doi: 10.1007/s10549-013-2607-x [DOI] [PubMed] [Google Scholar]
- 120. Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. (2009) 75:1374–9. doi: 10.1124/mol.108.054163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. (2013) 31:577–. doi: 10.1038/nbt0713-577 [DOI] [PubMed] [Google Scholar]
- 122. Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet. (2019) 10:478. doi: 10.3389/fgene.2019.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]