Abstract
An important regulator involved in oxygen-dependent gene expression is the transcription factor HIF (hypoxia-inducible factor), which is composed of an oxygen-sensitive α-subunit (HIF-1α or HIF-2α) and a constitutively expressed β-subunit. In normoxia, HIF-1α is destabilized by post-translational hydroxylation of Pro-564 and Pro-402 by a family of oxygen-sensitive dioxygenases. The three HIF-modifying human enzymes have been termed prolyl hydroxylase domain containing proteins (PHD1, PHD2 and PHD3). Prolyl hydroxylation leads to pVHL (von-Hippel-Lindau protein)-dependent ubiquitination and rapid proteasomal degradation of HIF-1α. In the present study, we report that human PHD2 and PHD3 are induced by hypoxia in primary and transformed cell lines. In the human osteosarcoma cell line, U2OS, selective suppression of HIF-1α expression by RNA interference resulted in a complete loss of hypoxic induction of PHD2 and PHD3. Induction of PHD2 by hypoxia was lost in pVHL-deficient RCC4 cells. These results suggest that hypoxic induction of PHD2 and PHD3 is critically dependent on HIF-α. Using a VHL capture assay, we demonstrate that HIF-α prolyl-4-hydroxylase capacity of cytoplasmic and nuclear protein extracts was enhanced by prolonged exposure to hypoxia. Degradation of HIF-1α after reoxygenation was accelerated, which demonstrates functional relevance of the present results. We propose a direct, negative regulatory mechanism, which limits accumulation of HIF-1α in hypoxia and leads to accelerated degradation on reoxygenation after long-term hypoxia.
Keywords: hypoxia-inducible factor, oxygen sensing, post-translational modification, prolyl hydroxylation, protein degradation, von-Hippel-Lindau protein
Abbreviations: aRP, acidic ribosomal protein; EGLN, egg-laying deficient nine-like protein; HIF, hypoxia-inducible factor; IVTT, in vitro transcription/translation; ODD, oxygen-dependent degradation domain; PHD, prolyl hydroxylase domain containing protein; pVHL, von-Hippel-Lindau protein; RNAi, RNA interference; RPTEC, human renal proximal tubular epithelial cells; RT, reverse transcriptase; siRNA, small interfering RNA
INTRODUCTION
The ability to adapt to acute and chronic hypoxia is crucial for survival of cells and organs under physiological and pathological conditions. This adaptation to hypoxia involves a precisely regulated system of altered gene expression. HIF-1 (hypoxia-inducible factor-1) is the main transcription factor responding to reduced oxygen availability. Target genes of HIF-1 regulate several metabolic processes including oxygen homoeostasis, energy metabolism, growth and differentiation [1,2]. HIF is composed of the constitutive nuclear protein ARNT (arylhydrocarbon receptor nuclear translocator), also termed HIF-1β, and an oxygen-sensitive α-subunit. HIF-1α is constantly synthesized but rapidly degraded under normoxic conditions, whereas reduced oxygen concentration results in stabilization of HIF-1α, translocation to the nucleus, heterodimerization with ARNT, DNA binding and promotion of target gene transcription.
The molecular mechanisms leading to normoxic degradation of HIF-1α have recently been elucidated (see [3] for a review). A family of newly discovered oxygen-dependent hydroxylases modify specific amino acid residues within HIF-α proteins [4,5]. Three distinct enzymes have been characterized [6] and named as prolyl hydroxylase domain-containing proteins (PHD), PHD1 is also called HPH3 (HIF-prolyl-hydroxylase 3) [7] and EGLN2 (egg-laying deficient nine-like protein 2) [8], PHD2 is identical with HPH2 [7] and EGLN1 [8] and PHD3 is identical with HPH1 [7] and EGLN3 [8]. These enzymes use molecular oxygen to hydroxylate the conserved Pro-564 and Pro-402 residues within the ODD (oxygen-dependent degradation domain) in HIF-1α [6,9]. This normoxic modification of the HIF-α subunit causes binding to the tumour suppressor pVHL (von-Hippel-Lindau protein), which is part of an E3-ubiquitin ligase and targets HIF-1α for subsequent proteasomal degradation [10]. An asparaginyl hydroxylase, termed factor inhibiting HIF (FIH-1), hydroxylates Asn-803 within the C-terminal transactivation domain of HIF-1α under conditions of normal oxygen tension. The hydroxylated C-terminal transactivation domain does not interact with transcriptional co-activators (p300/CREB-binding protein) and thus FIH-1 impairs normoxic transcription of HIF-1 target genes [11–13]. The HIF-α hydroxylases show a distinct pattern of subcellular localization [14,15] and seem to have different affinities for the proline residues in HIF-α subunits [16,17].
Although significant progress has been made, several aspects of the function and regulation of the HIF system are still incompletely understood. For example, normoxic activation of HIF by oncogenes [18], growth factors and cytokines [19] and small signalling molecules, such as NO [20,21], has been reported. Furthermore, pVHL has a stabilizing effect on microtubules, which has been shown to be unrelated to the HIF system [22]. Recent serial analysis of gene expression has shown that cells lacking HIF-1α can still express a number of genes in response to hypoxia [23]. Another still poorly understood observation relates to the fact that HIF-1α levels can decrease in cells during prolonged exposure to hypoxia [24]. This decrease indicates that a feedback mechanism may exist, which augments HIF-1α degradation under hypoxic conditions. Indeed, some studies have shown that PHD mRNA expression increases during hypoxia in human tumour cell lines [6,14,25].
In the present study, we analysed the transcriptional pathway leading to hypoxic induction of the human HIF-α prolyl hydroxylases PHD2 and PHD3. We show that PHD2 and PHD3 expression was induced by hypoxia in several primary and permanent cell lines and that the inducibility depended on functional VHL and HIF-1α. We propose that accumulation of PHD2 and PHD3 protein by hypoxia leads to limited steady-state levels of HIF-α under hypoxic conditions and to accelerated degradation after reoxygenation.
MATERIALS AND METHODS
Cell culture
Human osteosarcoma cells U2OS, hepatoma cells HepG2 and renal clear cell carcinoma cells RCC4 were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco, Karlsruhe, Germany) supplemented with 10% (v/v) foetal calf serum (Gibco), 2 mM glutamine (Gibco), 50 i.u./ml penicillin and 50 μg/ml streptomycin (Sigma, Deisenhofen, Germany). Primary RPTEC (human renal proximal tubular epithelial cells) were grown in DMEM/Ham's F12 medium (1:1 ratio), supplemented with insulin (5 μg/ml), transferrin (5 μg/ml), selenium (5 ng/ml), hydrocortisone (36 ng/ml), tri-iodothyronine (4 pg/ml) and epidermal growth factor (10 ng/ml; all from Sigma) and 15 mM Hepes buffer (BioWhittaker, Walkersville, MD, U.S.A.). Experiments were performed with subconfluent cultures. Hypoxic incubations were performed in an atmosphere of 1% O2, 5% CO2 and balance N2 in a hypoxia workstation (InvivO2, Ruskinn Technology, Leeds, U.K.). Normoxia was defined as incubation in a humidified atmosphere containing air plus 5% CO2. For reoxygenation experiments, dishes were removed from the 1% incubator and placed into a conventional incubator.
Protein extraction and analysis
Total cellular protein extracts for HIF-α Western analysis was prepared on ice using a denaturating lysis buffer containing 8 M urea, 1% SDS, 8.7% (v/v) glycerine and 10 mM Tris (pH 6.8). Protein samples for PHD Western blotting were prepared in a standard Nonidet P40 buffer supplemented with protease inhibitors (Protease Inhibitor Cocktail Set V, Calbiochem). Cultures were washed twice with ice-cold PBS, lysed and then sonicated. Equal amounts of cell lysates (40 μg of protein) were subjected to SDS/PAGE and blotted on to Protran membranes (Schleicher and Schuell, Dassel, Germany). Membranes were blocked with 5% skim milk in PBS overnight. HIF-1α was detected with a monoclonal anti-HIF-1α-antibody (BD-Transduction Laboratories, Heidelberg, Germany) diluted 1:2000 in skim milk for 2 h. A monoclonal HIF-2α antibody (Abcam, Acris GmbH, Bad Nauheim, Germany) was used in a dilution of 1:500 overnight. A polyclonal PHD2 antibody was obtained from NOVUS Biologicals (Littleton, CO, U.S.A.), diluted 1:500; incubations were for 2 h at room temperature (22 °C). The constitutively expressed transcription factor SP1 was detected with a monoclonal antibody (PEP2; Santa Cruz Biotechnology, Heidelberg, Germany), diluted 1:1000. After three washes with PBS, the membranes were incubated for another 2 h with an appropriate horseradish-peroxidase-coupled secondary antibody (Dako, Hamburg, Germany) diluted 1:2000 in skim milk. Binding of the antibodies was visualized using the ECL® Western Blotting Analysis system (Amersham Biosciences, Freiburg, Germany).
RNA extraction and real-time PCR
The total cellular RNA was extracted by the acid guanidinium thiocyanate–phenol–chloroform method [26]. In brief, the culture medium was removed and cells were immediately washed with ice-cold PBS. Subsequently, 1000 μl of TRIzol® Reagent (Invitrogen, Karlsruhe, Germany) was added and the total cellular RNA was extracted according to the manufacturer's instructions. The total RNA (1 μg) was used as template in a M-MLV RT (reverse transcriptase) reaction, which was performed according to the manufacturer's instructions (Invitrogen). The resulting cDNA (2 μl of RT reaction) was used as template for quantitative real-time PCR, which was performed on an ABI Prism 7000 (Applied Biosystems, Darmstadt, Germany) using qPCR core kit for SYBR Green I (Eurogentec, Cologne, Germany). PCRs were set up to a final volume of 25 μl. The established primers for PHD1-, PHD2-, PHD3-PCR were used at 10 pmol and for 60 S aRP-PCR (where aRP stands for acidic ribosomal protein) at 5 pmol respectively [14]. After initial activation of the DNA polymerase and double-strand dissociation at 95 °C for 10 min, a two-step PCR with 40 cycles of primer annealing and elongation at 60 °C for 1 min followed by denaturation at 95 °C for 15 s was performed. Specificity of the PCR product was confirmed by examination of the dissociation curve (60–95 °C) and by agarose gel electrophoresis. The purified PCR products served as internal standards. All samples were quantified in duplicate and normalized to 60 S aRP-cDNA.
Microarray procedures
cDNA microarrays were produced in-house. Human EST (expressed sequence tag) library (ResGen) was used as a template. PCR products (approx. 3700 genes) were spotted on to poly(L-lysine)-coated glass slides in triplicates. Relevant genes were sequence-verified. Probes for cDNA microarrays were generated using 20 μg of total DNase I (Qiagen, Hilden, Germany)-treated and purified RNA (Rneasy Mini column, Qiagen) from cells exposed to normoxia (21% O2) or hypoxia (1% O2, 4 h) in a standard RT reaction in which dTTP was partly (one-third) replaced with either 73 μM dUTP labelled with the fluorescent dye Cy3 (normoxia) or 73 μM dUTP labelled with the fluorescent dye Cy5 (hypoxia) (Amersham Biosciences). Probes were cleaned using the Microcon YM30 nucleotide removal kit (Millipore). Probes were combined and hybridized to the array overnight at 65 °C. Slides were washed, dried and scanned using ScanArray 5000 scanner (PerkinElmer Life Sciences) at 543 nm (Cy3) and 633 nm (Cy5) simultaneously. The images were analysed using ScanArrayExpress software. The raw data were first Lowes-normalized followed by calculation of background-subtracted median ratio value for each spot. Subsequently, each spot was corrected against internal β-actin control. Three replicate spots on each slide were averaged.
RNA interference (RNAi)
Oligonucleotides for RNAi were purchased from MWG-Biotech (Ebersberg, Germany). A sequence corresponding to nt 661–681 (GenBank® accession number NM 001530) of HIF-1α was chosen as siRNA (small interfering RNA) (5′-AACUAACUGGACACAGUGUGU-3′). Transfection of cells was performed using Oligofectamine® (Invitrogen) as the transfection reagent. For each dish, a solution of 450 pmol double-stranded RNA in 1500 μl of serum-free cell culture medium and 90 μl of Oligofectamine® in 360 μl medium was prepared in separate vials. After incubation for 7–10 min, both solutions were mixed and the medium was added after 25 min to a final volume of 3 ml/dish. Cells were transfected by dropwise addition of the siRNA-vehicle solution to the culture medium. Hypoxic incubation started after a 4 h normoxic period to allow degradation of preformed HIF-1α.
Protein interaction assays
U2OS cells were incubated in normoxia or in hypoxia for 1 or 24 h respectively. Cytoplasmic and nuclear extracts were prepared as described previously [16]. Briefly, the cells were washed and scraped into ice-cold PBS. After centrifugation, PBS was completely removed and cell membranes were lysed by incubation with cold hypotonic extraction buffer (20 mM Tris, pH 7.5/5 mM KCl/1.5 mM MgCl2/1 mM dithiothreitol) and vigorous mixing. Nuclei and debris were spun down, the supernatant was removed (designated cytoplasmic extract) and stored at −80 °C. Three volumes of hypertonic buffer [20 mM Tris, pH 7.5/420 mM KCl/1.5 mM MgCl2/20% (v/v) glycerol] were added to the nuclear pellet. The samples were incubated on ice. After centrifugation, the supernatant was stored at −80 °C (nuclear extract). Gal-HIF-1α549–582, PHD2 and 35S-labelled pVHL were prepared by coupled IVTT (in vitro transcription/translation) using TNT® reticulocyte lysate (Promega). Gal-HIF-1α549–582 was purified on agarose beads conjugated with antibodies raised against the DNA-binding domain of the yeast transcription factor Gal4 (Santa Cruz Biotechnology) and suspended in hypotonic extraction buffer supplemented with 1 mM ascorbate, 0.1 mM α-ketoglutarate and 20 μM FeCl2. Gal-HIF-1α549–582 was then incubated for 12 min either with cell extracts or with pcDNA3 programmed reticulocyte lysate serving as negative control or with IVTT-PHD2 as positive control. The reaction was terminated by the addition of 100 μM desferrioxamine. 35S-pVHL was added after washing the beads in wash buffer (150 mM NaCl/0.5 mM EDTA/0.5% Nonidet P40/50 mM Tris, pH 7.5) several times. The samples were incubated on an end-over-end rotator at 4 °C overnight. After removing unbound 35S-pVHL by washing, the beads were boiled in SDS/PAGE loading buffer. The supernatant was subjected to SDS/PAGE (15% gel); pVHL was detected by autoradiography. Due to an internal in-frame methionine at codon 54, two isoforms were regularly detected.
RESULTS
Hypoxia leads to induction of PHD2 and PHD3 in primary and permanent cell lines
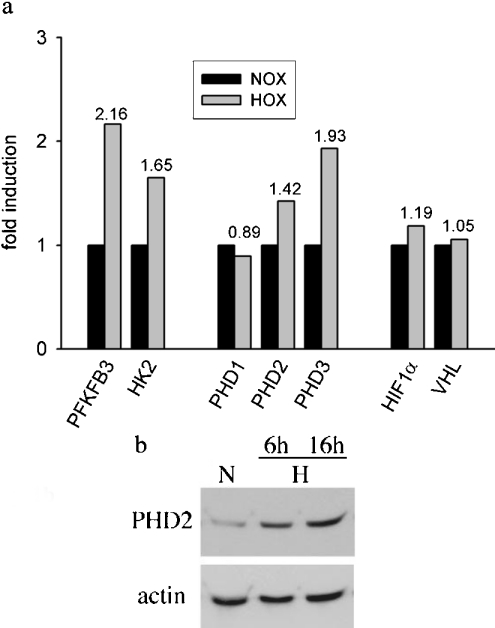
Our study was based on reports showing induction of PHDmRNA by hypoxia in HeLa cells [6,27], in U2OS cells [14], in C6 rat glioma cells [28] and in cells of cardiovascular origin [29]. We assessed PHD induction in HepG2 cells by real-time PCR and microarray analysis. In line with previous reports, we found that PHD2 and PHD3 were hypoxia inducible, whereas transcription of PHD1 appeared constitutive. Reassuringly, the PCR results were confirmed by microarray data (Figure 1a). In HepG2 cells, induction levels after 4 h of hypoxic incubation were comparable with 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 and hexokinase, both of which are known HIF-1 target genes [1]. Hypoxic induction of PHD2 was also confirmed by Western blotting (Figure 1b). Next, we tried to establish that regulation in primary human cell lines resembles regulation in permanent cell lines. To this end, untransformed RPTEC were subjected to hypoxia. This incubation resulted in an increase in PHD2- and PHD3mRNA as quantified by real-time PCR (Figure 2). In line with published results for cancer cells, PHD1 gene expression did not seem to be regulated by hypoxia. Interestingly, short-term hypoxia (1 h) did not induce PHD mRNA, whereas hypoxic incubation overnight had a striking effect.
Figure 1. PHD induction in HepG2 cells by hypoxia.
(a) Microarray analysis. Cells were incubated in an atmosphere of 1% O2 for 4 h. cDNA probes were generated as described in the Materials and methods section. Normoxic (NOX) and hypoxic (HOX) probes were combined and hybridized to the array overnight at 65 °C. Three replicate spots on each slide were averaged. (b) PHD2 Western blot. Cells were treated as for microarrays. Whole cell-protein extracts were subjected to SDS/PAGE and immunoblot analysis.
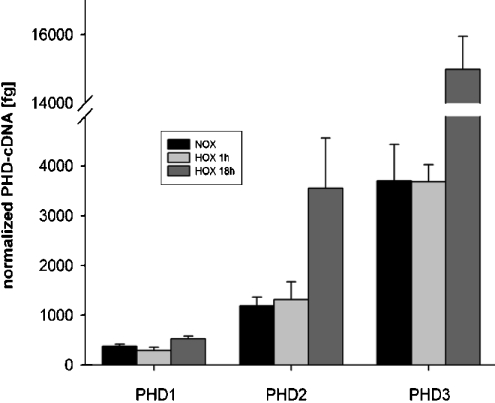
Figure 2. PHD induction in renal proximal tubule cells by hypoxia.
Real-time quantitative RT–PCR was performed for PHD1, PHD2, PHD3 of normoxic (NOX), short-term hypoxic (HOX 1 h) and long-term hypoxic (HOX 18 h) RPTEC cultures. Bars indicate means±S.D. of three samples from one experiment. Results are given normalized to 60 S aRP mRNA as a housekeeping gene. These results are representative of two independent experiments.
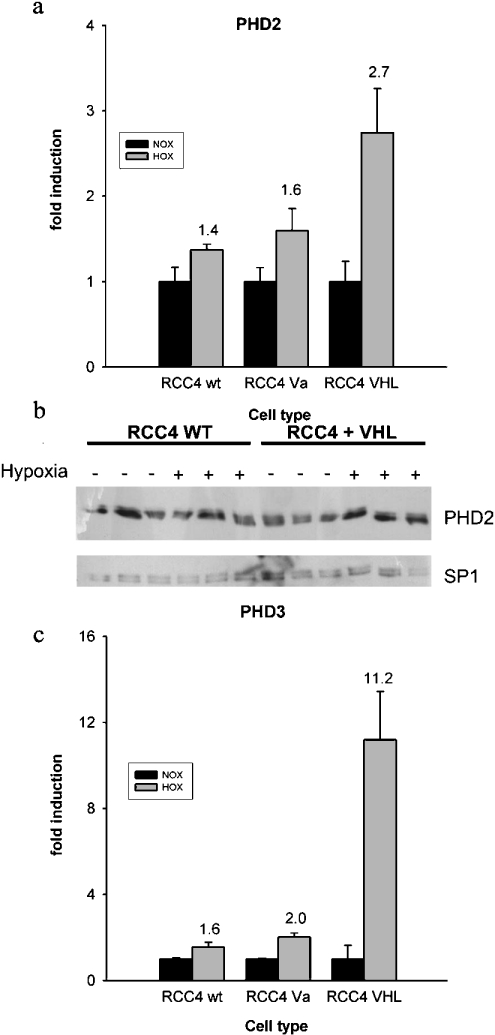
Hypoxic induction of PHD2 and PHD3 is VHL-dependent
pVHL is an essential component in the HIF-α degradation pathway as it is the substrate recognizing subunit of an E3-ubiquitin ligase. VHL-negative RCC4 cells display high normoxic levels of HIF-α. Recompletion by stable transfection with a VHL expression vector re-establishes normal HIF-α regulation in these cells. We studied whether the dysregulation of HIF-α is associated with the loss of hypoxic induction of PHD2 and PHD3 mRNA. Although we found that PHD induction by hypoxia was substantially reduced when compared with VHL recompleted cells, some induction was still present in wild-type RCC4 cells (Figures 3a and 3c). Western blotting indicated that PHD2 protein levels are somewhat variable in RCC4 cells. PHD2 bands were quantified by densitometry and correlated to SP1 expression, which is not hypoxia-inducible. This analysis showed that PHD2 was induced 1.4-fold in pVHL-positive cells, whereas induction was abolished in RCC4 wild-type cells.
Figure 3. Normoxic and hypoxic PHD levels in VHL-deficient cells.
Quantitative RT–PCR of different RCC4 sublines was performed. cDNA levels were corrected for 60 S aRP. Results are presented for (a) PHD2 and (c) PHD3 in wild-type (wt), mock-transfected (Va) and VHL-recompleted (VHL) RCC4 cells. Bars indicate means±S.D. of three independent samples from one experiment. (b) Samples were prepared from three independent cultures of both cell lines in normoxia and in hypoxia and analysed for PHD2 and SP1 protein expression. SP1 serves as a loading and transfer control.
Hypoxic induction of PHDmRNA depends on HIF-1α
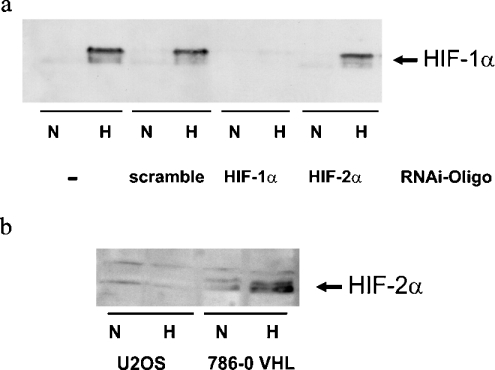
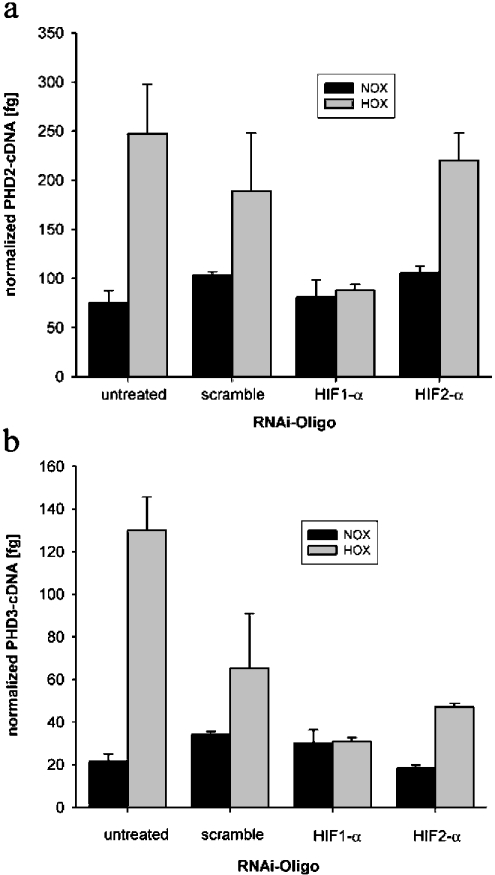
The role of HIF-1α in hypoxic PHD gene induction was assessed using RNAi. Double-stranded RNA oligonucleotides with sequence homology to endogenous mRNA lead to subsequent mRNA cleavage by a multienzyme complex [30]. Thus short double-stranded RNA derived from the HIF-1α sequence can be used to generate a temporal inhibition of HIF-1α function as reported recently [31]. Figure 4(a) shows efficiency of this procedure in U2OS cells as induction of HIF-1α protein was not detectable despite hypoxic incubation for 18 h. Western-blot analysis also demonstrated that HIF-2α was not present in these cells (Figure 4b). Real-time RT–PCR was performed after an overnight incubation in hypoxia with and without HIF-1α siRNA transfection. A 2-fold induction of PHD2 and PHD3 mRNA by hypoxia in mock-transfected cells was detected. This hypoxic induction was completely abrogated by HIF-1α siRNA transfection. As expected, transfection with HIF-2α RNAi-oligo-nucleotides did not suppress hypoxic PHD mRNA induction, showing that HIF-1α is sufficient to induce PHD expression (Figure 5).
Figure 4. Western-blot analysis of HIF-α in U2OS cells.
(a) Western-blot analysis of HIF-1α protein levels in U2OS cells treated with siRNA oligonucleotides. Cells were incubated with scramble or HIF-1α or HIF-2α oligonucleotides for 4 h and then subjected to hypoxia for 18 h. (b) Western-blot analysis of HIF-2α after 4 h of hypoxia demonstrating hypoxic induction of HIF-2α in VHL-transfected 786–0 cells but not in U2OS cells.
Figure 5. PHD expression in normoxia and in hypoxia after RNAi.
Real-time quantitative RT–PCR for (a) PHD2 and (b) PHD3 in U2OS cultures transfected with the indicated siRNA oligonucleotides. Bars indicate the normalized means±S.D. of three samples from one experiment.
Hypoxia increases cellular hydroxylation capacity
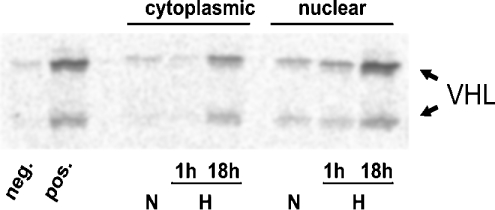
We assessed the hydroxylase capacity of cytoplasmic and nuclear protein extracts prepared from U2OS cells after normoxic or hypoxic incubation for 1 or 18 h respectively. As the protein extracts were used in vitro in the abundance of all substrates and cofactors, the results reflect maximal possible enzyme turnover rather than actual HIF hydroxylation in vivo. The in vitro protein interaction assay was performed as described previously [5]. Gal-HIF-1α549–582 fusion protein was exposed to the cell-protein extracts. Hydroxylation of the ODD by the extracts was determined by binding of [35S]methionine-labelled pVHL. Figure 6 shows increased hydroxylase capacity in U2OS cells after prolonged exposure to hypoxia. This result suggests that hypoxic induction of PHD2 and PHD3 led to an increased enzyme activity in the cytoplasm as well as in the cell nucleus.
Figure 6. In vitro hydroxylase activity assay.
Equal amounts of nuclear and cytoplasmic protein extracts from normoxic (N), short-term hypoxic (H, 1 h) and long-term hypoxic (H, 18 h) U2OS cultures hydroxylated Gal-HIF-1α549–582 bound to agarose beads. After stopping hydroxylation by adding desferrioxamine, 35S-labelled IVTT-pVHL was added and incubated overnight. Supernatants were subjected to SDS/PAGE and 35S-pVHL was detected by autoradiography. Captured 35S-VHL indicates hydroxylase activity of the protein extracts modifying the HIF-ODD. IVTT-pVHL appears in two isoforms due to an internal in-frame start codon. ‘neg.’ and ‘pos.’ indicate hydroxylase activity of unprogrammed reticulocyte lysate and IVTT-PHD2 respectively.
Hypoxia causes a decrease in the half-life of HIF-1α after reoxygenation
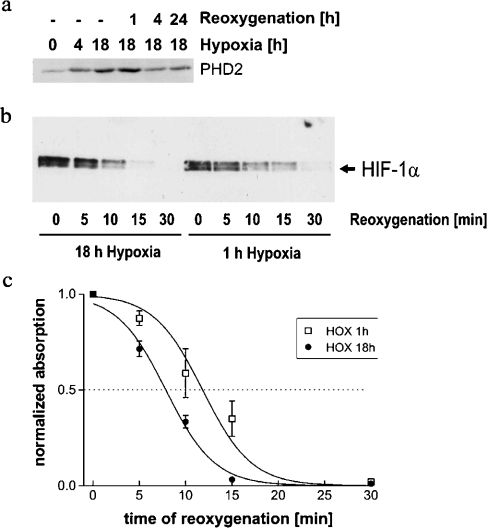
Increased PHD2 and PHD3 levels are expected to result in accelerated degradation of HIF-1α protein on reoxygenation after long-term hypoxia. In U2OS cells, maximal induction levels remained constant from 4 to 48 h after onset of hypoxia (results not shown). Figure 7(a) shows that PHD2 is strongly induced by overnight incubation in a hypoxic atmosphere in U2OS cells. After reoxygenation, PHD2 levels decline but remain increased even after return to normoxia for 24 h. Interestingly, induced PHD2 levels are constant after the first hour of reoxygenation, which may indicate that PHD2 protein is relatively stable. HIF-1α protein levels in reoxygenated whole cell lysates of U2OS cells were assessed after short and prolonged hypoxic periods (Figure 7b). Four independent hypoxia/reoxygenation experiments were performed following the same method. HIF-1α bands were quantified by densitometry. Means were calculated and plotted against time after reoxygenation (Figure 7c). This analysis demonstrated that reoxygenation after prolonged hypoxic incubation led to more rapid HIF-1α degradation, which is consistent with the induction of PHD2 and PHD3 under these conditions.
Figure 7. Western-blot analysis of PHD2 and HIF-1α after hypoxic incubation and reoxygenation.
U2OS cultures were lysed at the indicated time of hypoxia and reoxygenation after long-term (18 h) and short-term (1 h) hypoxia. SDS/PAGE was performed with a total of 40 μg protein loaded in each lane: (a) PHD2; (b) time course of HIF-1α degradation; (c) line plot showing levels of HIF-1α protein at the time points shown in (b). Data originate from four repetitions of the hypoxia/reoxygenation experiments. HIF-1α bands were analysed by densitometry; results are expressed as means±S.D. Lines were calculated by non-linear regression.
DISCUSSION
HIF-1 is the main transcription factor adapting gene expression to hypoxia [2]. HIF-1α is regulated by oxygen-sensitive hydroxylases, which modify distinct amino acid residues (see [3] and references therein). Hydroxylation of Pro-564 and Pro-402 leads to binding of pVHL, subsequent ubiquitination and proteasomal degradation. In the present study, we show that transcription of human PHD2 and PHD3 is hypoxia-inducible. Furthermore, we demonstrate that these enzymes, which initiate degradation of HIF-1α, are target genes of HIF-1 and thus are part of a direct negative feedback mechanism. A very recent report suggests that PHD2 is the main enzyme down-regulating HIF-1α in normoxia as RNAi for PHD2 is sufficient to up-regulate HIF-1α in normoxia [27]. Human PHD2 and PHD3 have been reported to be hypoxia-inducible in cancer cells [6,14,27], whereas only PHD3 was found to be hypoxia-inducible in cultured cardiovascular cells [29]. Our results confirm that PHD2 and PHD3 induction by hypoxia is a more general mechanism not restricted to cancer cells. Interestingly, PHD1 mRNA was not found to be hypoxia-inducible, which is in line with previous studies. Human PHD1 seems to differ in some way from its isoenzymes. First, it is the only HIF hydroxylase that is restricted to the cell nucleus and, secondly, it may have a lower efficiency in destabilizing endogenous HIF-1α when compared with PHD2 and PHD3 [15]. Besides these features, evidence exists for oestrogen inducibility of this gene in human mammary carcinoma [32] and for up-regulation in renal carcinoma compared with normal renal tubular epithelium [23].
It seemed important to us to define whether the up-regulation of PHD expression is HIF-1-dependent because manipulation of the HIF system is currently regarded as a potential therapeutic principle [33]. Having shown that PHD2 and PHD3 are induced by hypoxia in U2OS, HepG2 and untransformed renal proximal tubule cells, we analysed PHD regulation in pVHL-deficient renal carcinoma cells that show up-regulation of HIF-α in normoxia. While our paper was at the preparation stage, it was reported that these cells show loss of PHD2 and PHD3 induction in response to hypoxia [25]. In our experiments, hypoxic PHD induction was massively reduced but still present to a small extent. Western blotting for PHD2 indicated that hypoxic induction was blunted in pVHL-deficient cells, whereas pVHL recomplemented cells had regained normal induction. This result suggests that induction of PHD2 is dependent on normal regulation of HIF-1. We conducted a second series of experiments in which we used RNAi in U2OS cells to inhibit hypoxic activation of HIF-1α. This manoeuvre completely blunted PHD induction in hypoxia. These results suggest that PHD2 and PHD3 are indeed HIF-1 target genes although additional mechanisms may be involved in other tissues or species.
Many steps of the HIF processing are not under the control of oxygen, such as HIF-1α translation [34]. The activity of PHDs would offer a checkpoint to limit and fine-tune HIF-1 activity in hypoxia. Furthermore, a feedback for HIF would be required to allow fast limitation of its activity after reoxygenation. To demonstrate a negative feedback loop, it was important to show increased hydroxylase capacity of cytoplasmic and nuclear protein extracts prepared from the hypoxic cells. Interestingly, the induction of HIF-α prolyl hydroxylase activity closely paralleled mRNA and protein induction. Significant induction was achieved after overnight exposure to hypoxia in the cytoplasm and in the cell nucleus. This result is plausible because PHD2 is predominantly cytoplasmic, whereas PHD3 is present in cytoplasm and nucleus [14]. As overexpression of PHDs suppresses HIF-1α accumulation [9,14] it seems probable that induction of PHDs by hypoxia can limit HIF-1α protein abundance and gene activation in hypoxia. Furthermore, two observations that were published before the characterization of the PHDs are explained by our results: first, the half-life of HIF-1α is shortened by prolonged exposure to hypoxia ([35] and confirmed in the present study). Obviously, the induction of PHD2 and PHD3 would allow faster degradation after long periods of hypoxia. Secondly, it has been reported that HIF-1α and HIF-2α levels are maximal after 4 h of hypoxia and decline thereafter [24]. As significant PHD activity in hypoxia can be demonstrated by addition of prolyl hydroxylase inhibitors (P. Stengel, unpublished work), it seems possible that the induction of PHD activity in hypoxia is responsible for the down-regulation of HIF-1α abundance in a situation of prolonged hypoxia.
In conclusion, we have demonstrated HIF-dependent hypoxic induction of PHD2 and PHD3, which may in turn down-regulate HIF activity in hypoxia. Thus PHD induction forms a direct negative feedback mechanism. An open question is whether PHD induction by hypoxia leads to differential regulation of HIF activity in response to short- and long-term hypoxia. Clearly, further work is required to define hypoxia-responsive elements in the regulatory DNA sequences of the PHD2 and PHD3 genes.
Acknowledgments
We thank I. Jensen, C. Kauderer, R. Meuer, T. Svensson and B. Stier for excellent technical assistance. Dr M. Nitschke (Department of Internal Medicine I, University of Luebeck, Germany) is gratefully acknowledged for providing RPTEC cultures. This study was supported by a grant from DFG (SFB367-C8) to W. J. and E. M. and by grants from the Academy of Finland (200462), Sigrid Juselius Foundation and Finnish Cancer Foundation to P. J.
References
- 1.Wenger R. H. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 2002;16:1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- 2.Semenza G. L. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr. Opin. Genet. Dev. 1998;8:588–594. doi: 10.1016/s0959-437x(98)80016-6. [DOI] [PubMed] [Google Scholar]
- 3.Masson N., Ratcliffe P. J. HIF prolyl and asparaginyl hydroxylases in the biological response to intracellular O2 levels. J. Cell Sci. 2003;116:3041–3049. doi: 10.1242/jcs.00655. [DOI] [PubMed] [Google Scholar]
- 4.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 5.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 6.Epstein A. C. R., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell (Cambridge, Mass.) 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 7.Bruick R. K., McKnight S. L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 8.Taylor M. S. Characterization and comparative analysis of the EGLN gene family. Gene. 2001;275:125–132. doi: 10.1016/s0378-1119(01)00633-3. [DOI] [PubMed] [Google Scholar]
- 9.Bruick R. K., McKnight S. L. Oxygen sensing gets a second wind. Science. 2002;295:807–808. doi: 10.1126/science.1069825. [DOI] [PubMed] [Google Scholar]
- 10.Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature (London) 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 11.Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 12.Mahon P. C., Hirota K., Semenza G. L. FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hewitson K. S., McNeill L. A., Riordan M. V., Tian Y. M., Bullock A. N., Welford R. W., Elkins J. M., Oldham N. J., Bhattacharya S., Gleadle J. M., et al. Hypoxia inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 14.Metzen E., Berchner-Pfannschmidt U., Stengel P., Marxsen J. H., Stolze I., Klinger M., Huang W. Q., Wotzlaw C., Hellwig-Buergel T., Jelkmann W., et al. Intracellular localisation of human HIF-1α hydroxylases: implications for oxygen sensing. J. Cell Sci. 2003;116:1319–1326. doi: 10.1242/jcs.00318. [DOI] [PubMed] [Google Scholar]
- 15.Huang J., Zhao Q., Mooney S. M., Lee F. S. Sequence determinants in hypoxia inducible factor-1α for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J. Biol. Chem. 2002;277:39792–39800. doi: 10.1074/jbc.M206955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masson N., Willam C., Maxwell P. H., Pugh C. W., Ratcliffe P. J. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirsilä M., Koivunen P., Günzler V., Kivirikko K. I., Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 18.Chan D. A., Sutphin P. D., Denko N. C., Giaccia A. J. Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1α. J. Biol. Chem. 2003;277:40112–40117. doi: 10.1074/jbc.M206922200. [DOI] [PubMed] [Google Scholar]
- 19.Hellwig-Burgel T., Rutkowski K., Metzen E., Fandrey J., Jelkmann W. Interleukin-1β and tumor necrosis factor-α stimulate DNA binding of hypoxia-inducible factor-1. Blood. 1999;94:1561–1567. [PubMed] [Google Scholar]
- 20.Zhou J., Schmid T., Brüne B. Tumor necrosis factor-α causes accumulation of a ubiquitinated form of hypoxia inducible factor-1α through a nuclear factor-κB-dependent pathway. Mol. Biol. Cell. 2003;14:2216–2225. doi: 10.1091/mbc.E02-09-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metzen E., Zhou J., Jelkmann W., Fandrey J., Brune B. Nitric oxide impairs normoxic degradation of HIF-1α by inhibition of prolyl hydroxylases. Mol. Biol. Cell. 2003;14:3470–3481. doi: 10.1091/mbc.E02-12-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hergovich A., Lisztwan J., Barry R., Ballschmieter P., Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat. Cell Biol. 2003;5:64–70. doi: 10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Y., Zhang W., Kondo K., Klco J. M., Martin, St T. B., Dufault M. R., Madden S. L., Kaelin W. G., Nacht M. Gene expression profiling in a renal cell carcinoma cell line: dissecting VHL and hypoxia-dependent pathways. Mol. Cancer Res. 2003;1:453–462. [PubMed] [Google Scholar]
- 24.Wiesener M. S., Turley H., Allen W. E., Willam C., Eckardt K. U., Talks K. L., Wood S. M., Gatter K. C., Harris A. L., Pugh C. W., et al. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1α. Blood. 1998;92:2260–2268. [PubMed] [Google Scholar]
- 25.del Peso L., Castellanos M. C., Temes E., Martin-Puig S., Cuevas Y., Olmos G., Landazuri M. O. The von Hippel Lindau/hypoxia inducible factor (HIF) pathway regulates the transcription of the HIF-proline hydroxylase genes in response to low oxygen. J. Biol. Chem. 2003;278:48690–48695. doi: 10.1074/jbc.M308862200. [DOI] [PubMed] [Google Scholar]
- 26.Gassmann M., Hennet T. From genetically altered mice to integrative physiology. News Physiol. Sci. 1998;13:53–57. doi: 10.1152/physiologyonline.1998.13.2.53. [DOI] [PubMed] [Google Scholar]
- 27.Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Angelo G., Duplan E., Boyer N., Vigne P., Frelin C. Hypoxia up regulates prolyl hydroxylase. A feedback mechanism that limits HIF-1 responses during reoxygenation. J. Biol. Chem. 2003;278:38183–38187. doi: 10.1074/jbc.M302244200. [DOI] [PubMed] [Google Scholar]
- 29.Cioffi C. L., Liu X. Q., Kosinski P. A., Garay M., Bowen B. R. Differential regulation of HIF-1α prolyl-4-hydroxylase genes by hypoxia in human cardiovascular cells. Biochem. Biophys. Res. Commun. 2003;303:947–953. doi: 10.1016/s0006-291x(03)00453-4. [DOI] [PubMed] [Google Scholar]
- 30.Couzin J. Breakthrough of the year. Small RNAs make big splash. Science. 2003;298:2296–2297. doi: 10.1126/science.298.5602.2296. [DOI] [PubMed] [Google Scholar]
- 31.Krishnamachary B., Berg-Dixon S., Kelly B., Agani F., Feldser D., Ferreira G., Iyer N., LaRusch J., Pak B., Taghavi P., et al. Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res. 2003;63:1138–1143. [PubMed] [Google Scholar]
- 32.Seth P., Krop I., Porter D., Polyak K. Novel estrogen and tamoxifen induced genes identified by SAGE (serial analysis of gene expression) Oncogene. 2002;21:836–843. doi: 10.1038/sj.onc.1205113. [DOI] [PubMed] [Google Scholar]
- 33.Huang L. E., Bunn H. F. Hypoxia-inducible factor and its biomedical relevance. J. Biol. Chem. 2003;278:19575–19578. doi: 10.1074/jbc.R200030200. [DOI] [PubMed] [Google Scholar]
- 34.Lang K. J., Kappel A., Goodall G. J. Hypoxia-inducible factor-1α mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berra E., Richard D. E., Gothie E., Pouyssegur J. HIF-1-dependent transcriptional activity is required for oxygen-mediated HIF-1α degradation. FEBS Lett. 2001;491:85–90. doi: 10.1016/s0014-5793(01)02159-7. [DOI] [PubMed] [Google Scholar]