Abstract
In a previous study [Li, Wagner, Friesen and Borst (2003) Gen. Comp. Endocrinol. 134, 147–155], we showed that the MO (mandibular organ) of the lobster Homarus americanus has high levels of HMGR (3-hydroxy-3-methylglutaryl-CoA reductase) and that most (approx. 75%) of the enzyme activity is soluble. In the present study, we report the biochemical and molecular characteristics of this enzyme. HMGR had two forms in the MO: a more abundant soluble form (66 kDa) and a less abundant membrane-bound form (72 kDa). Two cDNAs for HMGR were isolated from the MO. A 2.6-kb cDNA encoded HMGR1, a 599-amino-acid protein (63 kDa), and a 3.2-kb cDNA encoded HMGR2, a 655-amino-acid protein (69 kDa). These two cDNAs had identical 3′-ends and appeared to be products of a single gene. The deduced amino acid sequences of these two proteins revealed a high degree of similarity to other class I HMGRs. Hydropathy plots indicated that the N-terminus of HMGR1 lacked a transmembrane region and HMGR2 had a single transmembrane segment. Recombinant HMGR1 expressed in Sf9 insect cells was soluble and had kinetic characteristics similar to native HMGR from the MO. Treatment with phosphatase did not affect HMGR activity, consistent with the observation that neither HMGR1 nor HMGR2 has a serine at position 490 or 546, the position of a conserved phosphorylation site found in class I HMGR from higher eukaryotes. Other lobster tissues (i.e. midgut, brain and muscles) had low HMGR activities and mRNA levels. MO with higher HMGR activities had higher HMGR mRNA levels, implying that HMGR is regulated, in part, at the transcription level.
Keywords: differential splicing, 3-hydroxy-3-methylglutaryl-CoA reductase, mandibular organ, methyl farnesoate, phosphorylation, transcription regulation
Abbreviations: ESA, eyestalk ablated; GSP, gene-specific primer; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; HMGR, HMG-CoA reductase; M-MLV, Moloney-murine-leukaemia virus; MO, mandibular organ; ORF, open reading frame; RACE, rapid amplification of cDNA ends; rec-HMGR1, recombinant HMGR1
INTRODUCTION
Mevalonic acid is the central substrate for the production of isoprenoids. A key enzyme in its synthesis is 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) reductase (HMGR) (EC 1.1.1.34). In many species, this enzyme is the rate-limiting step in the production of mevalonic acid and the activity of HMGR is an important factor controlling the production of isoprenoids. In mammalian cells, the most abundant compound produced from mevalonic acid is cholesterol, and the regulation of HMGR has received considerable attention as a means of controlling levels of this steroid. However, other isoprenoid products of mevalonic acid, although less abundant, are equally important for cellular function [1,2].
Arthropods do not synthesize cholesterol from mevalonic acid [3]. Nevertheless, the corpora allata of insects has high levels of HMGR that produce the mevalonic acid used in the synthesis of the sesquiterpene, juvenile hormone [4]. Similarly, the MO (mandibular organ) of decapod crustaceans has high levels of HMGR for the production of a juvenile hormone-related compound, methyl farnesoate [5,6], which appears to have physiological functions similar to those of juvenile hormone in insects [7,8].
A comparison of the amino acid sequences of HMGR proteins from different species reveals two distinct classes of this enzyme. Class I enzymes are found in eukaryotes. These proteins are membrane-bound through an N-terminal membrane domain and have an approximate molecular mass of 95 kDa. Class II enzymes are found in prokaryotes, are soluble, lack a membrane domain and have an approximate molecular mass of 55 kDa. The amino acid sequence of the C-terminal catalytic domain of HMGR is well conserved within each class, although there are considerable differences in this domain between class I and class II enzymes. Class I enzymes have a short linker region between the membrane domain and catalytic domain. The membrane domain of class I HMGR enzymes shows considerable diversity [9–11].
Whereas class I HMGR in eukaryotes is typically membrane-bound, soluble HMGR (approx. 50–56 kDa, approx. half the size of intact HMGR) is often found in homogenates of eukaryotic tissues. This activity is usually supposed to represent catalytic domains of the enzyme that have been proteolytically released from the membrane domain during isolation [11,12]. However, several insect species have substantial amounts of soluble HMGR implying that some of the enzyme in these species may be soluble in situ. For example, in the cockroach Diploptera punctata, more than half of HMGR activity in the corpora allata is soluble [4]. In the cockroach Blattella germanica, HMGRs in the fat body have smaller masses (58 and 66 kDa) [13,14] than predicted from the sequence of the cDNA (93 kDa) [15].
In a previous study, we showed that the lobster MO contains abundant HMGR activity and that approx. 75% of the activity is soluble [16]. In the present study, we extend these observations with a biochemical and molecular characterization of the soluble and membrane-bound forms of HMGR, providing a molecular basis for the presence of these forms of this enzyme in the lobster MO. In addition, we investigate the mechanisms used to regulate the activity of this enzyme.
EXPERIMENTAL
Animals
Lobsters were kept in artificial seawater at 13 °C. Some lobsters were ESA (eyestalk ablated) by severing each eyestalk at its base approx. 6 weeks before use.
Chemicals and reagents
Antisera against the recombinant HMGR of Pseudomonas mevalonii (Psm-HMGR) and purified Psm-HMGR were given by Dr V. W. Rodwell (Purdue University, West Lafayette, IN, U.S.A.). Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA, U.S.A.). ResourceQ anion-exchange column, Superdex 200 HR 10/30 gel filtration column, Hybond N membrane and Rapid Hybridization Buffer were from Amersham Biosciences (Piscataway, NJ, U.S.A.). TALON metal affinity resin was obtained from ClonTech Laboratories (Franklin Lakes, NJ, U.S.A.). Immobilon™-P transfer membrane, Centricon centrifugal filter devices and syringe filters were from Millipore (Bedford, MA, U.S.A.).
Enzyme preparation and HMGR assay
Tissue samples were prepared as described previously [16]. Briefly, MO tissue was homogenized in HMGR buffer (250 mM NaCl/30 mM EDTA/1.0 mM dithiothreitol/50 mM KH2PO4/0.1% BSA/0.1 mM leupeptin/1 mM PMSF/10 mM 2-mercaptoethanol/0.3 M sucrose, pH 7.4) at 4 °C. Some of the whole homogenate was centrifuged at 100000 g for 60 min and the pellet and supernatant were collected. HMGR activity was measured using 3R,S-hydroxy-[3-14C]methylglutaryl-CoA (1.9 GBq/mmol; PerkinElmer Life and Analytical Sciences, Boston, MA, U.S.A.) as described previously [16]. 3R,S-[5-3H]Mevalonic acid (1.4 TBq/mmol; PerkinElmer Life and Analytical Sciences) was added to each reaction tube to determine the recovery of mevalonic acid (typically >95%).
Electrophoresis and Western blotting
Samples with HMGR activities were separated on a 10% SDS/polyacrylamide gel, followed by Coomassie Brilliant Blue staining (for MO tissue fractions or Sf9 cell homogenate) or silver staining (for chromatographic fractions) [17].
After separation by SDS/PAGE, some samples were transferred on to Immobilon™-P membranes and analysed by Western blotting. After blocking with 1% BSA in PBS overnight at 4 °C, the membranes were incubated at room temperature (25 °C) with Psm-HMGR antiserum (1:5000 in PBS) for 2 h followed by peroxidase-conjugated anti-rabbit IgG (Sigma, St. Louis, MO, U.S.A.; 1:10000 in PBS) for an additional 2 h. After each step, the blots were washed three times for 10 min in PBS with 0.05% Tween 20. The blots were then incubated with 2 ml of SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL, U.S.A.) and exposed to X-ray film [17].
Purification of the soluble form of lobster HMGR from the MO
Proteins in the 100000 g MO supernatant were passed through a 0.2 μm syringe filter and separated by anion-exchange chromatography (ResourceQ column, 1 ml) using a NaCl gradient (0–1 M) in 50 mM Tris (pH 8.2). Fractions with HMGR activity were pooled and concentrated using a Centricon centrifugal filter. The concentrated sample was separated further using gel filtration (Superdex 200 HR 10/30 column) eluted with 50 mM phosphate buffer (pH 7.0) containing 150 mM NaCl. Fractions with highest HMGR activity were pooled and concentrated as above.
Cloning and sequencing of HMGR cDNAs
MO were dissected from ESA lobsters and immediately homogenized in Tri Reagent (Sigma) to isolate total RNA. First-strand cDNA was synthesized from total RNA using M-MLV (Moloney-murine-leukaemia virus) and Random Primers (Promega, Madison, WI, U.S.A.). A 312-bp cDNA fragment of lobster HMGR was amplified by PCR from the first-strand cDNA using two degenerate primers (ORS1, 5′-CCNATGGCNACNACNGARGG-3′ and ORS2, 5′-CATRTTCATNCCCATNGCRTCNCC-3′), which were designed from highly conserved amino acid sequences of the catalytic domain of class I HMGRs [15]. The PCR conditions used in the present study and elsewhere were 94 °C, 1 min; 50–65 °C, 1 min; 72 °C, 1–3 min; 30–35 cycles; 20 μl of reaction mixture including 0.5 unit of Taq DNA polymerase (Sigma). The PCR product was subcloned into the pCR®2.1 vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA, U.S.A.) and sequenced using the BigDye sequencing kit (Applied Biosystems, Foster City, CA, U.S.A.).
The 3′-RACE (rapid amplification of cDNA ends) System (Invitrogen) was used to amplify the 3′-end of the HMGR cDNA. First-strand cDNA was synthesized from total RNA using M-MLV and the Adapter Primer from the 3′-RACE System. Two GSPs (gene-specific primers; GSP1, 5′-CAGAGCTGTACATGTGGACAAG-3′ and GSP2, 5′-CGTCTGCAGGATTTGCAAGTG-3′) were designed from the 312-bp cDNA fragment. These primers were used sequentially with the Universal Amplification Primer to PCR-amplify the 3′-end of the HMGR cDNA with the first-strand cDNA. Similarly, the 5′-RACE System (Invitrogen) was used to amplify the 5′-end of the HMGR cDNA. GSP3 (5′-CTTGTCCACATGTACACCTCTG-3′) and GSP4 (5′-ACAACCACGATTGGTGGAAGC-3′) were designed from the inverse sequence of the 312-bp cDNA fragment. First-strand cDNA was synthesized from total RNA using M-MLV and GSP3 and then tailed with terminal deoxynucleotide transferase and dCTP. GSP4 and the Abridged Anchored Primer from the kit were used to PCR-amplify the 5′-end of the HMGR cDNAs with the dC-tailed first-strand cDNA. PCRs contained an additional 0.1 unit of Pfu DNA polymerase (Stratagene, La Jolla, CA, U.S.A.). PCR products from the 3′- and 5′-RACE reactions were subcloned and sequenced as above.
To confirm the nucleotide sequence of the cDNA for the soluble form of enzyme (HMGR1), its entire ORF (open reading frame) was amplified using GSP5 (5′-CCGGAATTCATGGCAGGCATCGGTCCC-3′; EcoRI site underlined) and GSP6 (5′-TACGTACCGCTCGAGCTACCTATCTCTAG-3′; XhoI site underlined). Similarly, GSP7 (5′-CCGGAATTCATGTATAGGTTGTTGAATG-3′; EcoRI site underlined) and GSP6 were used to amplify the entire ORF of the membrane-bound form of the enzyme (HMGR2). In these amplifications, an additional 0.1 unit of Pfu DNA polymerase and 0.1 μl of Perfect Match PCR Enhancer (Stratagene) were added to the PCR.
Northern blotting
A 563-bp fragment (HMGR1 and HMGR2 probe), common to both HMGR cDNAs (1595–2157 bp in HMGR1 cDNA and 2140–2702 bp in HMGR2 cDNA), was amplified with primers GSP8 (5′-TAGGTGGAGGCACTGGACTTG-3′) and GSP9 (5′-ACCCTTGATTGCAACCATCG-3′). A 308-bp fragment (HMGR1 probe) specific for the 5′-end of HMGR1 cDNA (30–337 bp, a sequence not found in HMGR2 cDNA) was produced using primers GSP10 (5′-CAAGTACAGTACTGTAATCCTC-3′) and GSP11 (5′-GCCTGCCATGAAGGTTAGTAG-3′). Finally, a 441-bp fragment (HMGR2 probe) specific for the 5′-end of the HMGR2 cDNA (442–853 bp) was amplified using the primers GSP12 (5′-CAACAGAGTTCAATCACCACG-3′) and GSP13 (5′-GAACATCCTGAGAGCGCTGAAG-3′). Alignment of the 308 bp HMGR1 and the 441 bp HMGR2 probe revealed no significant similarity. These three cDNA fragments were individually radiolabelled using [α-32P]dCTP (111 TBq/mmol; ICN Radiochemicals, Costa Mesa, CA, U.S.A.) and the Prime-a-Gene Labeling System (Promega). The 32P-labelled cDNA fragments were used as probes for Northern and Southern blotting [17].
Total RNA was prepared from MO and other lobster tissues using Tri Reagent. mRNA was prepared from MO using the Micro-FastTrack 2.0 kit (Invitrogen). Samples of total RNA (usually 4 μg) or mRNA (0.1 μg) were separated on 1% agarose/formaldehyde gels, transferred on to Hybond N membranes and cross-linked to the membranes with UV light. The membranes were then incubated in 3 ml of Rapid Hybridization buffer for 30 min at 65 °C and hybridized to approx. 3×106 c.p.m. of one of the radiolabelled probes for 4 h at 65 °C. After hybridization, membranes were washed under conditions of increasing stringency to remove non-specifically bound probe and then exposed to X-ray film.
Southern blotting
Genomic DNA was isolated from lobster muscle using the Wizard Genomic DNA Purification kit (Promega). The lobster genomic DNA (10 μg) was digested with approx. 20 units of BamHI, EcoRI, HindIII or NotI restriction endonucleases (New England Biolabs, Boston, MA, U.S.A.) respectively in 20 μl of the appropriate buffer at 37 °C overnight. DNA fragments were separated on 1% agarose gels, transferred on to Hybond N membranes and cross-linked to the membrane with UV light. The membranes were hybridized to the 32P-labelled 563-bp HMGR1 and HMGR2 probes as in Northern blotting.
Expression and purification of the recombinant HMGR1
GSP5 and GSP6 were used to amplify the 1.8-kb ORF of the 2.6-kb cDNA encoding HMGR1. The PCR product (approx. 1.8 kb) was subcloned into the pCR®2.1 vector and its sequence confirmed. It was then subcloned into the donor plasmid pFASTBAC HTa using the EcoRI and XhoI restriction sites in the multiple cloning sites. Cloning into pFASTBAC HTa appends an additional 29 amino acids (MSYYHHHHHHDYDIPTTENLYFNGAMGSH) to the N-terminus of the protein that includes a His6 tag. The sequence of the cDNA construct was verified.
Spodoptera frugiperda (Sf9) cells were grown in Sf900II cell medium (Invitrogen) in suspension culture at 28 °C and 135 rev./min. Cells were maintained at densities between 5×105 and 5×106 cells/ml. Cell viability was determined by Trypan Blue (Sigma) staining. Production of recombinant Bacmid DNA and its transfection of Sf9 cells to produce recombinant baculovirus were performed as described in the Bac-to-Bac baculovirus expression manual (Invitrogen). The recombinant baculovirus containing HMGR1 cDNA was collected and used to infect 50 ml of Sf9 cells at 1×106 cells/ml. The culture medium of these cells was harvested 6 days post-infection to collect recombinant baculovirus and used for protein expression.
Sf9 cells (400 ml) at 1×106 cells/ml were infected with the above recombinant baculovirus and grown in suspension to a density of 1×106 cells/ml. Cells were collected 48 h later by centrifugation, resuspended in buffer A (50 mM Tris/50 mM sodium phosphate/300 mM NaCl, pH 7.5) and centrifuged at 100000 g for 30 min. The supernatant was loaded on to a Co2+ affinity column (ClonTech Laboratories), equilibrated with buffer A containing 20 mM imidazole. After washing the column with 10 column volumes of buffer A containing 20 mM imidazole, the His6-tagged recombinant HMGR1 (rec-HMGR1) was eluted from the column with buffer A containing 150 mM imidazole. The purified rec-HMGR1 was tested for purity by SDS/PAGE and Western blotting and analysed using gel filtration and anion-exchange chromatography.
Biochemical characterization of rec-HMGR1
Time course, protein dependence, Km for HMG-CoA and NADPH, KI for lovastatin and mevalonic acid and substrate specificity of the purified rec-HMGR1 were determined as described previously [16].
Phosphatase and ATP treatment
Rec-HMGR1 (5 μg of protein) or MO fractions (10 μg of protein) in 50 μl of HMGR buffer without KH2PO4 were incubated alone (Control), with lambda protein phosphatase (1, 3 and 10 units; New England Biolabs) with ATP (1×10−8 to 10−5 M; Sigma) or with 1×10−8 M ATP and approx. 100 μg of rat liver whole homogenate for 30 min. EDTA was removed from the buffer used for treatments with ATP [18,19]. Psm-HMGR [18] was used as a negative control and the rat liver whole homogenate [19] as a positive control for all treatments.
RESULTS
Biochemical analysis of lobster HMGR
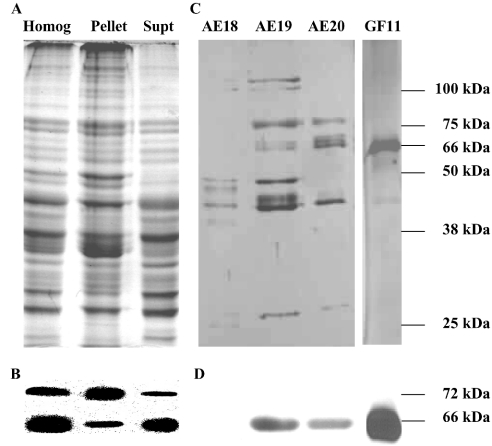
MO tissue was homogenized and separated into three fractions (whole homogenate, 100000 g pellet and 100000 g supernatant). Since freezing and thawing of mammalian tissues has been shown to increase the proteolytic cleavage of membrane bound HMGR into a soluble fragment [11], no freeze–thaw steps were used in the preparation of these MO fractions. After centrifugation, they were separated immediately on SDS/polyacrylamide gels (Figure 1A; Homog, Pellet and Supt) and then analysed by Western blotting with the Psm-HMGR antiserum. All the three fractions contained two immunoreactive protein bands (approx. 66 and 72 kDa) (Figure 1B). The 66 kDa band was most abundant in the supernatant and the 72 kDa band was most abundant in the pellet. These results indicated that the MO contained two proteins that were antigenically similar to HMGR.
Figure 1. Lobster MO has two HMGR proteins.
(A) Samples (approx. 20 μg of protein) of a whole MO homogenate (Homog) and its 100000 g pellet (Pellet) and supernatant (Supt) were prepared from freshly dissected tissue, and analysed immediately by SDS/PAGE (Coomassie Blue staining). (B) The SDS/polyacrylamide gels in (A) were analysed with Western blots using a Psm-HMGR antiserum and showed the presence of two immunoreactive protein bands. The more abundant 66 kDa protein was found predominantly in the Supt fraction, whereas the less abundant 72 kDa protein was predominantly in the Pellet. (C) Proteins in the supernatant were purified by anion-exchange chromatography and the HMGR activity was detected in fractions 18–20 (AE18–AE20). These fractions were pooled and further purified by gel filtration and HMGR activity was highest in fraction 11 (GF11). Aliquots of fractions AE18–AE20 and GF11 were analysed by SDS/PAGE (silver staining). (D) Only the 66-kDa protein was detected by Western blotting in the fractions separated. Molecular masses of the marker proteins are shown on the right-hand side of the SDS/polyacrylamide gel and the calculated masses of the immunoreactive bands are given for the Western blots. The Figure is representative of the results obtained in approx. ten experiments.
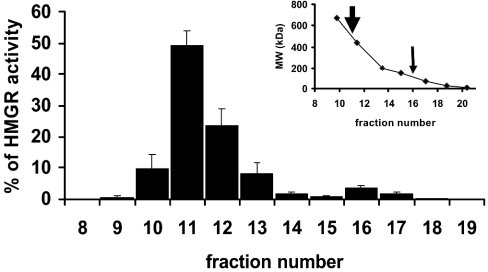
HMGR in the 100000 g MO supernatant (approx. 500 μg of protein) was purified by anion-exchange chromatography (0–1 M NaCl). HMGR activity eluted between 150 and 170 mM NaCl. The fractions with HMGR activity were concentrated and analysed by SDS/PAGE (Figure 1C; AE18–AE20). Western blots showed the presence of a single immunoreactive band (approx. 66 kDa) in the two fractions that contained approx. 98% of the total HMGR activity (Figure 1D; AE19–AE20). Fractions with the highest activity (e.g. AE19–AE20) were pooled from several anion-exchange separations, concentrated and analysed by gel filtration. The HMGR activity eluted in a major peak (approx. 500 kDa) and a minor peak (approx. 120 kDa) (Figure 2). The peak fractions of the major peak were pooled, concentrated and analysed by SDS/PAGE. They contained a major protein band with a molecular mass of approx. 66 kDa (Figure 1C, GF11). A single band of immunoreactive material of the same mass was detected in these gels by Western blotting (Figure 1D). When the 100000 g MO supernatant was analysed by gel filtration, its HMGR activity eluted with a profile similar to the material purified by anion exchange and the pooled fractions of the major peak had a single immunoreactive band of approx. 66 kDa (results not shown).
Figure 2. Soluble form of lobster HMGR has a molecular mass of approx. 500 kDa.
The 100000 g supernatant was analysed by gel filtration (Superdex 200 HR 10/30 column, 1 ml fraction) and HMGR activity was detected in several fractions, with the highest activity eluting in fraction 11 (estimated molecular mass approx. 500 kDa) and a minor peak in fraction 16 (estimated molecular mass approx. 120 kDa). Inset: elution volumes of the molecular standards and the peaks of HMGR activity (arrows). This Figure is representative of the results obtained in five experiments.
Isolation of lobster HMGR cDNAs
We used two degenerate primers (ORS1 and ORS2) [15] to amplify a 312-bp PCR fragment with the first-strand cDNA made from MO mRNA. The deduced amino acid sequence of this cDNA fragment (104 amino acid residues) had high identity to other HMGRs. Two nested pairs of GSPs (GSP1 and GSP2 for 3′-RACE and GSP3 and GSP4 for 5′-RACE) were then designed based on the sequence of the 312-bp HMGR cDNA fragment. Using these primers, we recovered the entire HMGR cDNA. The two sequential amplifications of the 3′-RACE yielded a single 1.5-kb PCR fragment containing a 1059-bp ORF encoding 352 amino acid residues with high identity to other HMGRs and a 452-bp 3′-untranslated region. The 5′-RACE produced two cDNA fragments (approx. 1.0 and 1.5 kb). The smaller (approx. 1.0 kb) PCR product contained a 655-bp ORF encoding 218 amino acid residues and a 328-bp 5′-untranslated region. The larger (approx. 1.5 kb) product contained an 823-bp ORF encoding 274 amino acid residues and a 705-bp 5′-untranslated region. Both the ORFs had high degrees of identity with other HMGR cDNAs.
The above results suggested that the lobster MO contains two mRNA transcripts with different 5′-ends and identical 3′-ends. This prediction was tested using the GSP5 and GSP6 to amplify the 1800 bp ORF of HMGR1 and GSP7 and GSP6 to amplify the 1998 bp ORF of HMGR2. The nucleotide sequences of these PCR products were identical with those determined above. Furthermore, the sequences of both the 5′-untranslated regions were confirmed using several alternative pairs of GSPs and the results were identical with 5′-RACE.
The sequence of the smaller mRNA indicated that it would produce a 2580 bp cDNA with a 1800 bp ORF, beginning at nt 329 and terminating at 2128 (GenBank® accession number AY292876). The location of the start codon in this cDNA was based on the presence of a stop codon at nt 314 (which is 5 codons upstream). The deduced amino acid sequence of this protein (HMGR1) had 599 amino acids with a calculated molecular mass of approx. 63 kDa. The larger mRNA would produce a 3125 bp cDNA with a 1998 bp ORF, beginning at nt 706 and terminating at 2673 (GenBank® accession number AY292877). The location of the start codon for translation-initiation site for this cDNA was based on the presence of a stop codon at nt 682 (which is 8 codons upstream). The deduced amino acid sequence of this protein (HMGR2) had 655 amino acids and a calculated molecular mass of approx. 69 kDa.
Sequence analysis of lobster HMGR1 and HMGR2
Although HMGR1 and HMGR2 differed at their N-termini, the amino acid sequences of their last 597 amino acids (HMGR13–599 and HMGR259–665) were identical (Figure 3). Both lobster HMGR sequences contained all three signature patterns common to other class I HMGRs (residues 264–278, 420–427 and 474–487 of HMGR1; residues 320–334, 476–483 and 530–543 of HMGR2). The last region contained the histidine residue (His-484 in HMGR1 and His-540 in HMGR2), whch has been shown to be involved in catalysis in other HMGRs [10,11]. In contrast, HMGR1 and HMGR2 were unusual in that both proteins had a valine residue (Val-490 in HMGR1 and Val-546 in HMGR2) in the position occupied by serine in other class I HMGRs. This serine in class I HMGR from higher eukaryotes regulates enzyme activity by reversible phosphorylation [10,11]. Another unusual feature of the two lobster HMGRs is their molecular masses, which were smaller than class I HMGRs (approx. 95 kDa) and larger than class II HMGRs (approx. 55 kDa).
Figure 3. Amino acid sequences of lobster HMGR1 and HMGR2 are closely related to other class I HMGR proteins.
The sequences of lobster HMGR1 and HMGR2 (lob-HMGR1 and HMGR2) were compared with the HMGRs of the cockroach B. germanica [15] and the beetle Ips paraconfusus [21], the fruitfly D. melanogaster [24] and the human Homo sapiens [36]. Only residues 1–80 of lob-HMGR1 are shown; the rest of its sequence is identical with residues 138–655 of lob-HMGR2. Conserved residues are shown as white letters with a black background, similar residues as white letters with a light grey background, non-conserved residues as black letters with a white background and gaps inserted to optimize the alignment are shown by a broken line. The putative transmembrane region of HMGR2 (residues 4–27) is shown with a bar and the putative PEST region (residues 549–585 of HMGR2) is shown with a broken line. The conserved catalytic histidine (residue 540 in HMGR2) is marked by an *. A valine (residue 546 in HMGR2) that replaces the serine that is phosphorylated in class I HMGRs from other higher eukaryotes is marked by #.
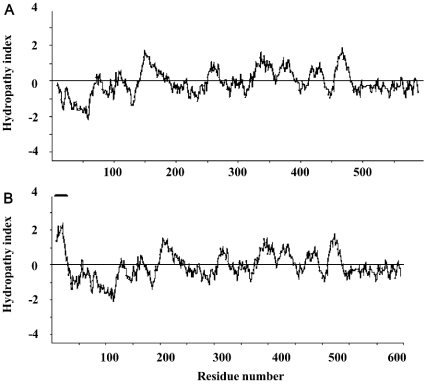
Hydropathy plots were used to identify potential membrane-spanning segments of the lobster HMGR proteins [20]. These plots suggest that HMGR1 does not possess a transmembrane segment. However, HMGR2 has a longer N-terminal domain (residues 1–66) and one region of this domain (residues 4–28) has a hydropathy value that is >2.2 (Figure 4), suggesting that it is a transmembrane segment. The sequence of this hydrophobic segment is well conserved in the final (8th) transmembrane segment of class I HMGRs in insects (Figure 3) [15,21–24]. The lobster HMGR proteins contain identical hydrophilic linker regions (residues 11–113 for HMGR1 and 67–169 for HMGR2) and C-terminal catalytic domains (residues 114–599 for HMGR1 and 170–665 for HMGR2). Both of these features are conserved in other class I HMGRs. The amino acid sequences of the lobster HMGRs and other class I HMGRs have extensive sequence conservation (45–59% identity). Sequence alignments (Figure 3) show that the catalytic domains of HMGR1 and HMGR2 have the highest sequence similarity to other class I HMGRs, whereas their linker region and the membrane domain of HMGR2 have moderate sequence similarity. An unusual approx. 80 amino acid region is found at the C-terminus of both lobster HMGRs. This region has not been observed in other HMGRs. This region contains a putative PEST sequence, and hence it may play a role in the turnover of this protein.
Figure 4. Hydropathy plots suggest that HMGR1 is soluble and that HMGR2 has a single transmembrane segment.
Plots were created using the program Kyte–Doolittle Hydropathy Plots (http://fasta.virgina.edu/o-fasta/grease.htm). Other hydropathy plot programs (such as Hopp–Woods Hydropathy plots, http://bioinformatics.weizmann.ac.il/hydroph/ and Dense Alignment Surface Methods, http://www.sbc.su.se/~miklos/DAS/) revealed the same results. The transmembrane segment of HMGR2 is indicated by a solid bar. Window size is 19.
Northern- and Southern-blot analyses
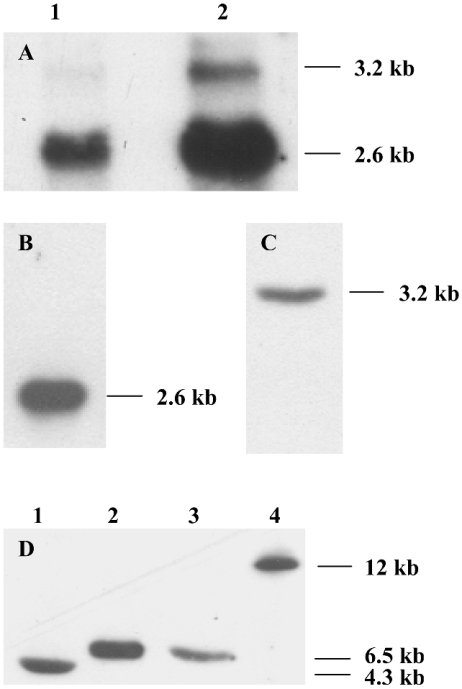
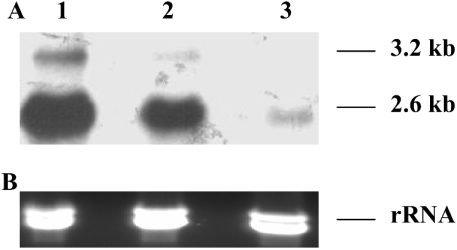
Northern blots of MO RNA revealed two HMGR transcripts, a more abundant 2.6 kb transcript and a less abundant 3.2 kb transcript. Both transcripts were detected in blots of total RNA from the MO using the 563-bp HMGR1 and HMGR2 probes (Figure 5A) or other probes containing regions common to both HMGR cDNAs (results not shown). Since the 308-bp HMGR1 and 441-bp HMGR2 probes cross-reacted with several bands in Northern blots of total MO RNA, we analysed blots using MO mRNA. In these blots, the 308-bp HMGR1 probe (which has no sequence common to HMGR2 cDNA) detected only the 2.6 kb transcript (Figure 5B) and the 441-bp HMGR2 probe detected only the 3.2-kb HMGR transcript (Figure 5C). Thus we conclude that the HMGR1 cDNA was produced from the 2.6 kb transcript and the HMGR2 cDNA from the 3.2-kb HMGR transcript.
Figure 5. Lobster MO contains two HMGR transcripts transcribed from a single gene.
(A) Northern-blot analysis using the 563-bp HMGR1 and HMGR2 probes showed the presence of a more abundant 2.6 kb transcript and a less abundant 3.2 kb transcript. Lanes 1, total RNA 1 μg; lane 2, total RNA 4 μg. (B) The 308-bp HMGR1 probe detected only the 2.6 kb transcript from 0.1 μg of mRNA. (C) The 441-bp HMGR2 probe detected only the 3.2-kb HMGR transcript from 0.1 μg of mRNA. (D) Southern-blot analysis. DNA (10 μg) was digested with 20 units of BamHI, EcoRI, HindIII or NotI (lanes 1–4 respectively) and probed with the 563-bp HMGR1 and HMGR2 probes. Each enzyme yielded one detectable DNA fragment, indicating that the putative HMGR gene is present as a single copy in the lobster genome. The transcript sizes are given on the right side of each blot. The Figure is representative of the results obtained in three experiments.
Southern blots of lobster genomic DNA digested with one of four restriction endonucleases and analysed with the 563-bp HMGR1 and HMGR2 probes had fragment patterns indicating that the HMGR gene was present as a single copy in the lobster genome (Figure 5D). This observation is consistent with the production of the two HMGR transcripts by differential splicing.
Characterization of rec-HMGR1
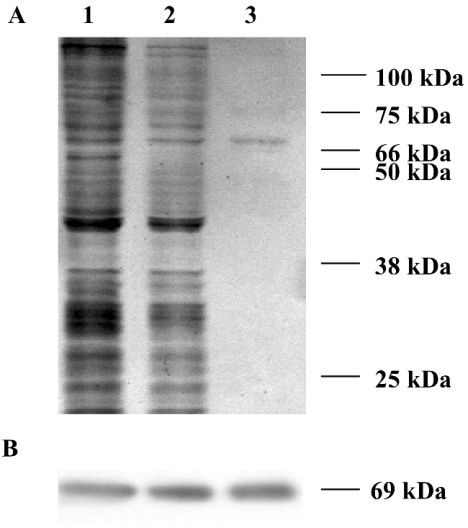
Bacterial expression of HMGR1 using the pET system (Novagen, Madison, WI, U.S.A.) produced a protein that was insoluble and inactive (results not shown). Therefore recombinant HMGR1 (rec-HMGR1) was expressed in Sf9 insect cells using the Bac-to-Bac baculovirus expression system. The HMGR activity of infected Sf9 cells [approx. 60 μmol·(mg of protein)−1·min−1] was approx. 100000 times higher than the activity of control cells. Aliquots of the infected Sf9 cell homogenates, their 100000 g supernatants and affinity-purified His-tagged rec-HMGR1 protein were analysed on 10% SDS/polyacrylamide gels (Figure 6A; lanes 1–3 respectively). A band with a molecular mass of approx. 69 kDa was observed in each lane. Western blots also showed one immunoreactive band with a molecular mass of approx. 69 kDa in each lane (Figure 6B; lanes 1–3). The molecular mass of rec-HMGR1 was slightly greater than the native HMGR1 due to the presence of the His tag and the linker (approx. 3 kDa). The activity of the purified rec-HMGR1 [5.5±0.1 mmol·(mg of protein)−1·min−1 (n=4; S.E.M.)] was approx. 50 times greater than the activity in the 100000 g supernatant of the infected Sf9 cells. Purified rec-HMGR1 eluted from anion exchange in the same volume as the native soluble HMGR from the MO. Analysis by gel filtration indicated a major peak corresponding to a molecular mass of approx. 540 kDa and a minor peak of approx. 140 kDa, also similar to native soluble HMGR. Similarly, the biochemical characteristics of the rec-HMGR1 (optimum pH, temperature dependence and effect of protein) were identical with the native enzyme [16]. Furthermore, the Km values of rec-HMGR1 for HMG-CoA and NADPH were 14.5 (±0.6 S.E.M.; n=3 determinations) and 14.0 (±0.5; n=3) μM respectively. These values were similar to those reported for the native enzyme [16]. The KI values of rec-HMGR1 for lovastatin and mevalonic acid (0.45±0.05 nM, n=3 and 80.5±13.9 μM, n=3 respectively) were similar to the values obtained with the native enzyme [16]. As observed with the native enzyme [16], other compounds (e.g. methyl farnesoate and farnesoic acid) had no effect on the activity of rec-HMGR1.
Figure 6. Recombinant HMGR1 (rec-HMGR1) is expressed in Sf9 cells.
Cells were infected with a recombinant baculovirus containing HMGR1 cDNA. The rec-HMGR1 was purified from the cell homogenates by affinity chromatography using the His tag. (A) Aliquots of the infected Sf9 cell homogenates, the 100000 g supernatant and the His-tag purified protein were analysed by SDS/PAGE (lanes 1–3 respectively). (B) Western blot of a similar gel using Psm-HMGR antiserum showed one immunoreactive protein band of 69 kDa. Molecular masses of the protein standards are shown on the right side of the SDS/polyacrylamide gel; the calculated masses of the immunoreactive bands are given for the Western blots. The Figure is representative of the three analyses of these fractions.
Effects of phosphatase and ATP on lobster HMGR activity
The replacement of serine residue with valine in lobster HMGR1 (Val-490) and HMGR2 (Val-546) suggests that this enzyme is not regulated by reversible phosphorylation. However, one potential cAMP- and cGMP-dependent protein kinase phosphorylation site, 11 potential protein kinase C phosphorylation sites and seven potential casein kinase II phosphorylation sites were found in both HMGR1 and HMGR2 (residues not shown).
To test the hypothesis that lobster HMGR is not regulated by phosphorylation, we measured the effects of lambda protein phosphatase on lobster HMGR activity. Treatment of rec-HMGR1 with lambda phosphatase (1 unit) had no effect (P>0.1; ANOVA) on its activity (Table 1). Similarly, treatment of 100000 g supernatants of MO from intact male lobsters, ESA male lobsters and intact female lobsters were unaffected by the phosphatase (P>0.1; ANOVA). Phosphatase treatment also did not affect the activity of purified Psm-HMGR (P>0.1; ANOVA); however, the HMGR activity of rat liver whole homogenate was significantly increased by phosphatase treatment (P<0.001; ANOVA). Treatment with higher concentrations of phosphatase (3 and 10 units) gave similar results (results not shown).
Table 1. Effects of phosphatase and ATP on HMGR activity.
Tissue and recombinant protein samples with HMGR activity (5–10 μg of protein in each sample) were incubated for 30 min alone in HMGR buffer without KH2PO4 (control), with 1 unit of lambda protein phosphatase (Phosphatase), with 1×10−8 M ATP (ATP) or with 1×10−8 M ATP plus approx. 100 μg of rat liver whole homogenate (ATP+rat liver homogenate). For both treatments containing ATP, EDTA was removed from the HMGR buffer. HMGR activity [nmol·(mg of protein)−1·min−1, ±S.E.M., n=4 determinations] was measured after treatment. N.D., not determined.
| HMGR activity [nmol·(mg of protein)−1·min−1] | ||||
|---|---|---|---|---|
| HMGR source | Control | Phosphatase | ATP | ATP+rat liver homogenate |
| Purified rec-HMGR1 | 5520±140 | 5548±182 | 5508±104 | 5550±88 |
| Intact male MO supernatant | 66.5±14.1 | 66.4±12.8 | 67.2±14.8 | 64.8±10.2 |
| ESA male MO supernatant | 246.1±34.5 | 252.3±32.6 | 261.8±35.2 | 249.6±15.9 |
| Intact female MO supernatant | 25.4±4.1 | 24.8±4.5 | 24.5±1.9 | 26.8±2.4 |
| Purified Psm-HMGR | 3.6±0.1 | 3.7±0.1 | 3.6±0.2 | 3.5±0.2 |
| Rat liver whole homogenate | 5.6±0.3 | 9.3±0.2 | 0.1±0.0 | N.D. |
We also attempted to stimulate HMGR phosphorylation by endogenous kinases by adding ATP (10−8 M) to these various fractions (Table 1). Since kinases require Mg2+ for activity, EDTA was removed from the HMGR buffer used with ATP [11,19]. As anticipated, addition of ATP to purified rec-HMGR1 and Psm-HMGR (which would be unlikely to have endogenous kinase activity) had no effect (P>0.1; ANOVA) on their activity. Similarly, addition of ATP to the 100000 g supernatants of MO from intact male lobsters, ESA male lobsters and intact female lobsters were not affected (P>0.1; ANOVA). However, the addition of ATP to rat liver whole homogenates caused a decline in HMGR activity to nearly undetectable levels (P<0.001; ANOVA), indicating the presence of a HMGR kinase in the rat liver. Treatment with higher concentrations of ATP (10−7–10−5 M) gave similar results to the above (results not shown). Finally, co-incubation of purified rec-HMGR1, Psm-HMGR and the 100000 g MO supernatants with 10−8 M ATP plus approx. 100 μg of rat liver whole homogenate (Table 1) had no effect on their HMGR activity (P>0.1; ANOVA). Thus the kinases present in rat liver homogenate (including the HMGR kinase that inhibits rat HMGR) did not affect lobster HMGR activity.
Tissue distribution and transcription regulation of HMGR in the lobster MO
HMGR activity was measured in different tissues from ESA male lobsters (n=4), intact male lobsters (n=4) and intact female lobsters (n=3) (Table 2). HMGR activity in MO of ESA male lobsters was 4-fold higher than that of the intact male lobsters and 10-fold higher than that of the intact female lobsters. This is consistent with our previous results showing that HMGR activity in the lobster MO is regulated by an eyestalk factor [16]. In addition, HMGR activity was detected in all other tissues of each animal tested (midgut, stomach, eyestalk, brain, central nervous cord, hepatopancreas, gonad and muscles). The level of HMGR activity in MO tissue [20–250 nmol·(mg of protein)−1·min−1] was 102–104 times higher than the levels measured in other tissues. Some tissues (midgut, stomach, eyestalk and brain) had modest levels of HMGR activity [0.08–0.21 nmol·(mg of protein)−1·min−1], whereas other tissues (central nervous cord, hepatopancreas, testis and ovary) had low levels of HMGR activity [0.02–0.07 nmol·(mg of protein)−1·min−1]. The HMGR activity of these tissues was similar in the three groups of lobsters. In contrast, HMGR activity in muscle tissues from intact female lobsters was relatively high [0.25±0.03 nmol·(mg of protein)−1·min−1] compared with the levels measured in both the intact and ESA males [<0.01 nmol·(mg of protein)−1·min−1; P<0.001, ANOVA].
Table 2. HMGR activity in tissues from female, male and ESA male lobsters.
HMGR activity [nmol·(mg of protein)−1·min−1, ±S.E.M.] was measured in the 100000 g supernatants of the indicated tissues. N.D., not determined.
| HMGR activity [nmol·(mg of protein)−1·min−1] | |||
|---|---|---|---|
| Tissue | ESA male lobsters (n=4) | Intact male lobsters (n=4) | Intact female lobsters (n=3) |
| MO | 246.1±34.5 | 66.5±14.1 | 25.4±4.1 |
| Midgut | 0.21±0.04 | 0.17±0.02 | 0.17±0.01 |
| Stomach | 0.12±0.02 | 0.16±0.01 | 0.14±0.01 |
| Eyestalk | N.D. | 0.10±0.01 | 0.08±0.01 |
| Brain | 0.15±0.01 | 0.13±0.01 | 0.17±0.04 |
| Central nervous cord | 0.06±0.01 | 0.07±0.01 | 0.05±0.02 |
| Hepatopancreas | 0.09±0.01 | 0.05±0.01 | 0.08±0.01 |
| Gonad | 0.02±0.01 | 0.02±0.01 | 0.02±0.01 |
| Muscles | 0.01±0.01 | 0.006±0.003 | 0.25±0.03 |
Northern blots using the 563-bp HMGR1 and HMGR2 probes detected both HMGR transcripts in total RNA prepared from the MO of the three lobster groups, although the larger transcript was often not detectable in female animals. In each sample, the 2.6 kb transcript was much more abundant than the 3.2 kb transcript. HMGR transcripts were more abundant in MO from ESA male lobsters when compared with that in the MO from intact male and female lobsters (Figure 7A; lanes 1–3 respectively). These observations indicate that at least some of the increased HMGR activity observed in MO from ESA animals is due to increased levels of mRNA. Northern blots of RNA from tissues other than the MO showed either extremely low levels or no HMGR transcripts, consistent with the low level of HMGR activity detected in these tissues (results not shown).
Figure 7. Eyestalk ablation increases the level of HMGR transcripts.
(A) Northern blot of total MO RNA (4 μg/lane) from ESA males (n=4; 6 weeks after eyestalk ablation), intact males (n=4) and intact females (n=3; lanes 1–3 respectively). The 2.6 kb transcript was always more abundant than the 3.2 kb transcript. Both transcripts were most abundant in ESA males and least abundant in intact females, which often lacked a detectable 3.2 kb transcript. These results mirror changes in HMGR activity in the same animals (see Table 1), suggesting that the HMGR activity is regulated at least partly at the level of transcription. (B) The amount of rRNA in 4 μg of total RNA of each sample (lanes 1–3) was similar. The actual sizes of the transcripts are given on the right side of the blot. The blot of one animal from each group is shown.
DISCUSSION
Results of the present study describe the molecular basis for the unique characteristics of HMGR in the lobster. In a previous study, we demonstrated that HMGR activity in homogenates of the lobster MO is found in two forms. Most (approx. 75%) of the activity was found in the soluble fraction, whereas the remainder (approx. 25%), was in the membrane fraction [16]. The current study confirms and extends this previous observation. First, we confirmed the existence of two forms of HMGR with a Psm-HMGR antiserum [11]. This antiserum labelled two immunoreactive protein bands in lobster MO homogenates and its subcellular fractions. On the basis of their distribution in these fractions, the more abundant 66-kDa protein appears to be soluble (HMGR1), and the less abundant 72 kDa protein to be membrane-bound (HMGR2). The molecular masses of lobster HMGR1 and HMGR2 were smaller than the class I (eukaryotic) HMGRs but larger than the class II (prokaryotic) HMGRs [10,11]. Since the native mass (determined by gel filtration) of the lobster HMGR is approx. 500 kDa, it appears that the native form is a multimeric complex, similar to observations for HMGR in other species [25,26].
Secondly, we cloned and sequenced two distinct cDNAs for HMGR from MO RNA. The deduced amino acid sequences of these two cDNAs indicate that the two lobster HMGR proteins have different N-terminal amino acids (HMGR11–2 and HMGR21–58) but identical C-terminal amino acids (HMGR13–599 and HMGR259–665). The deduced amino acid sequences of these two cDNAs strongly suggest that these proteins are class I HMGRs (see below).
Thirdly, the deduced amino acid sequences of HMGR1 and HMGR2 strongly support the identification of HMGR1 as the soluble form of the enzyme and HMGR2 as the membrane-bound form. Perhaps, the most compelling evidence for this conclusion is provided by the hydropathy plot of their amino acid sequences. Hydropathy plots represent the average hydrophobic character along the amino acid sequence of a protein. These plots can be used to identify clusters of hydrophobic amino acids, which may indicate a segment of the protein that is sufficiently hydrophobic to either interact with or reside in a membrane. When a window size of 19 residues is used to calculate hydropathy plots, the hydrophobic membrane-spanning segments stand out rather clearly, having values of at least 1.6 on the Kyte–Doolittle hydropathy scale at the midpoint of the region [20]. Hydropathy plots of HMGR1 suggest that this protein lacks an N-terminal transmembrane region, consistent with its designation as the soluble form of HMGR. In contrast, hydropathy plots of HMGR2 suggest that it has one N-terminal transmembrane segment (residues 4–28 with a maximum hydropathy value of >2.2), consistent with its designation as the membrane-bound form. This conclusion is supported by the molecular masses of HMGR1 and HMGR2 calculated from their deduced amino acid sequences, which closely match the molecular masses of the native proteins determined by Western blotting.
Fourthly, Northern blots of RNA from the MO showed the presence of two HMGR transcripts. One of these (2.6 kb) was more abundant than the other (3.2 kb), consistent with the observed abundance of the soluble and membrane-bound forms of HMGR proteins in the MO. Northern blots of MO mRNA using probes specific for HMGR1 and HMGR2 showed that the HMGR1 cDNA was produced from the 2.6 kb transcript and the HMGR2 cDNA was produced from the 3.2-kb HMGR transcript. Finally, expression of the HMGR1 cDNA in Sf9 cells produced infected cells with high levels of HMGR activity and this recombinant enzyme (rec-HMGR1) was in the soluble fraction. When separated by anion exchange and gel filtration, rec-HMGR1 had the same characteristics as the native HMGR1, including Km and KI values. When analysed by SDS/PAGE, rec-HMGR1 was slightly larger (approx. 3 kDa) than the native enzyme due to the presence of the His tag added by the Baculovirus system. In short, rec-HMGR1 has identical properties with native HMGR1, including its solubility.
Finally, our results indicate that the mRNAs encoding HMGR1 and HMGR2 result from the differential transcript splicing of a single gene. This conclusion is supported by the nucleotide sequences of the cDNAs of HMGR1 and HMGR2, which are identical for the last 2248 bp at the 3′-ends. In addition, Southern blots showed that the lobster HMGR gene was present as a single gene copy. Together, these results strongly support the conclusion that the two HMGR forms result from the differential splicing of a single HMGR gene to produce two transcripts, one for a soluble protein that lacks a transmembrane region and one for a membrane-bound protein that contains a single transmembrane segment.
Both lobster HMGRs contain all three signature patterns found in other class I HMGRs. Comparison of the amino acid sequences of the lobster HMGR1 and HMGR2 indicate that the catalytic (C-terminal) domain of this protein is closely related to the catalytic domains of other class I HMGRs (Figure 3). The amino acid sequences of HMGR1 and HMGR2 have the highest similarity to mammalian HMGRs. HMGRs of both the hamster, Mesocricetus auratus [27], and the mouse, Mus musculus (GenBank® accession number XP_127496), have 73% amino acid similarity and 59% amino acid identity to lobster HMGRs. High levels of similarity are also observed between the amino acid sequences of lobster HMGR1 and HMGR2 and insect HMGRs. The amino acid sequence of HMGR in beetles Ips pini (GenBank® accession number AAL09351), Ips paraconfusus [21], Dendroctonus jeffreyi [22], the cockroach, B. germanica [15], the moth, Agrotis ipsilon [23], and the fruitfly, Drosophila melanogaster [24], have 61–67% similarity and 45–50% identity to the amino acid sequence of lobster HMGR. In contrast, lobster HMGR has a lower level of similarity (<35%) to class II HMGRs (i.e. Psm-HMGR). In conclusion, the lobster HMGRs are members of class I HMGR.
The amino acid sequence of the lobster HMGR contains unusual features that distinguish them from other class I HMGRs. Foremost among these features is that the predominant form of the enzyme (HMGR1) appears to lack a membrane-bound region, whereas the less-abundant form (HMGR2) has a single transmembrane segment. Class I HMGRs typically have two (in plants), seven (in yeast) or eight (in insects and vertebrates) transmembrane spans [10,11]. Nevertheless, the single transmembrane region in HMGR2 (residues 4–28) is well conserved with the last (8th) transmembrane segment found in insect HMGRs. Although this segment contains 17 hydrophobic residues, it also contains an unusually high number of charged residues (Asp-10, His-11 and Lys-24). Although these charged residues are conserved in insect HMGRs, their importance is unclear. However, it seems probable that the presence of charged residues in the single transmembrane domain of lobster HMGR2 would decrease its stability in the membrane, which may account for the appearance of some of this form in the cytosol fraction (see Figure 1). In addition, the lobster HMGRs contain an additional approx. 80 amino acids at their C-terminal end. The presence of a PEST sequence suggests that this region controls degradation of the enzyme. However, the significance of this C-terminal segment in HMGR function will probably only be clarified by expressing a truncated rec-HMGR1. Finally, lobster HMGRs lack a conserved serine found in class I HMGRs. Since this serine is involved in the regulation of HMGR by reversible phosphorylation, its absence in lobster HMGRs suggests that they are regulated in other ways (see below).
The occurrence of multiple HMGR transcripts and proteins is not unique to the lobster and has been observed in several mammals and insects. In Chinese-hamster ovary cells, 16 different HMGR mRNAs are produced by differential splicing [28]. Humans and Syrian hamsters also have multiple HMGR transcripts [29]. In contrast with the lobster, the multiple mRNAs found in these mammals vary in their 5′-untranslated regions, so these multiple mRNAs produce the same HMGR. Drosophila pupae and adults also have two HMGR transcripts (approx. 3.2 and 4 kb) that are transcribed from a single HMGR gene [24]. A recent analysis of the Drosophila expressed sequence tag database [30] suggests that there are multiple alternative HMGR transcripts in the testes, but the structure of the encoded proteins is unclear since the expressed sequence tags might represent incomplete cDNAs. Whether the smaller transcripts encode shorter HMGRs lacking some or all of the transmembrane segments needs further clarification. Soluble forms of HMGR have also been observed in several insect species. For example, more than half of HMGR protein activity is found to be soluble in the corpora allata of D. punctata [4]. Most of the HMGR protein seems to be soluble or have fewer transmembrane spans in the fat body of B. germanica [13,14]. However, these observations do not preclude the proteolytic release of a soluble fragment from the membranes during tissue preparation. Nevertheless, the apparent tendency of the arthropod HMGRs to be soluble or have fewer transmembrane spans may reflect the lack of cholesterol synthesis in these species [3].
The activity of class I HMGR from higher eukaryotes is attenuated by HMGR kinase, which phosphorylates a serine located six residues from the catalytic histidine of the enzyme ([11,19,31–34], but see [33a]), a spacing that is conserved in other class I HMGRs from higher eukaryotes [18,32]. After phosphorylation, complete restoration of catalytic activity requires dephosphorylation of the HMGR by a protein phosphatase [35]. Lobster HMGRs contain the catalytic histidine but lack the target serine. Although multiple potential phosphorylation sites are found in this enzyme, lobster HMGR activity does not appear to be regulated by reversible phosphorylation. This conclusion is supported by several observations. First, treatment of rec-HMGR1 and native HMGR in MO fractions with phosphatase had no effect on their activity, although this treatment caused a substantial (60%) increase in HMGR activity in rat liver homogenates. Similarly, addition of ATP (to stimulate phosphorylation by an endogenous HMGR kinase) had no effect on lobster HMGR, although it decreased the HMGR activity in rat liver homogenates to a nearly undetectable level. Additionally, mixing experiments showed that the HMGR kinase and other kinases present in the rat liver homogenates did not affect the lobster HMGR. Taken together, these results strongly support the view that lobster HMGR is not regulated by reversible phosphorylation, a mechanism that is commonly used to regulate other class I HMGRs from higher eukaryotes [11].
In summary, lobster HMGR has unusual molecular and regulatory characteristics. Lobster HMGR is the first class I HMGR identified that has both soluble and membrane-bound forms. These forms arise from the differential splicing of a single gene to produce two transcripts. The larger transcript encodes the membrane-bound form that has a single transmembrane segment on its N-terminus. The smaller transcript encodes the soluble form, which lacks this N-terminal sequence. The other unusual feature of lobster HMGR is its lack of regulation by reversible phosphorylation. Class I HMGRs from higher eukaryotes can be inhibited by the phosphorylation of a conserved serine located near the active site, whereas this serine is missing in lobster HMGR. Although other potential phosphorylation sites are present in lobster HMGR, they do not appear to be used to regulate its activity.
Acknowledgments
D.W.B. was supported by grants NIH R15 HD37953-01 and NSF IBN 0240903. We thank Dr K. Edwards and Dr C. Gatto for technical assistance and suggestions. We also thank Ms E. Ostrowski for help in maintaining lobsters.
References
- 1.Goldstein J. L., Brown M. S. Regulation of the mevalonate pathway. Nature (London) 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein J. L., Brown M. S. The low-density lipoprotein pathways and its relation to atherosclerosis. Annu. Rev. Biochem. 1997;46:897–930. doi: 10.1146/annurev.bi.46.070177.004341. [DOI] [PubMed] [Google Scholar]
- 3.Beenakers A. M. T., Van der Horst D. J., Van Marrewijk W. J. Insect lipids and lipoproteins, and their role in physiological processes. Prog. Lipid Res. 1985;24:19–67. doi: 10.1016/0163-7827(85)90007-4. [DOI] [PubMed] [Google Scholar]
- 4.Feyereisen R., Farnsworth D. E. Characterization and regulation of HMG-CoA reductase during a cycle of juvenile hormone synthesis. Mol. Cell. Endocrinol. 1987;53:227–238. doi: 10.1016/0303-7207(87)90178-x. [DOI] [PubMed] [Google Scholar]
- 5.Borst D. W., Laufer H., Landau M., Chang E. S., Hertz W. A., Baker F. C., Schooley D. A. Methyl farnesoate and its role in crustacean reproduction and development. Insect Biochem. 1987;17:1123–1127. [Google Scholar]
- 6.Laufer H., Borst D. W., Baker F. C., Carrasco C., Sinkus M., Rueter C. C., Tsai L. W., Schooley D. A. Identification of a juvenile hormone-like compound in a crustacean. Science. 1987;325:202–205. doi: 10.1126/science.235.4785.202. [DOI] [PubMed] [Google Scholar]
- 7.Homola E., Chang E. S. Methyl farnesoate: crustacean juvenile hormone in search of functions. Comp. Biochem. Physiol. 1997;117B:347–356. [Google Scholar]
- 8.Borst D. W., Ogan J., Tsukimura B., Claerhout T., Holford K. C. Regulation of the crustacean mandibular organ. Amer. Zool. 2001;41:430–441. [Google Scholar]
- 9.Hampton R., Dimpster-Denk D., Rine J. The biology of HMG-CoA reductase: the pros of contra-regulation. Trends Biochem. Sci. 1996;21:140–145. [PubMed] [Google Scholar]
- 10.Bochar D. A., Stauffacher L. V., Rodwell V. W. Sequence comparison reveal two classes of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol. Genet. Metab. 1999;66:122–127. doi: 10.1006/mgme.1998.2786. [DOI] [PubMed] [Google Scholar]
- 11.Bochar D. A., Friesen J. A., Stauffacher C. V., Rodwell V. W. Biosynthesis of mevalonic acid from acetyl-CoA. In: Cane D., editor. Isoprenoids Including Caretenoids and Steroids, vol. 2, Comprehensive Natural Products Chemistry. Oxford: Pergamon Press; 2000. pp. 15–44. [Google Scholar]
- 12.Kleinsek D. A., Dugan R. E., Baker T. A., Porter J. W. 3-Hydroxy-3-methylglutaryl-CoA reductase from rat liver. Methods Enzymol. 1981;71C:462–479. doi: 10.1016/0076-6879(81)71057-7. [DOI] [PubMed] [Google Scholar]
- 13.Casals N., Buesa C., Piulachs M. D., Cabano J., Marrero P. F., Belles X., Hegardt F. Coordinated expression and activity of 3-hydroxy-3-methylglutaryl coenzyme A synthase and reductase in the fat body of Blattera germanica (L.) during vitellogenesis. Insect Biochem. Mol. Biol. 1996;26:837–843. doi: 10.1016/s0965-1748(96)00044-6. [DOI] [PubMed] [Google Scholar]
- 14.Casals N., Martin D., Buesa C., Piulachs M. D., Hegardt F., Belles X. Expression and activity of 3-hydroxy-3-methylglutaryl coenzyme A synthase and reductase in the fat body of ovariectomized and allatectomized Blattera germanica. Physiol. Entomol. 1997;22:6–12. [Google Scholar]
- 15.Martinez-Gonzalez J., Buesa C., Piulachs M.-D., Belles X., Hegardt F. G. Molecular cloning, developmental pattern and tissue expression of 3-hydroxy-3-methylglutaryl coenzyme A reductase of the cockroach Blattera germanica. Eur. J. Biochem. 1993;213:233–241. doi: 10.1111/j.1432-1033.1993.tb17753.x. [DOI] [PubMed] [Google Scholar]
- 16.Li S., Wagner C. A., Friesen J. A., Borst D. W. 3-Hydroxy-3-methylglutaryl coenzyme A reductase in the lobster mandibular organ: regulation by the eyestalk. Gen. Comp. Endocrinol. 2003;134:147–155. doi: 10.1016/s0016-6480(03)00246-6. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J., Fritsch E. F., Maniatis T. Molecular Cloning. In: Nolan C., editor. 2nd edn. Plainview, New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Friesen J. A., Rodwell V. W. Identification of elements critical for phosphorylation of 3-hydroxy-3-methylglutaryl coenzyme A reductase by adenosine monophosphate-activated protein kinase: protein engineering of the naturally nonphosphorylatable 3-hydroxy-3-methylglutaryl coenzyme A reductase from Pseudomonas mevalonii. Biochemistry. 1997;36:1157–1162. doi: 10.1021/bi962104l. [DOI] [PubMed] [Google Scholar]
- 19.Brown M. S., Goldstein J. L., Dietschy J. M. Active and inactive forms of 3-hydroxy-3-methylglutaryl coenzyme A reductase in the liver of the rat. J. Biol. Chem. 1979;254:5144–5149. [PubMed] [Google Scholar]
- 20.Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Tittiger C., Blomquist G. J., Ivarsson P., Borgeson C. E., Seybold S. J. Juvenile hormone regulation of HMG-R gene expression in the dark beetle Ips paraconfusus (Coleoptera: Scolytidae): implications for male aggregation pheromone biosynthesis. Cell. Mol. Life Sci. 1999;55:121–127. doi: 10.1007/s000180050275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tittiger C., Barkawi L. S., Bengoa C. S., Blomquist G. J., Seybold S. J. Structure and juvenile hormone-mediated regulation of the HMG-CoA reductase gene from the Jeffrey pine beetle, Dendroctonus jeffreyi. Mol. Cell. Endocrinol. 2003;199:11–21. doi: 10.1016/s0303-7207(02)00358-1. [DOI] [PubMed] [Google Scholar]
- 23.Duportets L., Belles X., Rossignol F., Couillaud F. Molecular cloning and structural analysis of 3-hydroxy-3-methylglutaryl conenzyme A reductase of the moth Agrotis ipsilon. Insect Mol. Biol. 2000;9:385–392. doi: 10.1046/j.1365-2583.2000.00200.x. [DOI] [PubMed] [Google Scholar]
- 24.Gertler F. B., Chiu C.-Y., Richter-Mann L., Chin D. Developmental and metabolic regulation of the Drosophila melanogater 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol. Cell. Biol. 1988;8:2713–2721. doi: 10.1128/mcb.8.7.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence C. M., Rodwell V. W., Stauffacher C. V. Crystal structure of Pseudomonas mevalonii HMG-CoA reductase at 3.0 Å resolution. Science. 1995;268:1758–1762. doi: 10.1126/science.7792601. [DOI] [PubMed] [Google Scholar]
- 26.Istvan E. S., Palnitkar M., Buchanan S. K., Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 2000;19:819–830. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin D. J., Gil G., Russell D. W., Liscum L., Luskey K. L., Basu S. K., Okayama H., Berg P., Goldstein J. L., Brown M. S. Nucleotide sequence of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase, a glycoprotein of endoplasmic reticulum. Nature (London) 1984;308:613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- 28.Reynolds G. A., Goldstein J. L., Brown M. S. Multiple mRNAs for 3-hydroxy-3-methylglutaryl coenzyme A reductase determined by multiple transcription initiation sites and intron splicing sites in the 5′-untranslated region. J. Biol. Chem. 1985;260:10369–10377. [PubMed] [Google Scholar]
- 29.Luckey K. L. Conservation of promoter sequence but not complex intron splicing pattern in human and hamster genes for 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol. Cell. Biol. 1987;7:1881–1893. doi: 10.1128/mcb.7.5.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubin G. M., Hong L., Brokstein P., Evans-Holm M., Frise E., Stapleton M., Harvey D. A. A Drosophila complementary DNA resource. Science. 2000;287:2222–2224. doi: 10.1126/science.287.5461.2222. [DOI] [PubMed] [Google Scholar]
- 31.Clarke P. R., Hardie D. G. Regulation of HMG-CoA reductase: identification of the site phosphorylated by the AMP-active protein kinase in vitro and in intact rat liver. EMBO J. 1990;9:2439–2446. doi: 10.1002/j.1460-2075.1990.tb07420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato R., Goldstein J. L., Brown M. S. Replacement of serine-871 of hamster 3-hydroxy-3-methylglutaryl coenzyme A reductase prevents phosphorylation by AMP-activated kinase and blocks inhibition of sterol synthesis induced by ATP depletion. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9261–9265. doi: 10.1073/pnas.90.20.9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omkumar R. V., Darnay B. G., Rodwell V. W. Modulation of Syrian hamster 3-hydroxy-3-methylglutaryl-CoA reductase by phosphorylation. Role of Ser871. J. Biol. Chem. 1994;269:6810–6816. [PubMed] [Google Scholar]
- 33a.Erratum. J. Biol. Chem. 1994;269:16518. [PubMed] [Google Scholar]
- 34.Omkumar R. V., Rodwell V. W. Phosphorylation of Ser871 impairs the function of His865 of Syrian hamster 3-hydroxy-3-methylglutaryl-CoA reductase. J. Biol. Chem. 1994;269:16862–16866. [PubMed] [Google Scholar]
- 35.Hardie D. Regulation of fatty acid and cholesterol metabolism by the AMP-activated protein kinase. Biochim. Biophys. Acta. 1992;1123:231–238. doi: 10.1016/0005-2760(92)90001-c. [DOI] [PubMed] [Google Scholar]
- 36.Luskey K. L., Stevens B. Human 3-hydroxy-3-methylglutaryl coenzyme A reductase. Conserved domains responsible for catalytic activity and sterol-regulated degradation. J. Biol. Chem. 1985;260:10271–10277. [PubMed] [Google Scholar]