Abstract
PtdSer (phosphatidylserine) synthesis in mammalian cells occurs through the exchange of L-serine with the base moieties of phosphatidylcholine and phosphatidylethanolamine, which is catalysed by PSS (PtdSer synthase) 1 and 2 respectively. PtdSer synthesis in intact cells and an isolated membrane fraction was inhibited by exogenous PtdSer, indicating that feedback control is involved in the regulation of PtdSer biosynthesis. PSS 1 and 2 are similar in amino acid sequence, with an identity of 32%; however, due to a lack of homology with other known enzymes, their amino acid sequences do not provide information on their catalytic and regulatory mechanisms. In the present study, to identify amino acid residues crucial for the activity and/or regulation of PSS 1, we systematically introduced mutations into a Chinese hamster PSS 1 cDNA clone; namely, each of the 66 polar amino acid residues common to PSS 2 was replaced with an alanine residue. On analysis of Chinese hamster ovary cells transfected with each of the alanine mutant clones, we identified eight amino acid residues (His-172, Glu-197, Glu-200, Asn-209, Glu-212, Asp-216, Asp-221 and Asn-226) as those crucial for the enzyme reaction or the maintenance of the correct structure required for serine base-exchange activity. Among these residues, Asn-209 was suggested to be involved in the recognition and/or binding of free L-serine. We also identified six amino acid residues (Arg-95, His-97, Cys-189, Arg-262, Gln-266 and Arg-336) as those important for regulation of PSS 1. In addition, we found that the alanine mutations at Tyr-111, Asp-166, Arg-184, Arg-323, and Glu-364 affected the production and/or stability of PSS 1 in Chinese hamster ovary cells.
Keywords: phospholipid, phosphatidylserine synthase, phospholipid biosynthesis, site-directed mutagenesis
Abbreviations: CHO, Chinese hamster ovary; PSS, phosphatidylserine synthase; PtdCho, phosphatidylcholine; PtdEtn, phosphatidylethanolamine; PtdSer, phosphatidylserine; R95K, etc., a site-directed mutant bearing a replacement of Arg-95 with lysine, etc
INTRODUCTION
PtdSer (phosphatidylserine) is an essential phospholipid for the growth of mammalian cells [1], comprising approx. 10% of the total phospholipids of various mammalian tissues and cultured cells. PtdSer is known to interact with various proteins, such as protein kinase C [2], myristoylated alanine-rich C kinase substrate [3], coagulation factor V [4], synaptotagmin [5], Raf-1 protein kinase [6], nitric oxide synthase [7], dynamin GTPase [8] and diacylglycerol kinase [9,10]. In addition, PtdSer functions as a precursor phospholipid in the formation of lysophosphatidylserine, which is believed to act as a lipid mediator under various pathophysiological conditions [11]. In the plasma membrane, PtdSer is localized almost exclusively in the cytoplasmic inner leaflet of the lipid bilayer, but it appears outside, for example, during apoptosis, where it serves as a signal for the clearance of apoptotic cells through phagocytosis. Moreover, PtdSer is thought to serve as some kind of scaffold for proteins recruited to membranes. Thus PtdSer plays many important physiological roles, and therefore the synthesis, degradation and localization of PtdSer appear to be strictly regulated.
The PtdSer formation in mammalian cells occurs through the exchange of L-serine with the choline moiety of PtdCho (phosphatidylcholine) or the ethanolamine moiety of PtdEtn (phosphatidylethanolamine) [12,13], whereas in bacteria and yeast, PtdSer is formed from CDP-diacylglycerol and L-serine [14,15]. The serine exchange in CHO (Chinese hamster ovary) cells is catalysed by at least two enzymes named PSS (PtdSer synthase) 1 and 2 [12,13], which are encoded by the pssA and pssB genes respectively [16,17]. PSS 1 and 2 are responsible for the conversion of PtdCho and PtdEtn, respectively, into PtdSer.
PtdSer biosynthesis in CHO-K1 cells is remarkably inhibited on the addition of PtdSer to the culture medium [18], indicating that feedback control is involved in the regulation of PtdSer biosynthesis. The activities of PSSs in homogenates of CHO-K1 cells grown with or without exogenous PtdSer are essentially the same [18]. In addition, PtdSer inhibits PSS 1 and 2 in an isolated membrane fraction of CHO-K1 cells [19–21]. These observations suggest that the feedback control of PtdSer biosynthesis occurs not through regulation of the expression of the PSS 1 and 2 genes, but through inhibition of PSSs by PtdSer.
A CHO cell mutant named ‘29’, whose PtdSer biosynthesis is highly resistant to inhibition by exogenous PtdSer, has been isolated from CHO-K1 cells [19]. This mutant has been shown to carry a point mutation in the pssA gene, which results in the replacement of Arg-95 of the gene product PSS 1 with a lysine residue [20]. Experiments involving mutant strain 29 and an Arg-95→Lys (R95K) mutant pssA cDNA clone [19,20] have indicated that the inhibition of PSS 1 by PtdSer is critical for the maintenance of a normal PtdSer level in CHO-K1 cells, and that Arg-95 of PSS 1 is an essential residue for normal control of PSS 1 activity. Furthermore, it has been shown that the activity of PSS 2, as well as that of PSS 1, is controlled through PtdSer-mediated inhibition, and that Arg-97 of PSS 2 is a critical residue for this feedback control [21].
Both Chinese hamster PSS 1 and 2 are predicted to be transmembrane proteins with nine or ten membrane-spanning segments from their cDNA sequences [16,17]. The two predicted synthases are similar in amino acid sequence, their identity being 32% [16,17]. However, PSS 1 and 2 lack significant homology with other well-characterized enzymes, including the PSSs of bacteria and yeast, and so characteristic motifs or domain structures of known function are missing in PSS 1 and 2. In addition, the membrane-bound properties of PSS 1 and 2 make it difficult to purify them and to analyse their three-dimensional structures. Therefore the crucial amino acid residues, such as those forming the catalytic or regulatory sites of PSS 1 and 2, remain unknown, except for Arg-95 of PSS 1 and Arg-97 of PSS 2, which are critical for enzyme regulation. Thus the details of the molecular mechanisms underlying PtdSer formation by, and the regulation of, PSS 1 and 2 are currently unknown.
In the present study, to identify amino acid residues crucial for the activity and/or regulation of PSS 1, we have systematically introduced site-directed mutations into a Chinese hamster PSS 1 cDNA clone; namely, each of the 66 polar amino acid residues common to PSS 2 was replaced with an alanine residue. On analysis of the 66 alanine mutants, we found 19 mutations that markedly disrupt PSS 1 function.
EXPERIMENTAL
Materials
L-[U-14C]Serine, [methyl-14C]choline and [2-14C]ethanolamine hydrochloride were purchased from Amersham Biosciences. An anti-PSS 1 N-terminal peptide antibody was raised by immunization of rabbits with a multiple antigen peptide corresponding to amino acid residues 1–17 of Chinese hamster PSS 1. The antigen-specific antibody was purified from the antiserum by affinity chromatography with antigen peptide-coupled resin.
Strains and culture conditions
Strain CHO-K1 was obtained from the American Type Culture Collection, and maintained in plastic flasks in Ham's F-12 medium supplemented with 10% (v/v) newborn calf serum, penicillin G (100 units/ml) and streptomycin sulphate (100 μg/ml) in a 5% CO2 atmosphere at 100% humidity and 37 °C. Mutant PSB-2 cells [22] were maintained under the same conditions, except that the growth medium was supplemented with 30 μM PtdSer liposomes, prepared as described previously [18].
Site-directed mutagenesis and plasmids
Alanine mutants were constructed with a QuikChange™ Site-Directed Mutagenesis Kit (Stratagene), according to the manufacturer's instructions. Plasmid pSVpssA/neo2 [20] was used as the template for all mutagenesis reactions. The resultant alanine mutant cDNAs were verified to have the intended mutations, but not extraneous mutations, by DNA sequencing (ABI PRISM™ 310; PerkinElmer).
DNA transfection of CHO cells
Cells (2.5×106 for CHO-K1; 3×106 for PSB-2) were seeded into 100 mm dishes containing 12 ml of cell culture medium. After overnight cultivation, the cells were transfected with plasmids using LIPOFECTAMINE Plus™ reagent (Invitrogen™ Life Technologies) as recommended by the manufacturer. Briefly, cells were incubated with a complex of DNA (the amount used in each experiment is given in the respective figure legend) and the reagents (20 μl of Plus™ reagent and 30 μl of LIPOFECTAMINE™ reagent) in 6.5 ml of Opti-MEM I (Invitrogen™ Life Technologies) at 37 °C for 5 h. Subsequently, 6.5 ml of cell culture medium and 650 μl of newborn calf serum were added to bring the final concentration of the serum and the volume of the medium to normal levels.
Metabolic labelling of PtdSer with [14C]serine
At 24 h after the start of transfection, the transfected cells (approx. 3×105) were seeded into the wells of a 24-well plate in Ham's F-12 medium supplemented with 10% newborn calf serum, followed by incubation at 37 °C. After 1 day, the cells were incubated in fresh growth medium with or without exogenous 100 μM PtdSer at 37 °C for 2 h, and then labelled with L-[U-14C]serine (0.5 μCi/ml) for 3 h at 37 °C in the corresponding growth medium with or without exogenous PtdSer. Phospholipids in the labelled cells were extracted by the method of Bligh and Dyer [23] and separated by one-dimensional TLC [24], and then the radioactivity of PtdSer was determined with a bioimage analyser (Fujix BAS 2000). The numbers of cells per well in parallel unlabelled cultures were determined with a Coulter model ZB1 counter, and used to standardize the results.
Enzyme assays for base-exchange reactions of phospholipids with serine, choline and ethanolamine in cell lysates
To measure the base-exchange activities in cell lysates, at 48 h after the start of transfection, cells were washed with PBS, harvested by scraping, resuspended in a sonication buffer consisting of 0.25 M sucrose, 10 mM Hepes, pH 7.5, and 1 mM EDTA, and then disrupted on ice with a W-225R Ultra Sonic disrupter (Heat System Ultrasonics) equipped with a No. 419 micro tip. A typical assay mixture (0.1 ml) for the base-exchange reactions comprised 50 mM Hepes, pH 7.5, 5 mM CaCl2 and either 0.2 mM L-[U-14C]serine (10 μCi/μmol), 0.2 mM [methyl-14C]choline (20 μCi/μmol) or 0.2 mM [2–14C]ethanolamine (10 μCi/μmol). The reaction was initiated by the addition of 50–200 μg of cell protein. For the experiments shown in Figure 4(B), the cell lysates were pre-incubated with or without PtdSer liposomes in 50 mM Hepes, pH 7.5, for longer than 30 min on ice, and then the reaction was initiated by the addition of CaCl2 and L-[U-14C]serine (10 μCi/μmol). The mixture was incubated at 37 °C for 20 min, and the reaction was terminated by adding 50 μl of 20 mM EDTA containing either 0.2 M L-serine, 0.2 M choline or 0.2 M ethanolamine. Then, 2.5 ml of chloroform/methanol (2:1, v/v) was added to the mixture, and lipids were extracted as described by Saito et al. [25]. A 1 ml portion of the lower chloroform phase was allowed to dry out, and then the radioactivity was measured by liquid scintillation spectrometry.
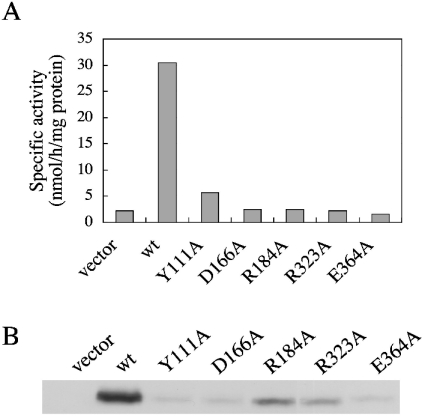
Figure 4. Five alanine mutations affecting the production and/or stability of PSS 1.
CHO-K1 cells were transiently transfected with 6 μg of each of the control empty vector plasmid (vector) or plasmids that encoded wild-type (wt) or Y111A, D166A, R184A, R323A and E364A mutant PSS 1. (A) Cell lysates of the transfectants were assayed for serine base-exchange activity. Three independent experiments gave similar results; the values shown are for one of the three independent experiments, and are the averages of duplicate determinations with a variation of <12% between duplicates. (B) Cell lysates (10 μg of protein) of the transfectants were separated by SDS/PAGE, and then analysed by Western blotting with an anti-PSS 1 N-terminal peptide antibody.
Western blotting
Proteins were incubated in SDS sample buffer consisting of 62.5 mM Tris/HCl, pH 6.8, 2% (w/v) SDS, 5% (v/v) 2-mercaptoethanol, 10% (v/v) glycerol and 0.001% (w/v) Bromophenol Blue at 37 °C for 30 min, separated by electrophoresis in 10% (w/v) polyacrylamide slab gels containing 0.1% SDS, as described previously [26], and then electroblotted on to PVDF membranes. The blots were incubated with the anti-PSS 1 N-terminal peptide antibody, and then with anti-rabbit IgG linked to horseradish peroxidase (Amersham Biosciences, Little Chalfont, Bucks., U.K.). Cross-reactive proteins were detected with an enhanced chemiluminescence kit (Amersham Biosciences), as recommended by the manufacturer.
Determination of protein concentrations
Protein concentrations were measured with a Pierce bicinchoninic acid protein assay kit, with BSA as a standard.
RESULTS
Systematic construction of alanine mutants of PSS 1
Alignment of the amino acid sequences of Chinese hamster PSS 1 and 2 revealed that 138 amino acid residues are conserved in these two enzymes. Among these conserved residues, 66 are polar ones. Although the phospholipid substrates used by PSS 1 and 2 for PtdSer formation are different [22,27], both PSS 1 and 2 catalyse the exchange of L-serine with the base moieties of pre-existing phospholipids for PtdSer formation. Furthermore, both enzymes are inhibited by exogenous PtdSer [19–21]. Thus the amino acid residues conserved in PSS 1 and 2 are candidates for critical amino acid residues for the activity and/or regulation of these two enzymes. Therefore, to identify such critical amino acid residues, we constructed a set of site-directed mutant cDNA clones, i.e. 66 different mutant clones that each encoded a PSS 1 mutant with a replacement of one of the conserved 66 polar amino acid residues with alanine.
Identification of eight alanine mutations that affect serine base-exchange activity but not the expression level of PSS 1
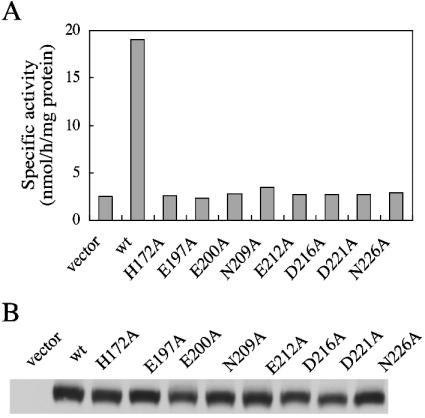
To examine the effect of the mutations on PSS 1 function, CHO-K1 cells were transiently transfected with plasmids encoding the 66 alanine mutants of PSS 1 and wild-type PSS 1, and a vector plasmid. Then a cell homogenate of each transfectant was subjected to assaying of PSS (serine base-exchange) activity. The serine base-exchange activity in the homogenate of the wild-type PSS 1 cDNA-transfected cells was remarkably higher than that found in the vector plasmid-transfected cells (Figure 1A), indicating overproduction of wild-type PSS 1 in the transfectants. As for the wild-type cDNA clone, most mutant cDNA clones led to remarkable increases in serine base-exchange activity in cell homogenates (results not shown). However, among the 66 mutant cDNA clones, eight caused no significant increase in the serine base-exchange activity in cell homogenates (Figure 1A), whereas the PSS 1 protein expression levels in the eight mutant cDNA-transfected cells were found to be comparable with that in the wild-type cDNA-transfected cells when examined by Western blotting with an anti-PSS 1 N-terminal peptide antibody (Figure 1B). Thus the eight mutations (H172A, E197A, E200A, N209A, E212A, D216A, D221A and N226A) carried by these mutant cDNA clones appeared to cause almost complete inactivation of the serine base-exchange activity.
Figure 1. Serine base-exchange activity of PSS1, and PSS 1 protein levels, in CHO-K1 cells transfected with the wild-type or the H172A, E197A, E200A, N209A, E212A, D216A, D221A and N226A mutant PSS 1 cDNA clones.
CHO-K1 cells were transiently transfected with 6 μg of each of the control empty plasmid (vector), and plasmids that encoded wild-type (wt) or H172A, E197A, E200A, N209A, E212A, D216A, D221A and N226A mutant PSS 1. (A) Cell lysates of the transfectants were assayed for serine base-exchange activity. Three independent experiments gave similar results; the values shown are for one of three independent experiments and are the averages of duplicate determinations with a variation of <12% between duplicates. (B) Cell lysates (10 μg of protein) of the transfectants were separated by SDS/PAGE, and then analysed by Western blotting with an anti-PSS 1 N-terminal peptide antibody.
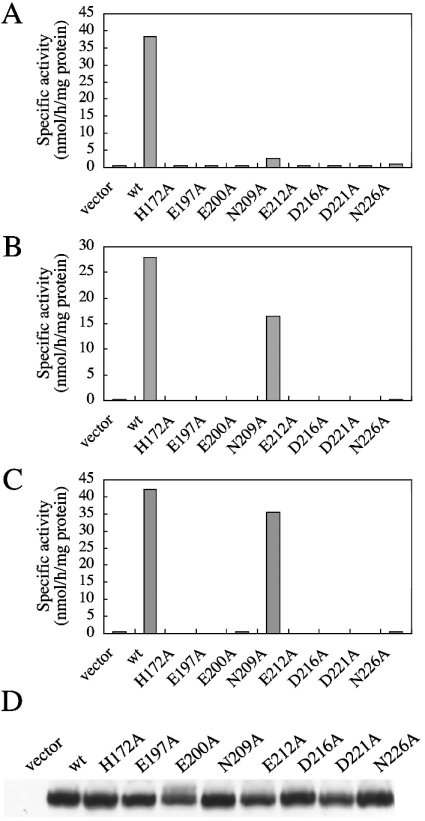
To confirm this, we transfected the PSS 1 cDNA clones carrying these eight mutations into a mutant strain of CHO-K1, PSB-2, which exhibits very low serine base-exchange activity because of defects in both PSS 1 and 2. As shown in Figure 2(A), the serine base-exchange activities in the cell homogenates of all the mutant cDNA transfectants except for the N209A mutant transfectant were much lower than that of the wild-type cDNA transfectant, similar to that of the vector plasmid transfectant. The N209A mutant cDNA clone caused a very slight increase in serine base-exchange activity (Figure 2A). The PSS 1 protein levels in all the mutant cDNA transfectants were comparable with that in the wild-type cDNA transfectant (Figure 2D). These results confirmed the inactivation of serine base-exchange activity by the alanine mutations at His-172, Glu-197, Glu-200, Asn-209, Glu-212, Asp-216, Asp-221 and Asn-226.
Figure 2. Serine, choline and ethanolamine base-exchange activities of PSS 1, and PSS 1 protein levels, in PSB-2 cells transfected with the wild-type or H172A, E197A, E200A, N209A, E212A, D216A, D221A and N226A mutant PSS 1 cDNA clones.
PSB-2 cells were transiently transfected with 6 μg of each of the control empty plasmid (vector), and plasmids that encoded the wild-type (wt) or H172A, E197A, E200A, N209A, E212A, D216A, D221A and N226A mutant PSS 1. Cell lysates of the transfectants were assayed for serine (A), choline (B), and ethanolamine (C) base-exchange activity. For the results shown in (A–C), two independent experiments gave similar results; the values shown are for one of the two independent experiments and are the averages of duplicate determinations with a variation of <5% (A), <0.4% (B) and 1% (C) between duplicates. (D) Cell lysates (10 μg of protein) of the transfectants were separated by SDS/PAGE, and then analysed by Western blotting with an anti-PSS 1 N-terminal peptide antibody.
PSS 1 is capable of catalysing choline and ethanolamine base-exchange for the formation of PtdCho and PtdEtn in vitro [1,16,28], in addition to serine base-exchange for PtdSer formation, although the major components of PtdCho and PtdEtn synthesis in vivo do not involve the base-exchange pathway [1,13,22,29,30]. Therefore we also examined the effects of these eight mutations causing almost complete inactivation of the serine base-exchange activity on the choline and ethanolamine base-exchange activities of PSS 1. Upon transfection into PSB-2 cells, the H172A, E197A, E200A, E212A, D216A, D221A and N226A mutant cDNA clones were incapable of inducing significant increases in either choline or ethanolamine base-exchange activity in cell homogenates (Figures 2B and 2C), indicating that these seven mutations affected the choline and ethanolamine base-exchange activities in addition to serine base-exchange activity. However, the homogenate of PSB-2 cells transfected with the N209A mutant cDNA clone showed striking increases in the choline and ethanolamine exchange activities, which were comparable with the increases induced by transfection with the wild-type PSS 1 cDNA clone (Figures 2B and 2C). Thus the N209A mutation was shown to specifically affect the serine base-exchange activity.
Identification of six amino acid residues crucial for the regulation of PSS 1
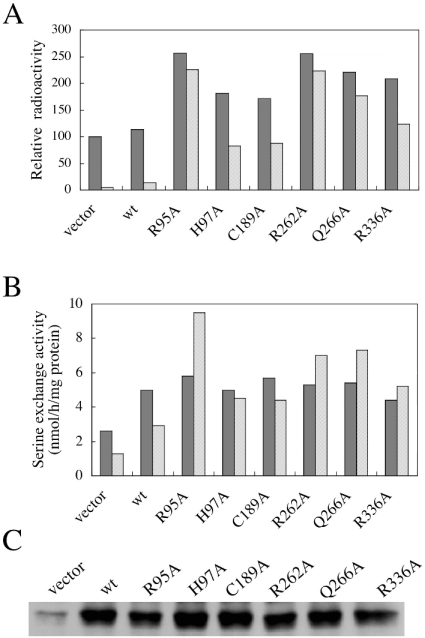
We previously found that replacement of Arg-95 of PSS 1 with a lysine residue affects regulation of this enzyme, indicating that Arg-95 of PSS 1 is a critical amino acid residue for the regulation of PSS 1 [19,20]. To confirm this by investigating a mutant carrying a replacement of Arg-95 with an amino acid residue other than lysine, and to identify amino acid residues involved in the enzyme regulation other than Arg-95, we examined the effects of the 66 different alanine mutations mentioned above, including the R95A mutation, on the PtdSer-mediated feedback control of PSS 1. CHO-K1 cells were transiently transfected with the plasmids encoding the 66 alanine mutants of PSS 1 and wild-type PSS 1, and a vector plasmid as a control, and then the PtdSer biosynthetic rates in the transfectants were determined in medium supplemented with or without 100 μM PtdSer. When the vector plasmid and the plasmid encoding wild-type PSS 1 were transfected, the PtdSer biosynthetic rates in the two transfectants were essentially the same in the medium without exogenous PtdSer, and were strikingly decreased on the addition of PtdSer to the medium (Figure 3A), confirming the PtdSer-mediated feedback control of PtdSer biosynthesis. Most of the mutant cDNA clone transfectants examined synthesized PtdSer at almost the same rate as the vector plasmid and wild-type cDNA clone transfectants in the medium supplemented with or without exogenous PtdSer. However, the six transfectants harbouring the R95A, H97A, C189A, R262A, Q266A and R336A mutant cDNA clones exhibited remarkable (1.7–2.6-fold) increases in the PtdSer biosynthetic rate in the medium lacking exogenous PtdSer compared with the vector plasmid and wild-type cDNA clone transfectants (Figure 3A). Furthermore, PtdSer biosynthesis in the transfectants harbouring the R95A, R262A or Q266A mutant cDNA clones was inhibited by only 10–20% on the addition of PtdSer to the culture medium (Figure 3A), whereas that by the wild-type PSS 1 cDNA transfectant was inhibited by approx. 90%. When cultivated with exogenous PtdSer, the PtdSer biosynthetic rate in these mutant clone transfectants was more than 10-fold that in the wild-type cDNA transfectant (Figure 3A). PtdSer biosynthesis in the transfectants harbouring the H97A, C189A and R336A mutant cDNA clones was also more resistant to inhibition by exogenous PtdSer than that in the wild-type cDNA transfectant (Figure 3A): the PtdSer biosynthetic rates in these mutant cDNA transfectants cultivated in the medium supplemented with PtdSer were approx. 50% of those in the respective transfectants cultivated without exogenous PtdSer, and more than 5-fold that in the wild-type cDNA transfectant cultivated with exogenous PtdSer (Figure 3A).
Figure 3. Six alanine mutations affecting the regulation of PSS 1.
(A) CHO-K1 cells were transiently transfected with 6 μg of each of the control empty plasmid (vector), and plasmids that, respectively, encoded wild-type (wt) or R95A, H97A, C189A, R262A, Q266A and R336A mutant PSS 1. The transfectants were metabolically labelled with [14C]serine for 3 h at 37 °C in the growth medium supplemented with (hatched bars) or without (filled bars) 100 μM PtdSer, and then the radioactivity of cellular PtdSer was determined. Three independent experiments gave similar results; the values shown are for one of three independent experiments and are the averages of duplicate determinations with a variation of <7% between duplicates. (B and C) Samples (0.25 μg) of each of the same plasmids as those used in the experiments described in (A) were transfected into CHO-K1 cells. (B) Cell lysates of the transfectants were assayed for serine base-exchange activity in the presence (hatched bars) or absence (filled bars) of 100 μM PtdSer. Two independent experiments gave similar results; the values shown are for one of the two independent experiments, and are the averages of duplicate determinations with a variation of <11% between duplicates. (C) Cell lysates (10 μg of protein) of the transfectants were separated by SDS/PAGE, and then analysed by Western blotting with an anti-PSS 1 N-terminal peptide antibody.
Next, we examined the effect of exogenous PtdSer on the serine base-exchange activity in homogenates of CHO-K1 cells transfected with each of the R95A, H97A, C189A, R262A, Q266A and R336A mutant cDNA clones. The specific activities of serine base-exchange in the cell homogenates of these transfectants were essentially the same as that found in the wild-type cDNA transfectant (Figure 3B). In addition, the PSS 1 protein levels in the cell homogenates of the mutant cDNA transfectants were comparable with that in that of the wild-type cDNA transfectant (Figure 3C). However, the serine base-exchange activities of all the mutant cDNA-transfected cells were either enhanced or not significantly inhibited upon the addition of 100 μM PtdSer to the reaction mixture, whereas those of the vector plasmid- and wild-type cDNA-transfected cells were inhibited by approx. 50% upon the addition of PtdSer (Figure 3B).
These results, taken together, indicate that, in addition to Arg-95, His-97, Cys-189, Arg-262, Gln-266 and Arg-336 are crucial residues for the regulation of PSS 1 activity.
Five alanine mutations affect PSS 1 production and/or stability
During the course of investigating which amino acid residues were crucial for enzyme activity, we found that some mutant clones other than the eight mutant clones described above were also incapable of inducing significant increases in serine base-exchange activity in transfectants, and that the PSS 1 protein levels in these mutant clone-transfectants were much lower than that in the wild-type cDNA-transfected cells (results not shown). To confirm that decreases in enzyme activities and protein levels were due to mutations in PSS 1 cDNA, and not to extraneous mutation(s) in the promoter region of the mutant PSS 1 expression plasmids, the PSS 1 cDNA inserts of the five plasmids incapable of yielding reasonable levels of PSS 1 production were cut out and each re-inserted into a fresh mammalian expression plasmid with an authentic promoter. When the five re-constructed mutant plasmids (Y111A, D166A, R184A, R323A and E364A) were transfected into CHO-K1 cells, the serine base-exchange activity in the homogenate of each transfectant was again found to be almost the same as that in the vector-transfected cells (Figure 4A), and the PSS 1 protein levels in the transfectants were also much lower than that in the wild-type PSS 1 cDNA-transfected cells (Figure 4B). Therefore the five PSS 1 mutations (Y111A, D166A, R184A, R323A and E364A) carried by these PSS 1 expression plasmids seemed to cause a lowering of PSS 1 production and/or stability.
DISCUSSION
PSS 1 in mammalian cells catalyses the exchange of free L-serine with the choline moiety of PtdCho for PtdSer formation [22,27]. A defect in PSS 1 causes cultured mammalian cells to become PtdSer auxotrophs [1]. In addition, the loss of normal feedback control of PSS 1 activity leads to the accumulation of cellular PtdSer and its decarboxylation product, PtdEtn [19,20]. Thus PSS 1 is a critical enzyme for the metabolism of the amino-glycerophospholipids PtdCho, PtdSer and PtdEtn in mammalian cells. However, the details of the molecular mechanisms underlying PtdSer synthesis by, and regulation of, PSS 1 are currently unknown. To address this issue, we attempted to identify amino acid residues crucial for the activity and/or regulation of Chinese hamster PSS 1 through systematic alanine mutagenesis.
We constructed a set of site-directed mutant cDNA clones, i.e. 66 different mutant clones that each encode a PSS 1 mutant with replacement of one of the 66 polar amino acid residues conserved in PSS 1 and PSS 2 with an alanine residue. On analysis of the 66 alanine mutants of PSS 1, we found that: (1) the H172A, E197A, E200A, N209A, E212A, D216A, D221A and N226A mutations caused almost complete abolition of the serine base-exchange activity; (2) the R95A, H97A, C189A, R262A, Q266A and R336A mutations disturbed the regulation of PSS 1 activity; and (3) the Y111A, D166A, R184A, R323A and E364A mutations lowered the production and/or stability of PSS 1 in CHO-K1 cells. Thus many amino acid residues conserved in PSS 1 and 2 were required for PSS 1 function, probably being involved in the catalytic action, regulation, adequate protein folding, targeting to an appropriate membrane, stability, or production of PSS 1. Interestingly, no mutation that affected both enzyme activity and its regulation was found in this study.
As the H172A, E197A, E200A, N209A, E212A, D216A, D221A and N226A mutations led to almost complete abolition of the serine base-exchange activity of PSS 1, each of the replaced amino acid residues (His-172, Glu-197, Glu-200, Asn-209, Glu-212, Asp-216, Asp-221 and Asn-226) appeared to participate in the recognition and/or binding of a substrate, the catalytic reaction or the maintenance of the normal structure required for the serine base-exchange activity. Furthermore, we found that the N209A mutation had a very specific effect: namely, it caused almost complete abolition of the serine base-exchange activity, but did not affect the choline or ethanolamine base-exchange activity of PSS 1. Thus, Asn-209 is probably involved in the recognition and/or binding of free L-serine.
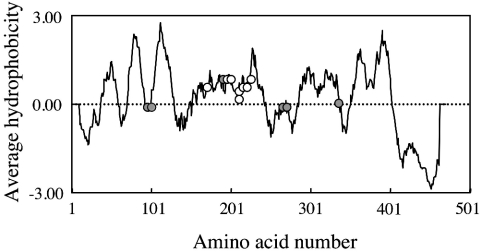
It is noteworthy that the eight amino acid residues, alanine mutations of which have caused abolition of serine base-exchange activity, are clustered in the middle region of the PSS 1 polypeptide (Figure 5). This region is relatively hydrophobic (Figure 5), and has been predicted to form multiple membrane-spanning segments by several programs for analysis of the topology of membrane proteins. As a phospholipid substrate of PSS 1 is supposed to be integrated into the membrane lipid bilayer, these findings raised the possibility that transmembrane segments within this middle region of PSS 1 polypeptide are involved in the recognition and/or binding of the phospholipid substrate in the membrane. In addition, it seems reasonable that Asn-209, which was suggested to be involved in the recognition and/or binding of the water-soluble substrate L-serine, was also found in this middle region of PSS 1 polypeptide and predicted to be located near or at the end, not in the middle, of a transmembrane segment by all the above programs. Thus we speculated that this middle region of the PSS 1 polypeptide forms an active site, where the binding of the phospholipid substrate and L-serine, and subsequent PtdSer formation, occur, although further investigations, such as analysis of the three-dimensional structure, are required to confirm this idea.
Figure 5. Hydrophobicity plots for the PSS 1 protein predicted from the cDNA sequence, and the positions of alanine mutations that affect the PSS 1 function.
The average hydrophobicity of a nanodecapeptide composed of amino acids n−9 to n+9 is plotted against n, the amino acid number. The positions of alanine mutations that cause almost complete abolition of the serine base-exchange activity (open circles) and those that disrupt the regulation of PSS 1 activity (grey circles) are indicated.
When the R95A, H97A, C189A, R262A, Q266A and R336A mutant PSS 1s were individually overproduced in CHO-K1 cells, the PtdSer synthesis in the medium without exogenous PtdSer was remarkably enhanced, although overproduction of the wild-type PSS 1 did not induce such enhancement. Furthermore, these six mutants of PSS 1 were shown to be highly resistant to inhibition by exogenous PtdSer compared with the wild-type PSS 1 in experiments involving intact cells and cell homogenates. These results indicated that Arg-95, His-97, Cys-189, Arg-262, Gln-266 and Arg-336 of PSS 1 are crucial residues for the regulation of PSS 1, and participate in the exogenous PtdSer-mediated inhibition of PSS 1.
All of the mutations identified as those disturbing the regulation of PSS 1 had the following two simultaneous effects: enhancement of PtdSer biosynthesis in the medium without exogenous PtdSer upon overproduction and conferment of resistance to inhibition by exogenous PtdSer (dysfunction of feedback control in intact cells and PtdSer-resistant PtdSer synthesis in cell homogenates). So far, a mutation that has only one of these effects has not been identified. Thus it appeared that the control of the activity of overproduced PSS 1 and the inhibition of PSS 1 by exogenous PtdSer occurred at least in part through the same molecular mechanism.
The PtdSer-mediated inhibition of PtdSer synthesis by another PSS, PSS 2, was recently suggested to occur through the direct interaction of PtdSer with PSS 2 [31]. From the analogy between the inhibition of PSS 1 and 2 by exogenous PtdSer, the inhibition of PSS 1 by exogenous PtdSer appears to occur through the direct interaction of PtdSer with PSS 1. However, whether the inhibition occurs through simple product inhibition (the binding of PtdSer to an active site) or allosteric regulation (the binding of PtdSer to a regulatory site) remains unknown. In the primary structure, five out of the six amino acid residues identified as those involved in the PtdSer-mediated inhibition were located away from the middle region of PSS 1, where alanine mutations that cause abolition of serine base-exchange activity are clustered (Figure 5). In addition, considering the activity in the homogenates of the mutant cDNA-transfected cells, the six regulatory mutants of PSS 1 seemed to exhibit normal serine base-exchange activity, implying that these regulatory mutations were located at a site other than the active site. Therefore allosteric regulation appears to be more plausible than simple product inhibition for the inhibition of PSS 1 by PtdSer.
PtdSer synthesis by the R95A mutant of PSS 1 was markedly enhanced on the addition of PtdSer to the reaction mixture. The reason for the enhancement is currently unknown. However, there are several possible explanations. When the wild-type PSS 1 was overproduced in CHO-K1 cells, the serine base-exchange activity in the cell homogenate was remarkably enhanced on the addition of PtdEtn (O. Kuge, unpublished work), which is not a substrate for PSS 1 in intact cells [22]. This observation implied that some phospholipids are able to activate overproduced PSS 1 in a non-specific manner. Therefore the enhancement of the R95A mutant PSS 1 activity by PtdSer might be due to a non-specific interaction between PtdSer and the mutant PSS 1 that is not regulated by the specific interaction with PtdSer. Another possible explanation is that, when the Arg-95 is mutated, the binding of PtdSer to a putative regulatory site causes activation instead of inhibition through, for example, stabilizing enzyme conformation.
The PSS 1 protein levels in CHO-K1 cells transfected with each of five mutant PSS 1 cDNA clones (Y111A, D166A, R184A, R323A and E364A) were much lower than that in the wild-type cDNA-transfected CHO-K1 cells. The low level expression of PSS 1 in the five mutant cDNA transfectants was reproducibly observed in transient expression experiments involving the control wild-type cDNA clone. The uptake of plasmid DNA by CHO-K1 cells in the transient expression system was supposed not to be affected by a few changes in the nucleotide sequence. Therefore it was not likely that the low level expression was due to the low efficiency of the incorporation of cDNA clones into CHO-K1 cells.
Although the expression levels were very low, substantial amounts of the mutant PSS 1 proteins were probably produced in the CHO-K1 cells transfected with the Y111A, D166A, R184A, R323A and E364A mutant cDNA clones, judging from the results of Western blot analysis with an anti-PSS 1 N-terminal peptide antibody; under the experimental conditions used in this study, we could detect no band of endogenous PSS 1, i.e. PSS 1 in the vector plasmid transfectant, but each homogenate prepared from the mutant cDNA transfectants gave a clearly visible band of PSS 1. Nevertheless, the serine base-exchange activities for PtdSer formation in the cell homogenates of these five mutant cDNA transfectants were almost the same as that in the homogenate of the vector plasmid transfectant. Therefore the Y111A, D166A, R184A, R323A and E364A mutations lowered the production and/or stability of PSS 1, and probably affected the serine base-exchange activity of PSS 1.
The recent advances in genome research and accumulation of DNA data have revealed that many model organisms, including Homo sapiens, Danio rerio, Drosophila melanogaster, Caenorhabditis elegans and Arabidopsis thaliana, have a gene encoding a putative protein homologous with Chinese hamster PSS 1, though it is currently unknown whether these genes are translated into functional proteins that possess serine base-exchange activity like Chinese hamster and murine PSS 1 and 2. These putative PSS 1-like proteins mentioned above showed various extents of amino acid sequence homology with Chinese hamster PSS 1, from 37% to 97%, but, interestingly, either all, or 18 out of 19, of the amino acid residues that had been identified as crucial for the Chinese hamster PSS 1 function in the present study were conserved in these putative proteins and murine PSS 1 and 2. This selective conservation supported the idea that the 19 amino acid residues were crucial for the PSS 1 function and implied that the PSS 1-like genes found in many organisms were functional and contributed to PtdSer synthesis through the serine base-exchange mechanism.
In summary, we found 19 amino acid residues crucial for PSS 1 function through systematic alanine mutagenesis, one of which was suggested to be specifically involved in L-serine binding and/or recognition. These findings, together with the results of future structural studies on PSS 1, such as ones on the membrane topology, and secondary and tertiary structures of PSS 1, will be of critical importance for elucidating the molecular mechanisms of PtdSer synthesis by, and regulation of, PSS 1, which is a key enzyme in the metabolism of amino-glycerophospholipids in mammalian cells.
Acknowledgments
We thank all the members of this laboratory for their helpful discussions. This work was supported in part by the Human Sciences Basic Research Project and Integrated Study Projects on Drug Innovation Science of the Japan Health Sciences Foundation, by Special Co-ordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and by Grants-in-Aid for General Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1.Kuge O., Nishijima M., Akamatsu Y. Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. II. Isolation and characterization of phosphatidylserine auxotrophs. J. Biol. Chem. 1986;261:5790–5794. [PubMed] [Google Scholar]
- 2.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 3.Taniguchi H., Manenti S. Interaction of myristoylated alanine-rich protein kinase C substrate (MARCKS) with membrane phospholipids. J. Biol. Chem. 1993;268:9960–9963. [PubMed] [Google Scholar]
- 4.Mann K. G., Jenny R. J., Krishnaswamy S. Cofactor proteins in the assembly and expression of blood clotting enzyme complexes. Annu. Rev. Biochem. 1988;57:915–956. doi: 10.1146/annurev.bi.57.070188.004411. [DOI] [PubMed] [Google Scholar]
- 5.Perin M. S., Fried V. A., Mignery G. A., Jahn R., Südhof T. C. Phospholipid binding by a synaptic vesicle protein homologous to the regulatory region of protein kinase C. Nature (London) 1990;345:260–263. doi: 10.1038/345260a0. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh S., Xie W. Q., Quest A. F., Mabrouk G. M., Strum J. C., Bell R. M. The cysteine-rich region of raf-1 kinase contains zinc, translocates to liposomes and is adjacent to a segment that binds GTP-ras. J. Biol. Chem. 1994;269:10000–10007. [PubMed] [Google Scholar]
- 7.Calderon C., Huang Z. H., Gage D. A., Sotomayor E. M., Lopez D. M. Isolation of a nitric oxide inhibitor from mammary tumor cells and its characterization as phosphatidyl serine. J. Exp. Med. 1994;180:945–958. doi: 10.1084/jem.180.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuma P. L., Stachniak M. C., Collins C. A. Activation of dynamin GTPase by acidic phospholipids and endogenous rat brain vesicles. J. Biol. Chem. 1993;268:17240–17246. [PubMed] [Google Scholar]
- 9.Kato M., Takenawa T. Purification and characterization of membrane-bound and cytosolic forms of diacylglycerol kinase from rat brain. J. Biol. Chem. 1990;265:794–800. [PubMed] [Google Scholar]
- 10.Yada Y., Ozeki T., Kanoh H., Nozawa Y. Purification and characterization of cytosolic diacylglycerol kinases of human platelets. J. Biol. Chem. 1990;265:19237–19243. [PubMed] [Google Scholar]
- 11.Bruni A., Monastra G., Bellini F., Toffano G. Autacoid properties of lysophosphatidylserine. Prog. Clin. Biol. Res. 1988;282:165–179. [PubMed] [Google Scholar]
- 12.Kuge O., Nishijima M. Phosphatidylserine synthase I and II of mammalian cells. Biochim. Biophys. Acta. 1997;1348:151–156. doi: 10.1016/s0005-2760(97)00137-9. [DOI] [PubMed] [Google Scholar]
- 13.Kuge O., Nishijima M. Biosynthetic regulation and intracellular transport of phosphatidylserine in mammalian cells. J. Biochem. (Tokyo) 2003;133:397–403. doi: 10.1093/jb/mvg052. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto K. Phosphatidylserine synthase from bacteria. Biochim. Biophys. Acta. 1997;1348:214–227. doi: 10.1016/s0005-2760(97)00110-0. [DOI] [PubMed] [Google Scholar]
- 15.Yamashita S., Nikawa J. Phosphatidylserine synthase from yeast. Biochim. Biophys. Acta. 1997;1348:228–235. doi: 10.1016/s0005-2760(97)00102-1. [DOI] [PubMed] [Google Scholar]
- 16.Kuge O., Nishijima M., Akamatsu Y. A Chinese hamster cDNA encoding a protein essential for phosphatidylserine synthase I activity. J. Biol. Chem. 1991;266:24184–24189. [PubMed] [Google Scholar]
- 17.Kuge O., Saito K., Nishijima M. Cloning of a Chinese hamster ovary (CHO) cDNA encoding phosphatidylserine synthase (PSS) II, overexpression of which suppresses the phosphatidylserine biosynthetic defect of a PSS I-lacking mutant of CHO-K1 cells. J. Biol. Chem. 1997;272:19133–19139. doi: 10.1074/jbc.272.31.19133. [DOI] [PubMed] [Google Scholar]
- 18.Nishijima M., Kuge O., Akamatsu Y. Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. I. Inhibition of de novo phosphatidylserine biosynthesis by exogenous phosphatidylserine and its efficient incorporation. J. Biol. Chem. 1986;261:5784–5789. [PubMed] [Google Scholar]
- 19.Hasegawa K., Kuge O., Nishijima M., Akamatsu Y. Isolation and characterization of a Chinese hamster ovary cell mutant with altered regulation of phosphatidylserine biosynthesis. J. Biol. Chem. 1989;264:19887–19892. [PubMed] [Google Scholar]
- 20.Kuge O., Hasegawa K., Saito K., Nishijima M. Control of phosphatidylserine biosynthesis through phosphatidylserine-mediated inhibition of phosphatidylserine synthase I in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4199–4203. doi: 10.1073/pnas.95.8.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuge O., Saito K., Nishijima M. Control of phosphatidylserine synthase II activity in Chinese hamster ovary cells. J. Biol. Chem. 1999;274:23844–23849. doi: 10.1074/jbc.274.34.23844. [DOI] [PubMed] [Google Scholar]
- 22.Saito K., Nishijima M., Kuge O. Genetic evidence that phosphatidylserine synthase II catalyses the conversion of phosphatidylethanolamine to phosphatidylserine in Chinese hamster ovary cells. J. Biol. Chem. 1998;273:17199–17205. doi: 10.1074/jbc.273.27.17199. [DOI] [PubMed] [Google Scholar]
- 23.Bligh E. G., Dyer W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 24.Kuge O., Yamakawa Y., Nishijima M. Enhancement of transport-dependent decarboxylation of phosphatidylserine by S100B protein in permeabilized Chinese hamster ovary cells. J. Biol. Chem. 2001;276:23700–23706. doi: 10.1074/jbc.M101911200. [DOI] [PubMed] [Google Scholar]
- 25.Saito M., Bourque E., Kanfer J. N. Studies on base-exchange reactions of phospholipids in rat brain particles and a “solubilized” system. Arch. Biochem. Biophys. 1975;169:304–317. doi: 10.1016/0003-9861(75)90345-8. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Kuge O., Nishijima M., Akamatsu Y. Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. III. Genetic evidence for utilization of phosphatidylcholine and phosphatidylethanolamine as precursors. J. Biol. Chem. 1986;261:5795–5798. [PubMed] [Google Scholar]
- 28.Kuge O., Nishijima M., Akamatsu Y. Isolation of a somatic-cell mutant defective in phosphatidylserine biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 1985;82:1926–1930. doi: 10.1073/pnas.82.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esko J. D., Raetz C. R. Autoradiographic detection of animal cell membrane mutants altered in phosphatidylcholine synthesis. Proc. Natl. Acad. Sci. U.S.A. 1980;77:5192–5196. doi: 10.1073/pnas.77.9.5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voelker D. R. Phosphatidylserine functions as the major precursor of phosphatidylethanolamine in cultured BHK-21 cells. Proc. Natl. Acad. Sci. U.S.A. 1984;81:2669–2673. doi: 10.1073/pnas.81.9.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuge O., Hasegawa K., Ohsawa T., Saito K., Nishijima M. Purification and characterization of Chinese hamster phosphatidylserine synthase 2. J. Biol. Chem. 2003;278:42692–42698. doi: 10.1074/jbc.M307270200. [DOI] [PubMed] [Google Scholar]