Abstract
HtrA1, a member of the mammalian HtrA (high temperature requirement A) serine protease family, has a highly conserved protease domain followed by a PDZ domain. Accumulating evidence has indicated that PDZ domains regulate protease activity of HtrA proteins. We searched for binding partners of the PDZ domain of mouse HtrA1 by yeast two-hybrid screening, and isolated proteins that were recognized by the HtrA1 PDZ domain through their C-terminal ends with a core consensus Φ-X-Φ-[V/L/F/A]-COOH sequence (where Φ is a hydrophobic/non-polar amino acid). C-propeptides of fibrillar collagens were most frequently isolated. Type III procollagen α1 C-propeptide, which was used as a model protein, was digested by HtrA1. HtrA1 cleavage of the collagen C-propeptide was enhanced by reductive denaturation of the C-propeptide and partly inhibited by removal of the C-terminal four amino acids from the C-propeptide, suggesting that the substrate recognition was facilitated by the binding of the free C-terminal ends of substrates to the PDZ domain of HtrA1. The synthetic oligopeptide (GM130Pep) that fitted the consensus recognition sequence bound to HtrA1 with a high affinity (Kd=6.0 nM). GM130Pep stimulated HtrA1 protease activity 3- to 4-fold, but did not efficiently stimulate the activity of an HtrA1 mutant lacking the PDZ domain, supporting the notion that the PDZ domain enhances protease activity upon ligand binding. The peptide derived from Type III collagen α1 C-propeptide specifically stimulated protease activity of HtrA1, but did not stimulate nor significantly bind to HtrA2, suggesting that the collagen C-propeptide is a specific physiological regulator of HtrA1.
Keywords: C-propeptide, collagen, HtrA1, HtrA2/Omi, PDZ domain, serine protease
Abbreviations: 3-AT, 3-amino-1,2,4-triazole; CAST, CAZ-associated structural protein; Cdyl, chromodomain protein, Y chromosome-like; Col1a1, Col2a1 and Col3a1; Types I, II and III procollagen α1 respectively; CoxVa, cytochrome c oxidase subunit Va; -C-Pro, -C-propeptide; DTT, dithiothreitol; F171D, Phe171→Asp; GM130, Golgi auto-antigen golgin, subfamily a,2; HtrA, high temperature requirement A; LRP9, low-density-lipoprotein-receptor-related protein 9; Ni-NTA, Ni2+-nitrilotriacetate; OMP, outer-membrane porin; PAR6B, partitioning defective 6 homologue β; SPR, surface plasmon resonance; ssrA, small stable RNA; TGF-β, transforming growth factor-β; Thlx, triple helical region; Trx, thioredoxin, Tsp, tail-specific protease
INTRODUCTION
PDZ domains are protein–protein interaction modules that often play a central role as organizers of multimeric signalling complexes. The primary targets of these domains are specific motifs, 4–6 amino acid in length, which occur at the C-terminus of target proteins or structurally related internal motifs [1,2]. In addition to the assembly of supramolecular signalling complexes, several PDZ-domain-containing proteins play an important role in the regulation of activity of cell surface receptor proteins or in protein trafficking [2]. Studies have revealed that PDZ domains also participate in the specific substrate recognition of Escherichia coli Tsp (tail-specific protease) [3,4] and regulation of protease activity of HtrA (high temperature requirement A) serine proteases [5].
HtrA is a highly conserved family of serine proteases, which are characterized by the combination of a trypsin-like catalytic domain with at least one C-terminal PDZ domain [6]. Bacterial HtrA is essential for survival at high temperatures and has a molecular chaperone activity at low temperatures and a serine protease activity that degrades misfolded proteins at high temperatures [7]. The crystal structure of a typical E. coli HtrA protein, DegP, has revealed that DegP is a hexamer formed by staggered association of trimer rings [8]. The structure of DegP suggested that the PDZ domains were responsible for substrate binding and subsequent translocation into the catalytic cavity [8]. The PDZ domain of another E. coli HtrA, DegS, specifically recognizes the C-terminal short sequence motifs (-YYF-COOH) of misfolded OMPs (outer-membrane porins) [9]. Binding of this peptide motif activates DegS cleavage of a transmembrane protein, RseA, that normally binds to and inhibits an envelope stress-response σ factor.
The human or mouse genome contains at least four HtrA family genes. HtrA2/Omi is localized in mitochondria where it is auto-processed to form the mature protein by removing the first 133 amino acids. HtrA2 is released into the cytosol upon apoptotic stimuli and promotes apoptosis through a caspase-dependent pathway [10] and a caspase-independent pathway. The latter pathway depends on the protease activity of HtrA2 [11]. Like the DegP trimer ring unit, HtrA2 is a trimer with the PDZ domains restricting the access of substrates to the inner catalytic cavity [12]. The trimeric structure is essential for its protease activity. The optimal HtrA2 PDZ-domain-binding peptide (-GQYYFV-COOH) was selected from a random peptide mixture by binding to the HtrA2 PDZ domain [5]. This peptide was able to facilitate the protease activity of HtrA2. These results from bacterial and mammalian HtrA serine proteases have revealed a new function of PDZ domains as ligand-dependent regulators of enzyme activity.
In contrast with HtrA2, mammalian HtrA1 and its close family members, HtrA3 and HtrA4, are secretory proteins and contain an insulin-like growth-factor-binding domain, and a Kazal-type serine-protease inhibitor domain in the N-terminal region upstream of the HtrA homology region [6,13,14]. HtrA1 was originally isolated as a gene repressed in SV40-transformed human fibroblasts [13]. HtrA1 expression is also down-regulated in several other cancers [15–17]. Stable over-expression of HtrA1 inhibits proliferation of metastatic melanoma cell lines [16] and induces cell death in ovarian cancer cells [17]. HtrA1 is therefore a candidate for a tumour suppressor gene. The tumour suppressive activity of HtrA1 was largely dependent on the protease activity. HtrA1 was also isolated as a gene up-regulated in cartilage of human osteoathritic patients [18].
Based on the finding that HtrA1 is characteristically expressed in distinct mouse embryo tissues, differentiation of which is mainly regulated by TGF-β (transforming growth factor-β) signalling, we have proposed that HtrA1 functions as an inhibitor of TGF-β signalling. We showed that HtrA1 was able to bind to and inhibit signalling of a wide range of TGF-β family proteins [19]. Surprisingly, the inhibitory activity of HtrA1 on the TGF-β signalling depended totally on the protease activity of HtrA1. Deletion of the C-terminal PDZ domain facilitated both the protease and the TGF-β inhibition activities. It is thus predicted that the C-terminal PDZ domain may function as a regulator of the protease activity of HtrA1. To understand the regulatory mechanisms of HtrA1, we have screened for PDZ-binding proteins using a yeast two-hybrid assay. In the present study, we investigated interactions of the HtrA1 PDZ domain with isolated partner proteins and the potential role of these interactions on the protease activity of HtrA1.
EXPERIMENTAL
DNA constructs and plasmids
Human HtrA2/Omi cDNA was given generously by A. S. Zervos (Biomolecular Science Center, University of Central Florida, Florida, U.S.A.). The full-length mouse HtrA1 and its various mutants, either tagged with the Myc epitope or not, were cloned into pcDNA3 mammalian expression vector (Invitrogen Corp.), as described previously [19]. The F171D (Phe171→Asp) HtrA1 mutant was constructed by changing the phenylalanine residue at amino acid 171 to aspartic acid by PCR mutagenesis. To make a bait plasmid for yeast two-hybrid assay, an HtrA1 fragment with a deletion in the signal peptide sequence (ΔSS-HtrA1, encoding amino acids 23–480) was amplified by PCR and the fragment was inserted into pGBT9 at the EcoRI and SalI sites. The encoding regions of other truncated HtrA1 fragments were; C-HtrA1, amino acids 132–480; ΔPDZ-HtrA1, amino acids 23–384; SP-HtrA1, amino acids 132–384; PDZ-HtrA1, amino acids 369–480. A bacterial expression vector for an active, N-terminally truncated HtrA protein, ΔN-HtrA1 (amino acids 151–480) or its mutant defective in protease activity, ΔNS328A-HtrA1, was constructed as follows. The full-length cDNA for HtrA1 or S328A HtrA1 [19] was digested with PstI at the position corresponding to amino acid 151 and SmaI in the 3′ non-coding region. The fragment was cloned into BSKS to add an EcoRI site at the 5′ end and a NotI site at the 3′ end. An EcoRI–NotI fragment was removed from this plasmid and inserted into the pET28a vector. To construct pET28a-ΔNΔPDZ-HtrA1, a PstI–BamHI fragment of HtrA1 cDNA encompassing amino acids 151–384 was inserted into BSKS. An EcoRI–NotI fragment was removed from this plasmid and cloned into pET28a. To construct a bacterial expression vector for an active, N-terminally truncated human HtrA2 (ΔN-HtrA2, amino acids 131–458), an EagI–BamHI fragment of HtrA2 cDNA was cloned into pET28a.
To construct prey plasmids for yeast two-hybrid assay, various cDNAs encoding PDZ-binding proteins were cloned in pACT2 vector. The Type III collagen α1 (Col3a1) triple helical region (Thlx) (amino acids 138–400) was amplified by PCR using the longest Col3a1 clone (clone M-220) isolated from two-hybrid screening as template. Among various fragments of Col3a1 C-propeptide (Col3a1-C-Pro) cloned into pACT2, two fragments designated as Col3a1-C-Pro(1–259) and Col3a1-C-Pro(59–259) were derived from yeast clones isolated from the two-hybrid screening, and encoded 259 and 201 C-terminal amino acids of Col3a1-C-Pro respectively. Col3a1-C-Pro(1–191) and Col3a1-C-Pro(1–236) were constructed by deleting the C-terminal 68 and 23 amino acids respectively from Col3a1-C-Pro(1–259) using EcoRI and EcoRV. The C-terminal four amino acid deletion mutant, Col3a1-C-Pro(1–255), and a central fragment, Col3a1-C-Pro(59–145), which lacked the N-terminal 58 and C-terminal 144 amino acids of Col3a1-C-Pro(1–259), were produced by PCR amplification using pACT2-Col3a1-C-Pro(1–259) as a template with the appropriate primers. Among the two-hybrid clones encoding a PDZ-binding protein [GM130 (Golgi auto-antigen golgin, subfamily a, 2), LRP9 (low-density-lipoprotein-receptor-related protein 9), CoxVa (cytochrome c oxidase subunit Va), Cdyl (chromodomain protein, Y chromosome-like), mKIAA0769 (mouse KIAA0769), CAST (CAZ-associated structural protein) or PAR6B (partitioning defective 6 homologue β)], one clone with a similar insert size was selected for further analysis (each encoded the C-terminal 143, 142, 117, 131, 108, 117 or 154 amino acids respectively). The C-terminal four amino acid deletion mutants of these clones (GM130Δ4aa, LRP9Δ4aa, CoxVaΔ4aa, CdylΔ4aa, mKIAA0769Δ4aa, CASTΔ4aa and PAR6BΔ4aa) were produced by PCR amplification. All of the PCR amplified fragments were inserted into pACT2 at the EcoRI and XhoI sites. To produce histidine-tagged (His6) proteins of Col3a1-C-Pro, Col1a1-C-Pro, GM130 or their C-terminal four amino acid deletion mutants in bacteria, the cDNA fragment was excised from the corresponding pACT-2 clone using BamHI and XhoI and inserted into pET28c.
Yeast two-hybrid screening for PDZ-binding proteins
The CG1945 yeast strain was transformed with pGBT9-ΔSS-HtrA1 or pGBT9-PDZ-HtrA1. The yeast cells were then transformed with a cDNA library and two-hybrid screening was carried out according to the ClonTech manual. A mouse adult brain Matchmaker library in pACT2 vector (ClonTech) was screened using PDZ-HtrA1 as bait. A mouse 17-day embryo Matchmaker cDNA library (ClonTech) was first screened using ΔSS-HtrA1 as bait, and individual clones isolated from the embryo library were next examined for PDZ binding using pGBT9-PDZ-HtrA1 transformed yeast. Yeast clones growing on His− plates in the absence of 3-AT (3-amino-1,2,4-triazole) were picked up and analysed for β-galactosidase activity according to the ClonTech filter-assay protocol. Clones positive for β-galactosidase activity were used for further analyses. Plasmid DNA was recovered from yeast, transformed into E. coli HB101 cells to select for pACT2 plasmid, and purified and processed for DNA sequencing using an ABI 310 sequencer.
For the quantitative binding assay, three independent transformants were measured for β-galactosidase activity using 2-nitrophenol-β-D-galactopyranoside (Wako) as the substrate according to the ClonTech liquid culture assay protocol. Results are presented in Miller units as the means±S.D. for three independent assays.
Preparation of HtrA1 in bacteria
E. coli BL21(DE3) (Novagen) cells were transformed with pET28a vector containing ΔN-HtrA1, ΔNS328A-HtrA1, ΔNΔPDZ-HtrA1 or ΔN-HtrA2. The transformed bacteria were cultured in 1 litre of Luria–Bertani medium containing 1% glucose at 37 °C until the D600 reached 0.6–0.8, and 1.0 mM isopropyl β-D-thiogalactoside was added to induce protein production. After incubation at 20 °C overnight, the cells were harvested and disrupted by sonication in 25 ml of TN buffer [20 mM Tris/HCl (pH 7.5)/500 mM NaCl]. The soluble fraction was applied to a Ni-NTA (Ni2+-nitrilotriacetate)–agarose (Qiagen) column. Bound HtrA protein was eluted with TN buffer containing 100 mM imidazole, and then dialysed against TN buffer.
Production and purification of PDZ-binding proteins
BL21(DE3) cells were transformed with pET28c-Col3a1-C-Pro and pET28c-Col3a1-C-ProΔ4aa. His6–Col3a1-C-Pro and His6–Col3a1-C-ProΔ4aa proteins were produced in insoluble forms after induction with 1.0 mM isopropyl β-D-thiogalactoside. Solubilization of proteins was performed by two methods. In the first method, the insoluble fraction was dissolved in a washing buffer [50 mM Tris/HCl (pH 7.5), 125 mM NaCl, 10% glycerol and 4 M urea] and applied to a Ni-NTA–agarose (Qiagen) column. The bound proteins were eluted with the washing buffer containing 100 mM imidazole, dialysed against 5 mM HCl, and concentrated by Centricon (Millipore). This protein preparation was used for the solid-phase binding assay. In the second method, the insoluble fraction was dissolve in a urea buffer [50 mM Tris/HCl (pH 8.0), 125 mM NaCl, 2mM DTT (dithiothreitol), 1 mM EDTA and 8 M urea] and chilled on ice for 2 h. The re-folding was achieved by adding 50 vol. of a folding buffer [50 mM Tris/HCl (pH 8.0), 125 mM NaCl, 2 mM DTT, 2 mM GSH, 1 mM GSSG and 1.8% Hepes] and incubation for 72 h at 4 °C. The solubilized proteins were purified with Ni-NTA–agarose (Qiagen) columns, dialysed against the folding buffer and used as substrates in protease assays.
GM130 and GM130Δ4aa proteins expressed in bacteria by the pET28c vector were produced in the soluble fraction. These proteins were purified with a Ni-NTA–agarose (Qiagen) column. The eluted proteins were dialysed against TN buffer.
Cell culture and transfection
HEK-293T cells were cultured in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum, in 10-cm diameter dishes to 60% confluence. Cells were transfected with 2 μg/dish pcDNA3-HtrA1-myc or its mutants (S328A-HtrA1, F171D-HtrA1 or ΔPDZ-HtrA1) by a calcium phosphate method [19]. Culture supernatant was collected 72 h after transfection.
Solid-phase binding assay
Human recombinant Col3a1-C-Pro protein produced by insect cells was a generously given by D.J.S. Hulmes (Institut de Biologie et Chimie des Proteines, Lyon, France). Bacterially produced Col3a1-C-Pro in 5 mM HCl was neutralized with a final concentration of 50 mM Tris/HCl (pH 7.5) just before use. ELISA plate (NUNC) wells were coated with 50 μl of Col3a1-C-Pro (100 ng per well) at 4 °C overnight. Coating with control Trx (thioredoxin)–His6, various collagen C-propeptide preparations and other PDZ-binding proteins was carried out similarly. After extensive washing with TBS [20 mM Tris/HCl (pH 7.6), 150 mM NaCl and 0.05% Tween 20], the wells were blocked with 0.1% skimmed milk in TBS. The Myc-tagged HtrA1 protein in the HEK-293T culture supernatant was serially diluted with fresh culture medium containing 10% fetal bovine serum, added to the wells (100 μl/well) and incubated at 4 °C overnight. After extensive washing with TBS, the wells were blocked with 0.5% BSA in TBS. The anti-Myc monoclonal antibody (9E10) in 4% BSA in TBS was added to the wells and incubated at 4 °C overnight. After washing with TBS, alkaline phosphatase-conjugated anti-mouse IgG antibody in TBS containing 4% BSA was added to the wells and incubated for 5 h at 25 °C. The bound alkaline phosphatase activity was detected using p-nitrophenyl phosphate (2.6 mM) in 100 μl of 10 mM diethanolamine buffer (pH 9.5) containing 0.5 mM MgCl2. After the reaction was stopped by adding 50 mM EDTA (disodium salt), the A405 was measured using Thermomax microplate reader (Molecular Devices). The amount of HtrA1 protein in the culture supernatant was measured by Western blot analysis using a purified bacterially produced HtrA1 fragment as standard.
Assay of protease activity
The protease activity was assayed by incubating HtrA1 or its derivatives with substrate proteins (5–10 μg of β-casein, BSA or other proteins) in a 20 μl mixture containing 50 mM Tris/HCl (pH 7.6) either with or without 1.5 mM DTT for 0.5–8 h at 37 °C. To examine effects of PDZ-binding peptides, bacterially produced protein (0.06–2 μg) or a synthetic peptide (obtained from BioSynthesis, Lewisville, TX, U.S.A.); see the structures in Figure 5) at 1–500 μM was included in the reaction mixture. The reaction was terminated by the addition of 20 μl of SDS loading buffer [50 mM Tris/HCl (pH 6.8), 2% SDS, 100 mM DTT, 0.1% Bromophenol Blue and 10% glycerol]. The mixture was boiled for 5 min, separated by SDS gel electrophoresis (10% polyacrylamide) and stained with Coomassie Blue. The stained gel was scanned using an Epson GT9500 scanner, and the intensity of protein bands was measured using Image Gauge V4.0 software (Fuji Photo Film Co.). The protease assays were carried out reveral times under the same or similar conditions, and representative data are presented in the Figures.
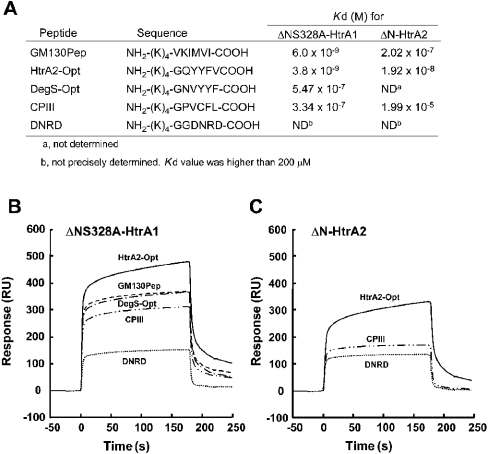
Figure 5. SPR assay for interaction of HtrA1 with synthetic peptides derived from C-terminal ends of PDZ-binding proteins.
(A) The sequences of the synthetic peptides and their dissociation constants for binding to ΔNS328A-HtrA1 and ΔN-HtrA2 proteins, as determined by SPR. The peptides were designed to include four lysine residues, (K)4, at the N-terminal region followed by six specific amino acid residues. The control peptide, Asp-Asn-Arg-Asp (DNRD), was designed not to contain hydrophobic amino acids. The specific sequence in GM130Pep was derived from rat GM130 protein. The binding of synthetic peptides to ΔNS328A-HtrA1 (B) and ΔN-HtrA2 (C). Concentration of peptides injected was 5 μM. SPR assay also confirmed that ΔNΔPDZ-HtrA1 lacking the PDZ domain did not interact with GM130Pep (results not shown). RU. resonance unit.
Western blotting
Rabbit polyclonal antibody that recognized a C-terminal fragment of HtrA1 [19] and C-propeptide of human Col3a1 (kindly given by L. Fisher, National Institute of Dental and Craniofacial Reasearch, National Institutes of Health, Bethesda, MD, U.S.A.) [20] were used as primary antibody. Protein samples were resolved on an SDS/12% polyacrylamide gel and then electrotransferred on to Protran nitrocellulose membrane (Schleicher and Schuell). The membrane was blocked with TBST [50 mM Tris/HCl (pH 7.5), 100 mM NaCl and 0.1% Tween 20] containing 5% skimmed milk for 2 h at 25 °C or overnight at 4 °C. The membrane was incubated with a rabbit polyclonal antibody diluted in TBST containing 4% BSA for 1 h at 25 °C. After washing extensively with TBST, membranes were incubated with horseradish peroxidase-conjugated anti-rabbit IgG antibody (Amersham Biosciences). The bound peroxidase was detected with ECL® Plus (Amersham Biosciences) using X-ray film (Fuji Photo Film Co.).
SPR (surface plasmon resonance) measurements
Interactions of ΔNS328A-HtrA1 and ΔN-HtrA2 with synthetic peptides were monitored by SPR measurements using the BIAcore™ 3000 (BIAcore AB). Purified His6-tagged ΔNS328A-HtrA1 or ΔN-HtrA2 was immobilized on an Ni-NTA sensor chip in an eluent buffer [10 mM Hepes (pH 7.4), 0.15 M NaCl, 50 μM EDTA and 0.005% Tween 20]. Because of the low-molecular masses of peptides used as analytes, the HtrA proteins in the range of 2000 or more resonance units were immobilized on the chips. Various concentrations of peptides (1 nM–100 μM) in the eluent buffer were injected at a flow rate of 10 μl/min. Dissociation of peptides was achieved with the eluent buffer containing 1 M NaCl. The specific changes in the experimental sensorgram were measured by subtracting the values of the reference cell containing no protein. Kinetic analysis was performed using BIAevaluation 3.0 software (BIAcore AB) according to the manufacturer's instructions.
RESULTS
Isolation of PDZ-binding proteins
In order to explore the regulatory mechanisms of HtrA1 protease, we searched for target proteins of the PDZ domain of HtrA1 (PDZ-HtrA1) via yeast two-hybrid screen. After screening of 3.5×106 clones each from a mouse embryonic cDNA library and an adult brain cDNA library, we isolated 71 clones that represented eleven known genes (Table 1). The most frequently isolated gene was Col3a1. Other fibrillar procollagen genes, type I α1 and type II α1 (Col1a1 and Col2a1 respectively), were also isolated. All collagen clones were obtained from the embryonic cDNA library. The second most frequently isolated gene encoded a cis-Golgi matrix golgin subfamily a protein (GM130). GM130 clones were obtained from both the embryonic and adult brain cDNA libraries. Among the eight isolated genes, other than the three collagen genes, only one gene, a cell surface LRP9, encoded a protein that had an extracellular segment. All of the three isolated LRP9 clones encoded only the C-terminal intracellular domain of this receptor molecule. Because HtrA1 is believed to be extracellular, collagens are the only possible candidates for physiological partners of PDZ-HtrA1.
Table 1. Genes isolated from yeast two-hybrid screening.
The day-17 mouse embryo cDNA library and the adult mouse brain cDNA library, 3.5×106 clones each, were screened for PDZ-binding proteins by yeast two-hybrid system as described in the Experimental section. The identity of WAC (WW domain-containing adapter with a coiled-coil region) has been obtained only recently and therefore this clone was not analysed in this study.
| Gene | Number of isolated clones | C-terminal sequence | Accession number |
|---|---|---|---|
| Col3a1 | 29* | -GPVCFL-COOH | AK019448 |
| Col1a1 | 5* | -GPACFV-COOH | BC003198 |
| Col2a1 | 2* | -GPVCFL-COOH | NM_031163 |
| GM130 | 15*† | -VKIMVV-COOH | BC011407 |
| Cdyl | 6† | -RKIDEF-COOH | NM_009881 |
| LRP9 | 3* | -DEPLLA-COOH | AB042200 |
| CoxVa | 3* | -LGLDKV-COOH | NM_007747 |
| CAST | 3† | -EEGIWA-COOH | AK122265 |
| mKIAA0769 | 3† | -VEITLV-COOH | AB018312 |
| PAR6B | 1* | -GTIITL-COOH | NM_021409 |
| WAC | 3† | -QNSFMF-COOH | AF320996 |
* Clones isolated from the embryo library.
† Clones isolated from the adult brain library.
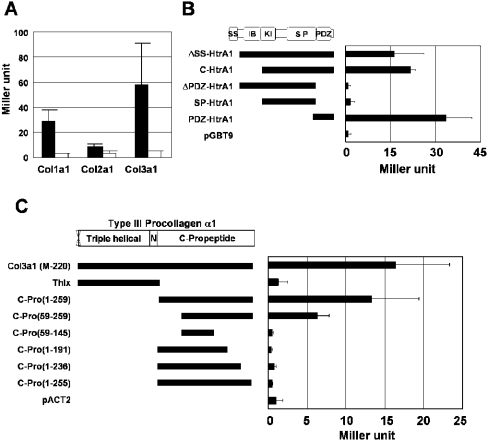
Among the three collagen clones, Col3a1 induced the highest β-galactosidase activity in quantitative yeast two-hybrid assay, followed by Col1a1 and then Col2a1 (Figure 1A). The C-terminal 12 amino acid sequence is identical for Col3a1 and Col2a1. It is likely that regions other than the C-terminal end are involved at least in part in the binding of collagens to HtrA1. In the following study, we used Col3a1 for further characterization of the interaction with PDZ-HtrA1.
Figure 1. Interaction of collagen C-propeptides with the PDZ domain of HtrA1.
(A) Interaction of Type I, II and III collagen with HtrA1. One clone encoding almost the same size of Type I, II or III collagen α1 molecule (each encoding C-terminal 365, 359 or 345 amino acids respectively) was selected from the pACT2 collagen clones isolated by yeast two-hybrid screening. They were analysed for interaction with HtrA1 by the β-galactosidase liquid assay of yeast two-hybrid using pGBT9-ΔSS-HtrA1 (closed bar) or the empty pGBT9 vector (open bar) as bait. (B) Specific interaction of Col3a1 with the PDZ domain of HtrA1. Various constructs of HtrA1 in pGBT9 as indicated were analysed for interaction with the Col3a1 clone in pACT2, as described in (A). (C) Interaction of the C-terminal end of Col3a1-C-Pro with HtrA1. The various constructs of Col3a1 in pACT2 as indicated were analysed for interaction with HtrA1 using pGBT9-ΔSS-HtrA1 as bait, as described in (A). SS, signal sequence; IB, IGF-binding-protein-like domain; KI, Kazal-type protease inhibitor domain; SP, serine protease domain; N, non-helical region of mature collagen.
First, we tested the interaction of various HtrA1 fragments with Col3a1 (Figure 1B). Only the HtrA1 fragments which contained the PDZ domain showed interaction with Col3a1, indicating that the PDZ domain was the sole region that was able to interact with Col3a1.
The longest Col3a1 clone (clone M-220) encompassed 151 amino acids of the triple helical region, the non-helical region and the entire 261 amino acid C-propeptide region (Figure 1C). A short Col3a1 clone [Col3a1-C-Pro(1–259)] encoded 259 amino acids of the C-propeptide. The yeast clones containing these long and short Col3a1 cDNA fragments exhibited comparable β-galactosidase activity (Figure 1C). Furthermore, the N-terminal half of Col3a1(M-220), Thlx, which contained the triple helical and non-helical regions, did not interact with HtrA1 (Figure 1C). Therefore, the C-propeptide of Col3a1 (Col3a1-C-Pro) was the sole region necessary for the interaction with PDZ-HtrA1. Four amino acids at the C-terminus were revealed to be essential for the interaction with HtrA1 (Figure 1C). On the other hand, an N-terminal truncation of 58 amino acids compromised the binding considerably, but still retained significant binding activity (Figure 1C). This is consistent with the finding that PDZ domains generally recognize specific 4–6 residue motifs at the C-terminus [1].
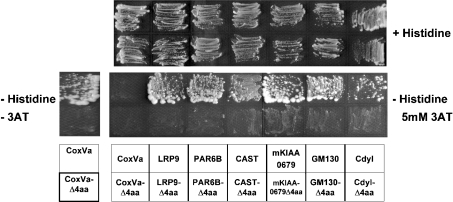
To verify the role of C-terminal ends of other isolated clones, we deleted their C-terminal four amino acids. All of the yeast clones harbouring the truncated prey plasmids did not grow on His− plates either in the presence or in the absence of 3-AT (Figure 2). These results indicated that interaction of all these clones with PDZ-HtrA1 was mediated through their C-terminal ends. The C-terminal six amino acid sequences of these clones are listed in Table 1. The isolated proteins terminated in leucine, valine, phenylalanine and alanine residues (C-terminal amino acid position was defined as follows: position 0, the C-terminal residue; position −1, the amino acid residue next to the C-terminus, and so on). We also observed strong selectivity for hydrophobic/non-polar amino acids at positions −1 (with preference for phenylalanine and leucine) and −3 (with preference for valine and isoleucine). Position −5 also showed weak selectivity for hydrophobic/non-polar amino acids. It is likely that the consensus motif recognized by HtrA1-PDZ is (Φ)-X-Φ-X-Φ-[L/V/F/A]-COOH, where (Φ) represents weak preference for hydrophobic/non-polar amino acids, Φ represents hydrophobic/non-polar amino acids and X represents any amino acid. The C-terminal sequences in Col3a1, Col2a1, GM130 and mKIAA0769 match the consensus recognition sequence ([L/I]-X-Φ-[I/L/V]-COOH) for the PDZ domain of HtrA2 (PDZ-HtrA2), as reported by Junqueira et al. [21].
Figure 2. Interaction of HtrA1 with C-terminal ends of PDZ-binding proteins.
Yeast cells were co-transformed with pGBT9-ΔSS-HtrA1 and pACT2 vector containing cDNA for CoxVa, LRP9, PAR6B, CAST, mKIAA0769, GM130, Cdyl (upper lane of each panel) or their mutants that lacked the C-terminal 4 amino acids (lower lane of each panel). The transformants were streaked on an agar plate with (upper panels) or without (lower panels) histidine, containing either 5 mM (right-hand panels) or no (left hand panels) 3-AT. All yeast clones transformed with these wild-type prey cDNA required the presence of the bait HtrA1 cDNA for growth on His− plates (results not shown).
Semi-quantitative filter β-galactosidase assay and growth assay in the presence of various concentrations of 3-AT (0–30 mM) indicated that PDZ-HtrA1 bound to GM130 with the greatest strength (results not shown). PDZ-HtrA1 bound to Col3a1 and mKIAA0769 strongly, followed by moderate binding to CAST and Col1a1, and a weak binding to LRP9, PAR6B, Cdyl and CoxVa (results not shown). C-terminal sequences of PAR6B, Cdyl and CoxVa contain polar/charged amino acids (threonine, glutamic acid and lysine respectively) at position −1 and significantly deviated from the above mentioned consensus.
Interaction of Col3a1-C-pro with HtrA1
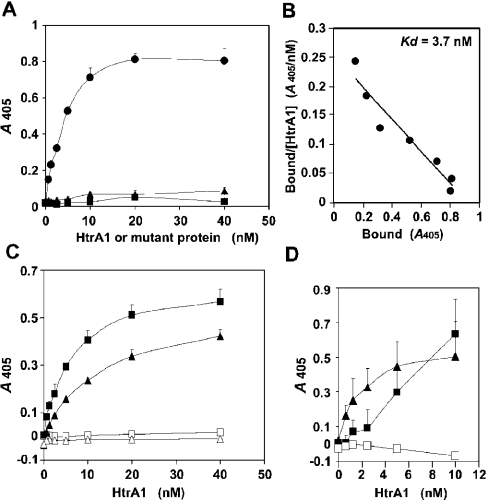
To analyse more quantitatively the interaction of Col3a1-C-Pro with PDZ-HtrA1, we employed a solid-phase binding assay using His6–Col3a1-C-Pro protein and HtrA1 protein produced by bacteria and HEK-293T cells (Figure 3). HtrA1 bound to Col3a1-C-Pro showing a typical saturation binding curve after subtraction of non-specific binding to control Trx–His6 (Figure 3A). In this assay, the Kd of HtrA1 for Col3a1-C-Pro was estimated to be 3.7 nM (Figure 3B). This binding affinity was comparable with the reported affinities of various PDZ domains to their specific partner proteins [2,22]. Consistent with the results of two-hybrid assay, the ΔPDZ-HtrA1 mutant which lacked the PDZ domain did not bind to Col3a1-C-Pro in this solid-phase binding assay. The requirement for the C-terminal sequence of Col3a1-C-Pro was also confirmed; a Col3a1-C-Pro mutant with a four amino acid deletion at the C-terminus (Col3a1-C-ProΔ4aa) was completely devoid of interaction with HtrA1 (Figure 3C). Similarly, interaction of HtrA1 with GM130 (Figure 3C) and LRP9 (results not shown) required their C-terminal four amino acids. Surprisingly, an HtrA1 mutant with mutation at amino acid position 171 (F171D-HtrA1) did not interact with Col3a1-C-Pro (Figure 3A). F171D-HtrA1 has a phenylalanine→aspartic acid mutation in a small region between the Kazal-type inhibitor domain and the protease homology domain. This mutant is defective in efficient trimer formation (M. Yano, Y. Ueta, C. Oka and M. Kawaichi, unpublished work). This finding indicates that the higher order structure of HtrA1 is necessary for recognition of the PDZ-binding proteins. It is known that the Gal4 DNA-binding domain that is commonly used in the yeast two-hybrid assay can mediate dimer formation [23]. This may explain our success in isolation of the PDZ-binding proteins by yeast two-hybrid screening.
Figure 3. Solid-phase binding assay for PDZ-binding proteins.
(A) Binding of HtrA1 and its mutants to immobilized Col3a1-C-Pro. The bacterially produced His6–Col3a1-C-Pro protein or control Trx–His6 protein was immobilized on the well walls and the binding of Myc-tagged proteins of wild-type HtrA1 (•), ΔPDZ-HtrA1 (▴) and F171D-HtrA1 (▪) were assayed. The values obtained with the Col3a1-C-Pro wells were subtracted from the values obtained with Trx wells and presented in this Figure. (B) Scatchard plot analysis for binding of HtrA1 to Col3a1-C-Pro using data in (A). (C) The C-terminal four amino acids of PDZ-binding proteins were essential for interaction with HtrA1. The bacterially produced His6-fusion proteins of Col3a1-C-Pro (▪), Col3a1-C-ProΔ4aa (□), GM130 (▴) or GM130Δ4aa (▵) were immobilized in the wells. The binding of wild-type HtrA1 were assayed as described in (A). (D) The native trimeric Col3a1-C-Pro did not interact with HtrA1. The well walls were coated with the native Col3a1-C-Pro produced in insect cells in the absence (□) or presence (▪) of 1.5 mM DTT. Binding of HtrA1 was assayed as described in (A), except that 1.5 mM DTT was included in the binding reaction of reduced Col3a1-C-Pro. HtrA binding to His6–Col3a1-C-Pro was monitored simultaneously as a reference (▴).
The naturally occurring collagen C-propeptides are in trimer forms [24], whereas bacterially produced Col3a1-C-Pro exists mostly as a monomer, as judged from the electrophoretic pattern under non-reducing conditions (results not shown). We next examined whether native Col3a1-C-Pro trimer molecules bound to HtrA1. The human recombinant Col3a1-C-Pro produced by insect cells has a trimeric structure almost indistinguishable from that of native Col3a1-C-Pro [24]. In contrast with bacterially produced Col3a1-C-Pro, this recombinant trimeric Col3a1-C-Pro did not bind to HtrA1 (Figure 3D). In the native Col3a1-C-Pro trimer, three subunits are held together by interchain disulphide bonds and several C-terminal cysteine residues are involved in intrachain disulphide linkages [25]. When the Col3a1-C-Pro trimer was reduced with 1.5 mM DTT, a sufficient concentration to disrupt the trimeric structure, the reduced Col3a1-C-Pro began to bind to HtrA1 (Figure 3D). These results indicate either that the monomeric C-propeptide is the actual ligand for the HtrA1 PDZ domain, or that the intrachain disulphide bonds prevent the C-terminus from associating with the PDZ domain, or both. The latter idea is consistent with the requirement for a free C-terminus of target proteins in interaction with PDZ domains [22].
Roles of the C-terminal PDZ recognition sequence in the protease reaction of HtrA1
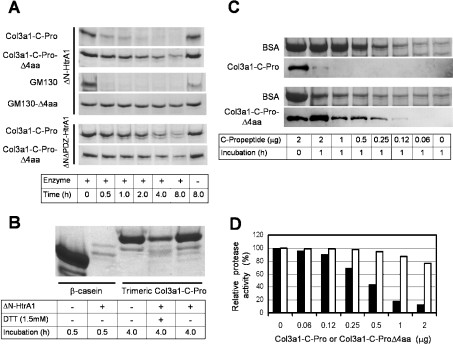
Some PDZ domains function in substrate recognition and tethering [3,4]. We therefore next examined whether collagens and their C-propeptides serve as substrates for HtrA1. For this purpose and for further biochemical analysis, we produced an N-terminally truncated form of HtrA1 (ΔN-HtrA1) in bacteria. ΔN-HtrA1 showed approx. 3-fold enhanced protease activity toward β-casein as compared with intact HtrA1 (results not shown), suggesting a negative regulatory role of the N-terminal region on the protease activity. ΔN-HtrA1 bound to various PDZ-binding proteins and its binding activity was comparable with that of full-length HtrA1 in the solid-phase binding assay (results not shown). ΔN-HtrA1 did not degrade collagen preparations conventionally used for collagenase assay (results not shown), but digested bacterially produced His6–Col3a1-C-Pro (Figure 4A). As compared with β-casein, Col3a1-C-Pro was a poor substrate; Col3a1-C-Pro was digested at rates 1/20 or less than β-casein under the conditions tested (results not shown). The Col3a1-C-ProΔ4aa mutant was also digested by HtrA1 (Figure 4A). However, its digestion rate was less than 1/16 of that observed for Col3a1-C-Pro, because comparable amounts of proteins were digested by ΔN-HtrA1 after 30 min digestion of Col3a1-C-Pro and after 8 h digestion of Col3a1-C-ProΔ4aa (Figure 4A). GM130 produced in bacteria was also quickly digested by ΔN-HtrA1, but GM130Δ4aa mutant lacking the C-terminal 4 amino acids was rather resistant to ΔN-HtrA1 digestion (Figure 4A). The native Col3a1-C-Pro trimer was barely digested by ΔN-HtrA1 (Figure 4B). However, it was digested after reduction with DTT. Taken together, these results suggest that effective digestion of a protein by HtrA1 requires binding of that protein to the HtrA1 PDZ domain. To confirm this conclusion, digestion by the ΔNΔPDZ-HtrA1 mutant that lacked the PDZ domains was analysed. The protease activity of ΔNΔPDZ-HtrA1 was enhanced 1.5-fold (β-casein as substrate) and approx. 5-fold (BSA as substrate) as compared with ΔN-HtrA1 (results not shown), supporting a suppressive function of the unengaged PDZ domain on the protease activity. ΔNΔPDZ-HtrA1 degraded Col3a1-C-Pro and Col3a1-C-ProΔ4aa mutant at almost the same rate (Figure 4A). This result showed that the PDZ domain was responsible for the differential digestion between Col3a1-C-Pro and Col3a1-C-ProΔ4aa observed with ΔN-HtrA1.
Figure 4. Digestion of Col3a1-C-Pro and GM130 by HtrA1.
(A) Efficient digestion by HtrA1 required interaction with the PDZ domain. His6–Col3a1-C-Pro, His6–GM130, His6–Col3a1-C-ProΔ4aa and His6–GM130Δ4aa (3 μg of each) were digested either with ΔN-HtrA1 (0.1 μg) or ΔNΔPDZ-HtrA1 (0.02 μg) for the indicated times. The digests were separated by SDS gel electrophoresis and detected by Coomassie Blue staining. (B) Native trimeric Col3a1 was resistant to HtrA1 digestion. Col3a1 (2 μg) produced by insect cells was digested with ΔN-HtrA1 (0.1 μg) for 4 h in the presence or absence of 1.5 mM DTT. This amount of ΔN-HtrA1 was sufficient to completely digest 5 μg of β-casein in 30 min (first and second lanes). (C) Col3a1-C-Pro interfered with the digestion of reduced BSA by ΔN-HtrA1. BSA (7.5 μg) was incubated with ΔN-HtrA1(0.1 μg) in a reaction mixture containing 1.5 mM DTT in the presence of various amounts of Col3a1-C-Pro (upper two panels) or Col3a1-C-ProΔ4aa (lower two panels), as indicated, for 1 h. The digests were separated by gel electrophoresis in two gels. One gel was stained with Coomassie Blue (first and third panels) and the other was used for Western-blot analysis with anti-(human Col3a1-C-Pro) polyclonal antibody, which cross-reacted with mouse Col3a1-C-Pro (second and fourth panels). (D) Relative protease activity toward reduced BSA in the presence of Col3a1-C-Pro (closed bar) or Col3a1-C-ProΔ4aa (open bar). The intensity of BSA bands in the lanes from (C) was quantified by densitometry. The amount of digested BSA in 1 h was defined as protease activity. Protease activity relative to that observed in the absence of C-propeptides was shown.
We next examined whether Col3a1-C-Pro modulated the protease activity of HtrA1. Col3a1-C-Pro had little effect on the protease activity of HtrA1 toward β-casein. Since β-casein is an extremely favourable substrate for most proteases, it may not require further activation of HtrA1 for efficient digestion. Besides, bovine β-casein has a C-terminal sequence similar to a putative PDZ-binding peptide (-FPIIV-COOH). It has been suggested that this C-terminal sequence leads to HtrA2/Omi activation thus masking the effect of other activators [5]. We therefore decided to use BSA as substrate in the following experiments. HtrA1 did not digest native BSA, haemoglobin or lysozyme, but could digest these proteins when they were reduced by DTT. Reduced BSA was degraded by ΔN-HtrA1 at a much slower rate than β-casein, but at a significantly higher rate than Col3a1-C-Pro; approx. 70% of BSA and Col3a1-C-Pro were digested after 15 min and 45 min incubation of the respective proteins with the same amount of ΔN-HtrA1 in the presence of DTT (results not shown). Digestion of reduced BSA by ΔN-HtrA1 was inhibited by the addition of relatively small amounts of Col3a1-C-Pro (Figures 4C and 4D); 50% inhibition of ΔN-HtrA1 (0.1 μg) protease activity was obtained with Col3a1-C-Pro at an amount between 0.25–0.5 μg that was 1/40–1/20 of that of BSA (10 μg). In the presence of 2 μg of Col3a1-C-Pro, the digestion of BSA was 90% inhibited (Figure 4D), whereas almost all of Col3a1-C-Pro was digested (Figure 4C, second panel). It seemed therefore that Col3a1-C-Pro was preferentially digested in the presence of BSA despite the fact that Col3a1-C-Pro was a poorer substrate for ΔN-HtrA1 than BSA when they were digested separately with ΔN-HtrA1. Col3a1-C-ProΔ4aa mutant, on the other hand, showed only a weak inhibition on the BSA cleavage by ΔN-HtrA1 (Figures 4C and 4D) and 80% or more of Col3a1-C-ProΔ4aa protein remained after reaction with ΔN-HtrA1 (Figure 4C, fourth panel). The inhibition of BSA digestion was not simply due to an addition of a preferable substrate, because β-casein did not inhibit BSA digestion (results not shown).
These data can be interpreted that Col3a1-C-Pro is preferentially taken up into the catalytic cavity of HtrA1 through the interaction with the PDZ domain, stays in the cavity for an extended time because it is a poor substrate for HtrA1, and thereby interferes with access and digestion of reduced BSA. Therefore the apparent inhibition of BSA digestion may not necessarily mean that Col3a1-C-Pro suppresses the protease activity of HtrA1.
Effects of C-terminal peptides on the protease activity of HtrA
The above mentioned characteristics of Col3a1-C-Pro as a substrate and a PDZ-binding partner hampered precise analysis of effects of PDZ-binding factors on the protease activity of HtrA1. To circumvent this problem, we synthesized oligopeptides derived from C-terminal sequences of representative PDZ-binding proteins (Figure 5A). The synthetic peptide, GM130Pep, contained the C-terminal sequence derived from GM130, which showed the strongest interaction with the PDZ domain of HtrA1 in the yeast two-hybrid assay. SPR measurements demonstrated a high-affinity interaction of GM130Pep with ΔNS328A-HtrA1 immobilized on a Ni-NTA chip (Figure 5B). The Kd for the binding of GM130Pep to ΔNS328A-HtrA1 was calculated to be 6.0 nM. This affinity of GM130Pep is comparable with those obtained by the same method for the binding of ssrA (small stable RNA) peptide to E. coli or Salmonella typhimurium HtrA (Kd=4.9 or 3.2 nM respectively) [4], and for the binding of the optimal peptide to mouse Dlg-2 protein (Kd=42 nM) [1]. HtrA2-Opt, which was derived from the reported optimal binding sequence for PDZ-HtrA2 [5], showed a slightly higher affinity for ΔNS328A-HtrA1 (Kd=3.8 nM). CPIII (procollagen III C-propeptide trimer) which contained the C-terminal sequence of Col3a1-C-Pro and DegS-Opt which was derived from the optimal binding sequence for bacterial DegS PDZ domain [9], showed Kd for PDZ-HtrA1 (334 nM and 547 nM respectively) nearly two orders of magnitude higher than that observed with GM130Pep.
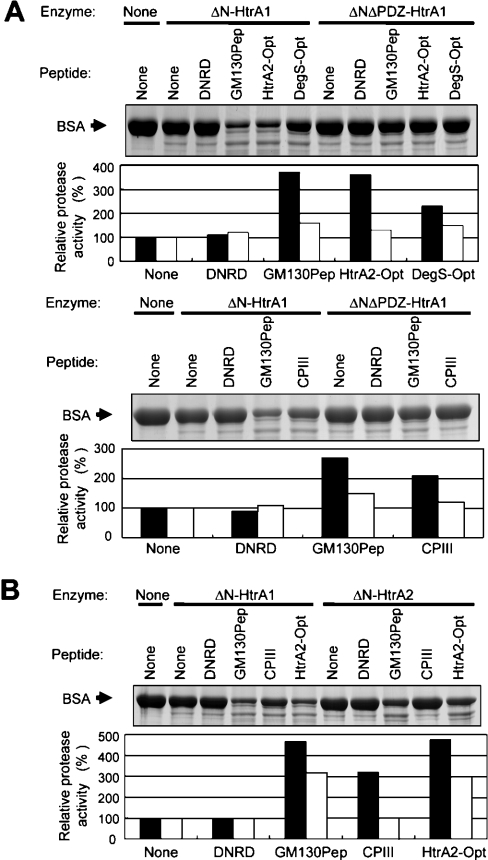
Next, we examined effects of these synthetic peptides on the protease activity of ΔN-HtrA1 using reduced BSA as a substrate (Figure 6A). GM130Pep and HtrA2-Opt peptides stimulated the ΔN-HtrA1 protease activity more than 3-fold. DegS-Opt and CPIII peptides stimulated the protease activity two-fold or more. These peptides had a similar stimulatory effect on the activity of ΔN-HtrA1 toward reduced haemoglobin (results not shown). The stimulatory effects of peptides on the ΔN-HtrA1 protease activity depended largely on the presence of the PDZ domain, because the protease activity of ΔNΔPDZ-HtrA1 was stimulated only slightly by these peptides (Figure 6A). The control peptide with hydrophilic C-terminal amino acids (Asp-Asn-Arg-Asp) did not show any stimulatory effects on ΔN-HtrA1, nor did it show significant binding to ΔNS328A-HtrA1 (Kd was higher than 200 μM and could not be determined precisely).
Figure 6. Effects of PDZ-binding peptides on protease activity of HtrA1 and HtrA2.
(A) Stimulation of protease activity of HtrA1 by PDZ-binding peptides. BSA (20 μg) was incubated with ΔN-HtrA1 (0.1 μg) or ΔNΔPDZ-HtrA1 (0.02 μg) in a mixture containing 1.5 mM DTT and 0.5 mM of indicated peptides for 1 h. The digests were separated by gel electrophoresis and detected by Coomassie Blue staining (upper panels). Relative protease activity (lower panels) was calculated as described in Figure 4. Closed and open bars indicate protease activity of ΔN-HtrA1 and ΔNΔPDZ-HtrA1 respectively. The amount of ΔNΔPDZ-HtrA1 (0.02 μg) was chosen so that it gave the same protease activity toward reduced BSA as that obtained with 0.1 μg of ΔN-HtrA1. (B) CPIII specifically activated protease activity of HtrA1. BSA (20 μg) was incubated with ΔN-HtrA1(0.1 μg) or ΔN-HtrA2 (0.1 μg) in a mixture containing 1.5 mM DTT and 0.5 mM of indicated peptides for 1 h. Other procedures were as described in (A). Relative protease activity of ΔN-HtrA1 (closed bar) and ΔN-HtrA2 (open bar) are shown in the lower panel.
Interestingly, CPIII peptide showed stimulatory effect specifically on HtrA1 (Figure 6B). This peptide did not enhance the activity of ΔN-HtrA2, whereas GM130Pep and HtrA2-Opt peptides were able to stimulated ΔN-HtrA2 activity efficiently. The SPR assay demonstrated that the binding of CPIII peptide to HtrA2 was very weak (Kd=19.9 μM), whereas binding of HtrA2-Opt and GM130Pep to ΔN-HtrA2 showed high affinity (Kd=19.2 nM and 202 nM respectively) (Figure 5B). Therefore, binding to PDZ domains correlated with the enhancement of protease activity by peptides.
DISCUSSION
HtrA1 PDZ-binding proteins
Consensus sequences for recognition by PDZ domains of HtrA serine proteases have been conventionally searched for by selective binding of a peptide library to purified HtrA proteins [5,9]. In the present study we employed yeast two-hybrid system to isolate binding proteins of the PDZ domain of mouse HtrA1, expecting that by this method we could obtain not only information on the consensus recognition sequence, but also the best-fit peptide sequence in its physiological context in a naturally occurring protein. We have isolated 11 proteins that interacted with the PDZ domain of mHtrA1 through their extreme C-terminal regions, which are characteristically rich in hydrophobic and non-polar amino acids. The consensus recognition sequence predicted for PDZ-HtrA1 {(Φ)-X-Φ-X-Φ-[L/V/F/A]-COOH} appears to be very similar to that observed for the PDZ domain of HtrA2 by Junqueira et al. [21] ([L/I]-X-Φ-[I/L/V]-COOH]), or by Martins et al. [5] (-Φ-X-Φ-Φ-Φ-COOH, where Φ represents hydrophobic amino acids with a preference for aromatic amino acids). In fact, two synthetic peptides, GM130Pep, which was the best recognition sequence for PDZ-HtrA1, and HtrA2-Opt, which was optimal for binding to PDZ-HtrA2, displayed similar high affinities (Kd=6.0 nM and 3.8 nM respectively) for PDZ-HtrA1. On the other hand, HtrA2 showed differential binding toward these optimal sequences; HtrA2 bound to GM130Pep at a Kd value 10 times higher than that observed for HtrA2-Opt. A highly specific binding of PDZ-HtrA2 was also reported for its binding to Mxi2; PDZ-HtrA1 was not able to interact with Mxi2 [26]. We found that PDZ-HtrA1, but not PDZ-HtrA2, specifically interacted with CPIII peptide, although the C-terminal amino acid sequence of CPIII (-GPVCFL-COOH) appeared to match the consensus sequence for HtrA2.
The selective binding of CPIII to PDZ-HtrA1 raises the possibility that collagen C-propeptides are physiological targets of the PDZ domain of HtrA1. In fact, C-propeptides of fibril-forming Type I, II and III pro-collagens were isolated frequently in our two-hybrid screening. We showed in this report that the bacterially-produced Col3a1-C-Pro monomer bound to PDZ-HtrA1 at a very low Kd (3.7 nM) and was digested by HtrA1. The efficient digestion depended on the presence of four amino acids at the C-terminus of Col3a1-C-Pro. In contrast, the native trimeric Col3a1-C-Pro protein produced in insect cells was not recognized by PDZ-HtrA1 and was resistant to the digestion by HtrA1. It was only after reduction with DTT that the native Col3a1-C-Pro bound to PDZ-HtrA1 and served as a substrate of HtrA1. All together, these results imply that the HtrA1 PDZ domain functions in substrate recognition.
Regulation of HtrA1 protease activity by the PDZ domain
The binding of ligand to PDZ domains has been reported to elevate protease activity of several PDZ-domain-containing proteases [5,9]. The enhancement of protease activity could be ascribed to at least two factors; increase in protease activity itself and facilitation of substrate recognition. The former example may be the increased proteolysis of RseA by DegS upon binding of OMP to the DegS PDZ domains [9]. On the contrary, the PDZ domain of E. coli Tsp is involved in specific substrate recognition. The Tsp PDZ domain specifically interacts with ssrA-encoded peptide that is tagged at the C-terminal ends of misfolded proteins as a degradation signal, thus facilitating their degradation [3]. We showed in the present report that the protease activity of HtrA1 toward denatured BSA was stimulated by oligopeptides that bound to PDZ-HtrA1. The degree of stimulation roughly correlated to the affinity of peptides to PDZ-HtrA1. For example, CPIII that bound to PDZ-HtrA1 fairly well, but only weakly to PDZ-HtrA2, was able to increase the protease activity of HtrA1 more than 3-fold, but not the activity of HtrA2. High concentrations of peptide (500 μM) were required to clearly demonstrate the enhancement of the protease activity, as reported for HtrA2 [5]. Probably, efficient stimulation of the protease activity requires the whole structure or a large mass of the physiological partner protein.
Col3a1-C-Pro binding to PDZ-HtrA1 may represent both of the two different consequences; facilitation of the substrate transport into the catalytic cavity and stimulation of the proteolytic activity. To clearly distinguish these two processes, we need to analyse enzyme kinetics quantitatively under various conditions using model substrates, such as the GST (glutathione S-transferase) protein tagged with recognition peptides or fluorescence-labelled peptide substrates [5].
Physiological implications
The present results imply a possible physiological function of HtrA1 in metabolism of fibrillar collagen C-propeptides. Other fibril-forming procollagens, Type V procollagen α2 and Type XI procollagen α1, also contain C-terminal end sequences that fit well into the consensus sequence for PDZ-HtrA1. C-propeptides mediate assembly of procollagen molecules into a triple-helical structure and keep the assembled procollagen as a soluble form inside the secretory pathways [27], and eventually it is cleaved from the helical region leading to the maturation of collagen fibrils outside the cell. In analogy with the bacterial HtrA proteins, mammalian HtrA1 might function in a surveillance system for quality control of collagens. HtrA1 may recognize denatured procollagen through the exposed C-terminal ends and may digest misfolded procollagen molecules in the secretory pathways.
An alternative idea is that C-propeptides released from the triple-helical region into the extracellular space may be the target for HtrA1. The released C-propeptides are known to have diverse biological activities. Chondrocalcin, the C-propeptide of Col2a1, is localized in the calcifying cartilage and is implicated in the creation of nucleation sites for the cartilage calcification during endochondral bone formation [28]. The C-propeptide of Col1a1, with an apparent native molecular mass of 35 kDa, the size of a monomer, was reported to induce growth arrest and spontaneous differentiation of clonally derived Schwann cells through an autocrine loop [29]. HtrA1 may negatively regulate biological activities of C-propeptides by degrading them.
We have shown that HtrA1 is expressed in embryonic tissues where cell differentiation is mainly regulated by TGF-β family proteins [19]. We have also demonstrated that HtrA1 inhibits signalling of a wide range of TGF-β proteins. TGF-β signalling has a close and mutual correlation with the extracellular matrix; TGF-βs regulate extracellular matrix synthesis [30] and TGF-β signalling is profoundly modulated by extracellular matrix proteins. The expression of HtrA1 is up-regulated in the osteo-arthritic cartilage [18] and down-regulated during malignant transformation and metastasis of several types of cancers [15–17]. The same fundamental phenomena of synthesis and degradation of extracellular matrix proteins underlie these seemingly different processes. All these findings imply important roles of HtrA1 in the metabolism of collagens and other extracellular matrix proteins.
Acknowledgments
We thank Dr Antonis S. Zervos, Dr David J. S. Hulmes and Dr Larry W. Fisher for the gifts of cDNA, recombinant proteins and antibodies, and Dr Seiji Takayama for his advice in SPR analysis. We are very grateful to Mrs Sachiko Iida, Choryo Syuzo Company, for her constant encouragement and support for Murwantoko during this work. This work was supported by research grants from the Foundation for the Nara Institute of Science and Technology, and the Inamori Foundation, and by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology, Japan. Murwantoko was a recipient of the fellowship for Indonesian students from the Iida Foundation.
References
- 1.Harris B. Z., Lim W. A. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 2.Hung A. Y., Sheng M. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- 3.Beebe K. D., Shin J., Peng J., Chaudhury C., Khera J., Pei D. Substrate recognition through a PDZ domain in Tail-specific protease. Biochemistry. 2000;39:3149–3155. doi: 10.1021/bi992709s. [DOI] [PubMed] [Google Scholar]
- 4.Spiers A., Lamb H. K., Cocklin S., Wheeler K. A., Budworth J., Dodds A. L., Pallen M. J., Maskell D. J., Charless I. G., Hawkins A. R. PDZ domains facilitate binding of high temperature requirement protease (HtrA) and tail-specific protease (Tsp) to heterologous substrates through recognition of small stable RNA A (ssrA)-encoded peptide. J. Biol. Chem. 2002;277:39443–39449. doi: 10.1074/jbc.M202790200. [DOI] [PubMed] [Google Scholar]
- 5.Martins L. M., Turk B. E., Cowling V., Borg A., Jarrell E. T., Cantley L. C., Downward J. Binding specificity and regulation of the serine protease and PDZ domains of HtrA2/Omi. J. Biol. Chem. 2002;278:49417–49427. doi: 10.1074/jbc.M308659200. [DOI] [PubMed] [Google Scholar]
- 6.Clausen T., Southan C., Ehrmann M. The HtrA family of proteases: Implications for protein composition and cell fate. Mol. Cell. 2002;10:443–455. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 7.Spiess C., Beil A., Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 8.Krojer T., Garrido-Franco M., Huber R., Ehrmann M., Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature (London) 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 9.Walsh N. P., Alba B. M., Bose B., Gross C. A., Sauer R. T. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell. 2003;113:61–71. doi: 10.1016/s0092-8674(03)00203-4. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki Y., Imai Y., Nakayama H., Takahashi K., Takio K., Takahashi R. A serine protease, HtrA2, is released from mitochondria and interacts with XIAP, inducing cell death. Mol. Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q., Chruch-Hajduk R., Ren J., Newton M. L., Du C. Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis. Genes Dev. 2003;17:1487–1496. doi: 10.1101/gad.1097903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W., Srinivasula S. M., Chai J., Li P., Wu J., Zhang Z., Alnemri E. S., Shi Y. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 2002;9:436–441. doi: 10.1038/nsb795. [DOI] [PubMed] [Google Scholar]
- 13.Zumbrun J., Trueb B. Primary structure of putative serine protease specific for IGF-binding. FEBS Letter. 1996;398:187–192. doi: 10.1016/s0014-5793(96)01229-x. [DOI] [PubMed] [Google Scholar]
- 14.Nie G. Y., Hampton A., Li Y., Findlay J. K., Salamonsen L. A. Identification and cloning of two isoforms of human high-temperature requirement factor A3 (HtrA3), characterization of its genomic structure and comparison of its tissue distribution with HtrA1 and HtrA2. Biochem. J. 2003;371:39–48. doi: 10.1042/BJ20021569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shridhar V., Sen A., Chien J., Staub J., Avula R., Kovats S., Lee J., Lillie J., Smith D. I. Identification of underexpressed genes in early- and late-stage primary ovarian tumors by suppression subtraction hybridization. Cancer Res. 2002;62:262–270. [PubMed] [Google Scholar]
- 16.Baldi A., De Luca A., Morini M., Battista T., Felsani A., Baldi F., Catricala C., Amantea A., Noonan D. M., Albini A., et al. The HtrA1 serine protease is down-regulated during human melanoma progression and repress growth of metastatic melanoma cells. Oncogene. 2002;21:6684–6688. doi: 10.1038/sj.onc.1205911. [DOI] [PubMed] [Google Scholar]
- 17.Chien J., Staub J., Hu S., Erickson-Johnson M. R., Couch F. J., Smith D. I., Crowl R. M., Kaufmann S. H., Shridhar V. A candidate tumor suppressor HtrA1 is downregulated in ovarian cancer. Oncogene. 2004;23:1636–1644. doi: 10.1038/sj.onc.1207271. [DOI] [PubMed] [Google Scholar]
- 18.Hu S., Carozza M., Klein M., Nantermet P., Luk D., Crowl R. M. Human HtrA, an evolutionarily conserved serine protease identified as differentially expressed gene product in osteoarthritic cartilage. J. Biol. Chem. 1998;273:34406–34412. doi: 10.1074/jbc.273.51.34406. [DOI] [PubMed] [Google Scholar]
- 19.Oka C., Tujimoto R., Kajikawa M., Kashiba-Takeuchi K., Ina J., Yano M., Tsuciya A., Ueta Y., Soma A., Kanda H., et al. HtrA1 serine protease inhibits signaling mediated by TGF-β family proteins. Development. 2004;131:1041–1053. doi: 10.1242/dev.00999. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein E. F., Fisher L. W., Li K., LeBaron R. G., Tan E. M., Uitto J. Differential expression of the versican and decorin genes in photoaged and sun-protected skin. Comparison by immunohistochemical and northern analyses. Lab. Invest. 1995;72:662–669. [PubMed] [Google Scholar]
- 21.Junqueira D., Cilenti L., Musumei L., Sedivy J. M., Zervos A. S. Random mutagenesis of PDZ Omi domain and selection of mutants that specifically bind the Myc proto-oncogene and induce apoptosis. Oncogene. 2003;22:2772–2781. doi: 10.1038/sj.onc.1206359. [DOI] [PubMed] [Google Scholar]
- 22.Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., Cantley L. C. Recognition of unique carboxyl-terminus motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 23.Marmorstrein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein–DNA complex. Nature (London) 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 24.Bernocco S., Finet S., Ebel C., Eichenberger D., Mazzorana M., Farjanel J., Hulmes D. J. Biophysical characterization of the C-propeptide trimer from human procollagen III reveals a tri-lobed structure. J. Biol. Chem. 2001;276:48930–48936. doi: 10.1074/jbc.M108611200. [DOI] [PubMed] [Google Scholar]
- 25.Lees J. F., Bulleid N. J. The role of cysteine residues in the folding and association of the COOH-terminal propeptide of type I and III procollagen. J. Biol. Chem. 1994;269:24354–24360. [PubMed] [Google Scholar]
- 26.Faccio L., Fusco C., Chen A., Martinotti S., Bonventre J. V., Zervos A. S. Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J. Biol. Chem. 2000;275:2581–2588. doi: 10.1074/jbc.275.4.2581. [DOI] [PubMed] [Google Scholar]
- 27.Hulmes D. J. Building collagen molecules, fibrils, and suprafibrillar structures. J. Struct. Biol. 2002;137:2–10. doi: 10.1006/jsbi.2002.4450. [DOI] [PubMed] [Google Scholar]
- 28.Poole A. R., Pidoux I., Reiner A., Choi H., Rosenberg L. C. Association of an extracellular protein (chondrocalcin) with the calcification of cartilage in endochondral bone formation. J. Cell Biol. 1984;98:54–65. doi: 10.1083/jcb.98.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rushton J., Schmitz S., Gunn-Moore F., Sherman D., Pappas C. A., Ritchie J. M., Haynes L. W. Growth arrest and spontaneous differentiation are initiated through an autocrine loop in clonally derived Schwann cells by α1-procollagen I C-propeptide. J. Neurochem. 1999;73:1816–1827. [PubMed] [Google Scholar]
- 30.Kresse H., Schonherr E. Proteoglycans of the extracellular matrix and growth control. J. Cell Physiol. 2001;189:266–274. doi: 10.1002/jcp.10030. [DOI] [PubMed] [Google Scholar]