Abstract
Glutamine transport into the human hepatoma cell line HepG2 is catalysed primarily by an ASCT2-type transporter identical in sequence with that cloned previously from JAR cells. An antibody raised against the C-terminus of the ASCT2 protein was shown to recognize ASCT2 on Western blots. Using this antibody, it was found that variation in cell growth rate did not affect ASCT2 expression, but both growth rate and ASCT2 expression were significantly reduced by glutamine deprivation. Expression of a number of other proteins was shown to be unaffected under these conditions. The sequence of the 5′-flanking region of the ASCT2 gene was derived from the human genome database. A 907 bp fragment of this sequence was directionally ligated into a luciferase reporter vector and was shown to exhibit promoter activity when transfected into HepG2 cells. Promoter activity was greatly reduced when transfection was performed in glutamine-free medium and was restored when glutamine was added post-transfection. The absence of other essential amino acids did not affect promoter activity, and glutamine deprivation did not affect the MCT1 (monocarboxylate transporter 1) promoter. These results indicate that both ASCT2 promoter activity and ASCT2 protein expression in these cells are dependent on glutamine availability.
Keywords: ASCT2, glutamine, hepatoma, promoter, regulation, transporter
Abbreviations: AP1, activator protein 1; DMEM, Dulbecco's modified Eagle's medium; MCT1, monocarboxylate transporter 1
INTRODUCTION
Hepatoma cells transport and utilize glutamine at a much higher rate than do normal liver cells [1]. Glutamine is also an essential requirement for tumour cell growth. Expression of the cell-membrane glutamine transporter has been suggested to be an integral event in hepatocellular transformation, and this transporter has been considered as a potential target in approaches aimed at attenuating the growth of tumour cells. In some more differentiated human hepatoma cell lines, competitive inhibition of glutamine transport has been shown to decrease cell proliferation under certain conditions (see [2]).
Glutamine transport across mammalian cell membranes is mediated by a number of transport systems (see [3]). In human [4] and rat [5] hepatoma cell lines, the predominant Na+-dependent glutamine transporter has properties similar to, but not identical with, proteins of the classical amino acid transport System ASC (alanine/serine/cysteine-preferring). cDNAs encoding ASC-like transporters have been cloned and sequenced from a number of different sources, and these have been termed ASCT2 [5–7] or ATB0 [8–10]. Although there are minor species differences in the amino acid specificity of these transporters, it is clear from sequence similarity that they are all closely related members of the same protein family now denoted as SLC1A5 (see [11] for a recent review of transporter nomenclature).
The importance of glutamine transport in hepatoma cell proliferation has been demonstrated recently by Fuchs et al. [12], who showed that attenuation of ASCT2 expression by inducible antisense RNA in a number of human hepatoma cell lines led to cell death via apoptosis. In the present paper, it is shown that both the expression of the glutamine transporter ASCT2 and the activity of the ASCT2 promoter in the human hepatoma cell line HepG2 are dependent upon glutamine availability.
EXPERIMENTAL
Cell culture
The human hepatoma cell line HepG2 and the monkey kidney cell line COS were maintained in DMEM (Dulbecco's modified Eagle's medium; Gibco BRL) supplemented with 10% FBS (foetal bovine serum; Gibco BRL), 100 units/ml penicillin G and 0.1 mg/ml streptomycin (Sigma). In some experiments, glutamine-free DMEM (Gibco BRL), leucine-free DMEM (ICN) or methionine-free DMEM (ICN) replaced the standard medium. In growth-rate assays, cells were grown in the appropriate medium for 72 h. Multiple cell counts using a haemocytometer were taken every 24 h for each condition.
Antibodies
In order to produce an ASCT2-specific antibody, the C-terminal peptide CPTGDSSATFEKESVM was synthesized and coupled to keyhole limpet haemocyanin via the terminal cysteine residue. The conjugate was injected into rabbits following a standard protocol, and the unfractionated serum was used in Western blotting experiments. The anti-calreticulin antibody used in some experiments was produced to a GST (glutathione S-transferase)-fusion protein as described in [13]. The glutamate dehydrogenase antibody was produced by injecting the commercially available purified enzyme into rabbits.
Western blot analysis
Cells were trypsinized, sedimented and then solubilized in the detergent N-decanoylmethylglucamide (2% final concentration). Cell protein concentration was measured by the method of Bradford [14]. Each sample (30 μg) was separated by SDS/PAGE and proteins were transferred on to nitrocellulose, which was then blocked by incubation with 5% (w/v) dried skimmed milk. Blots were probed with the appropriate primary antibody, followed by a suitable anti-rabbit secondary antibody and developed by ECL® (enhanced chemiluminescence; Amersham Biosciences). The intensity of bands were quantified using ImageQuant (Molecular Dynamics).
Verification of specificity of the anti-ASCT2 antibody
The full-length H4-ASCT2 cDNA in the pGem vector [5] was excised by EcoRI digestion and subcloned into the mammalian expression vector pCI-neo (Promega). Clones were checked for the correct orientation by PCR using a sequence-specific forward primer and T3 as the reverse primer. COS cells were transfected with 2 μg of the construct using FuGENE 6 (Roche) according to the manufacturer's protocol. Cells were harvested at 24 h and 48 h time points for Western blot analysis.
Cloning of the HepG2 ASCT2 promoter region
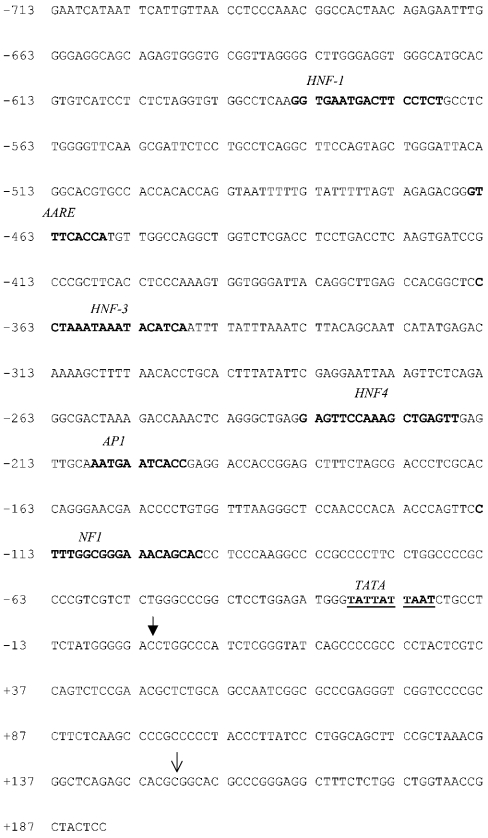
Genomic DNA was isolated from HepG2 cells using the Nucleospin Tissue Kit (Clontech) according to the manufacturer's instructions. The following primer pair was designed using sequence information from the human genome database (forward primer corresponds to bases −713 to −689 and the reverse primer corresponds to +194–+167 of the sequence shown in Figure 6). The forward primer contained a KpnI restriction site and the reverse primer contained a BglII site: forward (ASCT2 F1), 5′-GCTACCGGAATCATAATTCATTGTTAACCTCC-3′; reverse (ASCT2 R1), 5′-AGATCTGGAGTAGCGGTTACCAGCCAGAGAAAG-3′.
Figure 6. Promoter region of the human ASCT2 gene.
The human ASCT2 gene was derived from the human genome database as described in the text. ↓ shows the 5′ end of the published cDNA sequence [9] and ↓ shows the predicted transcriptional start site, which is designated nucleotide 0. The TATA box (underlined) and putative consensus binding sites for transcription factors commonly involved in liver gene expression are in bold. AARE, amino acid response element; HNF, hepatocyte nuclear factor; NF, nuclear factor.
PCR was performed using genomic DNA as a template as follows: 35 cycles of 94 °C for 40 s, 65 °C for 40 s and 72 °C for 1.5 min.
The single 907 bp band was gel extracted and cloned into the pGEM T-Easy vector (Promega). Sequencing was performed by MWG Biotech using M13 forward and reverse primers. The promoter insert was then ligated into pGL3-basic vector after cutting both the vector and the insert with KpnI and BglII to allow the correct orientation.
Determination of promoter activity
HepG2 cells were co-transfected with 1.5 μg of pGL3-basic vector containing the ASCT2 promoter insert and 20 ng of pRL CMV vector as transfection control, using Tfx™-50 reagent (Promega) according to the manufacturer's protocol. Cell samples were taken at 24 and 48 h time points. The optimum time point used for promoter activity was 48 h. The activity of the promoter was measured using a standard luciferase assay (Promega) and expressed as the ratio of firefly luminescence to Renilla luminescence. Light emission was measured using a luminometer.
The pGL3-MCT1 (containing the monocarboxylate transporter 1) promoter construct used in some experiments was a gift from Professor A. P. Halestrap (Department of Biochemistry, University of Bristol).
RESULTS
Glutamine transport into HepG2 cells
The transport of glutamine into HepG2 cells was found to be Na+-dependent, did not tolerate the substitution of Li+ for Na+, and was inhibited by excess concentrations of serine, cysteine and asparagine, but not by N-methyl aminoisobutyrate (results not shown). These properties are identical with those of the ASCT2 transporter characterized previously in the rat hepatoma cell line H4-IIE and expressed in oocytes [5]; they are also identical with kinetic data reported previously for HepG2 cells by Bode et al. [4].
In order to determine the cDNA sequence encoding the HepG2 transporter, reverse transcription-PCR was performed using primers corresponding to conserved regions of the ASCT2 transporter family.
Individual PCR products were sequenced and overlapping sequences were used to assemble the ASCT2 cDNA sequence. This was found to be 100% identical with the ASCT2/B0 transporter cloned from the human choriocarcinoma cell line JAR [9] (GenBank® accession number U53347).
Characterization of an antibody against human ASCT2
Previous investigations of the expression of ASCT2 transporters have been limited by the lack of a suitable specific antibody. Alignment of the last 15 amino acids from the mouse, rat and human ASCT2/B0 amino acid sequences show that they all contain the C-terminal sequence EKESVM, although there is variation between the amino acids preceding this sequence: human (JAR cells), GPAGDATVASEKESVM [9]; mouse testis, GPTGDSSATFEKESVM [6]; and rat hepatoma H4-IIE, GPAGDA-AACEKESVM [5].
As described in the Experimental section, an antibody was made to the C-terminus of mouse ASCT2, which was the only sequence available at the time. It was therefore necessary to determine whether or not the antibody recognized the human ASCT2 transporter.
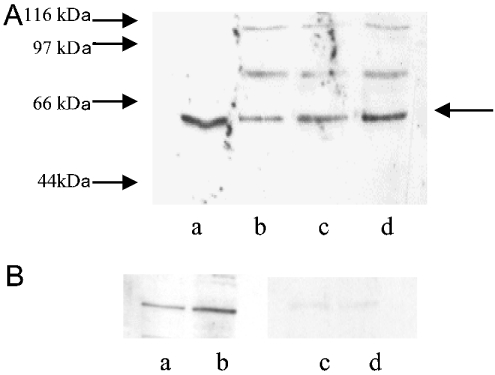
COS cells were transfected with full-length H4-ASCT2 cloned into the expression vector pCI-neo, and cell protein was probed with the ASCT2 antibody on Western Blots. Figure 1(A) shows that the antibody recognized a protein band of 58–60 kDa in non-transfected COS cells, together with two minor bands at approx. 70 kDa and 100 kDa. The intensity of the major band increased with time post-transfection. In four separate experiments, the intensity of the bands at 24 h and 48 h post-transfection were 3.9±0.8- and 13.8±4.2-fold that in non-transfected cells respectively. The expected molecular mass of the ASCT2 protein calculated from the derived amino acid sequence was 57 kDa. The antibody recognized a single band of identical molecular mass in HepG2 cell extracts. Since the band recognized by the antibody in HepG2 cells corresponded to the band which increased in the transfected cells, it was concluded that this antibody is specific for the C-terminus of the HepG2 transporter. It appears that COS cells endogenously express an ASCT2-like protein. Figure 1(B) shows that the 57 kDa band was eliminated when the blot was incubated with the peptide to which the antibody was raised.
Figure 1. Antibody specificity.
(A) Representative Western blot of cell extracts probed with the anti-ASCT2 antibody. Lane a, HepG2 cells; lane b, untransfected COS cells; lane c, COS cells at 24 h post-transfection; lane d, COS cells at 48 h post-transfection. The arrow indicates the band of interest. Molecular-mass sizes are indicated in kDa to the left of the blot. (B) Blot showing peptide blocking of the ASCT2 band in HepG2 cell extract. Lane a, control (25 μg of cell extract); lane b, control (35 μg of cell extract); lane c, blot as in lane a, blocked with 10 μg/ml immunogenic peptide; lane d, blot as in lane b, blocked with 10 μg/ml immunogenic peptide.
ASCT2 protein expression in relation to cell growth rate in HepG2 cells
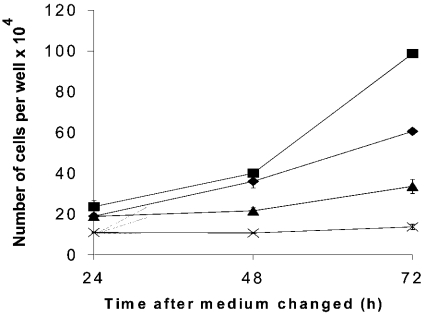
This antibody was used to explore the relationship between transporter expression and cell growth rate. The growth rate of HepG2 cells was altered by growing the cells under different conditions. PMA and novobiocin are known to affect the rate of growth of hepatoma cells, and PMA has been shown to inhibit glutamine uptake in SK-Hep cells via the protein kinase C pathway [15]. Cells were incubated in normal glutamine-containing medium (control), medium+PMA, medium+novobiocin and normal medium lacking glutamine. Cells were counted over a 72 h time period (Figure 2). Novobiocin and medium without glutamine both reduced the cell growth rate, while the addition of PMA increased the rate of growth.
Figure 2. Effects of culture medium on cell growth rate.
Cells were seeded at approx. 105 cells/well in normal medium and grown for 24 h. The medium was then changed to control medium (♦), medium supplemented with 0.1 μM PMA (▪), medium supplemented with 0.1 mM novobiocin (×) or medium lacking glutamine, but otherwise identical with control medium (▴). At 24, 48 and 72 h after the medium change, cells were trypsinized, suspended in 1 ml of medium and counted. Results are means±S.E.M. of four determinations.
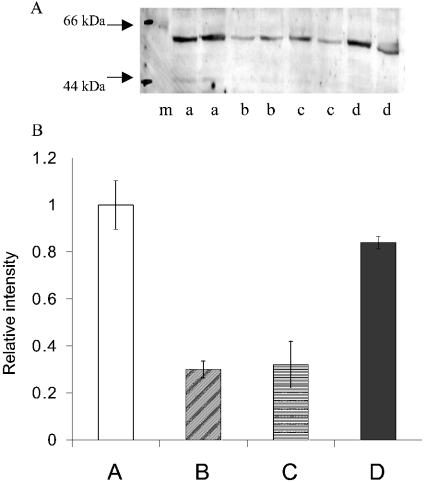
Western blot analysis for the ASCT2 transporter was then performed on extracts from cells grown under these different conditions (Figure 3). The levels of ASCT2 protein expressed differed significantly. Cells growing at a reduced rate in the absence of glutamine showed a large reduction in ASCT2 expression. However, ASCT2 expression in the cells growing slowly in the presence of novobiocin was similar to that in cells growing in normal medium. Cells growing at a higher rate in the presence of PMA always showed a significantly lower expression of ASCT2. The results show that ASCT2 expression is not simply a function of growth rate. It appears that ASCT2 expression is dependent rather on glutamine availability. Figure 4 shows an experiment in which cells were deprived of glutamine for 7 days, and glutamine was then added in the presence and absence of the transcription-inhibitor actinomycin D or the translation-inhibitor cycloheximide. Expression of ASCT2 protein increased over time, and after 24 h had attained a higher level than that in cells grown in normal glutamine-containing medium. The increase in protein expression was blocked by both inhibitors, and hence involved changes in transcription as well as translation.
Figure 3. Western blot showing ASCT2 levels in HepG2 cells grown under the conditions shown in Figure 2.
(A) Representative blot. Lane m, molecular-mass markers (sizes in kDa are indicated to the left of the blot); lanes a, control medium; lanes b, medium+PMA; lanes c, glutamine-free medium; lanes d, medium+novobiocin. In each case, 35 μg of cell extract was used. (B) Relative intensity of ASCT2 bands in four separate experiments similar to that shown above estimated using Imagequant. The mean value for cells in control medium was taken as 1, and results are means±S.E.M. A, control medium; B, medium+PMA; C, glutamine-free medium; D, medium+novobiocin.
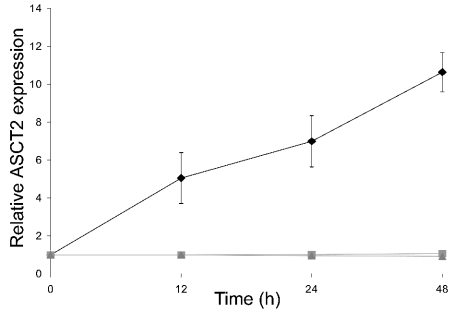
Figure 4. Time course of ASCT2 protein expression in response to added glutamine.
HepG2 cells were initially cultured in glutamine-free medium for 7 days. Subsequently, glutamine (5 mM) was added and 30 μg samples of cell extract were analysed by Western blotting as in Figure 3 above. The value of ASCT2 protein content at zero time was taken as 1, and values at other time points are expressed relative to this. The results are means±S.E.M. of four determinations. The relative value for ASCT2 expression in cells grown in standard glutamine-containing medium for a minimum of 7 days in a separate experiment was 4.5. ♦, No inhibitors; ▴, +10 μg/ml actinomycin D; ▪, +10 μg/ml cycloheximide.
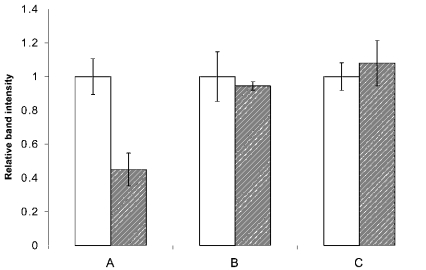
In order to determine whether or not the effect of glutamine deprivation on ASCT2 was due to a non-specific increase in protein degradation, the expression of other proteins for which antibodies were readily available was also measured by Western blotting. Figure 5 shows the expression levels of ASCT2, the mitochondrial protein glutamate dehydrogenase and the endoplasmic reticulum protein calreticulin for cells grown in the presence and absence of glutamine. Glutamine availability did not affect the expression of these other two proteins.
Figure 5. Effects of glutamine availability on protein expression.
HepG2 cells were grown in the presence and absence of glutamine. Three replicate blots of cell protein were then probed with antibodies against ASCT2 (A), calreticulin (B) or glutamate dehydrogenase (C). The entire experiment was repeated on four separate cell cultures; bands were quantified using Imagequant. The mean intensity of bands for cells in the presence of glutamine was separately set to 1 for each antibody. Open bars, glutamine present; hatched bars, glutamine absent.
Effects of glutamine on ASCT2 promoter activity
The 5′ end of human ASCT2 cDNA sequence cloned previously [9] was used to run a BLAST search of the human genome database. The ASCT2 (SLC1A5) gene is contained within a 31 Mb genomic contig of chromosome 19 (GenBank® accession number NT_011109.15). Figure 6 shows a length of DNA sequence immediately preceding the 5′ end of the cDNA (bp 19560031–19560932 of the genomic contig). This sequence is expected to contain the ASCT2 promoter. The 5′ end of published cDNA sequence [9] is at base +149 in the numbering used in Figure 6. Promoter-identification software (Berkeley Drosophila Genome Project Promoter Prediction database; http://www.fruitfly.org/) predicted a transcriptional start site at base 0 and a putative TATA box starting at −20. Putative transcription-factor-binding sites for a number of proteins commonly involved in liver gene regulation (hepatocyte nuclear factors 1, 3 and 4, and nuclear factor 1) were identified using MatInspector software (http://www.genomatix.de/software_services/software/MatInspector/matinspector.html) and are indicated. The sequence also contains a putative amino-acid-regulatory element and a consensus site for binding of the transcription factor AP1 (activator protein 1).
The DNA sequence shown in Figure 6 was generated by PCR using HepG2 genomic DNA as a template, as described in the Experimental section, ligated into the cloning vector pGem-T-Easy, amplified and sequenced. The 907 bp product obtained was identical in sequence with that shown in Figure 6. The insert was directionally subcloned into the pGL3-basic vector (Promega). The vector contains cDNA that encodes a modified firefly luciferase, but lacks a promoter. This allows the promoter activity of a DNA insert to be measured by determination of luciferase activity following transfection of the vector–insert construct into a suitable cell system. The cells were co-transfected with the pRL CMV vector as a transfection control. This vector contains cDNA encoding Renilla luciferase and a constitutive CMV promoter.
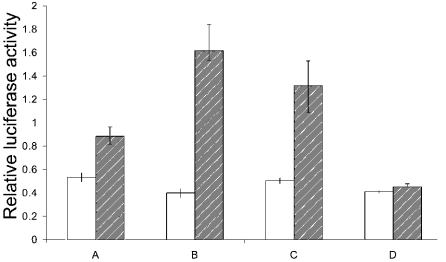
Figure 7 shows an experiment in which cells were transfected and grown for 48 h in media containing no glutamine or with glutamine present, and promoter activity was measured after 24 h and 48 h. In parallel, cells were transfected and grown without glutamine for 24 h and then supplemented with glutamine for a further 24 h. Luciferase activity in HepG2 cells transfected with this pGL3-promoter construct increased with time, indicating that the cloned DNA sequence contained an active promoter. In addition, these results show that although the promoter is active to some extent when no glutamine is present, the activity increases significantly when glutamine is supplied. Addition of glutamate did not mimic the effect of glutamine.
Figure 7. Luciferase activity in extracts of HepG2 cells transfected with the pGL3-basic promoter construct.
HepG2 cells were co-transfected with the pGL3 construct and pRL-CMV vector and grown in different media. A, cells transfected at zero time and then grown without glutamine; B, cells transfected at zero time and grown in the presence of glutamine; C, cells transfected at zero time and grown without glutamine for 24 h, followed by the addition of glutamine and growth for a further 24 h; D, cells transfected at zero time and grown without glutamine for 24 h, then grown for a further 24 h with 5 mM glutamate. Results are means±S.E.M. for six separate transfection experiments and are expressed as a ratio of firefly luminescence to Renilla luminescence. Promoter activity was determined 24 h (open bars) and (hatched bars) 48 h post-transfection.
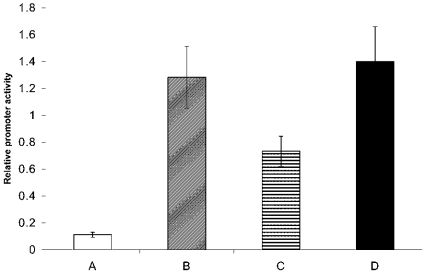
In order to determine whether or not the promoter activity responded specifically to glutamine, transfection was performed in media lacking the essential amino acids leucine or methionine. Figure 8 shows that lack of leucine or methionine did not greatly affect promoter activity. The same experiment was performed with a construct containing the MCT1 promoter. In this case, promoter activity was not affected by removal of glutamine, methionine or leucine (results not shown). These results show that glutamine itself in some way activates the ASCT2 promoter and increases ASCT2 expression.
Figure 8. Luciferase activity of HepG2 cells transfected with the pGL3-basic vector in various media.
Cells were co-transfected with the pGL3 construct and pRL-CMV vector in various media and cultured in the same media for 48 h post-transfection. Luminescence readings were taken at the 48 h time point. A, medium lacking glutamine; B, standard medium containing all amino acids; C, medium lacking leucine; D, medium lacking methionine.
DISCUSSION
The results of this paper characterize some factors which affect expression of the ASCT2 transporter in the human hepatoma cell line HepG2. Deprivation of glutamine reduces both the growth rate of cells and ASCT2 expression. PMA increased the growth rate, but decreased the expression of ASCT2 protein. This decreased protein expression may account for the decreased rate of glutamine uptake by PMA observed previously in SK-Hep cells [15,16]. Novobiocin decreased the growth rate, but had no effect on protein expression. Glutamine is not acting as the sole energy source of these cells, since all media contained 5 mM glucose. Similarly, glutamine is not exerting its effects by simply acting as a source of glutamate, since low levels of glutamate are routinely present in the media and addition of 5 mM glutamate did not increase promoter activity. Therefore it appears that the expression of ASCT2 is directly related to the availability of glutamine itself. The effect of glutamine deprivation is not merely due to a general increase in protein degradation, since a number of other proteins are unaffected. It cannot be excluded, however, that the ASCT2 is one of a group of proteins with rapid turnover that may be affected by glutamine.
It is also shown that glutamine enhances the activity of the cloned ASCT2 promoter, again under conditions where it is not acting as a sole energy source. The effect of glutamine deprivation on promoter activity was not mimicked by the removal of the essential amino acids methionine or leucine, and may therefore be relatively specific. Glutamine removal did not affect cell transfection or the activity of a different promoter (MCT1), indicating that the response is not a general effect on promoter function.
Glutamine is known to affect gene expression in other systems. Glutamine stimulates the proliferation of intestinal epithelial cells, and this is associated with activation of both the ERK (extracellular-signal-regulated kinase) and JNK (c-Jun N-terminal kinase) signalling pathways resulting in a 4-fold increase in activator protein-dependent gene transcription [17,18]. These effects were not observed with a number of glutamine metabolites, suggesting that glutamine itself can act as a signal indirectly influencing gene expression. It is possible that a similar mechanism involving activation of signalling pathways is responsible for activation of the ASCT2 transporter. Further analysis of the ASCT2 promoter is required to determine the site of action of glutamine and the involvement of specific transcription factors. In particular, investigation of the possible role of the putative amino-acid-response element and AP1-binding site will be of importance.
Activation of ASCT2 expression by glutamine is a novel example of nutrient-gene regulation. Removal of amino acid substrates has been shown to increase the expression of certain other proteins, such as amino acid transport System A (e.g. see [19]), the renal epithelial glutamate transporter EAAC1 [20] and asparagine synthetase [21]. To our knowledge, this is the first example of decreased expression of a transport protein by deprivation of an amino acid substrate in a mammalian cell system.
Acknowledgments
This work was funded by the BBSRC (Biotechnology and Biological Sciences Research Council).
References
- 1.Souba W. W. Glutamine and cancer. Ann. Surg. 1993;218:715–728. doi: 10.1097/00000658-199312000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bode B. P., Fuchs B. C., Hurley B. P., Conroy J. L., Suetterlin J. E., Tanabe K. K., Rhoads D. B., Abcouwer S. F., Souba W. W. Molecular and functional analysis of glutamine uptake in human hepatoma and liver-derived cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2002;283:G1062–G1073. doi: 10.1152/ajpgi.00031.2002. [DOI] [PubMed] [Google Scholar]
- 3.Bode B. P. Recent molecular advances in mammalian glutamine transport. J. Nutr. 2001;131(Suppl.):2475S–2487S. doi: 10.1093/jn/131.9.2475S. [DOI] [PubMed] [Google Scholar]
- 4.Bode B. P., Kaminski D. L., Souba W. W., Li A. P. Glutamine transport in isolated human hepatocytes and transformed liver cells. Hepatology. 1995;21:511–520. [PubMed] [Google Scholar]
- 5.Pollard M., Meredith D., McGivan J. D. Identification of a plasma membrane glutamine transporter from the rat hepatoma cell line H4-IIE-C3. Biochem. J. 2002;368:371–375. doi: 10.1042/BJ20020982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Utsunomiya-Tate N., Endou H., Kanai Y. Cloning and functional characterization of a System ASC-like Na+-dependent neutral amino acid transporter. J. Biol. Chem. 1996;271:14883–14890. doi: 10.1074/jbc.271.25.14883. [DOI] [PubMed] [Google Scholar]
- 7.Broer A., Brookes N., Ganapathy V., Dimmer K. S., Wagner C. A., Lang F., Broer S. The astroglial ASCT2 amino acid transporter as a mediator of glutamine efflux. J. Neurochem. 1999;73:2184–2194. [PubMed] [Google Scholar]
- 8.Kekuda R., Torres-Zamorano V., Fei Y. J., Prasad P. D., Li H. W., Mader L. D., Leibach F. H., Ganapathy V. Molecular and functional characterization of intestinal Na+-dependent neutral amino acid transporter B0. Am. J. Physiol. 1997;272:G1463–G1472. doi: 10.1152/ajpgi.1997.272.6.G1463. [DOI] [PubMed] [Google Scholar]
- 9.Kekuda R., Prasad P. D., Fei Y. J., Torres-Zamorano V., Sinha S., Yang-Feng T. L., Leibach F. H., Ganapathy V. Cloning of the sodium-dependent, broad-scope, neutral amino acid transporter Bo from a human placental choriocarcinoma cell line. J. Biol. Chem. 1996;271:18657–18661. doi: 10.1074/jbc.271.31.18657. [DOI] [PubMed] [Google Scholar]
- 10.Pollard M., Meredith D., McGivan J. D. Characterisation and cloning of a Na+-dependent broad-specificity neutral amino acid transporter from NBL-1 cells: a novel member of the ASC/B0 transporter family. Biochim. Biophys. Acta. 2002;1561:202–208. doi: 10.1016/s0005-2736(02)00346-2. [DOI] [PubMed] [Google Scholar]
- 11.Kanai Y., Hediger M. A. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflügers Arch. 2004;447:469–479. doi: 10.1007/s00424-003-1146-4. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs B. C., Perez J. C., Suetterlin J. E., Chaudhry S. B., Bode B. P. Inducible antisense RNA targeting amino acid transporter ATB0/ASCT2 elicits apoptosis in human hepatoma cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286:G467–G478. doi: 10.1152/ajpgi.00344.2003. [DOI] [PubMed] [Google Scholar]
- 13.Heal R., McGivan J. D. Induction of calreticulin expression in response to amino acid deprivation in Chinese hamster ovary cells. Biochem. J. 1998;329:389–394. doi: 10.1042/bj3290389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 15.Pawlik T. M., Souba W. W., Sweeney T. J., Bode B. P. Amino acid uptake and regulation in multicellular hepatoma spheroids. J. Surg. Res. 2000;91:15–25. doi: 10.1006/jsre.2000.5888. [DOI] [PubMed] [Google Scholar]
- 16.Bode B. P., Reuter N., Conroy J. L., Souba W. W. Protein kinase C regulates nutrient uptake and growth in hepatoma cells. Surgery. 1998;124:260–265. [PubMed] [Google Scholar]
- 17.Rhoads J. M., Argenzio R. A., Chen W., Rippe R. A., Westwick J. K., Cox A. D., Berschneider H. M., Brenner D. A. L-Glutamine stimulates intestinal cell proliferation and activates mitogen-activated protein kinases. Am. J. Physiol. 1997;272:G943–G953. doi: 10.1152/ajpgi.1997.272.5.G943. [DOI] [PubMed] [Google Scholar]
- 18.Rhoads J. M., Argenzio R. A., Chen W., Graves L. M., Licato L. L., Blikslager A. T., Smith J., Gatzy J., Brenner D. A. Glutamine metabolism stimulates intestinal cell MAPKs by a cAMP-inhibitable, Raf-independent mechanism. Gastroenterology. 2000;118:90–100. doi: 10.1016/s0016-5085(00)70417-3. [DOI] [PubMed] [Google Scholar]
- 19.McGivan J. D., Pastor-Anglada M. Regulatory and molecular aspects of mammalian amino acid transport. Biochem. J. 1994;299:321–334. doi: 10.1042/bj2990321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGivan J. D., Nicholson B. Regulation of high-affinity glutamate transport by amino acid deprivation and hyperosmotic stress. Am. J. Physiol. 1999;277:F498–F500. doi: 10.1152/ajprenal.1999.277.4.F498. [DOI] [PubMed] [Google Scholar]
- 21.Hutson R. G., Kitoh T., Moraga Amador D. A., Cosic S., Schuster S. M., Kilberg M. S. Amino acid control of asparagine synthetase: relation to asparaginase resistance in human leukemia cells. Am. J. Physiol. 1997;272:C1691–C1699. doi: 10.1152/ajpcell.1997.272.5.C1691. [DOI] [PubMed] [Google Scholar]