Abstract
Numerous invertebrate species belonging to several phyla cannot synthesize sterols de novo and rely on a dietary source of the compound. SCPx (sterol carrier protein 2/3-oxoacyl-CoA thiolase) is a protein involved in the trafficking of sterols and oxidation of branched-chain fatty acids. We have isolated SCPx protein from Spodoptera littoralis (cotton leafworm) and have subjected it to limited amino acid sequencing. A reverse-transcriptase PCR-based approach has been used to clone the cDNA (1.9 kb), which encodes a 57 kDa protein. Northern blotting detected two mRNA transcripts, one of 1.9 kb, encoding SCPx, and one of 0.95 kb, presumably encoding SCP2 (sterol carrier protein 2). The former mRNA was highly expressed in midgut and Malpighian tubules during the last larval instar. Furthermore, constitutive expression of the gene was detected in the prothoracic glands, which are the main tissue producing the insect moulting hormone. There was no significant change in the 1.9 kb mRNA in midgut throughout development, but slightly higher expression in the early stages. Conceptual translation of the cDNA and a database search revealed that the gene includes the SCP2 sequence and a putative peroxisomal targeting signal in the C-terminal region. Also a cysteine residue at the putative active site for the 3-oxoacyl-CoA thiolase is conserved. Southern blotting showed that SCPx is likely to be encoded by a single-copy gene. The mRNA expression pattern and the gene structure suggest that SCPx from S. littoralis (a lepidopteran) is evolutionarily closer to that of mammals than to that of dipterans.
Keywords: cholesterol, ecdysone, lipid metabolism, prothoracic glands, steroid
Abbreviations: FAM, carboxyfluorescein; GST, glutathione S-transferase; RACE, rapid amplification of cDNA ends; RP49, ribosomal protein 49; SCP2, sterol carrier protein 2; SCPx, sterol carrier protein 2/3-oxoacyl-CoA thiolase; TAMRA, carboxytetramethylrhodamine
INTRODUCTION
Inability to synthesize sterols is apparently universal within the Arthropoda. Within this phylum the most detailed work on dietary sterol requirements and metabolism has been undertaken in insects. The majority of insect species investigated require some cholesterol, or a sterol which is convertible into cholesterol, for satisfactory growth, development and reproduction [1].
SCP2 (sterol carrier protein 2), which is encoded by a part of a fusion gene encoding SCPx (sterol carrier protein 2/3-oxoacyl-CoA thiolase) in many species, has multiple proposed functions in lipid metabolism and intracellular movement of cholesterol and other lipids in mammals [2,3]. SCP2 protein is expressed in various tissues, but is particularly abundant in organs involved in cholesterol absorption, transport or metabolism.
In vertebrate peroxisomes there are three 3-oxoacyl-CoA thiolases. While all three thiolases are involved in fatty acid oxidation, SCPx plays an exclusive role in branched-chain fatty acid oxidation and in the oxidation of the branched side chain of cholesterol to form bile acids. This is because the two other thiolases are inactive with branched-chain fatty-acyl-CoAs as substrates, but SCPx can use them [2,4]. Bile acids are essential to biliary cholesterol secretion, which involves bile acids, cholesterol and phospholipid. Bile acid formation involves β-oxidation of the branched alkyl side chain of 24-oxo-3α,7α,12α-trihydroxy-5β-cholestanoyl-CoA to cleave off propionyl-CoA. The SCPx exhibits 3β-oxothiolase activity to convert 24-oxo-3α,7α,12α-trihydroxy-5β-cholestanoyl-CoA into cholyl-CoA, which is subsequently converted into cholic acid and glycinated to glycocholic acid. Also, the SCPx shows 3β-oxothiolase activity to convert 24-oxo-3α,7α,12α-trihydroxy-5β-cholestanoyl-CoA into chenodeoxycholyl-CoA, which is the precursor of chenodeoxycholic acid [4–6]. Mice in which the gene was knocked out exhibited inhibition of β-oxidation in bile acid synthesis, based on the accumulation of 3α,7α,12α-trihydroxy-27-nor-5β-cholestane-24-one, which is a known bile alcohol derivative of the cholic acid synthesis intermediate 3α,7α,12α-trihydroxy-24-oxo-cholestanoyl-CoA, in bile and serum [7].
SCPx belongs to the SCP2 gene family, which at present includes four proteins: SCP2, SCPx, 17β-hydroxysteroid dehydrogenase IV and UNC-24 [8]. Apart from SCP2, the other homologues have SCP2 domains towards their C-termini. A single structural gene encodes both SCP2 and SCPx in humans [9], mice [10], rats [11] and chickens [12]. Alternative transcription initiation gives rise to two mRNAs encoding SCPx or SCP2 in these species. In humans, the SCPx mRNA encodes 547 amino acid residues representing a fusion protein of 3-oxoacyl-CoA thiolase (amino acid residues 1–404) and the SCP2 domain (amino acid residues 425–547). The product is post-translationally cleaved to the 46 kDa 3-oxoacyl-CoA thiolase and the 13 kDa SCP2. As mentioned above, SCP2 is also transcribed as a short mRNA which encodes 143 amino acid residues identical with the C-terminal 143 amino acids of SCPx. On the basis of a database search, the fused SCPx gene can be traced back to the fruitfly Drosophila melanogaster, whereas in yeast and the nematode worm Caenorhabditis elegans, the thiolase and SCP2 are not fused, but encoded by two separate genes [13,14].
Here we report the molecular cloning and characterization of the cDNA encoding SCPx from Spodoptera littoralis (cotton leafworm). The results suggest that SCPx may be involved in sterol absorption and synthesis of insect moulting hormones (ecdysteroids).
EXPERIMENTAL
Protein sequence
SCPx was isolated during the purification of the enzymes involved in the 3-epimerization of ecdysteroids as described previously [15]. The protein co-migrated closely with the second form of 3-dehydroecdysone 3α-reductase throughout the chromatographic purification steps. The purified protein was subjected to SDS/10%-(w/v)-PAGE, electrotransferred to ProBlott™ membrane (Applied Biosystems, Warrington, Cheshire, U.K.) and visualized by Coomassie Brilliant Blue staining. A single band was observed, which was excised and sequenced by an automated pulsed liquid-phase sequencer (Applied Biosystems 471A), giving the N-terminal amino acid sequence PRKVFVVGVGMTNFI.
cDNA cloning and sequencing
A PCR-based cloning strategy was used to isolate a cDNA fragment encoding this protein. Two degenerate sense primers were synthesized. Primers based on adjacent parts of the N-terminal amino acid sequence (primer 1: 5′-CCN MGI AAR GTI TTY GTN GTN GG, where N represents A/T/C/G, M is A/C, I is inosine, R is A/G, Y is C/T; primer 2, 5′-GGN GTN GGN ATG ACI AAY TTY AT).
Total RNA was extracted using TRIzol (Life Technologies, Ltd.) from midgut dissected from larvae at 18 h into the last larval instar. First-strand cDNA was reverse-transcribed from the total RNA using a 1st Strand cDNA Synthesis Kit (Roche Molecular Biochemicals) with QT adapter primer: 5′-CCA TCA GTG CTA GAC AGC TAA GCT TGA GCT CGG ATC C(T)17 (modified from [16]). cDNA synthesized with QT primer served as template for PCR in which the above degenerate primers were combined in turn with the adapter Q0 primer: 5′-CCA TCA GTG CTA GAC AGC T (modified from [16]). PCR was carried out as follows: one cycle of 94 °C for 3 min, and 35 cycles of 94 °C for 1 min, 50 °C for 1 min, 72 °C for 3 min and one cycle of 72 °C for 7 min using primer 1 and Q0. This PCR product was used as template for the nested PCR, which was carried out as follows: one cycle of 94 °C for 3 min, and 30 cycles of 94 °C for 1 min, 53 °C for 1 min, 72 °C for 3 min and one cycle of 72 °C for 7 min. The nested PCR with primer 2 and Q1–2 {5′-TAA GCT TGA GCT CGG A (modified from [16])} yielded a product of approx. 1.8 kb.
The purified PCR product was cloned into pGEM®-T Easy Vector (Promega). Transformants were screened by colony PCR using M13 forward and reverse primers (5′-GTA AAA CGA CGG CCA G and 5′-CAG GAA ACA GCT ATG AC respectively), and those showing the correct size of inserts were propagated in Luria–Bertani broth containing 100 #x3BC;g/ml ampicillin, and plasmid DNA was purified after 16 h incubation at 37 °C. Double-stranded DNA sequencing was performed by the dideoxy termination method using Sequenase Version 2.0 (usb™; Amersham Pharmacia Biotech Ltd.). The sequences of three independent clones were compared to detect errors that could have occurred during the reverse transcription and PCR amplification.
5′-RACE (5′ rapid amplification of cDNA ends)
5′-RACE was carried out to obtain the 5′-end of the cDNA using the solid-phase CapFinder™ (Clontech, Cowley, Oxford, U.K.) approach [17]. mRNA was isolated from the total RNA using a Dynabeads® mRNA Purification Kit (Dynal Biotech UK, Bromborough, Wirral, Cheshire, U.K.), and used to synthesize a solid-phase cDNA library with CapFinder™ Primer (5′-AAG CAG TGG TAT CAA CGC AGA GTG GCC ATT ATG GCC GGG). The RNA was then degraded with RNase H, and the immobilized cDNA was purified by using a magnet. The cDNA region corresponding to the 5′-end of the mRNA was amplified by two successive rounds of PCR using gene-specific primers 4 and 5 (5′-TAG AGT GCG TTG GAG CCA GT and 5′-GAT ACC AGT CAT GCC GAC CT respectively), together with the 5′-Primer (5′-AAG CAG TGG TAT CAA CGC AGA GT). The second round of PCR yielded a product of approx. 300 bp, which was cloned into pGEM®-T Easy Vector, and the nucleotide sequences of several clones determined.
In vitro translation of SCPx
The cDNA containing the entire open reading frame was amplified by PCR with the following gene specific primers: IVORF-F (5′-GGG AAC AGC CAC CAT GCC TAG AAA AGT GTT CGT TGT) and IVORF-R (5′-CCA CCG CCT CTA GAG CGC AGT TTG GAG CGG ATT GTG T). PCR was conducted as follows: one cycle of 94 °C for 3 min, 15 cycles of 94 °C for 1 min, 53 °C for 30 s, 72 °C for 2 min, and one cycle of 72 °C for 5 min. The resulting PCR product was used as the template for the nested PCR, which was carried out as follows: 1 cycle of 94 °C for 3 min, 30 cycles of 94 °C for 1 min, 53 °C for 30 sec, 72 °C for 2 min, and 1 cycle of 72 °C for 5 min. The primers for the nested PCR were as follows: T7IV primer (5′-GGA TCC TAA TAC GAC TCA CTA TAG GGA ACA GCC ACC ATG) and IVterminator primer (5′-TTT TTT TTT AGT GAT GGT GAT GGT GAT GGC CAC CGC CAG AAC CGG TGC CAC CGC CAG AAC CGC CAC CGC CTC TAG AGC G). These primers were designed to incorporate the T7 promoter into the 5′-end of the sense strand. The product was used as the template to produce the protein using TNT® T7 Quick for PCR DNA (Promega). The protein was expressed under the conditions recommended by the manufacturer. The translated products were resolved on an SDS/12%-(w/v)-polyacrylamide gel and visualized by autoradiography.
Northern- and Southern-blot analysis
Total RNA from various tissues was isolated using TRIzol® reagent (Invitrogen, Paisley, Renfrewshire, Scotland, U.K.). A 10 #x3BC;g portion of total RNA was fractionated on a formaldehyde/agarose gel, transferred to Electran® nylon membrane (Merck) and hybridized with a probe corresponding to the open reading frame of the SCPx cDNA. The probe was radiolabelled with [α-32P]dCTP using a Ready-To-Go™ DNA labelling kit (−dCTP; Amersham Pharmacia Biotech) and loading was normalized by probing with a mouse 18S ribosomal RNA probe. Prehybridization and hybridization were carried out using QuikHyb® (Stratagene) under the conditions recommended by the manufacturer. The blots were washed at high stringency [0.1×SSC (1×SSC is 0.15 M NaCl/0.015 M sodium citrate)/0.1% SDS, at 60 °C] and labelled bands visualized by autoradiography.
Genomic DNA was prepared from the fat body and Malpighian tubules of last larval instar S. littoralis using a modified phenol extraction method as described by Sambrook et al. [18]. Aliquots (10 #x3BC;g) of DNA were digested with BamHI, BglII, EcoRI or XbaI, fractionated on a 1%-agarose gel, transferred to a nylon membrane, and hybridized using radiolabelled probes corresponding to different regions of SCPx cDNA. Hybridization was carried out using QuikHyb® (Stratagene) under the conditions recommended by the manufacturer. The blot was washed at high stringency (0.1×SSC/0.1% SDS at 60 °C), and labelled bands were visualized by autoradiography.
Recombinant SCPx protein expression
The cDNA containing the entire open reading frame was amplified by PCR with the following gene specific primers: ORF-F (5′-CGG ATC CCC TAG AAA AGT GTT CGT T) and ORF-R (5′-CGA ATT CTC ACA GTT TGG AGC GGA TTG T). These primers were designed to incorporate BamHI and EcoRI sites into the 5′-end and 3′-end respectively of the sense strand. PCR was conducted as follows: one cycle of 94 °C for 3 min, 30 cycles of 94 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min, and one cycle of 72 °C for 10 min. The resulting PCR product was gel-purified and cloned into pGEM®-T Easy vector. The product was cut out with BamHI and EcoRI from the plasmid and transferred into pGEX-2T (Amersham Biosciences). The recombinant protein was expressed using the conditions recommended by the manufacturer. The whole-cell extract from the induced cells was used for Western-blot analysis.
Preparation of anti-SCP2 antibodies
To produce the recombinant SCP2 domain of SCPx protein, the corresponding cDNA region (positions 1381–1671; Figure 1A below) was amplified by PCR with the following gene-specific primers: SCP2F (5′-GGG ATC CCC GGG CAT CTA CGG ATT CAA AGT) and SCP2R (5′-GGA ATT CGT GAT GGT GAT GGT GAT GCA GTT TGG AGC GGA TTG T). The SCP2R primer was designed to incorporate a His6 (hexahistidine) tag into the C-terminus of the recombinant protein. The resulting PCR product was gel-purified and cloned into pGEM®-T Easy vector. The product was cut out with BamHI and EcoRI from the plasmid and transferred into pGEX-2T (Amersham Biosciences). The recombinant protein was expressed using the conditions recommended by the manufacturer. The GST (glutathione S-transferase)–SCP2 fusion protein was purified with glutathione–agarose (Sigma). The purified protein was digested with thrombin (Amersham Biosciences) to remove the GST tag and re-purified using Ni-CAM™ affinity resin (Sigma). The recombinant protein (70 #x3BC;g) was emulsified with complete Freund's adjuvant and injected into a rabbit. Booster injections were made 14 days later with 70 #x3BC;g of the protein.
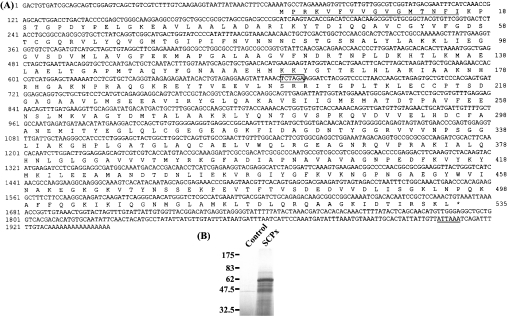
Figure 1. Nucleotide and deduced amino acid sequence of SCPx.
(A) The cDNA sequence is indicated on the top line, and the deduced amino acid sequence is on the second line. The putative polyadenylation signal is double-underlined, and the amino acid sequence obtained from the purified SCPx is underlined. An XbaI restriction site is shown in a box. Numbers on the right refer to the last amino acid residue on each line of the respective protein sequences. Numbers on the left refer to the first residue on each line of the respective nucleotide sequences. (B) In vitro translation of SCPx. Lane 1, no template; lane 2, DNA. The fragment containing the open reading frame of SCPx was used as template.
Western-blot analysis
Protein was extracted from various tissues in 20 mM Tris/HCl, pH 7.5. Protein samples (100 #x3BC;g) were resolved by SDS/PAGE and transferred on to nitrocellulose membrane. Escherichia coli cells expressing the recombinant SCPx protein were lysed in SDS sample buffer and protein extracts were subjected to SDS/PAGE. Membranes were incubated for 18 h at 4 °C in blocking solution [20 mM Tris/HCl (pH 7.5)/500 mM NaCl/5% (w/v) non-fat dry milk]. After washing with TBST solution [20 mM Tris/HCl (pH 7.5)/500 mM NaCl/0.05% Tween 20], the membranes were incubated with rabbit polyclonal antibody (1:1000 dilution) against C. elegans P-44 protein (equivalent to 3-oxoacyl-CoA thiolase; see Figure 2 below) [19] or rabbit antiserum (1:2000 dilution) against the SCP2 domain for 2 h. After washing with TBST solution, the membranes were incubated with horseradish-peroxidase-conjugated goat anti-rabbit IgG (Bio-Rad, 1:10000 dilution) for 1 h. The membranes were washed and developed with ECL Plus® Western-blotting detection reagents (Amersham Biosciences).
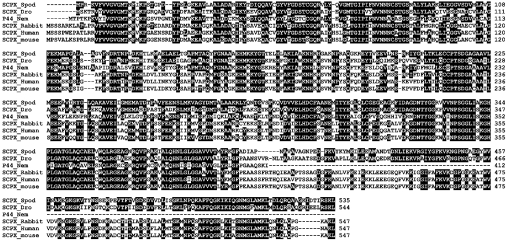
Figure 2. Alignment of the deduced amino acid sequences of SCPx and the most similar database proteins.
The deduced sequence of SCPx from S. littoralis (SCPX_Spod) was compared with all sequences in the database using the BLAST program. Only the amino acid sequences of the most similar proteins in the SCP2 gene family are shown in the alignment. Gaps introduced to optimize alignment are indicated by hyphens. Identical amino acids between SCPx from S. littoralis and at least one other sequence are indicated in black boxes. Numbers on the right refer to the last amino acid residue on each line of the respective protein sequences. The accession number and the percentage similarity to SCPx of S. littoralis (SCPX_Spod) of each protein are as follows: D. melanogaster SCPx (SCPX_Dro), AE003660, 63%; C. elegans P-44 (P44_Nem), D86473, 55%; rabbit SCPx (SCPX_Rabbit), AF051897, 55%; human SCPx (SCPX_Human), M75883, 54%; mouse SCPx (SCPX_mouse), M91458, 55%. The alignment was constructed by use of the CLUSTALW program.
Semi-quantitative real-time PCR
Quantification of SCPx expression in prothoracic glands derived from different times within the sixth instar was performed by an ABI Prism 7700 sequence detector (Applied Biosystems), a real-time PCR machine, using a TaqMan® probe for target DNA quantification. For this, batches of seven prothoracic glands were homogenized using a QIAshredder homogenizer (Qiagen), and RNAs were prepared using the RNeasy Mini kit (Qiagen). First-strand cDNA synthesis was performed using the Reverse-iT™ first-strand synthesis kit (ABgene, Epsom, Surrey, U.K.). The SCPx-specific primers used were 5′-AGG CAT CTA CGG ATT CAA AGT GAA (forward), 5′-CGC TGC TAT TGT ATG TGA CTT TGC (reverse) and 5′-FAM-CCC CAA CGG CGC GGA AGG-TAMRA (TaqMan® probe). RP49 (ribosomal protein 49)-specific primers were used as an internal control during quantification. RP49-specific primers used were 5′-ATC CTG ATG ATG CAG AAC AGG AA (forward), 5′-GAT GGT CTT GCG CTT CTT TGA G (reverse) and 5′-FAM-CGC AGA GAT CGC CCA CGG AGT CT-TAMRA (TaqMan® probe; FAM is carboxyfluorescein, and TAMRA is carboxytetramethylrhodamine). Taqman® probes labelled with FAM and TAMRA were supplied by Eurogentec (Romsey, Hants., U.K.). An ABI Prism 7700 sequence detector measured the fluorescent signal generated by the sequence-specific probes. The plate was scanned at 518 nm (FAM) and 582 nm (TAMRA). The relative expression level was determined using the threshold cycle, which is the cycle number at which the reporter fluorescence generated by the cleavage of the probe passed a fixed threshold above baseline. All samples were normalized to the RP49 value. The quantitative PCRs were done twice, with three replicates for each.
RESULTS
Cloning of the cDNA encoding SCPx
Using a combination of column-chromatographic approaches as described previously [15], we purified SCPx from Spodoptera midgut. SDS/PAGE analysis revealed that the apparent molecular mass of the purified SCPx was approx. 46 kDa and provided the material to obtain the N-terminal amino acid sequence.
A PCR-based cloning strategy, as detailed in the Experimental section, allowed us to obtain a cDNA fragment corresponding to the sequences between positions 70 and 1942 (Figure 1A). Gene-specific primers derived from this sequence were synthesized and used for 5′-RACE to obtain the 5′-end of the cDNA. 5′-RACE produced a cDNA clone of 285 bp, which contained a putative translation start site at position 67. Taken together, all overlapping cDNAs span a total of 1942 bp. The polyadenylation signal is located at position 1907. As shown in Figure 1(A), using the first ATG as the start codon, the full-length SCPx cDNA encodes a protein of 535 amino acids with a predicted molecular mass of 57450.13. The result of in vitro translation confirmed this prediction (Figure 1B). Comparison of this value with that of the purified protein (approx. 46 kDa), suggests that post-translational processing of the protein may occur. The protein is mildly basic, with an estimated pI of 8.02.
Similarity of deduced amino acid sequence to the proteins of the SCP2 gene family
The deduced amino acid sequence of the coding region showed similarity to members of the SCP2 gene family (Figure 2) such as D. melanogaster SCPx (63%), C. elegans P-44 (55%), Oryctolagus cuniculus (rabbit) SCPx (55%), Homo sapiens (human) SCPx (54%) and Mus musculus (mouse) SCPx (55%).
Tissue distribution and developmental expression of SCPx
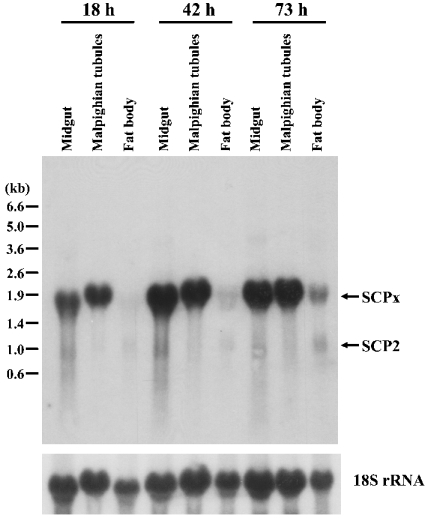
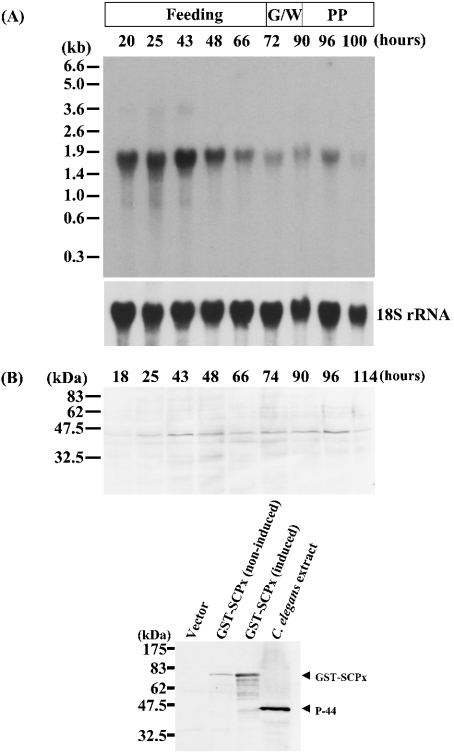
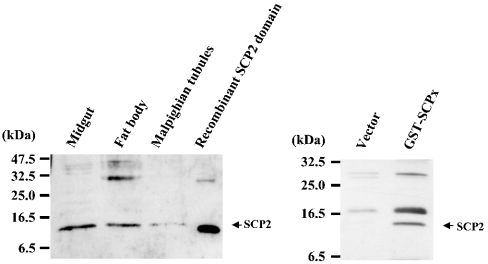
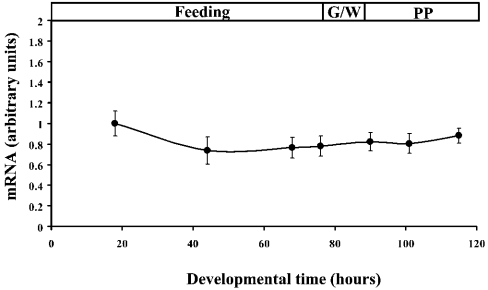
As demonstrated in Figure 3, a cDNA probe representing the protein coding region detected two transcripts of approximately 1.9 and 0.95 kb, although the 0.95 kb mRNA was expressed at a much lower level than the 1.9 kb transpcript. Presumably, these mRNAs encode SCPx and SCP2, since the probe corresponding to the SCP2 domain (1357–1671) was also able to hybridize to both bands (results not shown). The expression level of SCPx mRNA is much higher in midgut and Malpighian tubules than in fat body (Figure 3). Northern-blot analysis of mRNA isolated from midgut at different developmental stages of the last larval instar (Figure 4A) revealed that the expression of the SCPx mRNA was detected during all stages of the instar, but was higher during the feeding stage. The mRNA was readily detected at 20 h into the last larval instar and was strongly expressed until 48 h. It started to decrease from 66 h, and its expression level became very low before pupation. Western-blot analysis showed that protein levels reflected the transcription of the SCPx gene (Figure 4B, top panel). The major band detected by the antibody against C. elegans P-44 protein in Western blotting is approx. 46 kDa, and corresponds well to the molecular mass of the protein originally purified and suggests that SCPx is post-translationally processed. Figure 4(B) (bottom panel) showed that the antibody against the C. elegans P-44 protein specifically detects recombinant S. littoralis SCPx protein. On the other hand, the major band detected by the antiserum against the SCP2 domain of S. littoralis SCPx protein was approx. 13 kDa in various tissues. The cleaved product was also detected in E. coli cells (Figure 5). In the prothoracic gland, SCPx is constitutively expressed during the last larval instar, as determined by semi-quantitative PCR, where the result has been normalized to RP49 (Figure 6).
Figure 3. Northern-blot analysis of the tissue distribution and expression of SCPx.
A 10 #x3BC;g aliquot of total RNA from various tissues at different times (top) within the last larval instar was used. The blot was hybridized with a 32P-labelled SCPx cDNA probe corresponding to the whole protein-coding region. The same blot was stripped and reprobed with 18 S rRNA probe to normalize sample loading. The positions of RNA size markers are shown on the left.
Figure 4. The developmental profile of SCPx during the sixth instar.
(A) A 10 #x3BC;g portion of total RNA from midgut at various time points within the sixth larval instar, as indicated, was used. The blot was hybridized with a 32P-labelled SCPx cDNA probe corresponding to the open reading frame. The same blot was stripped and reprobed with 18S rRNA probe. G, W and PP refer to the stages ‘gut purge’, ‘wandering’and ‘prepupa’ respectively that occur before pupation. (B) Western-blot analysis of SCPx protein. Top panel: 100 #x3BC;g of protein from midgut at various time points within the sixth larval instar, as indicated, was used. Bottom panel: Western-blot analysis of the recombinant S. littoralis SCPx protein. Lane 1, proteins from E. coli carrying the pGEX-2T vector without the SCPx cDNA; lane 2, proteins from E. coli carrying the pGEX-2T vector containing the SCPx cDNA without induction of expression; lane 3, proteins from E. coli carrying the pGEX-2T vector containing SCPx cDNA with induction of expression; lane 4, whole extract from C. elegans. The antibody against C. elegans P-44 was used to detect the SCPx protein.
Figure 5. Western-blot analysis of the SCP2 domain.
Left panel: proteins from various tissues within the last larval instar were used. Lane 1, midgut; lane 2, fat body; lane 3, Malpighian tubules; lane 4, recombinant protein corresponding to the SCP2 domain of SCPx. The antibody against the SCP2 domain of S. littoralis SCPx protein was used. Right panel: Western-blot analysis of the recombinant S. littoralis SCPx protein using anti-(SCP2 domain) antibody. Lane 1, proteins from E. coli carrying the pGEX-2T vector without the SCPx cDNA; lane 2, proteins from E. coli carrying the pGEX-2T vector containing SCPx cDNA with induction of expression.
Figure 6. Developmental profile of SCPx in the prothoracic glands during the sixth instar.
The SCPx mRNA level in the prothoracic glands was measured by real-time PCR as described in the Experimental section. The arbitrary unit for mRNA was set as 1 at the 18 h time point and all samples were normalized relative to RP49. Results are means±S.E.M. for two experiments, with three replicates for each time point. G/W and PP are defined in the legend to Figure 4.
Genomic structure of SCPx
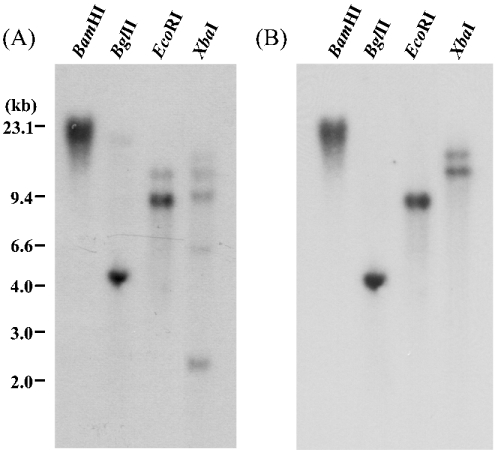
Genomic DNA was prepared from combined fat body and Malpighian tubules and digested with various restriction enzymes, including XbaI, which has a cleavage site in the SCPx cDNA (at position 667; Figure 1), whereas BamHI, BglII and EcoRI do not. The resultant DNA fragments were analysed by Southern blotting using a probe representing the coding region of the cDNA (67–1674; Figure 1). As shown in Figure 7(A), more than one band was observed in BglII-, EcoRI- and XbaI-digested samples. However, when the same blot was re-probed with a cDNA fragment corresponding to the N-terminus (67–667; Figure 1), only one band was observed in BamHI-, BglII- and EcoRI-digested DNA (Figure 7B). This result can be most simply interpreted if SCPx is encoded by a single gene in the S. littoralis genome. However, this point would need to be confirmed by performing a quantitative Southern blot, but this would require knowledge of the genome size of S. littoralis. Multiple bands detected using the full-length coding region may be due to hybridization to other members of the SCP2 gene family, but could also be caused by polymorphism, since this genomic DNA was prepared from ten individuals. The two bands in the XbaI digests in Figure 7(B) can only be due to polymorphism of the gene.
Figure 7. Southern-blot analysis of S. littoralis genomic DNA.
A 10 #x3BC;g portion of genomic DNA was digested with various restriction enzymes as indicated on the top of each panel. The blot was hybridized with 32P-labelled SCPx cDNA corresponding to the entire coding region (A) or a region from the start codon to the XbaI site (67–667; Figure 1) (B). DNA marker sizes are indicated on the left.
DISCUSSION
Using a reverse-transcriptase PCR-based cloning strategy, employing degenerate primers designed on the basis of the partial amino acid sequence of the purified protein, together with 5′-RACE, the complete cDNA encoding SCPx was isolated and sequenced. The full-length SCPx cDNA encodes a protein of 535 amino acids. In fact, the in vitro expression showed that this cDNA expressed an approx. 57 kDa product (Figure 1).
In humans, SCPx mRNA encodes a fusion protein, and the 58 kDa product is post-translationally processed to the 46 kDa thiolase and the 13 kDa SCP2. In contrast, the mRNA encoding P-44, which is homologous with SCPx in C. elegans, lacks the SCP2 domain near the 3′-end [14]. The situation concerning regulation of SCPx in Drosophila is intermediate between that of mammals and C. elegans. Drosophila produces two types of mRNA encoding SCPx: a fused mRNA and a SCP2-lacking mRNA, the latter being the major transcript in the larval stage [20]. In the case of S. littoralis, Northern blotting using the full-length coding region of SCPx detected two transcripts of approx. 1.9 and 0.95 kb (Figure 3), but the short transcript, which presumably encodes SCP2, was expressed at a much lower level than the 1.9 kb transcript (Figure 3). The SCPx mRNA lacking the SCP2 domain was not detected using Northern blotting or 3′-RACE (results not shown). On the other hand, the antibody against the 3-oxoacyl-CoA thiolase domain could detect a product of approx. 46 kDa (Figure 4B). By contrast, the antibody against the SCP2 domain could detect a product of approx. 13 kDa not only in vivo but also in E. coli cells which expressed the recombinant SCPx protein (Figure 5). These data suggest that S. littoralis SCPx is mainly transcribed as a fusion mRNA and the translation product is post-translationally cleaved into two proteins: 3-oxoacyl-CoA thiolase and SCP2. Accordingly, the lepidopteran system may be closer to that of mammals rather than that of dipterans.
Database searching revealed that the gene product has significant similarity to products of other members of the SCP2 gene family (Figure 2). Most importantly, a cysteine residue (Cys82), which has been suggested to be part of the active site in a bacterial (Zoogloea ramigera) 3-oxoacyl-CoA thiolase [21], is conserved in S. littoralis SCPx. Also, this protein contains Ser-Lys-Leu, a putative peroxisome targeting motif, at the end of the C-terminus. This means that SCPx can be transported to peroxisomes and processed in a similar manner to other species.
Northern blotting also revealed that the 1.9 kb mRNA is highly expressed in midgut and Malpighian tubules during the feeding stage of the last larval instar. In C. elegans, P-44 is mainly expressed in the larval intestine [19], whereas in Drosophila, SCPx is highly expressed in midgut epithelium during late embryogenesis [20]. These expression patterns are very similar to that of S. littoralis. These data suggest that SCPx may be involved in lipid transport and metabolism in the gut of invertebrate species. In particular, gut plays an important role for absorption of fatty acids and cholesterol in mammals, and most invertebrate species cannot synthesize sterols and depend on a dietary source. The high expression level in midgut is very consistent with this role. The lipid metabolism may include that of sterol/steroid, since the midgut is a major site of sterol dealkylation in insects [22] and the midgut and Malpighian tubules are also known to be tissues which strongly express steroid-metabolizing enzymes [23–26].
Warren and Gilbert [27] suggested that SCP2 may be involved in the transport of 7-dehydrocholesterol from the endoplasmic reticulum to the mitochondrial outer membrane during the early steps of ecdysteroid synthesis in the prothoracic glands. Our demonstration by semi-quantitative PCR that SCPx is constitutively expressed in prothoracic glands (Figure 6) lends support to this tenet. Furthermore, a recent report revealed that SCP2 from the yellow-fever mosquito (Aedes aegypti) is also highly expressed in the tissues which are involved in ecdysteroid synthesis [28]. These data suggest that this protein may have a role in sterol transport, including that associated with ecdysteroid synthesis.
Acknowledgments
The work was supported by a grant from the BBSRC (Biotechnology and Biological Sciences Research Council). We thank Mr S. G. Corrigan for culture of S. littoralis and Ms Andrea Davies for help with the semi-quantitative PCR experiments.
References
- 1.Rees H. H., Isaac R. E. Biosynthesis and metabolism of ecdysteroids and methods of isolation and identification of the free and conjugated compounds. Methods Enzymol. 1985;111:377–410. doi: 10.1016/s0076-6879(85)11024-4. [DOI] [PubMed] [Google Scholar]
- 2.Gallegos A. M., Atshaves B. P., Storey S. M., Starodub O., Petrescu A. D., Huang H., McIntosh A. L., Martin G. G., Chao H., Kier A. B., Schroeder F. Gene structure, intracellular localization, and functional roles of sterol carrier protein-2. Prog. Lipid Res. 2001;40:498–563. doi: 10.1016/s0163-7827(01)00015-7. [DOI] [PubMed] [Google Scholar]
- 3.Puglielli L., Rigotti A., Greco A. V., Santos M. J., Nervi F. Sterol carrier protein-2 is involved in cholesterol transfer from the endoplasmic reticulum to the plasma membrane in human fibroblasts. J. Biol. Chem. 1995;270:18723–18726. doi: 10.1074/jbc.270.32.18723. [DOI] [PubMed] [Google Scholar]
- 4.Ferdinandusse S., Denis S., van Berkel E., Dacremont G., Wanders R. J. A. Peroxisomal fatty acid oxidation disorders and 58 kDa sterol carrier protein X (SCPx): activity measurements in liver and fibroblasts using a newly developed method. J. Lipid Res. 2000;41:336–342. [PubMed] [Google Scholar]
- 5.Bun-ya M., Maebuchi M., Kamiryo T., Kurosawa T., Sato M., Tohma M., Jiang L. L., Hashimoto T. Thiolase involved in bile acid formation. J. Biochem. (Tokyo) 1998;123:347–352. doi: 10.1093/oxfordjournals.jbchem.a021943. [DOI] [PubMed] [Google Scholar]
- 6.Wanders R. J., Denis S., van Berkel E., Wouters F., Wirtz K. W. A., Seedorf U. J. Identification of the newly discovered 58 kDa peroxisomal thiolase SCPx as the main thiolase involved in both pristanic acid and trihydroxycholestanoic acid oxidation: implications for peroxisomal β-oxidation disorders. J. Inherited Metab. Dis. 1998;21:302–305. doi: 10.1023/a:1005349028853. [DOI] [PubMed] [Google Scholar]
- 7.Kannenberg F., Ellinghaus P., Assmann G., Seedorf U. Aberrant oxidation of the cholesterol side chain in bile acid synthesis of sterol carrier protein-2/sterol carrier protein-x knockout mice. J. Biol. Chem. 1999;274:35455–35460. doi: 10.1074/jbc.274.50.35455. [DOI] [PubMed] [Google Scholar]
- 8.Seedorf U., Ellinghaus P., Nofer J. R. Sterol carrier protein-2. Biochim. Biophys. Acta. 2000;1486:45–54. doi: 10.1016/s1388-1981(00)00047-0. [DOI] [PubMed] [Google Scholar]
- 9.Ohba T., Rennert S. M., Pfeifer Z. G., He R., Yamamoto R., Halt J. A., Billheimer J. F., Strauss J. F. The structure of the human sterol carrier protein-x sterol carrier protein 2 gene (scp2) Genomics. 1994;24:370–374. doi: 10.1006/geno.1994.1630. [DOI] [PubMed] [Google Scholar]
- 10.Seedorf U., Raabe M., Ellinghaus P., Kannenberg F., Fobker M., Engle T., Denis S., Wouters F., Wirtz K. W. A., Wanders R. J. A., Maeda N., Assmann G. Defective peroxisomal catabolism of branched fatty acyl coenzyme A in mice lacking the sterol carrier protein-2 sterol carrier protein-x gene function. Genes Dev. 1998;12:1189–1201. doi: 10.1101/gad.12.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seedorf U., Assmann G. Cloning, expression and nucleotide-sequence of rat-liver sterol carrier protein-2 cDNAs. J. Biol. Chem. 1991;266:630–636. [PubMed] [Google Scholar]
- 12.Pfeifer S. M., Sakuragi N., Ryan A., Johnson A. L., Deeley R. G., Billheimer J. T., Baker M. E., Strauss J. F. Chicken sterol carrier protein-2 sterol carrier protein-x – cDNA cloning reveals evolutionary conservation of structure and regulated expression. Arch. Biochem. Biophys. 1993;304:287–293. doi: 10.1006/abbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 13.Tan H., Okazaki K., Kubota I., Kamiryo T., Utiyama H. A novel peroxisomal nonspecific lipid-transfer protein from Candida tropicalis – gene structure, purification and possible role in β-oxidation. Eur. J. Biochem. 1990;190:107–112. doi: 10.1111/j.1432-1033.1990.tb15552.x. [DOI] [PubMed] [Google Scholar]
- 14.Bun-ya M., Maebuchi M., Hashimoto T., Yokota S., Kamiryo T. A second isoform of 3-oxoacyl-CoA thiolase found in Caenorhabditis elegans, which is similar to sterol carrier protein x but lacks the sequence of sterol carrier protein 2. Eur. J. Biochem. 1997;245:252–259. doi: 10.1111/j.1432-1033.1997.t01-1-00252.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen J.-H., Powls R., Rees H. H., Wilkinson M. C. Purification of ecdysone oxidase and 3-dehydroecdysone 3α-reductase from the cotton leafworm, Spodoptera littoralis. Insect Biochem. Mol. Biol. 1999;29:899–908. [Google Scholar]
- 16.Frohman M. A. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 17.Schramm G., Bruchhaus I., Roeder T. A simple and reliable 5′ RACE approach. Nucleic Acids Res. 2000;28:e96. doi: 10.1093/nar/28.22.e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J., Fritsch E. F., Maniatis T. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 19.Maebuchi M., Togo S. H., Yokota S., Ghenea S., Bun-ya M., Kamiryo T., Kawahara A. Type-II 3-oxoacyl-CoA thiolase of the nematode Caenorhabditis elegans is located in peroxisomes, highly expressed during larval stages and induced by clofibrate. Eur. J. Biochem. 1999;264:509–515. doi: 10.1046/j.1432-1327.1999.00655.x. [DOI] [PubMed] [Google Scholar]
- 20.Kitamura T., Kobayashi S., Okada M. Regional expression of the transcript encoding sterol carrier protein x-related thiolase and its regulation by homeotic genes in the midgut of Drosophila embryos. Develop. Growth Differ. 1996;38:373–381. doi: 10.1046/j.1440-169X.1996.t01-3-00005.x. [DOI] [PubMed] [Google Scholar]
- 21.Thompson S., Mayerl F., Peoples O. P., Masamune S., Sinskey A. J., Walsh C. T. Mechanistic study on β-oxoacyl thiolase from Zoogloea ramigera: identification of the active-site nucleophile as Cys89, its mutation to Ser89, and kinetic and thermodynamic characterization of wild-type and mutant enzymes. Biochemistry. 1989;28:5735–5742. doi: 10.1021/bi00440a006. [DOI] [PubMed] [Google Scholar]
- 22.Rees H. H. Biosynthesis of ecdysone. In: Kerkut G. A., Gilbert L. I., editors. Comprehensive Insect Physiology, Biochemistry and Pharmacology, vol. 7. Oxford: Pergamon Press; 1985. pp. 249–293. [Google Scholar]
- 23.Chen J.-H., Kabbouh M., Fisher M. J., Rees H. H. Induction of an inactivation pathway for ecdysteroids in larvae of the cotton leafworm, Spodoptera littoralis. Biochem. J. 1994;301:89–95. doi: 10.1042/bj3010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams D. R., Chen J.-H., Fisher M. J., Rees H. H. Induction of enzymes involved in molting hormone (ecdysteroid) inactivation by ecdysteroids and an agonist, 1,2-dibenzoyl-1-tert-butylhydrazine (RH-5849) J. Biol. Chem. 1997;272:8427–8432. doi: 10.1074/jbc.272.13.8427. [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi H., Chen J.-H., O'Reilly D. R., Rees H. H., Turner P. C. Regulation of ecdysteroid signalling: molecular cloning, characterization and expression of 3-dehydroecdysone 3α-reductase, a novel eukaryotic member of the short-chain dehydrogenases/reductases superfamily from the cotton leafworm, Spodoptera littoralis. Biochem. J. 2000;349:239–245. doi: 10.1042/0264-6021:3490239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi H., Chen J.-H., O'Reilly D. R., Turner P. C., Rees H. H. Regulation of ecdysteroid signaling: Cloning and characterization of ecdysone oxidase, a novel steroid oxidase from the cotton leafworm, Spodoptera littoralis. J. Biol. Chem. 2001;276:26819–26828. doi: 10.1074/jbc.M104291200. [DOI] [PubMed] [Google Scholar]
- 27.Warren J. T., Gilbert L. I. Metabolism in vitro of cholesterol and 25-hydroxycholesterol by the larval prothoracic glands of Manduca sexta. Insect Biochem. Mol. Biol. 1996;26:917–929. doi: 10.1016/s0965-1748(96)00058-6. [DOI] [PubMed] [Google Scholar]
- 28.Krebs K. C., Lan Q. Isolation and expression of a sterol carrier protein-2 gene from the yellow fever mosquito, Aedes aegypti. Insect Mol. Biol. 2003;12:51–60. doi: 10.1046/j.1365-2583.2003.00386.x. [DOI] [PubMed] [Google Scholar]