Abstract
The p160 co-activators, SRC1 (steroid receptor co-activator 1), GRIP1 (glucocorticoid-receptor-interacting protein 1) and ACTR (activator for thyroid hormone and retinoid receptors), have two ADs (activation domains), AD1 and AD2. AD1 is a binding site for the related co-activators, CBP (cAMP-response-element-binding protein-binding protein) and p300, whereas AD2 binds to another co-activator, co-activator-associated arginine methyltransferase 1 (CARM1). Here, we identified two CBP-interacting sites [amino acids 1075–1083 (site I) and 1095–1106 (site II)] in a so-called CBP-dependent transactivation domain (AD1; amino acids 1057–1109) of GRIP1. Site I was the major site for CBP-dependent AD1 transactivation activity of GRIP1 whereas, following the deletion of site II, full or partial transactivation activity was retained without the recruitment of CBP in yeast, HeLa, human embryonic kidney 293 and CV-1 cells. GRIP1 (with a deletion of site II) expressed stronger co-activator activity than that of wild-type GRIP1 in the TR (thyroid receptor) and the AR (androgen receptor), but not the ER (oestrogen receptor), systems in HeLa cells. We also demonstrated that these CBP-binding sites of GRIP1 are not the only functional domains for its AD1 function in TR, AR and ER systems in HeLa cells by the exogenous overexpression of one E1A mutant, which led to a lack of CBP-binding ability. Our results suggest that these two CBP-interacting sites in the GRIP AD1 domain not only determine its AD1 activity, but are also involved in its co-activator functions in some nuclear receptors.
Keywords: cAMP-responsive-element-binding-protein-binding protein (CBP), co-activator, E1A, glucocorticoid-receptor-interacting protein 1 (GRIP1), interacting site, nuclear receptor
Abbreviations: ACTR, activator for thyroid hormone and retinoid receptors; AD, activation domain; AF, activation function; AR, androgen receptor; β-gal, β-galactosidase; CARM1, co-activator-associated arginine methyltransferase 1; CBP, cAMP-responsive-element-binding protein-binding protein; DBD, DNA-binding domain; ER, oestrogen receptor; GR, glucocorticoid receptor; GRIP1, glucocorticoid-receptor-interacting protein 1; GST, glutathione S-transferase; HBD, hormone-binding domain; HEK, human embryonic kidney; LBD, ligand-binding domain; LUC, luciferase; MMTV, murine-mammary-tumour virus; NR, nuclear receptor; p/CAF, p300/CBP-associated factor; PR, progesterone receptor; SRC1, steroid receptor co-activator-1; TIF2, transcriptional mediator/intermediary factor 2; TR, thyroid receptor
INTRODUCTION
The NRs (nuclear receptors) constitute a large superfamily of ligand-regulated transcription factors that trigger complex events during development, differentiation and homoeostasis [1–4]. They control gene expression upon binding of small lipophilic hormones such as steroids, retinoids, thyroid hormones and some unidentified ligands. NRs stimulate transcription by promoting the local modification of chromatin structure and the recruitment of a pre-initiation complex containing RNA polymerase II to the promoter. Two transcriptional activation functions (AF1 and AF2) of NRs provide molecular surfaces for the recruitment of transcriptional co-activator proteins to achieve these purposes.
There are three families of NR co-activators: p300/CBP (cAMP-responsive-element-binding protein-binding protein), the p160 family and p/CAF (p300/CBP-associated factor) [3,5,6]. At least three distinct p160 proteins have been identified, including SRC1 (steroid receptor co-activator-1) [7], GRIP1 (glucocorticoid-receptor-interacting protein 1) [8] and its human homologue TIF2 (transcriptional mediator/intermediary factor 2) [9] and ACTR (activator for thyroid hormone and retinoid receptors) [10], also known as AIB1 (amplified in breast cancer 1; [11]), RAC3 (receptor-associated co-activator 3; [12] or TRAM1 (thyroid hormone receptor activator molecule 1; [13]), and its mouse homologue p/CIP [14]. The interaction of the p160 proteins with the LBD (ligand-binding domain) of NRs is mediated by the LXXLL (Leu-Xaa-Xaa-Leu-Leu) motif [15–19]. This sequence forms part of an amphipathic α-helix, which binds in a conserved hydrophobic cleft on the surface of liganded LBDs. The C-terminal region of the p160 co-activators is also able to interact with the AF1 domain of some NRs, and then enhance the AF1 activities of those NRs in the absence of ligands [20,21]. However, it is unclear whether these AF1s bind to specific motif(s) on the p160 co-activator similar to the LXXLL motif for AF2 binding. CBP, p300 and p/CAF have each been shown to possess histone acetyltransferase activities and directly interact with AF1 and AF2 [6,22]. The co-activator function of CBP/p300 probably involves a variety of mechanisms, because these proteins can bind to many transcriptional activators, NR co-activators and components of the basal transcriptional machinery [23–27]. These properties have prompted the suggestion that CBP/p300 serves as a scaffold or platform for the assembly of transcriptional co-activators and as a signal in multiple regulatory pathways [25,28–30].
Two ADs (activation domains), AD1 and AD2, in the p160 co-activator for the transmission of the activation signal from NRs have been identified in previous studies [21,31,32]. GRIP1 and CBP associate both in vivo and in vitro. The GRIP1 sequences required for this interaction have been mapped to amino acids 1010–1131 or 1057–1109 [21,31]. This region co-localizes with the potent transcriptional activation domain AD1, which has been shown to be CBP-dependent. A number of studies have emphasized the functional importance of p160–CBP interactions [10,33–36]. It has been shown that the deletion of amino acids 1018–1088 in ACTR, which includes the CBP-interaction domain AD1, negates the ability of ACTR to stimulate GR (glucocorticoid-receptor)-mediated transcription in transiently transfected cells. Similarly, it has been shown that a deletion in the region encoding SRC1 AD1 (amino acids 900–950) abrogated the ability of SRC1 to enhance ligand-dependent and -independent transcription by the ER (oestrogen receptor). CBP has been shown to acetylate ACTR at a lysine residue adjacent to an LXXLL motif, resulting in the dissociation of ACTR from the LBD. Thus it has been suggested that CBP may both facilitate and attenuate NR-mediated transcription.
A second AD located close to the C-termini of the p160s (AD2; amino acids 1122–1462 in GRIP1) has previously been shown to bind CARM1 (co-activator-associated arginine methyltransferase 1), a protein with arginine methyltransferase activity that methylates histones and CBP [32]. Both binding of p300 to AD1 and binding of CARM1 to AD2 are required for their respective ER co-activator functions, and for their synergy [37]. Furthermore, CARM1 can methylate CBP in vitro and in vivo, and control CBP co-activating activity in GRIP1-dependent transcriptional activation and in hormone-induced gene activation [38,39]. The goals of the present study were to map the CBP/p300-interacting sites in GRIP1 in detail, and to investigate the importance of these CBP-interacting sites of GRIP1 for its co-activation function.
MATERIALS AND METHODS
Plasmids
The pSG5.HA vector coding for full-length GRIP1 (codons 5–1462) has been described previously [32]. For other GRIP1s, codons 563–1121, 563–1094 and 563–1012 were cloned as an EcoRI–SalI fragment into the EcoRI and XhoI sites of vector pSG5.HA; for GRIP1, codons 1013–1121 were cloned as an EcoRI–XhoI fragment into the EcoRI and XhoI sites of vector pSG5.HA. Deletions of pSG5.HA.GRIP15-1462 and pSG5.HA. GRIP1563–1121, including codons 1075–1083 and 1095–1106, were performed using the Promega Gene Edit kit. The pSG5.HA vector coding for full-length CBP was cloned as a full-length CBP HindIII–BamHI fragment from pCDNA3b.Flag.CBP.HA (a gift from Dr. Tso-Pang Yao, Duke University, Durham, NC, U.S.A.) into the HindIII and BamHI sites of vector pSG5.HA modified by introducing a NotI–HindIII fragment into the EcoRI and XhoI sites. Various fragments of GRIP1s were constructed by inserting EcoRI–SalI fragments of the appropriate PCR-amplified GRIP1 cDNA into the EcoRI and SalI sites of the pM vector (ClonTech), a vector for the expression of GAL4 DBD (DNA-binding domain) fusion proteins from a constitutive SV40 (simian virus 40) early promoter. The expression vector (pM) for the GAL4 DBD fused to CBP2041-2240 and the Ad5 E1A(12S) and CBP-binding mutants have been described previously [5,40]. The reporter genes consisting of an MMTV (mouse-mammary-tumour virus) promoter that contains the LUC (luciferase)-coding region (MMTV-LUC), EREII-LUC (GL45), MMTV(TRE)-LUC and a reporter gene GK1 that contains five GAL4 response elements upstream of a minimal adenovirus E1B have been described previously [41,42]. For the expression of NRs in mammalian cells, vectors pSVAR0 for human AR (androgen receptor) [43], pHE0 for human ERα (Gly-400→Val) [44], pCMX.hTRβ1 for human TRβ1 (thyroid receptor β1) [18], pKSX for mouse GR [45], and pCMV.hPR-B for human PR (progesterone receptor) B [46], were used as described previously.
Yeast expression plasmids coding for fusion proteins of GAL4 DBD and p300 (codons 1856–2414) and ER HBD (hormone-binding domain) have been described previously [17,47]. Yeast expression plasmids coding for fusion proteins of GAL4 AD (or GAL4 DBD) and various GRIP1 fragments were constructed by inserting EcoRI–SalI PCR fragments into pGAD424 (or pGBT9 vectors) (ClonTech).
The bacterial expression vector for GST (glutathione S-transferase) fused to CBP (codons 2041–2240) was constructed by inserting the appropriate PCR fragment into pGEX-4T1 (Amersham Biosciences) via the EcoRI and XhoI sites.
Cell culture and transient transfection assays
For functional assays, HeLa, COS-7, HEK (human embryonic kidney)-293 and CV-1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% charcoal/dextran-treated foetal bovine serum. Transient transfections and LUC assays were performed as described previously [21] in 24-well culture dishes. Total DNA was adjusted to 1 μg by adding the necessary amount of pSG5.HA vector. Luciferase activity of the transfected cell extracts is presented as relative light units, and represents the mean and standard deviation for three transfected cultures. As the expression of many control vectors was used to monitor transfection efficiency influenced by co-activators, internal controls were not used. Instead, reproducibility of observed effects was determined in multiple independent transfection experiments.
Immunoblotting
Immunoblots were performed as described previously [21] using 30% of the extract from lysates for immunoprecipitation with monoclonal antibodies 3F10 (Roche) against the HA epitope and RK5C1 (Santa Cruz Biotechnology) against GAL4 DBD.
Protein–protein interaction assays
For GST pull-down assays, 35S-labelled proteins were produced with the TNT T7-coupled reticulocyte lysate system (Promega), and GST fusion proteins were produced in Escherichia coli strain BL21, eluted and analysed by gel electrophoresis, as described previously [17].
Yeast two-hybrid assays were performed as previously described [17]. Where indicated, yeast cultures were incubated for approx. 15 h before harvest with 100 nM oestradiol for ER. β-Galactosidase (β-gal) activity of extracts from the liquid yeast cultures is expressed as the mean and standard deviation of results from three independently transformed yeast colonies, and is representative of two or more independent experiments.
RESULTS
Two distinct CBP-interacting sites in the GRIP1 AD1 domain
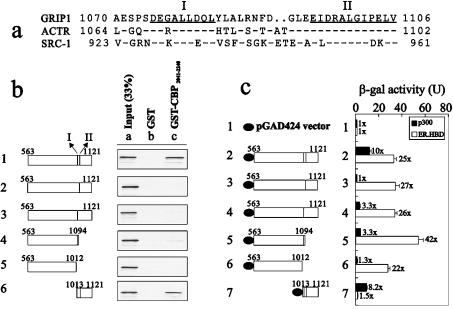
To study possible CBP-interacting sites in the GRIP1 AD1 domain, we analysed the AD1 domain of three p160 co-activator family proteins based on two previous studies [21,31], and found that there were two highly conserved regions, amino acids 1075–1083 (I) and 1095–1106 (II), in this AD1 domain (Figure 1a). We then constructed these two internally deleted fragments in GRIP1563–1121 to test whether these two regions were responsible for direct interaction with CBP. In the GST pull-down assay, any fragment deleted in these two regions lost the ability to bind the CBP C-terminus (amino acids 2041–2240) compared with the wild-type GRIP1 fragment (Figure 1b). Similar observations were also made in the yeast two-hybrid assay; however, the deletion of site II caused a weaker binding ability (approx. 30%) to p300 C-terminus (amino acids 1856–2414) to be retained than wild-type GRIP1 did (Figure 1c, compare histograms 1, 2, 4 and 5, closed columns). A smaller fragment containing these two regions, amino acids 1013–1121, was sufficient to interact with CBP or p300 C-terminus, whether analysed by GST pull-down or yeast two-hybrid assays (Figures 1b and 1c). All pGAD424.GRIP1 fragments, except for amino acids 1013–1121, were proven to retain their ability to interact directly with ER, which suggests that these GRIP1 fragments functioned normally through the intact LXXLL motif in cells (Figure 1c, open columns).
Figure 1. Two CBP-interacting sites in the GRIP1 AD1 are mapped to amino acids 1075–1083 (I) and 1095–1106 (II).
(a) Sequence alignment of the CBP-interaction domain in GRIP1 with the corresponding region in ACTR and SRC-1. Identical residues in all three proteins are indicated (−), as are gaps introduced for optimal sequence alignment (..). Sequences of the two sub-CBP-interaction domains are underlined and labelled I and II. (b) Interaction of the C-terminal region of CBP with two sub-CBP-binding domains of GRIP1 in vitro. Glutathione–Sepharose bound to GST or GST–CBP2041-2240 was incubated with 35S-labelled GRIP1 fragments translated in vitro from pSG5.HA vectors. GRIP1 fragments translated in vitro include the amino acids below. (c) Interaction of C-terminal region of p300 with two CBP-interacting sites of GRIP1 in yeast two-hybrid assays. pGAD424 (GAL4 AD) vectors, fused to the indicated GRIP1 fragments, were expressed in yeast strain SFY526, along with GAL4 DBD-p300 (amino acids 1856–2414) or GAL4 DBD-ER HBD fusion proteins; when ER HBD was present, yeast cells were grown in 100 nM oestradiol. Activation of the integrated β-gal reporter gene controlled by GAL4-binding sites was determined by measuring β-gal activity in cell extracts. Numbers beside the columns indicate fold activity relative to no added GRIP1 fragment. We observed a similar expression pattern (b) in two independent experiments. The data in (c) are based on at least three separate experiments.
CBP-dependent and -independent transactivation activity in the GRIP1 AD1 domain
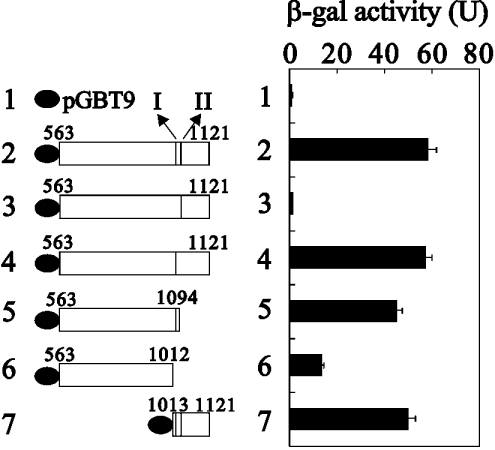
Results shown in Figure 1 demonstrate that there were two distinct CBP-interacting sites in the AD1 transactivation domain of GRIP1. To examine the functional role of these two CBP-interacting sites for GRIP1 AD1 activity in yeast strain SFY526, we used the pGBT9 vector, fused to a yeast GAL4 DBD and which could activate a β-gal reporter gene controlled by a GAL4 enhancer site in yeast, to express wild-type GRIP1563–1121, two internal truncated fragments (deletion in site I or II) in GRIP1563–1121, C-truncated GRIP1s (amino acids 563–1094 and 563–1012), and C-terminal GRIP1 (amino acids 1013–1121). Our data showed that the GRIP1 fragment with site I deleted completely lost its transactivation activity from the AD1 domain (Figure 2, compare histograms 2 and 3). However, following the deletion of site II, the same level of transactivation activity was retained as in wild-type GRIP1 (Figure 2, compare histograms 2 and 4). A similar result was observed in the C-truncated GRIP1 (amino acids 563–1094), which contained site I. Compared with the activity of wild-type GRIP1 (amino acids 563–1121), the region of amino acids 1013–1121 was able to express full transactivation activity in the AD1 domain (Figure 2, compare histograms 2 and 7). In contrast, one construct without these two CBP-interacting sites in amino acids 563–1012 expressed a weaker transactivation activity (Figure 2, compare histograms 1, 2 and 6).
Figure 2. The effect of the two CBP-interacting sites on GRIP1 AD1 activity in yeast strain SFY526.
pGBT9 (GAL4 DBD) fusion proteins of GRIP1 fragments (indicated by amino acid numbers) were expressed in yeast strain SFY526. Activation of the integrated β-gal reporter gene controlled by GAL4-binding sites was determined by measuring β-gal activity in cell extracts. The two sub-CBP-interaction domains are labelled I and II. The data are based on at least three separate experiments.
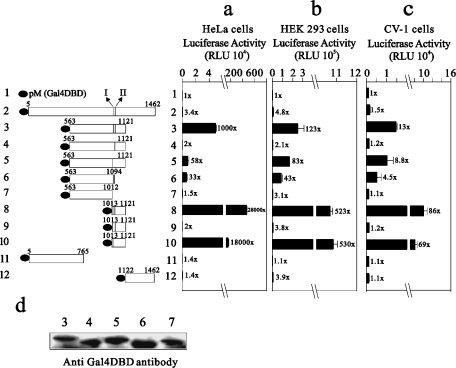
We also analysed the transactivation activities of these GRIP1 constructs in HeLa, HEK-293 and CV-1 cells (Figure 3). Our previous studies have demonstrated that the transactivation activity of the AD1 domain of GRIP1 could be repressed by its own C-terminal region (S.-M. Huang, W.-C. Hsu and T.-Y. Hsieh, unpublished work). Here, our observation showed that GRIP11013-1121 had the highest transactivation activity of all tested GRIP1 fragments (Figures 3a–3c, compare histograms 1–3 and 8); GRIP1563–1121 had a higher transactivation activity than full-length GRIP1 (GRIP15-1462; Figures 3a–3c, compare histograms 1, 2 and 3). All constructs with site I deleted expressed very low or no transactivation activity (Figures 3a–3c, compare histograms 4, 7 and 9), whereas following the site II deletion they retained part or all of the wild-type GRIP1 activity (Figures 3a–3c, compare histograms 3 and 5 or 8 and 10). The difference in activity in histograms 4 and 7 was not caused by the expression of these constructs in cells (Figure 3d). In contrast with the result from yeast (Figure 2, histogram 6), GRIP1563–1012 without these two CBP-interacting sites had no transactivation activity in HeLa and CV-1 cells (Figures 3a and 3c, compare histograms 1 and 7). In summary, our findings in yeast, HeLa, HEK-293 and CV-1 cells suggest that these two CBP-interacting sites in GRIP1 play different functional roles in its AD1 transactivation activity. However, site I was more important than site II for the AD1 transactivation activity of GRIP1, whether in mammalian or yeast cells.
Figure 3. The effect of the two CBP-interacting sites on GRIP1 AD1 activity in HeLa, HEK-293 and CV-1 cells.
HeLa cells (a), HEK-293 cells (b) and CV-1 cells (c) were transiently transfected with 0.5 μg of the expression vector pM or various pM–GRIP1 fragments, along with the GK1 reporter gene (0.5 μg), which encodes LUC and is controlled by GAL4 response elements. Luciferase activities of the transfected cell extracts were determined. RLU, relative light units. Numbers beside the columns indicate fold activity relative to no added GRIP1 fragment. The two sub-CBP-interaction domains are labelled I and II. (d) COS-7 cells were transiently transfected in 6-well plates with 1 μg of the indicated pM–GRIP1 fragments. Immunoblots were performed as described in the Materials and methods section using 30% of the extract from lysates for immunoprecipitation and monoclonal antibody RK5C1 (Santa Cruz Biotechnology) against GAL4 DBD. We observed a similar expression pattern (d) in two independent experiments. These data (a–c) are the average of three experiments (means±S.D.; n=3).
These two distinct CBP-interacting sites of GRIP1 are necessary for the enhancement of the transactivation activity of CBP
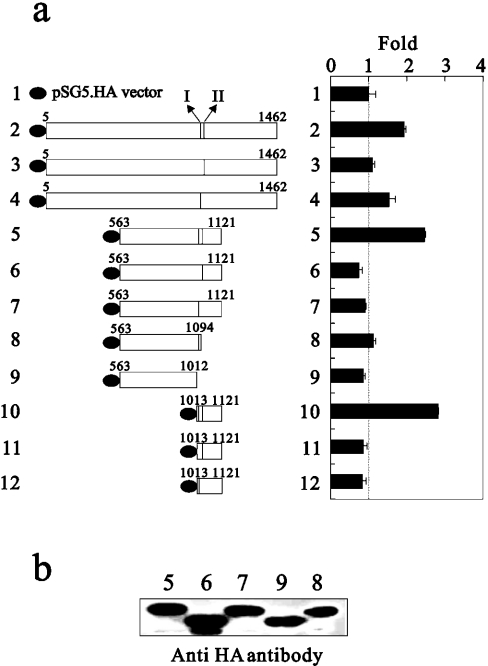
The fact of the physical interaction between CBP and GRIP1 suggests that GRIP1 might enhance CBP's primary co-activator functions through its effect on CBP transactivation activity. To test the ability of two CBP-interacting sites (I and II) of GRIP1 to enhance the transactivation activity of the C-terminal region of CBP, CBP amino acids 2041–2240, a primary region for binding with the AD1 region of GRIP1, was fused to the GAL4 DBD and tested in transiently transfected HeLa cells in the presence and absence of the two CBP-interacting sites of various GRIP1s. As shown in Figure 4(a), any GRIP1 fragments containing both sites I and II were able to enhance further CBP transactivation activity, whereas GRIP1 fragments lacking either CBP binding site I or II lost this ability. Thus we demonstrated the mutuality of GRIP1 and CBP through the CBP-interacting sites of GRIP1 (Figures 3a and 4a).
Figure 4. The effect of the two CBP-interacting sites on CBP putative transactivation activity in HeLa cells.
(a) HeLa cells were transiently transfected with 0.25 μg of pM.CBP2041-2240 and 0.25 μg of pSG5.HA or various pSG5.HA.GRIP1 fragments, along with the GK1 reporter gene (0.5 μg). LUC activities of the transfected cell extracts were determined. The basal activity of pM.CBP2041-2240 is indicated by the dotted line. The two sub-CBP-interaction domains are labelled I and II. (b) COS-7 cells were transiently transfected in six-well plates with 1 μg of the indicated pSG5.HA.GRIP1 fragments. Immunoblots were performed as described in the Materials and methods section using 30% of the extract from lysates for immunoprecipitation and monoclonal antibody 3F10 (Roche) against the HA tag. These data (a) are the average of three experiments (means±S.D.; n=3).
Differential use of these two GRIP1 CBP-interacting sites by TR
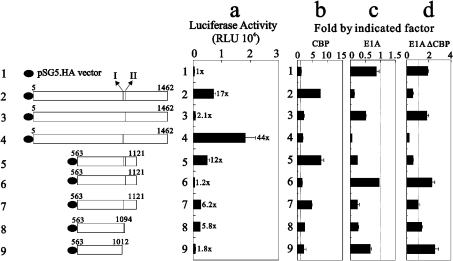
The study of Ma et al. [21] demonstrated that the two signal input domains (one that binds NR AF2 domains and one that binds AF1 domains of some, but not all, NRs) and the two signal output domains (AD1 and AD2) of p160 co-activators played different relative roles for two different NRs: AR and TR (Figures 5 and 6). We confirmed that the AD1 domain made a stronger contribution than the AD2 domain to the TR function through site I (Figure 5a, compare histograms 2, 3 and 5). In contrast, the deletion of site II, retaining AD1 activity, had a higher co-activation effect on TR function than wild-type GRIP1 (Figure 5a, compare histograms 2 and 4). These findings suggest that these two CBP-interacting sites in the GRIP1 AD1 region might simultaneously have positive and negative roles on TR functions.
Figure 5. The CBP-interacting site I of GRIP1 is required for its co-activator function in TR transcriptional activity.
(a) HeLa cells were transiently transfected with MMTV(TRE)-LUC reporter gene (0.25 μg), pCMX-hTRβ1 (0.01 μg) encoding hTRβ1, and 0.3 μg of pSG5.HA or various pSG5.HA-GRIP1 fragments in the presence of (b) pSG5.HA.CBP (0.25 μg) (c) E1A (0.25 μg) or (d) E1A(ΔCBP) (0.25 μg). Transfected cultures were grown in 100 nM 3,5,5′-tri-iodo-L-thyronine (T3), and LUC activities of the transfected cell extracts were determined. Numbers beside the columns indicate fold activity relative to that of hormone-activated TR with no added GRIP1. The actual LUC activities measured for TR activity were as follows: no added T3, 195±15 RLU (relative light units); T3 added, 411±20 RLU. Fold activations in (b), (c) and (d) indicate fold activity relative to that shown in (a). The dotted line is indicated as 1-fold activity with CBP (b), E1A (c) and E1A(ΔCBP) (d). The two sub-CBP-interaction domains are labelled I and II. These data (a–d) are the average of three experiments (means±S.D.; n=3).
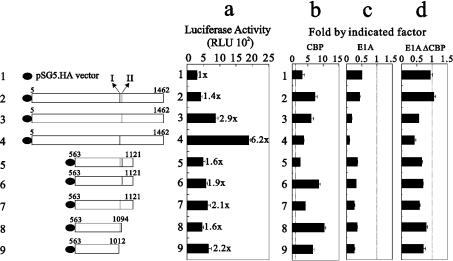
Figure 6. The CBP-interacting site II of GRIP1 exerts a negative role in its co-activator function in AR transcriptional activity.
(a) HeLa cells were transiently transfected with the MMTV-LUC reporter gene (0.25 μg), pSVAR0 (0.2 μg) encoding AR, and 0.3 μg of pSG5.HA, or various pSG5.HA-GRIP1 fragments in the presence of (b) pSG5.HA.CBP (0.25 μg) (c) E1A (0.25 μg) or (d) E1A(ΔCBP) (0.25 μg). Transfected cultures were grown in 100 nM dihydrotestosterone, and LUC activities of the transfected cell extracts were determined. Numbers beside the columns indicate fold activity relative to that of hormone-activated AR with no added GRIP1. The actual LUC activities measured for AR activity were as follows: no added dihydrotestosterone, 96±5 RLU (relative light units); dihydrotestosterone added, 323±30 RLU. Fold activations in (b), (c) and (d) indicate fold activity relative to that in (a). The dotted line shows 1-fold activity with CBP (b), E1A (c) and E1A(ΔCBP) (d). The two sub-CBP-interaction domains are labelled I and II. These data (a–d) are the averages of three experiments (means±S.D.; n=3).
The LUC activities of exogenously overexpressed CBP, E1A or an E1A mutant lacking the CBP-binding site [E1A(ΔCBP), which were co-transfected with all tested GRIP1 fragments, and their effect on TR function were calculated, and converted into the ‘fold value’ by dividing the activity observed when empty vector or specific GRIP1 was added with CBP (Figure 5b), E1A (Figure 5c) or E1A(ΔCBP) (Figure 5d) by that of the empty vector or specific GRIP1 alone. Our results demonstrated that CBP displayed higher enhancement effects on GRIP1 fragments containing site I, or sites I and II (Figure 5b, compare histograms 2, 5, 7 and 8). However, inhibition by the exogenous overexpression of E1A worked primarily on GRIP1 fragments containing site I (Figure 5c, compare histograms 2, 4, 5, 7 and 8). The application of the E1A mutant E1A(ΔCBP) to the TR system suggests that the inhibition of TR function by E1A is not only mediated through CBP, but is also mediated through other unknown factors in cells, because this E1A mutant still repressed the co-activator functions of GRIP1 fragments with an intact site I (Figure 5d, compare histograms 2, 4 and 5).
GRIP1 AD2 activity in AR functions is negatively regulated by its AD1 domain
Full-length GRIP1 fragments, with site I or II deleted, retained the AD2-dependent co-activator function in the AR system (Figure 6a, compare histograms 1–4), because AR is a GRIP1 AD2-dependent NR [21]. The higher AR co-activator function of the full-length GRIP1 fragment with site II deleted suggests that this site might be involved in the negative cross-talk between GRIP1 AD1 and AD2 regions (Figure 6a, compare histograms 1–4), because this effect was lost when GRIP1 fragments lacked the AD2 region (Figure 6a, compare histograms 4 and 7). The exogenous overexpression of CBP enhanced AR function alone and the effects of all tested GRIP1 fragments on AR function (Figure 6b). The inhibition by exogenous overexpression of E1A worked primarily on the AR itself and on the GRIP1 site I or II (Figure 6c). The effects of the E1A mutant E1A(ΔCBP) in the AR system suggest that the inhibition of the AR function by E1A is mediated primarily through CBP (Figure 6d). However, the deletion of site I or II might change the functional role of CBP in the AR-GRIP1 complex.
Cross-talk between GRIP1 AD1 and AD2 in ER functions
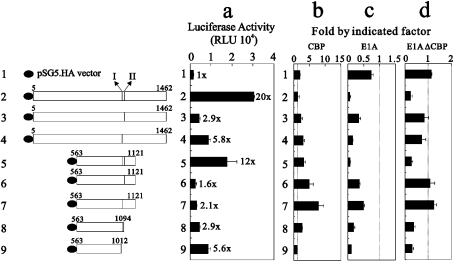
The study of Chen et al. [37] demonstrated that the co-activator functions of p300 and CARM1 depend on the co-expression of GRIP1. Co-expression of all three co-activators causes synergistic enhancement of ER function. Both AD1 and AD2 are required for their respective co-activator functions and for their synergy. Thus mutations in AD1 (site I or site II) might disrupt the cross-talk between AD1 and AD2. The enhancement of ER function by the full-length GRIP1 fragment, expressing the strongest co-activator function among truncated and mutant AD1 GRIP1 (Figure 7a, compare histograms 2–5), supports the suggestion that both AD1 and AD2 are required for its full co-activator roles for ER function. Effects by mutation in either site I or site II suggest that intact AD1 is important for GRIP1's co-activator function in the ER system (Figure 7a, compare histogram 2 with 3 and 4). However, AD1 played a more important role than AD2 (Figure 7a, compare histograms 2 and 5).
Figure 7. The CBP-interacting sites I and II of GRIP1 are required for its co-activator function in ER transcriptional activity.
(a) HeLa cells were transiently transfected with EREII-LUC reporter gene (0.25 μg), pHE0 (0.01 μg) encoding hERα, and 0.3 μg of pSG5.HA, or various pSG5.HA-GRIP1 fragments in the presence of (b) pSG5.HA.CBP (0.25 μg) (c) E1A (0.25 μg) or (d) E1A(ΔCBP) (0.25 μg). Transfected cultures were grown in 100 nM oestradiol, and LUC activities of the transfected cell extracts were determined. Numbers beside the columns indicate fold activity relative to that of hormone-activated ER with no added GRIP1. The actual LUC activities measured for ER activity were as follows: no added oestradiol, 242±15 RLU (relative light units); oestradiol added, 1515±130 RLU. Fold activations shown in (b), (c) and (d) indicate fold activity relative to those in (a). The dotted line shows 1-fold activity with CBP (b), E1A (c) and E1A(ΔCBP) (d). Two sub-CBP-interaction domains are labelled I and II. These data (a–d) are the average of three experiments (means±S.D.; n=3).
Exogenous CBP overexpression enhanced further the effect of all tested GRIP1 fragments, except for ER itself, full-length GRIP1 and the fragment derived from amino acids 563–1012, on ER function (Figure 7b, compare histograms 1, 2 and 9). However, inhibition by exogenous E1A overexpression worked primarily on the GRIP1 AD1 region (sites I and II; Figure 7c, compare histograms 2 and 5). The application of E1A(ΔCBP) to the ER system suggests that the inhibition by E1A on ER function is not only mediated through CBP, but is also mediated through other unknown factors in cells, because this E1A mutant still repressed the co-activator functions of GRIP1 fragments with an intact AD1 region (Figure 7d, compare histograms 2 and 5).
The specific CBP-interacting site preference for GRIP1 co-activator functions is dependent on the NR and on the cell type
The precise assembly of co-activators and components of co-activator complexes required for physiological NR functions are not fully understood. Since the functions of components of coactivator complexes appear to be distinct, it is likely that transcription factor-specific differences exist in their association with factors within cells [48]. Hence we checked the preference of these two CBP-interacting sites for GRIP1 co-activator functions on GR, PR and TR in HeLa, HEK-293 and CV-1 cells (Table 1). In HeLa cells, CBP-interacting site I was important for full-length GRIP1 co-activator function in the GR, PR and TR systems [Table 1, compare the data for full-length GRIP1 (amino acids 5–1462) with different statuses for sites I and II in HeLa cells]. A small region (amino acids 563–1012) without these two CBP-interacting sites retained co-activator function in the GR system in HeLa cells. GRIP1 expressed AD1- and AD2-dependent co-activator functions with the GR and PR in HeLa cells. In HEK-293 cells, CBP-interacting site I and AD2 of GRIP1 were necessary for GRIP1 co-activator function in the PR system [Table 1; for the importance of site I, compare the data for full-length GRIP1 (amino acids 5–1462) with different statuses for sites I and II or the difference between amino acids 563–1094 and 563–1012; for the importance of AD2, compare the data for wild-type GRIP1 (amino acids 5–1462 and 563–1121) in HEK-293 cells]. CBP-interacting site I was the sole determinant for GRIP1 co-activator function in the TR system in HEK-293 cells (Table 1, compare GRIP1 fragments with those that were internally deleted in site I). No CBP-interacting site was essential for GRIP1 co-activator function in the GR system in CV-1 cells (Table 1, compare amino acids 5–1462 and 563–1012). In CV-1 cells, the PR function was dependent on GRIP1 AD1 and AD2, whereas the fragment comprising amino acids 563–1012 without these two CBP-interacting sites retained 67% of the activity of the full-length GRIP1 co-activator (Table 1). Similarly to the functional role of GRIP1 in the TR system in HEK-293 cells, CBP-interacting site I was essential for GRIP1 co-activator function in CV-1 cells (Table 1; compare data for GRIP1 fragments with that of the fragment internally deleted in site I).
Table 1. Various functional roles of CBP-interacting sites I and II of GRIP1 with GR, PR and TR in HeLa, HEK-293 and CV-1 cell lines.
HeLa, HEK-293 and CV-1 cells were transfected with plasmids encoding NR (0.1 μg, except for 0.04 μg of TR in HeLa cells) and the GRIP1 fragment indicated (0.35 μg), which also contained the LUC reporter gene (0.25 μg), as shown. Transfected cultures were grown with the appropriate hormone (100 nM dexamethasone for GR; 10 nM progesterone for PR; 100 nM 3,3′,5-tri-iodothyronine for TR). LUC fold activation is expressed relative to that of samples containing NR, but no GRIP1. Similar activity profiles were observed in three independent experiments.
| Fold activation | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CBP-interacting site status | GR+MMTV-LUC | PR+MMTV-LUC | TR+MMTV(TRE)-LUC | ||||||||
| GRIP1 | I | II | HeLa | HEK-293 | CV-1 | HeLa | HEK-293 | CV-1 | HeLa* | HEK-293 | CV-1 |
| Vector | − | − | 1±0.2 | 1±0.1 | 1±0.2 | 1±0.2 | 1±0.1 | 1±0.1 | 1±0.1 | 1±0.2 | 1±0.01 |
| 5–1462 | + | + | 10±1 | 1±0.1 | 32±1 | 5±0.6 | 19±2 | 12±0.2 | 17±2 | 5±0.3 | 5±0.7 |
| 5–1462 | − | + | 6±0.7 | 1±0.1 | 18±1 | 2±0.5 | 5±0.4 | 3±0.3 | 2±0.3 | 0.5±0.05 | 0.5±0.02 |
| 5–1462 | + | − | 13±1 | 1±0.1 | 24±0.2 | 5±0.5 | 29±0.2 | 13±1 | 44±8 | 6±0.6 | 3±0.7 |
| 563–1121 | + | + | 7±1 | 0.6±0.01 | 48±5 | 2±0.1 | 6±0.1 | 7±0.3 | 12±1 | 7±0.6 | 13±0.5 |
| 563–1121 | − | + | 6±0.1 | 1±0.01 | 34±1 | 1±0.5 | 6±0.4 | 5±0.2 | 1±0.1 | 0.3±0.01 | 0.7±0.09 |
| 563–1121 | + | − | 7±0.7 | 0.8±0.01 | 31±0.6 | 2±0.7 | 5±0.3 | 6±0.9 | 6±0.1 | 6±0.3 | 4±0.2 |
| 563–1094 | + | − | 9±0.4 | 1±0.05 | 36±1 | 2±0.1 | 7±0.4 | 8±1 | 6±0.2 | 6±0.2 | 4±0.4 |
| 563–1012 | − | − | 7±0.2 | 1±0.02 | 31±6 | 2±0.1 | 1±0.1 | 8±1 | 2±0.3 | 0.6±0.05 | 1±0.2 |
* Fold values are calculated based on the relative data shown in Figure 5(a).
DISCUSSION
The functional role of sites I and II in the GRIP1 AD1 domain
Previous studies have suggested that GRIP1 AD1 activity appears to be mediated through CBP, because AD1 could not be separated from the CBP interaction domain by mutation [21,31]. However, these studies defined the AD1 domain to be either within amino acids 1010–1131, or a smaller fragment of amino acids 1057–1109 in GRIP1/TIF2. Our study has further identified two distinct CBP-interacting sites located in amino acids 1057–1109: amino acids 1075–1083 (site I) and 1095–1106 (site II). Following the deletion of site I, the ability to interact with CBP and the AD1 transactivation activity was lost, whereas following the deletion of site II, the fragment primarily lost the ability to interact with CBP, but retained part or all of the AD1 activity (Figures 2 and 3). Hence our findings reveal that AD1 transactivation activity of GRIP1 may or may not be mediated through CBP; that is, site I expresses CBP-dependent, and site II has CBP-independent, AD1 activity. However, the real mechanism for site II remains to be elucidated.
The functional roles of sites I and II depend on the NR and cell type
The definition of GRIP1 as a primary NR co-activator is based on its ability to interact directly with all NRs through its LXXLL motif [15–19]. Many papers currently suggest that GRIP1 AD1 and AD2 synergistically enhance NR functions through CBP/p300 and CARM1 respectively [21,32]. CBP/p300 and CARM1 may act synergistically in vitro and in vivo on GRIP1's NR co-activation via their distinct protein-modifying enzymic activities [32,37–39]. The question is which of these interactions is required for the co-activator function of GRIP1 towards NRs. In the present study, we have provided several lines of evidence to suggest that GRIP1 AD1 (major region through site II) has effects on the cross-talk between AD1 and AD2 functions in HeLa cells (Figures 5a and 6a). Previous studies suggested that the TR transcriptional system is primarily dependent on GRIP1 AD1 [21]. Hence the deletion of site I in GRIP1 totally removed its co-activator function in the TR system in HeLa cells (Figure 5a, compare histograms 1–3) or played dominant negative roles in HEK-293 and CV-1 cells (Table 1, shown by GRIP1 fragments with the fold activation value≤1). The deletion of site II in GRIP1, which led to the expression of a higher co-activator activity than for wild-type GRIP1 in the TR system, suggests that site II might play a negative role in AD1 activity in HeLa cells, but not in HEK-293 or CV-1 cells [Figure 5a (compare histograms 2 and 4) and Table 1], whereas we failed to observe a similar result when one smaller fragment (amino acids 563–1121, lacking the AD2 domain) was deleted in site II in HeLa cells (Figure 5a, compare histograms 5 and 7). A similar pattern of events was observed in the PR system in HEK-293 cells, but not in HeLa or CV-1 cells (Table 1). Similar to the findings of cell-specific AR co-activation and repression by a novel zinc-finger protein, Zac1 [49], these results suggest that GRIP1-specific differences exist in terms of its association with factors within three tested cell lines.
The enhancement of AR transcriptional activity by GRIP1 is primarily dependent on its AD2 region [21]. Hence, following the deletion of either site I or site II of GRIP1, it retained its NR co-activator function in our assays because of an intact AD2 region in these GRIP1 fragments in HeLa cells (Figure 6a, compare histograms 1–5). However, the deletion of site II still elicited a greater co-activator effect than that for wild-type GRIP1, whereas we also failed to observe a similar result for the smaller fragment (amino acids 563–1121) in HeLa and CV-1 (Figure 6a, compare histograms 5 and 7, and results not shown). Our NR co-activator function analysis reveals that site II in GRIP1 may serve as a domain to repress GRIP1 co-activator function in TR and AR systems in HeLa cells, and the repression mechanism may directly mediate the cross-talk between AD1 and AD2 domains or other unknown pathways. In the ER transcriptional system, both AD1 and AD2 are required for GRIP1's full enhancement because GRIP1563–1121 (lacking AD2) expressed a lower effect than full-length GRIP1 in HeLa, HEK-293 and CV-1 cells (Figure 7a, compare histograms 2 and 5, and results not shown). However, the functional role of site II in the ER system was different from that in the TR or AR system, and sites I and II were both required for GRIP1's co-activator function through its AD1 in the ER system (Figure 7a, compare histograms 2–7, and results not shown).
The introduction of exogenous CBP, E1A and E1A(ΔCBP) into GRIP1–NR complexes suggests that CBP might be an NR co-player, i.e. a primary or secondary NR co-activator, depending on the NR type (compare Figures 5b, 6b and 7b). However, some unknown factors, for example retinoblastoma or p/CAF, might also be important co-players in GRIP1–NR transcriptional complexes (compare Figures 5d and 7d). One GRIP1 fragment (amino acids 563–1012) has partial or full co-activator activity in the GR system in HeLa and CV-1 cells (Table 1), and has partial co-activator activity in the PR system in CV-1 cells (Table 1). The unknown factors (novel molecules or mechanisms) remain to be studied in the future.
In summary, our study further subdivides the AD1 domain into CBP-interacting sites I and II to provide a new definition of GRIP1 function domains: TR is dependent on the GRIP1 CBP-interacting site I; CBP-interacting site I is necessary for GRIP1 AD1 function in the PR system in HEK-293 cells, but not not CV-1 cells. Our study reveals the preferential role of the CBP-interacting site in GRIP1 for its co-activator functions in cells.
Acknowledgments
We thank Weijun Feng (University of California, San Francisco, CA, U.S.A.) for expression vectors and reporter genes for TR, Paul Webb and Peter J. Kushner (University of California) for expression vectors and reporter genes for ER, Albert O. Brinkmann (Erasmus University, Rotterdam, The Netherlands) for the AR expression vector, Dean P. Edwards (University of Colorado Health Sciences Center, Denver, CO, U.S.A.) for the PR expression vector, Tony Kouzarides (University of Cambridge, U.K.) for E1A and E1A(ΔCBP) expression vectors, and Tso-Pang Yao (Duke University, Durtham, NC, U.S.A.) for the pCDNA3b.Flag. CBP.HA expression vector. This work was supported by grants from the National Science Council and National Health Research Institute, Taiwan, Republic of China (NSC 92-2320-B-016-009 and NHRI-EX93-9224NC to S.-M.H.).
References
- 1.Moras D., Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr. Opin. Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 2.Lemon B. D., Freedman L. P. Nuclear receptor cofactors as chromatin remodelers. Curr. Opin. Genet. Dev. 1999;9:499–504. doi: 10.1016/s0959-437x(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 3.McKenna N. J., Lanz R. B., O'Malley B. W. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 4.Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 5.Swope D. L., Mueller C. L., Chrivia J. C. CREB-binding protein activates transcription through multiple domains. J. Biol. Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 6.Blanco J. C., Minucci S., Lu J., Yang X. J., Walker K. K., Chen H., Evans R. M., Nakatani Y., Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oñate S. A., Tsai S. Y., Tsai M. J., O'Malley B. W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 8.Hong H., Kohli K., Trivedi A., Johnson D. L., Stallcup M. R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4948–49452. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voegel J. J., Heine M. J., Zechel C., Chambon P., Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 10.Chen H., Lin R. J., Schiltz R. L., Chakravarti D., Nash A., Nagy L., Privalsky M. L., Nakatani Y., Evans R. M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 11.Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 12.Li H., Gomes P. J., Chen J. D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeshita A., Cardona G. R., Koibuchi N., Suen C. S., Chin W. W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J. Biol. Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 14.Torchia J., Rose D. W., Inostroza J., Kamei Y., Westin S., Glass C. K., Rosenfeld M. G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature (London) 1997;387:677–684. [Google Scholar]
- 15.Heery D. M., Kalkhoven E., Hoare S., Parker M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 16.Darimont B. D., Wagner R. L., Apriletti J. W., Stallcup M. R., Kushner P. J., Baxter J. D., Fletterick R. J., Yamamoto K. R. Structure and specificity of nuclear receptor–coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding X. F., Anderson C. M., Ma H., Hong H., Uht R. M., Kushner P. J., Stallcup M. R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol. Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 18.Feng W., Ribeiro R. C., Wagner R. L., Nguyen H., Apriletti J. W., Fletterick R. J., Baxter J. D., Kushner P. J., West B. L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 19.McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bevan C. L., Hoare S., Claessens F., Heery D. M., Parker M. G. The AF1 and AF2 domains of the androgen receptor interact with distinct regions of SRC1. Mol. Cell. Biol. 1999;19:8383–8392. doi: 10.1128/mcb.19.12.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma H., Hong H., Huang S. M., Irvine R. A., Webb P., Kushner P. J., Coetzee G. A., Stallcup M. R. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol. Cell. Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bannister A. J., Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 23.Trouche D., Cook A., Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao T. P., Ku G., Zhou N., Scully R., Livingston D. M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurokawa R., Kalafus D., Ogliastro M. H., Kioussi C., Xu L., Torchia J., Rosenfeld M. G., Glass C. K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–803. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 26.Scoggin K. E., Ulloa A., Nyborg J. K. The oncoprotein Tax binds the SRC-1-interacting domain of CBP/p300 to mediate transcriptional activation. Mol. Cell. Biol. 2001;21:5520–5530. doi: 10.1128/MCB.21.16.5520-5530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livengood J. A., Scoggin K. E., Van Orden K., McBryant S. J., Edayathumangalam R. S., Laybourn P. J., Nyborg J. K. p53 Transcriptional activity is mediated through the SRC1-interacting domain of CBP/p300. J. Biol. Chem. 2002;277:9054–9061. doi: 10.1074/jbc.M108870200. [DOI] [PubMed] [Google Scholar]
- 28.Chen H., Lin R. J., Xie W., Wilpitz D., Evans R. M. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 29.Goodman R. H., Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 30.Chan H. M., La Thangue N. B. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 31.Voegel J. J., Heine M. J., Tini M., Vivat V., Chambon P., Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 33.Sheppard H. M., Harries J. C., Hussain S., Bevan C., Heery D. M. Analysis of the steroid receptor coactivator 1 (SRC1)-CREB binding protein interaction interface and its importance for the function of SRC1. Mol. Cell. Biol. 2001;21:39–50. doi: 10.1128/MCB.21.1.39-50.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demarest S. J., Martinez-Yamout M., Chung J., Chen H., Xu W., Dyson H. J., Evans R. M., Wright P. E. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature (London) 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 35.Demarest S. J., Deechongkit S., Dyson H. J., Evans R. M., Wright P. E. Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB-binding protein. Protein Sci. 2004;13:203–210. doi: 10.1110/ps.03366504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H., Chen J. D. The receptor-associated coactivator 3 activates transcription through CREB-binding protein recruitment and autoregulation. J. Biol. Chem. 1998;273:5948–5954. doi: 10.1074/jbc.273.10.5948. [DOI] [PubMed] [Google Scholar]
- 37.Chen D., Huang S. M., Stallcup M. R. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 2000;275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 38.Chevillard-Briet M., Trouche D., Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daujat S., Bauer U. M., Shah V., Turner B., Berger S., Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr. Biol. 2002;12:2090–2097. doi: 10.1016/s0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- 40.Bannister A. J., Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umesono K., Evans R. M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 42.Paech K., Webb P., Kuiper G. G., Nilsson S., Gustafsson J., Kushner P. J., Scanlan T. S. Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- 43.Brinkmann A. O., Faber P. W., van Rooij H. C., Kuiper G. G., Ris C., Klaassen P., van der Korput J. A., Voorhorst M. M., van Laar J. H., Mulder E., et al. The human androgen receptor: domain structure, genomic organization and regulation of expression. J. Steroid Biochem. 1989;34:307–310. doi: 10.1016/0022-4731(89)90098-8. [DOI] [PubMed] [Google Scholar]
- 44.Green S., Issemann I., Sheer E. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milhon J., Kohli K., Stallcup M. R. Genetic analysis of the N-terminal end of the glucocorticoid receptor hormone binding domain. J. Steroid Biochem. Mol. Biol. 1994;51:11–19. doi: 10.1016/0960-0760(94)90110-4. [DOI] [PubMed] [Google Scholar]
- 46.Boonyaratanakornkit V., Melvin V., Prendergast P., Altmann M., Ronfani L., Bianchi M. E., Taraseviciene L., Nordeen S. K., Allegretto E. A., Edwards D. P. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol. 1998;18:4471–4487. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eckner R., Ewen M. E., Newsome D., Gerdes M., DeCaprio J. A., Lawrence J. B., Livingston D. M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 48.Xu J., Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol. Endocrinol. 2003;17:1681–1692. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- 49.Huang S. M., Stallcup M. R. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol. Cell. Biol. 2000;20:1855–1867. doi: 10.1128/mcb.20.5.1855-1867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]