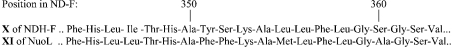Abstract
We have investigated the topologies of Ndh (a plastid complex with NADH dehydrogenase activity) and its NDH-F subunit in thylakoids by trypsin and proteinase V8 digestion of both intact and Triton X-100-permeabilized barley thylakoids and identification of the products with antibodies against specific sequences of the NDH-A, NDH-K and NDH-F subunits. Antibody binding and protection against proteinases were also assayed. The analysis of the digestion products of NDH-F by immunodetection and matrix-assisted laser-desorption ionization–time-of-flight allowed us to propose its membrane topology and to compare it with bioinformatic predictions and with that of the homologous subunit (ND5/NuoL/NQO12) of the respiratory complex I. Results indicate that the thylakoid Ndh complex may have an L-shaped structure, similar to that of respiratory complex I, with the hydrophilic arm orientated towards the stroma and the hydrophobic arm inserted into the thylakoid. NDH-A and NDH-K may be located at the bridge between the two arms. Similar to ND5/NuoL/NQO12 of complex I, NDH-F must be distally located in the hydrophobic arm. NDH-F would include up to 15 transmembrane helices and 14 hydrophilic regions. A conserved His-349 in the X transmembrane helix could be involved in H+ pumping. The conserved Thr-181 NDH-F, whose probable phosphorylation increases the activity of the Ndh complex, is located within the hydrophilic region between the V and VI transmembrane helices.
Keywords: chloroplast, chlororespiration, Ndh complex, NDH-F, topology
Abbreviations: Chl, chlorophyll; MALDI–TOF, matrix-assisted laser-desorption ionization–time-of-flight; Ndh, a plastid complex with NADH dehydrogenase activity
INTRODUCTION
Ndh (a plastid complex with NADH dehydrogenase activity), analogous to the NADH dehydrogenase or complex I (EC 1.6.5.3) of the mitochondrial respiratory chain, which catalyses the transfer of electrons from NADH to plastoquinone, has been purified from the pea [1] and barley [2]. A total of 11 NDH polypeptides of the Ndh complex, located in stromal thylakoids [3–5], are encoded by the respective ndh genes of plastid DNA [6]. The Ndh complex (providing electrons) together with thylakoid plastoquinol peroxidase [7], the Mehler reaction and superoxide dismutase (draining electrons) might poise the redox level of photosynthetic electron carriers. This poising mechanism (chlororespiration) would optimize cyclic photosynthetic electron transport under a variety of environmental conditions while scavenging the reactive oxygen species generated under continuous photo-oxidative stress or by the successive sunflecks and light gaps [2]. Accordingly, the amounts of NDH polypeptides and NADH dehydrogenase activity of the Ndh complex increase under photo-oxidative stress [2,8–10] and ndh mutants show higher sensitivity to photo-oxidative stress [11,12]. The increase in plastid-encoded NDH polypeptides under photo-oxidative stress is mediated by H2O2 [13]. The activity of the Ndh complex also increases through an H2O2-stimulated phosphorylation of the Thr-181 of the NDH-F subunit [14].
The molecular basis of Ndh complex activation needs to be investigated to elucidate key aspects of this enzymic complex, such as its transmembrane orientation, subunit composition, the presence and identity of nuclear-encoded polypeptides, proton-translocating properties and accessibility of subunits to modifying enzyme(s). In this respect, comparison with the better known mitochondrial and eubacterial complex I [15–17] is probably a useful tool. Complex I has an L-shaped structure consisting of one hydrophilic arm and one hydrophobic arm.
We have investigated the topology of the Ndh complex in the thylakoid membranes of barley, especially of the NDH-F polypeptide (homologous with mitochondrial ND5 and bacterial NuoL or NQO12 subunits of the respiratory complex I). We prepared antibodies against specific sequences of NDH-A, NDH-F and NDH-K subunits of the Ndh complex and performed assays of proteolytic cleavage, antibody binding and antibody protection against trypsin digestion with intact and permeabilized thylakoids. Detailed assays with the NDH-F polypeptide allowed us to propose its transmembrane orientation in thylakoids.
EXPERIMENTAL
Plant materials and thylakoid isolation
Barley (Hordeum vulgare cv Hassan) was grown on vermiculite under controlled conditions at 23±1 °C and a 16 h photoperiod of 80 μmol of photons·m−2·s−1 white light. Thylakoid membranes were isolated from 14 day-old primary leaves incubated for 20 h under 300 μmol of photons·m−2·s−1, as described in [2].
Preparation of the immunoaffinity matrix and immunopurification of the Ndh complex
Monospecific NDH-F antibody was produced by Sigma-GenoSys (Cambridge, U.K.) using a synthetic peptide as antigen, which was established by the protein sequence analysis of barley NDH-F polypeptide (DDBJ/EMBL/GenBank® accession number U22003). The amino acid sequence of the antigen peptide was WSKDEILSNSWLYS and corresponds to amino acids 415–428 of the NDH-F protein. NDH-F antibody was bound to a Protein A–Sepharose CL-4B (Sigma, St. Louis, MO, U.S.A.) matrix and then cross-linked with dimethyl pimelimidate [18].
The thylakoid membranes were solubilized with Triton X-100 using a Chl (chlorophyll)/detergent ratio of 1:20 (w/w) that solubilized the thylakoid lamellae and the Ndh complex [19]. The immunoaffinity matrix, previously equilibrated with 50 mM Tris/HCl (pH 8.3), 150 mM NaCl, 1 mM EDTA and 0.5% Triton X-100, was incubated batchwise with solubilized thylakoid samples at 4 °C for 1 h with gentle agitation. The matrix was pelleted by a microfuge short pulse, and washed twice with 10 vol. of 50 mM Tris/HCl (pH 8.3), 150 mM NaCl, 1 mM EDTA and 0.5% Triton X-100. The Ndh complex was eluted with 50 mM diethylamine (pH 11.5) containing 0.5% Triton X-100. The eluted samples were immediately neutralized with 1 M NaH2PO4.
Proteolysis of thylakoids and immunopurified Ndh complex
Freshly prepared thylakoids were treated with trypsin [360 units·(mg of Chl)−1] for the indicated time periods at 20 °C in the presence or absence of Triton X-100 [up to 8 mg·(mg of Chl)−1]. Immunopurified Ndh complex (0.5 μg) was incubated in 100 mM Tris/HCl (pH 7.5) and 10 units of trypsin for up to 16 min at 20 °C. In both cases, reactions were stopped by the addition of soya-bean trypsin inhibitor [75 ng·(μg of trypsin)−1] in SDS sample buffer [20] and then boiling for 10 min.
In other experiments, freshly isolated thylakoids were treated with proteinase V8 [0.2 unit·(mg of Chl)−1] at 30 °C in the presence or absence of Triton X-100 [1 mg·(mg of Chl)−1]. Reactions were stopped by the addition of 2 mM PMSF in SDS sample buffer and then boiling for 10 min.
Samples were subjected to continuous or gradient SDS/PAGE [20] or Tricine–SDS/PAGE [21], and transferred to PVDF membranes (Millipore). NDH-A and NDH-K polypeptides and their proteolytic fragments were immunodetected with antibodies raised against the synthetic peptides PFDLPEAEEELVAGY and DFDRYGLVPRSSPR, corresponding to amino acids 229–243 and 80–93 of the NDH-A (DDBJ/EMBL/GenBank® accession number AJ011848) and NDH-K (DDBJ/EMBL/GenBank® accession number AY243565) proteins respectively. NDH-F polypeptide and its proteolytic fragments were detected with the monospecific NDH-F antibody described above. D1 protein of photosystem II was immunodetected with a commercial antibody (AgriSera AB, Vannas, Sweden) raised against the C-terminal part of the protein. The ATPase α-subunit antibody was a gift from Dr A. Guéra (from our own laboratory). Phosphorylated polypeptides were immunodetected with mouse monoclonal antiphosphothreonine (Sigma). The different immunocomplexes were detected with the alkaline phosphatase Western-blot analysis system (Boehringer Mannheim, Barcelona, Spain).
MS analysis
Freshly isolated thylakoids were treated with proteinase V8 [0.2 unit·(mg of Chl)−1] at 30 °C for 60 min. Reactions were stopped with 2 mM PMSF and then centrifuged at 15000 g for 15 min. Water-soluble V8 degradation products coming from exposed regions of membranous proteins were collected in the super-natant and fractionated by reversed-phase HPLC on a Deltapak C18 column [300 Å(1 Å=0.1 nm), 5 μM; Waters, Milford, MA, U.S.A.] employing an HPLC HP1050 apparatus (Hewlett–Packard, Waldbron, Germany). Peptides were eluted with 0.08% trifluoroacetic acid in water for 20 min, followed by a 0–50% gradient of 80% (v/v) acetonitrile in water and 0.075% trifluoroacetic acid (solvent A) for 50 min and by a 50–80% gradient of solvent A for 20 min. Fractions were collected continuously every 2 min. Each fraction was dried down, resuspended in acetonitrile/water/trifluoroacetic acid (30:69.9:0.1, by vol.), and analysed by MS. The mass spectra were obtained automatically by MALDI–TOF (matrix-assisted laser-desorption ionization–time-of-flight) with a Bruker Reflex IV mass spectrometer (Bruker-Franzen Analytic, Bremen, Germany) in positive-reflection mode and were annotated automatically by employing the Auto-Xecute™ acquisition software. The mass spectrometer was previously calibrated with a protonated peptide standard mixture within the m/z range of 1000–4000. Reversed-phase chromatography and MALDI–TOF analysis were performed by the Proteomic Facility of the Centro Nacional de Biotecnología (CSIC, Madrid, Spain).
NDH-F antibody binding to the thylakoid membrane
Freshly isolated thylakoids, resuspended in a buffer containing 10 mM Tricine (pH 8.0), 5 mM MgCl2, 10 mM NaCl and 100 mM sorbitol (incubation medium) to a concentration of 0.5 μg of Chl·μl−1, were incubated with preimmune antiserum [100 μl·(mg of Chl)−1] for 20 min at 20 °C. Thereafter, thylakoids were washed twice with 10 mM Tricine (pH 8.0) and 140 mM NaCl (wash buffer), resuspended in incubation medium and incubated with anti-NDH-F (see above) for 20 min at 20 °C. After two washings with wash buffer and resuspension in incubation medium, thylakoids were incubated with goat anti-rabbit IgG conjugated with alkaline phosphatase (Sigma) for 20 min at 20 °C and then washed and resuspended in wash buffer and incubation medium respectively. The immunocomplexes were quantified by measuring alkaline phosphatase activity with p-nitrophenyl phosphate as described in [22]. In some cases, no NDH-F antibody was added (control of unspecific binding). In other cases, thylakoids were subjected to trypsin digestion [360 units·(mg of Chl)−1] for 20 min at 4 °C before adding the preimmune antiserum or after NDH-F antibody incubation.
Methods for the prediction of transmembrane topology
To predict potentially transmembrane-hydrophobic helices, we used the following informatic programs: PHDhtm [23], TMHMM [24], HMMTOP [25], TopPred [26], TMpred [27] and MEMSAT [28].
Other determinations
Total protein levels were determined as described in [29] using BSA as a standard. Chl content was measured as described in [30]. Editing of the 62nd nt (C) of barley mRNA was performed as described in [31] with the primers GGGTAATCCCTCTTCTCC and CACTCGCTGCAATTGGCCGT, spanning a 495 nt sequence starting from the 27th nt.
All experiments were repeated at least three times.
RESULTS
Cleavage of NDH-K and NDH-A by trypsin incubation
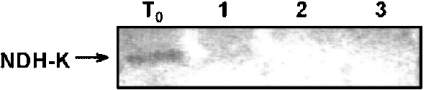
The orientation of NDH-K relative to the thylakoid membrane was studied by controlled trypsin digestion [360 units of trypsin·(mg of Chl)−1 for 20 min at 20 °C] of barley thylakoids. As shown in Figure 1, NDH-K was completely digested by trypsin in intact membrane vesicles. No tryptic peptide containing the selected antigenic sequence was observed, probably because of the large number of tryptic cleavage sites (more than 30 in a 248 amino acid sequence). The addition of either Triton X-100 or NaCl to the incubation medium did not alter the tryptic digestion of NDH-K, indicating that, within the whole Ndh complex, this polypeptide occupies an external position facing the stromal surface of thylakoids.
Figure 1. Tryptic degradation of NDH-K of the Ndh complex in barley thylakoids.
Freshly prepared thylakoids were incubated with trypsin [360 units·(mg Chl)−1] for 0 (T0) or 20 min at 20 °C in the absence (1) or in the presence of Triton X-100 [8 mg·(mg of Chl)−1] (2) or 140 mM NaCl (3). After SDS/PAGE (12% acrylamide) and blotting on to PVDF membranes, NDH-K polypeptide was revealed by a monospecific NDH-K antibody.
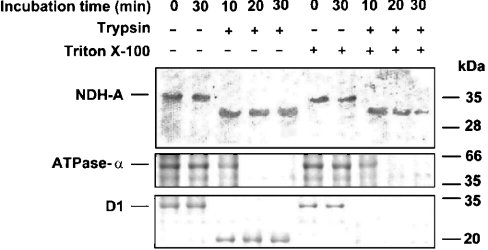
It should be pointed out that the procedure employed in the present study for isolating thylakoids renders mostly right-side-out vesicles. Moreover, even though some mechanical perturbations could produce a small amount of inside-out vesicles, these vesicles would originate from granal regions [19] that do not contain an Ndh complex [4]. In this context, Figure 2 shows controls of thylakoid intactness and orientation by digestion analysis of an ATP-ase α-subunit, which is a peripheral polypeptide (facing the stroma), and D1 protein, an integral polypeptide with five transmembrane helices. Tryptic degradation of the ATPase α-subunit was almost complete after 20 min incubation in either the presence or absence of Triton X-100, indicating that thylakoid preparations exposed all of the ATPase α-subunit, and hence the stromal surface, to the incubation medium. Moreover, D1 degradation in the absence of Triton X-100 was not complete, producing a polypeptide of approx. 20 kDa recognized by the D1 antibody. As stated above, this antibody was raised against the C-terminal part of D1, which is orientated towards the luminal surface of thylakoids and is therefore not exposed to trypsin in untreated membranes. When Triton X-100 (in subsolubilizing concentration) was added to the incubation medium, the degradation of D1 was complete. In summary, the control experiments demonstrated that (i) thylakoid vesicles were indeed sealed in right-side orientation, (ii) the luminal compartment was not accessible to proteinases and (iii) subsolubilizing levels of Triton X-100 rendered accessible the proteinase target sites of proteins orientated towards the lumen.
Figure 2. Effect of Triton X-100 on the tryptic degradation of NDH-A, ATPase α-subunit and D1 protein in barley thylakoids.
Freshly prepared thylakoids were incubated in the absence or presence of trypsin [360 units·(mg of Chl)−1] and Triton X-100 [4 mg·(mg of Chl)−1] for the indicated times at 20 °C. After SDS/PAGE (12% acrylamide) and blotting on to PVDF membranes, NDH-A, ATPase α-subunit, D1 and their tryptic derivatives were revealed by specific antibodies.
Controlled incubation [360 units of trypsin·(mg of Chl)−1, for up to 30 min at 20 °C] of intact thylakoids with trypsin (Figure 2) revealed the production of a stable 30.5 kDa peptide reacting with the NDH-A antibody. Evidently, this peptide was produced by trypsin cleavage of the 35 kDa NDH-A protein. Triton X-100 treatment enhanced the degradation of the 30.5 kDa peptide. These results suggest that NDH-A must have at least one trypsin cleavage site accessible from the stroma that produces the 30.5 kDa peptide and has additional trypsin sites in the luminal side of thylakoids. The NDH-A protein is homologous with the ND1/NuoH/NQO8 protein of respiratory complex I, and its amino acid sequence deduced from edited mRNA [31] suggests that it may include eight hydrophobic transmembrane helices. The results of NDH-A digestion with trypsin agree with the current model for its homologous protein in complex I [15], suggesting a limited exposure of NDH-A to the stromal side (probably by protection by the hydrophilic arm) and a wide exposure to the lumen, which allowed full digestion of the 30.5 kDa peptide in permeabilized thylakoids.
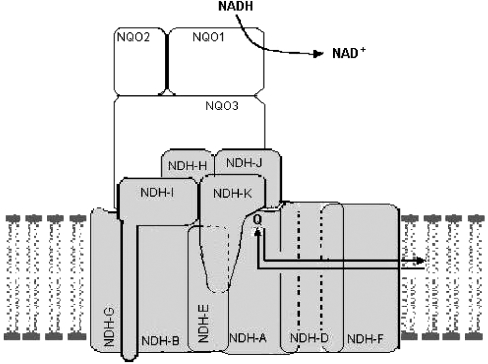
NDH-K and NDH-A trypsin-cleavage assays suggest that the thylakoid Ndh complex may have an L-shape structure, similar to respiratory complex I, with the hydrophilic arm orientated towards the stroma and the hydrophobic L-arm immersed in the thylakoid membrane. Based on current models proposed for respiratory complex I [32], Figure 3 shows the probable structural organization of the Ndh complex in thylakoid membranes. The conformation-prediction program PHD [23] applied to the amino acid sequence of NDH-A and NDH-K deduced from the corresponding barley genes predicts that NDH-A is a highly hydrophobic polypeptide containing eight transmembrane helices, whereas NDH-K contains a main hydrophilic core and two transmembrane helices. These predictions agree with our results and with the model shown in Figure 3, where NDH-K is a peripheral polypeptide of the Ndh complex anchored to the membrane (probably by the two hydrophobic helices). NDH-A is an integral membrane polypeptide, with some hydrophilic regions orientated towards the lumen and others towards the stroma. The latter may be partially protected from trypsin cleavages by components of the hydrophilic arm of the complex, among them NDH-K.
Figure 3. Schematic structural organization of the Ndh complex in thylakoid membranes, including chloroplast-encoded NDH-A/NDH-K subunits and the putative nuclear-encoded subunits homologous with NQO1, NQO2 and NQO3 of bacterial complex I.
Models for transmembrane topology of NDH-F
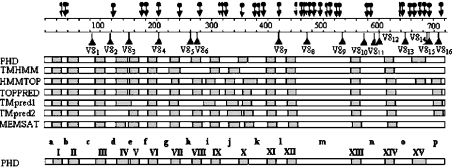
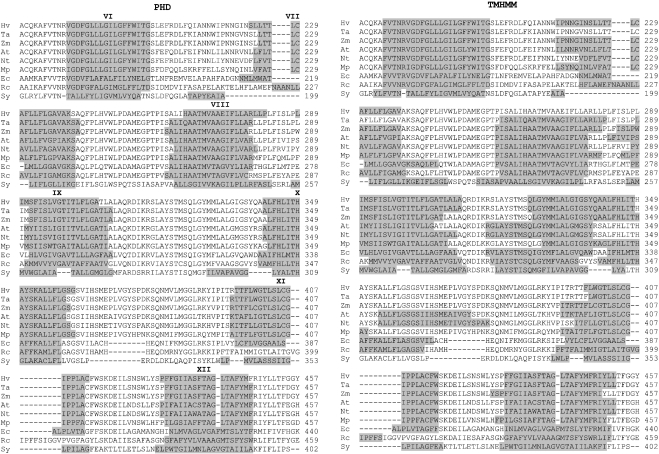
According to the model depicted in Figure 3, NDH-F (homologous with respiratory ND5/NQO12/NuoL protein) is an integral membrane protein connecting the distal polypeptides of the hydrophobic arm to the bridge between the two arms. To investigate the topology of NDH-F in thylakoids, the conformation prediction programs PHD, TMHMM, HMMTOP, TopPred, TMpred1, TMpred2 and MEMSAT were applied to the amino acid sequence of NDH-F deduced from the barley gene sequence, taking into account that the 62nd C is edited to U, therefore encoding Leu as the 21st amino acid (results not shown), as found in maize ndhF [6]. NDH-F is a highly hydrophobic protein for which the different programs predict between 13 and 17 potential transmembrane hydrophobic helices (Figure 4). All the seven programs predict nine consensus hydrophobic helices, which are, using the nomenclature described in Figure 4 for the PHD model: I, II, III, VI, VII, XI, XII, XIII and XIV. The main discrepancies among the seven predictions are between helices VII and XII. Since membrane topology must be similar in chloroplastic and respiratory NDH-F/NuoL/ND5 polypeptides from different organisms, we compared the transmembrane helix predictions of the different programs when applied to the NDH-F of six representative plants and NuoL of Escherichia coli, Rhodobacter capsulatus and Synechocystis. Figure 5 shows that, at least between the helices VI and XII represented, the PHD program predicted five equivalent transmembrane helices in NDH-F of the six plants tested and in NuoL of E. coli and Synechocystis. Only in NuoL of R. capsulatus, the PHD program did not predict helix XI. In contrast, helix predictions by other programs varied significantly among the six plants, even when no significant sequence divergences could be observed (e.g. TMHMM, Figure 5). Therefore, in the following, we shall use the helix prediction by the PHD program as the main reference for contrast with antibody binding and proteolytic data of NDH-F.
Figure 4. Transmembrane helices of the deduced sequence of barley chloroplast NDH-F polypeptide predicted by different bioinformatic programs.
According to the PHD model, hydrophobic helices (grey boxes) are labelled with roman numerals and hydrophilic regions (white boxes) are labelled with lower-case letters. Potential sites for trypsin and proteinase V8 digestion (according to the PeptideSort program, http://www.es.embnet.org/cgi-bin/w2h/) are indicated by arrows and triangles respectively.
Figure 5. Transmembrane helices predicted by PHD and TMHMM programs for NDH-F/NuoL of different organisms.
Predicted helices (roman numerals) are shown in grey for the aligned sequences from 174 to 457 amino acids (according to the barley polypeptide) of NDH-F of barley (Hv), wheat (Ta), Zea mays (Zm), Arabidopsis (At), tobacco (Nt) and Marchantia polymorpha (Mp) and NuoL of E. coli (Ec), R. capsulatus (Rc) and Synechocystis (Sy).
Binding of the anti-NDH-F antibody to thylakoids
In a first approach to test the different models of NDH-F topology in thylakoids, we assayed the capacity of freshly prepared thylakoids to bind a monospecific antibody raised against the sequence WSKDEILSNSWLYS (l) corresponding to amino acids 415–428. This sequence is included within the hydrophilic region l in the nomenclature of the PHD program (Figure 4). The amount of bound NDH-F antibody was estimated by assaying alkaline phosphatase activity conjugated with a secondary antibody (goat anti-rabbit IgG). Table 1 shows that intact thylakoids specifically bind NDH-F antibody, as indicated by a higher alkaline phosphatase activity of the thylakoids incubated with anti-NDH-F [7.1 and 8.0 units·(mg of Chl)−1] compared with that of thylakoid either incubated with preimmune serum alone [3.8 units·(mg of Chl)−1] or after trypsin treatment [3.6 units·(mg of Chl)−1]. These results indicated that the l region of NDH-F lies on the stromal side of intact thylakoid membranes.
Table 1. NDH-F antibody binding to thylakoid membranes.
Freshly isolated thylakoids (0.5 μg of Chl·μl−1) were incubated with preimmune antiserum for 20 min at 20 °C, washed twice with 10 mM Tricine (pH 8.0) and 140 mM NaCl (wash buffer) and then incubated with monospecific anti-NDH-F. After two washings with wash buffer, thylakoids were incubated with goat anti-rabbit IgG conjugated with alkaline phosphatase (AP–anti-rabbit IgG) for 20 min at 20 °C and then washed twice with the wash buffer. The immunocomplexes were quantified by measuring alkaline phosphatase activity conjugated with the goat anti-rabbit IgG. Where indicated, no NDH-F antibody was added (treatment 2) or thylakoids were subjected to trypsin digestion [360 units·(mg of Chl)−1] for 20 min at 4 °C before preimmune antiserum (treatment 3) or after NDH-F antibody (treatment 4) incubation. One unit of alkaline phosphatase hydrolyses 1 μmol of p-nitrophenyl phosphate per min at 37 °C. Values in parentheses represent S.D., n=4.
| Treatment | Phosphatase activity [units·(mg of Chl)−1] |
|---|---|
| 1. Preimmune serum→anti-NDH-F→AP–anti-rabbit IgG | 7.1 (0.5) |
| 2. Preimmune serum→AP–anti-rabbit IgG | 3.8 (0.1) |
| 3. Trypsin→preimmune serum→anti-NDH-F→AP–anti-rabbit IgG | 3.6 (0.3) |
| 4. Preimmune serum→anti-NDH-F→trypsin→AP–anti-rabbit IgG | 8.0 (0.3) |
Analysis of V8-generated peptides from NDH-F
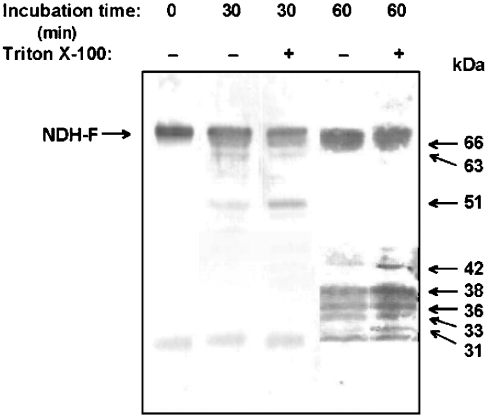
In this approach, we intend to establish the orientation of potential proteinase V8 target sites by analysing the peptides produced after incubation of intact and Triton X-100-permeabilized thylakoids with proteinase V8. Different Triton X-100 and proteinase V8 concentrations were assayed to select the partial digestion conditions yielding reproducible patterns of V8-generated peptides detectable with NDH-F antibody (raised against the l sequence). After 30 and 60 min incubations of intact and permeabilized thylakoids respectively with proteinase V8, digestion fragments were separated by SDS/PAGE and detected with NDH-F antibody, which would react only with peptides containing the antigenic l sequence. As Figure 6 shows, large-sized peptides of 51, 63 and 66 kDa were detected after 30 min incubations of intact thylakoids with proteinase V8. When incubations were increased to 60 min, most of these peptides and the original 70 kDa NDH-F were further degraded to produce mainly 33, 36 and 38 kDa peptides. With respect to intact thylakoids, 30 min incubations of permeabilized thylakoids with V8 produced an increase in the 51 kDa peptide in parallel to a decrease in the 63 kDa peptide. Incubations of Triton X-100-permeabilized thylakoids for 60 min with V8 increased the production of 33, 36 and 38 kDa peptides and of two peptides of 31 and 42 kDa, barely detected in 60 min-incubated intact thylakoids. An unspecific 30 kDa band was detected under all conditions and was not considered in the following peptide analysis. Although several potential V8 cleavage sites may not be accessible under the assay conditions, Triton X-100 treatment increases the accessibility of some cleavage sites, especially in the luminal side of thylakoids. Digestion peptides of 33, 36, 38, 51, 63 and 66 kDa must result from cleavages at V8 sites facing the stroma, whereas the production of 31 and 42 kDa peptides seems to require at least one additional cleavage at V8 sites facing the lumen.
Figure 6. Proteinase V8 fragments of NDH-F containing the WSKDEILSNSWLYS sequence from intact and detergent-treated thylakoids.
Freshly prepared thylakoids were incubated with proteinase V8 [0.2 unit·(mg of Chl)−1] for the indicated times at 30 °C in the absence or presence of Triton X-100 [1 mg·(mg of Chl)−1]. After SDS/PAGE (10–16% acrylamide) and blotting on to PVDF membranes, NDH-F and its V8 derivatives were revealed by a monospecific NDH-F antibody. The approximate molecular mass of V8 peptides is indicated on the right.
Theoretically, the size and identity of the peptides produced by proteinases could be determined by MS. However, when proteolytic peptides derive from a large number of proteins, like those of thylakoids, complex separation methods of highly hydrophobic peptides followed by MS identification of a large number of peptides are required to find among them those derived from NDH-F polypeptide. We could only use MS to identify small hydrophilic peptides produced when intact thylakoids were incubated with proteinase V8. Alternatively, the conventional approach used in the present study focuses on peptides carrying specific antibody-binding sites, even though their sizes can be determined only with limited accuracy by SDS/PAGE. However, the fact that NDH-F contains only 16 potential V8 digestion sites (numbered V81 to V816 in Figure 4) allows a tentative identification of the 33, 36 and 38 kDa l-bearing peptides shown in Figure 6 and, therefore, of some of the V8 sites exposed to the stromal side of thylakoids. It should be noted that proteinase V8 cleaves at the CO2H of glutamyl residues; however, peptide bonds involving prolyl residues are commonly not cleaved by the proteinase [33], thus excluding the Glu-368 (Figure 5) as a potential V8 digestion site. The antigenic l sequence (from amino acids 415–428) includes the V87 site (position 420), whose cleavage presumably produces peptides not detectable with the NDH-F antibody, as suggested by the lack of specific antibody binding to thylakoids treated with trypsin (which also cleaves within the antigenic l sequence). Peptides of 33, 36 and 38 kDa cannot be produced by single cleavages of NDH-F since the V86 (position 273) and V88 (position 470) sites are too far from, respectively, the C- and N-termini of NDH-F to produce such small mono-cleavage l-bearing peptides. Therefore only peptides resulting from pairs of cleavages (one at the left and the other at the right of the l sequence) could have the observed sizes. Table 2 shows the predicted number of amino acids of the peptides resulting from candidate combinations of one cleavage on the left (sites V83, V84, V85 and V86), with one cleavage on the right (sites V88, V89, V810, V811, V812 and V813) of the l sequence.
Table 2. Length (number of amino acids) of potential small-sized V8 peptides containing the l antigenic region.
The length of peptides that would be produced by cleavage at each pair of the indicated V8 sites is expressed as the number of amino acids. The position of each cleavage site is indicated within parentheses.
| No. of amino acids | ||||||
|---|---|---|---|---|---|---|
| From↓To→ | V88 (470) | V89 (536) | V810 (573) | V811 (591) | V812 (600) | V813 (647) |
| V83 (152) | 318 | 384 | 421 | 439 | 448 | 495 |
| V84 (203) | 267 | 333 | 370 | 388 | 397 | 444 |
| V85 (260) | 210 | 276 | 313 | 331 | 340 | 387 |
| V86 (273) | 197 | 263 | 300 | 318 | 327 | 374 |
Sites V810, V811 and V812 are within the same hydrophilic n region, flanked by hydrophobic transmembrane helices XIII and XIV (nomenclature referred to PHD model, Figure 4). In fact, all models predict the same four hydrophobic transmembrane helices (XI, XII, XIII and XIV) from approx. 400–650 amino acids of NDH-F (Figure 4). Similarly, sites V88 and V89 (in the hydrophilic m region) and V813 (in the hydrophilic o region) are orientated towards the same thylakoid side, which is opposite to the side faced by n. Therefore the 33, 36 and 38 kDa peptides may result from cleavages involving sites V88, V89 and V813 or V810, V811 and V812 (at the right of the l sequence), but not from any combination of the two groups of sites. Predicted peptide sizes (Table 2) strongly suggest that no combination of cleavages at sites V83, V84, V85 and V86 with cleavages at sites V88, V89 and V813 would produce the 33–38 kDa peptides. Cleavage pairs that could produce the 33, 36 and 38 kDa peptides (involving the minimum number of cleavage sites) are, respectively: V85/V810, V85/V811 and V85/V812 (peptides of 313, 331 and 340 amino acids respectively, Table 2). It must be noted that the expected number of peptides is the product of the number of cleavages on the left and the number of cleavages on the right of l. If more than one cleavage site is accessible on the left and right of l (e.g. V83 and V84 on the left, and V810 and V813 on the right of l), more than three peptide products would be detected. Therefore the three peptides of 33, 36 and 38 kDa must result from one cleavage at the left site and three cleavages at the right site of l or vice versa. From the results in Table 2, it seems obvious that any combination of three cleavages at V83 to V86 left sites with one cleavage on the right of l can hardly originate the three observed peptides. Sites V81, V82, V814, V815 and V816, not included in Table 2, are clearly outside the range of possibilities involved in the production of the 33–38 kDa polypeptides.
Predicted sizes of V85/V810, V85/V811 and V85/V812 peptides (34.5, 36.6 and 37.6 kDa respectively) are in good agreement with the observed migration of V8 products in SDS/PAGE: 33, 36 and 38 kDa. In fact, these polypeptides did not shift to higher or lower apparent molecular mass when subjected to urea-SDS/PAGE or by extended denaturation with SDS (up to 4%) before conventional SDS/PAGE. This absence of large hydrophobicity-related effects on peptide electrophoretic mobility could be due to the influence of the long (more than 90 amino acids) hydrophilic m sequence, probably held by all l-bearing peptides. In support of this contention, binding assays of NDH-F antibody (Table 1) showed that an antigenic l sequence faces the stromal side, which would be followed by the hydrophobic transmembrane helix XII and the m sequence, as predicted by all the models (Figure 4) in all organisms (Figure 5). Therefore the m sequence would be orientated towards the luminal side and all l-bearing peptides originated by proteinase attack to intact thylakoids must also bear the hydrophilic m sequence.
The deduction that V85/V810, V85/V811 and V85/V812 cleavages produced the 33, 36 and 38 kDa peptides recognized by anti-NDH-F was further confirmed by MALDI–TOF identification of small stromal-sided peptides produced by proteinase V8. These proposed cleavages predict that the V810/V811 (17 amino acids, 2.00509 kDa) and V811/V812 peptides (10 amino acids, 1.13247 kDa) must also be produced. In fact, when soluble digestion products were analysed by MALDI–TOF, two low-abundance but clearly distinguishable peptides of 2.0051 and 1.1325 kDa respectively were observed (Supplementary Figures 1 and 2 available as online data at http://www.BiochemJ.org/bj/382/bj3820145add.htm).
In summary, results from V8 digestion experiments probably match with the conclusion reached from binding assays of NDH-F antibody (Table 1) and, in agreement with the hydrophobic helices predicted by all models in the region from amino acid 400 to 650 (Figure 4), indicate that the antigenic l sequence is on the stromal side, sites V85, V810, V811 and V812 are orientated towards the stroma and sites V88, V89 and V813 are orientated towards the lumen. Since both the site V85 and the l sequence face the same stromal side, there must be an even number of hydrophobic transmembrane helices between V85 and l. This agrees with the PHD and TMpred1 models (four helices, although in different positions) but not with TMHMM, HMMTOP, TopPred, TMpred2 or MEMSAT (3, 5, 5, 5 and 5 helices respectively). According to the PHD consensus for the NDH-F of six plants, and assuming four hydrophobic transmembrane helices between the site V85 and the l sequence, the results suggest a precise topology for a large part of the NDH-F polypeptide in thylakoid membranes. Using the sequence region nomenclature indicated for the PHD model (Figure 4), hydrophilic f, h, j, l and n regions would be orientated towards the stroma and hydrophilic g, i, k, m and o regions towards the lumen. They span approximately from amino acid 170 to 650 and include nine transmembrane helices (VI–XIV of the PHD model, Figure 4). In agreement with this tentative orientation of NDH-F, the Thr-181 of the f region, which would face towards the stroma, has recently been proposed to be reversibly phosphorylated [14]. An alternative luminal orientation of the f region would hardly be compatible with a reversible phosphorylation.
When thylakoids were treated with Triton X-100, new V8 sites became accessible and several combinations of V8 sites could account for the peptides produced specifically from permeabilized thylakoids (Figure 6). The 31 kDa peptide could be produced by cleavages at sites V85 and V89, whereas the 42 kDa peptide (amino acids 370–390) could be produced by several pairs of cleavages (Table 2). The topology of f to o regions proposed above predicts that at least one cleavage of each of these pairs faces the lumen, which agrees with the idea that the 31 and 42 kDa peptides are produced only in Triton X-100-treated thylakoids.
The mature NDH-F polypeptide migrates in SDS/PAGE (Figure 6) with an apparent mass of 70 kDa [9], which (even considering a faster electrophoretic migration linked to its hydrophobic properties) suggests some processing of the primary translation product that must contain 713 amino acids. Uncertainties about the N- and C-termini of the mature NDH-F polypeptide open several possibilities for the origin of the 66, 63 and 51 kDa peptides that make it difficult to select among the different model predictions for the topology of regions upstream of position 170 and downstream position 650.
Tryptic peptide analysis of NDH-F
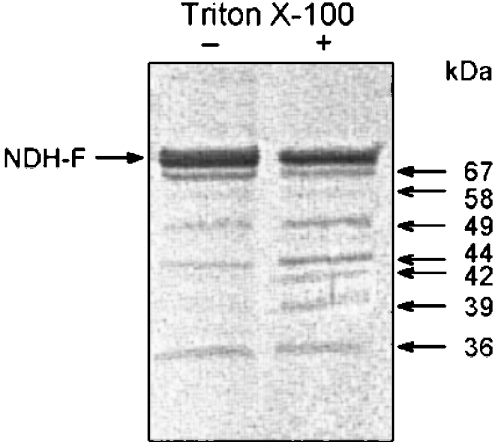
Controlled trypsin digestion of intact and detergent-treated thylakoids confirmed the topology proposed for the region from amino acid 170 to 650 and provided new insights. We assayed different Triton X-100 concentrations and trypsin incubation temperatures to select partial digestion conditions yielding reproducible patterns of tryptic peptides detectable with the NDH-F antibody after Tricine–SDS/PAGE and transfer to PVDF membranes. As expected, treatment of thylakoids with Triton X-100 over the range of 1–4 mg·(mg of Chl)−1 accelerated the digestion of NDH-F polypeptide within the assayed temperature range (results not shown), probably because several trypsin cleavage sites on the luminal side became accessible. Standard digestion conditions were finally established at 360 units of trypsin·(mg of Chl)−1 and 0 or 1 mg of Triton X-100·(mg of Chl)−1 for 20 min at 20 °C.
NDH-F contains many potential tryptic sites closely grouped within different regions (Figure 4). In the absence of Triton X-100, trypsin digestion produced four main peptides of 36, 44, 49 and 67 kDa and a less abundant peptide of 58 kDa (Figure 7) containing the l sequence detectable with NDH-F antibody. Despite uncertainties about the N- and C-termini of mature NDH-F polypeptide, the 67 kDa digestion peptide (which must contain from 600 to 630 amino acids) could be produced only by trypsin cleavage at positions downstream of amino acid 600 (obviously facing the stroma) combined with either a cleavage at the b region or the previous processing of the N-terminus. It must be noted that a single cleavage at a trypsin site within the d region (Lys-123) would hardly produce the 67 kDa peptide. V8 peptide analysis indicated the presence of a luminal region (o region) downstream from the hydrophobic helix XIV; however, the 67 kDa tryptic peptide indicated a stromal region downstream of helix XIV. The two conclusions are compatible if there is an additional hydrophobic helix downstream of the helix XIV as all models predict, with the exception of TMHMM and MEMSAT. Following the PHD model sequence nomenclature (Figure 4), we designated this transmembrane helix as XV and p, corresponding to the C-terminal hydrophilic stromal region of NDH-F. The production of the 66 kDa peptide requires at least cleavage at a trypsin site (e.g. Arg-679 or Lys-690) within or close to the p region, which the PHD model predicts for amino acids from 684 to 713 at the C-terminus.
Figure 7. Tryptic fragments of NDH-F containing the WSKDEILSNSWLYS sequence from intact and detergent-treated thylakoids.
Freshly prepared thylakoids were incubated with trypsin [360 units·(mg of Chl)−1] for 20 min at 20 °C in the absence or presence of Triton X-100 [1 mg·(mg of Chl)−1]. After Tricine-SDS/PAGE (10% acrylamide) and blotting on to PVDF membranes, NDH-F and its tryptic derivatives were revealed by a monospecific NDH-F antibody. The approximate molecular mass of tryptic peptides is indicated on the right.
The 36 kDa peptide is probably produced by trypsin cleavages at the stromal h and n regions. On the other hand, the 44, 49 and 58 kDa peptides may be produced in intact thylakoids through combinations of several trypsin cleavage sites, and therefore a selection among the different models for the topology of NDH-F between the N-terminus and the f region is not possible. Similarly, a large number of different luminal cleavages may explain the additional 39 and 42 kDa peptides produced in detergent-treated thylakoids.
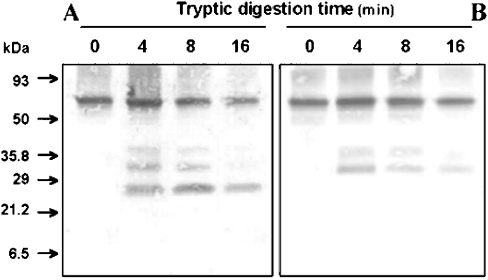
As described previously [14], phosphorylation, probably at the Thr-181 of NDH-F, regulates the activity of the Ndh complex and phosphorylated NDH-F can be detected with phospho-Thr antibody. To obtain additional information on the identity of tryptic peptides, they were tested with NDH-F and phospho-Thr antibodies. To avoid the interference of other thylakoid phospho-proteins [34] in Western-blot assays, in this approach, tryptic digestion was performed with purified Ndh complex where both stromal- and luminal-sided tryptic sites may be accessible. In the present case, the results are independent of the integrity of the purified Ndh complex, which might be affected by the purification procedure.
According to previous results, Figure 8 shows that both NDH-F and phospho-Thr antibodies recognized the 70 kDa NDH-F polypeptide. After 8 min incubation of purified Ndh complex with trypsin, two peptides of 39 and 31 kDa were produced that were similarly recognized by the two antibodies. One additional 25 kDa peptide was recognized by NDH-F but not by phospho-Thr antibody. Whereas the 25 kDa peptide persisted and even slightly increased after another 8 min (16 min incubation), the 39 and 31 kDa polypeptides were reduced. Obviously, the two larger tryptic peptides include the l antigenic sequence and probably the phosphorylated Thr-181. The shorter 25 kDa peptide is a fragment (probably derived from the former ones) still retaining the l sequence but not the phospho-Thr-181. This amino acid lies between the two potential and close trypsin sites (Lys-177 and Arg-183) of the stromal f region. The following closest, upstream potential trypsin site is Arg-123. On the other hand, there are many potential trypsin sites downstream of l (from Arg-448 to Lys-528) in the hydrophilic m region. Therefore the 31 kDa peptide (approx. 280 amino acids) recognized by the two antibodies must span from Lys-177 in the hydrophilic f region to one of the first trypsin sites (probably Lys-469) in the hydrophilic m region. Considering the involvement of a minimum number of cleavage sites, the 39 kDa peptide would derive from cleavages at Arg-123 (in the d region) and Lys-469 (in the m region). The 25 kDa peptide, containing the l sequence but not the phosphorylated Thr-181, may derive from the 31 and 39 kDa peptides by a further trypsin cleavage at Lys-240 at the beginning of the hydrophilic h region.
Figure 8. Tryptic digestion of phosphorylated NDH-F polypeptide in the immunopurified Ndh complex.
Ndh complex (0.5 μg) was immunopurified from freshly prepared thylakoids and incubated in 100 mM Tris/HCl (pH 7.5) and 10 units of trypsin at 20 °C. At the indicated times, reactions were stopped by the addition of soya-bean trypsin inhibitor [75 ng·(μg of trypsin)−1] in SDS sample buffer and then boiling for 10 min. Samples were subjected to Tricine-SDS/PAGE (10% acrylamide) and Western-blot analysis using a monospecific NDH-F antiserum (A) or a monoclonal phosphothreonine antibody (B).
Probably, the 39 and 42 kDa peptides produced after trypsin incubation of detergent-treated thylakoids (Figure 7) are also produced by cleavages at Arg-123 and in the luminal m region that would be accessible in the isolated complex and in permeabilized thylakoids. Although these results do not provide information on the stromal or luminal orientation of the d region, they strongly suggest the accessibility of the Arg-123 site to trypsin if the g region is exposed. With this in mind, we may conclude that the 58 kDa peptide produced by trypsin on intact thylakoids (Figure 7) could result from the cleavage pair at Arg-123 and Arg-679/Lys-690 (the trypsin site in the p region that is also required to produce the 66 kDa peptide). However, this interpretation suggests, not conclusively, that the d (as well as the f) region faces the stroma, which in turn requires that the short e region faces the lumen.
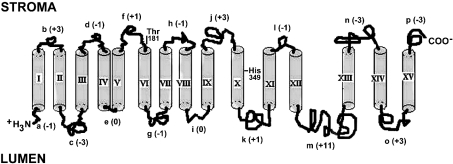
In summary, with minor uncertainties on the first 175 amino acid sequence, the results suggests the topology of NDH-F in thylakoids represented in Figure 9.
Figure 9. Transmembrane topology model of the NDH-F polypeptide.
Hydrophobic helices are labelled with roman numerals and hydrophilic regions are labelled with lower-case letters, according to the PHD model (see Figure 4). The net charge of hydrophilic regions is indicated within parentheses. The length of all transmembrane helices was 18 amino acids, except in IV, XI and XV, which have 20 amino acids, and XIII, which has 19 amino acids. The conserved Thr-181 (phosphorylation site) and His-349 (for H+ translocation) are indicated.
DISCUSSION
Polypeptides encoded by plastid ndh genes are homologous with those of the hydrophobic arm and the bridge connecting both arms of the L-shaped structure of respiratory complex I. Although genes and polypeptides of the hydrophilic arm have not yet been identified, the NADH:plastoquinone oxidoreductase activity [2] of the purified Ndh complex indicates that it must contain a hydrophilic arm composed, among others, of the NuoF/NQO1 NADH-binding polypeptide. Furthermore, a comparison of trypsin sensitivity of the NDH-A and NDH-K polypeptides in thylakoid membranes (Figures 1 and 2) with the structures proposed for complex I [15,35,36] suggests that the overall structures of complex I and the Ndh complex must be similar. NDH-A (the probable plastoquinone-binding subunit) and NDH-K would be at the connecting bridge of the Ndh complex. The Ndh polypeptide homologous with complex I NQO1 would be at the distal end of the hydrophilic arm, which would protrude towards the stroma (Figure 3), where the available NADH provides electrons to reduce plastoquinone. Functionally, the stromal side is to the Ndh complex as mitochondrial matrix and bacterial cytoplasm are to complex I, whereas the thylakoid lumen is equivalent to intermembranous mitochondrial and bacterial periplasmic spaces.
Digestion assays of intact and permeabilized thylakoids (Figures 6–8) with V8 and trypsin indicated an easy accessibility of several cleavage sites in the stromal and luminal sides of NDH-F, indicating that most (if not all) of the hydrophilic regions of NDH-F are not covered by the connecting bridge. Binding of NDH-F antibody to intact thylakoids (Table 1) also supports the accessibility of hydrophilic regions of NDH-F orientated towards the stroma. The results agree with the situation of the homologous ND5 polypeptide of complex I at the distal end of the membrane arm [32].
The topology of bacterial components of complex I has been investigated with genetic constructs encoding segments of the polypeptides fused to the alkaline phosphatase marker [17,37]. Recently, a similar approach has been used to investigate the topology of the PsaA subunit of photosystem I complex in Chlamydomonas chloroplasts transformed with genes encoding chimaeric polypeptides consisting of aminoglycoside adenyltransferase (spectinomycin resistance) fused to psaA fragments of different lengths [38]. However, the difficulties involved in transforming chloroplasts of higher plants with genetic chimaeras limit the use of this approach to investigate the membrane orientation of the components of the Ndh complex. The accessibility of stromal and luminal hydrophilic regions of NDH-F allowed us to investigate the topology of this polypeptide in thylakoid by analyses of antibody binding and proteinase cleavage peptides. The importance of investigating the NDH-F/ND5/NuoL/NQO12 topology is strengthened because sequence analyses indicate that it is a component of a proton-pumping mechanism [17].
The transmembrane topology of NDH-F (Figure 9) deduced from our experiments has similarities and differences with the membrane topology deduced by other experimental approaches for the homologous NuoL of R. capsulatus [17]. The transmembrane predictions (especially that of PHD, Figure 4) and sequences (alignments not shown) of the first 330 amino acids are rather similar in NDH-F and NuoL. Therefore the transmembrane topologies deduced for the first nine helices are the same for NDH-F and NuoL. The N-terminus extends towards the lumen and OUT (periplasm) respectively, and the successive hydrophilic regions alternately face towards the stroma (IN-cytoplasm) and lumen (OUT) until the j region, which follows transmembrane helix IX (Figure 9). The conserved Asp-90 and Glu-152, characteristic of antiporter proteins [17], are in or close to transmembrane helices III and V respectively near the luminal (in NDH-F, Figure 9) and OUT (in NuoL) sides. The stroma (IN)-faced hydrophilic f region between transmembrane helices V and VI is shorter in NuoL (12 amino acids) than in NDH-F (19 amino acids). The latter includes at its C-terminus the Thr-181 that is presumably reversibly phosphorylated and is conserved from Cyanobacteria to higher plants (Figure 5). In fact, the sequence surrounding the Thr-181 (ACQKAFVTNR) is conserved in NDH-F of land plants but not in NuoL, which lacks, among other residues, the Thr in this region. Sequence comparisons of the NDH-F polypeptides of higher plants indicate a high homology for two-thirds of the sequence at the N-terminus and more variability for the remaining one-third at the C-terminus [39].
Amino acid sequence alignments (Figure 10) indicate that transmembrane helix X that we deduced for NDH-F corresponds to hydrophobic helix XI proposed for NuoL [17]. Between transmembrane helices IX and X of NDH-F, the model in Figure 9 included the 33 amino acid hydrophilic region j, which contains sequences homologous with the hydrophobic helix X proposed for NuoL [17]. An equivalent hydrophobic helix is not predicted for NDH-F by the PHD model. In fact, there is no agreement among different model predictions (Figure 4) for the position and number of hydrophobic helices in this polypeptide segment. Despite this difference, our results and those for NuoL [17] agree in the membrane side location (stroma-IN) of this polypeptide region (j in NDH-F) since it is proposed that hydrophobic helix X of NuoL is not transmembrane but rather protrudes into the cytoplasm. However, the last six hydrophobic helices of NDH-F and NuoL (X, XI, XII, XIII, XIV and XV of the NDH-F and XI, XII, XIII, XIV, XV and XVI of the NuoL) show clear differences in the proposed topologies.
Figure 10. Alignment of amino acid sequences of the predicted hydrophobic arms X of NDH-F and XI of NuoL.
As pointed out in Figure 10, the hydrophobic helix X of NDH-F is homologous with the hydrophobic helix XI of NuoL: both contain the Phe and the His (positions 357 and 349 respectively in NDH-F) conserved in all the homologous plastid and respiratory polypeptides analysed (Figure 5). This His and probably another conserved His (position 345 in NDH-F) may be involved in the H+ pumping channel, as predicted by the Pfan collection of hidden Markov models and described for similar channels in H+ pumping transhydrogenases [40]. Accordingly, our results indicate that hydrophobic helix X of NDH-F is transmembrane; however, the homologous helix XI of NuoL was proposed [17] to protrude into the IN side (equivalent to stroma). The differences between the two results would affect predictions with respect to the role of NDH-F/NuoL in H+ translocation.
As a consequence of the different proposed locations of hydrophobic helices (X and XI of NDH-F and NuoL respectively), the transmembrane orientation of the following hydrophilic regions is also different. However, these differences are not surprising, considering the markedly low sequence homologies, e.g. of approx. the 390th amino acid to the C-terminus of NDH-F compared with the corresponding region in NuoL. Therefore the correspondence of transmembrane helices X, XI, XII, XIII, XIV and XV of NDH-F and transmembrane helices XI, XII, XIII, XIV, XV and XVI of NuoL is only numerical, and not in sequence or probably in function. The same may be argued in relation to the connecting hydrophilic regions. As a matter of fact, whereas region m between transmembrane helices XII and XIII of NDH-F contains more than 90 amino acids, the corresponding hydrophilic region between transmembrane helices XIII and XIV of NuoL contains approx. 25 amino acids. As mentioned above, the sequence variability in NDH-F of different plants is high in the last one-third and low in the first two-thirds of the protein [39].
As far as we know, a transmembrane helix number of 15 has not been seen thus far in the X-ray structure of membrane proteins. However, a similar number of helices (16) has been proposed for NuoL [17]. In any case, 15 transmembrane helices is a rather high and unusual number and further experiments are required to confirm the complete topology of the NHD-F polypeptide in thylakoids, especially for the first 170 amino acids (hypothetical helices I–V) and the last 60 amino acids (hypothetical helix XV).
At present, it is not possible to ascertain whether discrepancies between the models of transmembrane topology proposed for NDH-F and NuoL are due to the different methodologies used in each case or if they are genuine and related to specific functions. In any case, the antibody binding and proteinase cleavage approach used in this work to investigate the topology of NDH-F in thylakoids has been employed for the first time to investigate a subunit of the chloroplast Ndh complex and seems to be appropriate for other components. The comparison of these with homologous subunits of complex I would shed light on the similarities and evolution of the functional differences between the two complexes.
Online data
Acknowledgments
We thank Dr Alberto Paradela (Centro Nacional de Biotecnología, CSIC, Madrid, Spain) for advice on the interpretation of MALDI–TOF results. This work was supported by grants BFI2000-0781 and BMC2003-01261 of the Spanish DGICT (Ministerio Educación y Cultura). We thank Mrs P. H. Serrot for a fruitful discussion and suggestions. H.R.L. was supported by postdoctoral fellowships from Consejo Nacional de Investigaciones Científicas y Técnicas (Argentina), and from the Spanish Ministerio de Educación, Cultura y Deportes.
References
- 1.Sazanov L., Burrows P. A., Nixon P. J. The plastid ndh genes code for a NADH-specific dehydrogenase: purification and characterization of a mitochondrial-like complex I from pea thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1319–1324. doi: 10.1073/pnas.95.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casano L. M., Zapata J. M., Martín M., Sabater B. Chlororespiration and poising of cyclic electron transport: plastoquinone as electron transporter between thylakoid NADH dehydrogenase and peroxidase. J. Biol. Chem. 2000;275:942–948. doi: 10.1074/jbc.275.2.942. [DOI] [PubMed] [Google Scholar]
- 3.Berger S., Ellersiek U., Westhoff P., Steinmuller K. Studies on the expression of NDH-H, a subunit of the NAD(P)H-plastoquinone-oxidoreductase of higher plant chloroplasts. Planta. 1993;190:25–31. [Google Scholar]
- 4.Nixon P. Chlororespiration. Philos. Trans. R. Soc. London B. 2000;355:1541–1547. doi: 10.1098/rstb.2000.0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albertsson P.-A. A quantitative model of the domain structure of the photosynthetic membrane. Trends Plant Sci. 2001;6:349–354. doi: 10.1016/s1360-1385(01)02021-0. [DOI] [PubMed] [Google Scholar]
- 6.Maier R. M., Neckermann K., Igloi G. L., Kössel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 7.Zapata J. M., Sabater B., Martín M. Identification of a thylakoid peroxidase which oxidized hydroquinone. Phytochemistry. 1998;48:1119–1123. [Google Scholar]
- 8.Martín M., Casano L. M., Sabater B. Identification of the product of ndhA gene as a thylakoid protein synthesized in response to photooxidative treatment. Plant Cell Physiol. 1996;37:293–298. doi: 10.1093/oxfordjournals.pcp.a028945. [DOI] [PubMed] [Google Scholar]
- 9.Catalá R., Sabater B., Guéra A. Expression of the plastid ndhF gene product in photosynthetic and non-photosynthetic tissues of developing barley seedlings. Plant Cell Physiol. 1997;38:1382–1388. doi: 10.1093/oxfordjournals.pcp.a029133. [DOI] [PubMed] [Google Scholar]
- 10.Casano L. M., Martín M., Zapata J. M., Sabater B. Leaf age- and paraquat-dependent effects on the levels of enzymes protecting against photooxidative stress. Plant Sci. 1999;149:13–22. [Google Scholar]
- 11.Endo T., Shikanai T., Takabayashi A., Asada K., Sato F. The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett. 1999;457:5–8. doi: 10.1016/s0014-5793(99)00989-8. [DOI] [PubMed] [Google Scholar]
- 12.Horvath E. M., Peter S. O., Joet T., Rumeau D., Cournac L., Horvath G. V., Kavanagh T. A., Schafer C., Peltier G., Medgyesy P. Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 2000;123:1337–1349. doi: 10.1104/pp.123.4.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casano L. M., Martín M., Sabater B. Hydrogen peroxide mediates the induction of chloroplastic Ndh complex under photooxidative stress in barley. Plant Physiol. 2001;125:1450–1458. doi: 10.1104/pp.125.3.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lascano H. R., Casano L. M., Martín M., Sabater B. The activity of the chloroplastic Ndh complex is regulated by phosphorylation of the NDH-F subunit. Plant Physiol. 2003;132:256–262. doi: 10.1104/pp.103.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yano T., Yagi T. H(+)-translocating NADH–quinone oxidoreductase (NDH-1) of Paracoccus denitrificans. Studies on topology and stoichiometry of the peripheral subunits. J. Biol. Chem. 1999;274:28606–28611. doi: 10.1074/jbc.274.40.28606. [DOI] [PubMed] [Google Scholar]
- 16.Friedrich T., Scheide D. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 2000;479:1–5. doi: 10.1016/s0014-5793(00)01867-6. [DOI] [PubMed] [Google Scholar]
- 17.Mathiesen C., Hägerhäll C. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta. 2002;1556:121–132. doi: 10.1016/s0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- 18.Schneider C., Roland A., Newman D., Sutherland D. R., Asser U., Greaves M. F. A one-step purification of membrane protein using a high efficiency immunomatrix. J. Biol. Chem. 1982;257:10766–10769. [PubMed] [Google Scholar]
- 19.Morrisey P. J., McCauley S. W., Melis A. Differential detergent-solubilization of integral thylakoid membrane complexes in spinach chloroplasts. Localization of photosystem II, cytochrome b6/f complex and photosystem I. Eur. J. Biochem. 1986;160:389–393. doi: 10.1111/j.1432-1033.1986.tb09983.x. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Schägger H., Von Jagow G. Tricine-sodium dodecyl sulfate-polycrylamide gel electrophoresis for the separation of proteins in range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 22.Bergmeyer H.-U. Phosphatasen (Phosphoestearasen) In: Bergmeyer H.-U., editor. Methoden der Enzymatischen Analyse. Weinheim/Bergstr., Germany: Verlag Chemie, GMBH; 1962. pp. 783–785. [Google Scholar]
- 23.Rost B., Casadio R., Fariselli P., Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonnhammer E. L., von Heijne G., Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 25.Tusnady G. E., Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 26.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann K., Stoffel W. PROFILEGRAPH: an interactive graphical tool for protein sequence analysis. Comput. Appl. Biosci. 1992;8:331–337. doi: 10.1093/bioinformatics/8.4.331. [DOI] [PubMed] [Google Scholar]
- 28.Jones D. T., Taylor W. R., Thornton J. M. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 29.Bradford M. M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 30.Arnon D. I. Copper enzymes in isolated chloroplasts. Polyphenol oxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.del Campo E. M., Sabater B., Martín M. Transcripts of the ndhH-D operon of barley plastids: possible role of unedited site III in splicing of the ndhA intron. Nucleic Acids Res. 2000;28:1092–1098. doi: 10.1093/nar/28.5.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sazanov L. A., Walker J. E. Cryo-electron crystallography of two subcomplexes of bovine complex I reveals the relationship between the membrane and peripheral arms. J. Mol. Biol. 2000;302:455–464. doi: 10.1006/jmbi.2000.4079. [DOI] [PubMed] [Google Scholar]
- 33.Smith B. J. Enzymatic methods for cleaving proteins. In: Walker J. M., editor. Methods in Molecular Biology, vol. 32. Totowa, NJ, U.S.A.: Humana Press; 1994. pp. 289–309. [DOI] [PubMed] [Google Scholar]
- 34.Rintamäki E., Salonen M., Suoranta U.-M., Calberg I., Anderson B., Aro E.-M. Phosphorylation of light-harvesting complex II and photosystem II core protein shows different irradiance-dependent regulation in vivo. J. Biol. Chem. 1997;272:30476–30482. doi: 10.1074/jbc.272.48.30476. [DOI] [PubMed] [Google Scholar]
- 35.Finel M. Organization and evolution of structural elements within complex I. Biochim. Biophys. Acta. 1998;1364:112–121. doi: 10.1016/s0005-2728(98)00022-x. [DOI] [PubMed] [Google Scholar]
- 36.Friedrich T. The NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochim. Biophys. Acta. 1998;1364:134–146. doi: 10.1016/s0005-2728(98)00024-3. [DOI] [PubMed] [Google Scholar]
- 37.Roth R., Hägerhäll C. Transmembrane orientation and topology of the NADH:quinone oxidoreductase putative quinone binding subunit NuoH. Biochim. Biophys. Acta. 2001;1504:352–362. doi: 10.1016/s0005-2728(00)00265-6. [DOI] [PubMed] [Google Scholar]
- 38.Franklin J. L., Zhang J., Reding K. Use of aminoglycoside adenyltransferase translational fusions to determine topology of thylakoid membrane proteins. FEBS Lett. 2003;536:97–100. doi: 10.1016/s0014-5793(03)00034-6. [DOI] [PubMed] [Google Scholar]
- 39.Ki-Joong K., Jansen R. K. ndhF sequence evolution and the major clades in the sunflower family. Proc. Natl. Acad. Sci. U.S.A. 1995;92:10379–10383. doi: 10.1073/pnas.92.22.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bizouarn T., Meuller J., Axelsson M., Rydstrom J. The transmembrane domain and the proton channel in proton-pumping transhydrogenases. Biochim. Biophys. Acta. 2000;1459:284–290. doi: 10.1016/s0005-2728(00)00163-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.