Abstract
BthTx-I (bothropstoxin-I) is a myotoxic Lys49-PLA2 (phospholipase A2 with Lys49) isolated from Bothrops jararacussu venom, which damages liposome membranes by a Ca2+-independent mechanism. The highly conserved Phe5/Ala102/Phe106 motif in the hydrophobic substrate-binding site of the Asp49-PLA2s is substituted by Leu5/Val102/Leu106 in the Lys49-PLA2s. The Leu5/Val102/Leu106 triad in BthTx-I was sequentially mutated via all single- and double-mutant combinations to the Phe5/Ala102/Phe106 mutant. All mutants were expressed as inclusion bodies in Escherichia coli, and the thermal stability (Tm), together with the myotoxic and Ca2+-independent membrane-damaging activities of the recombinant proteins, were evaluated. The far-UV CD profiles of the native, wild-type recombinant and the L106F (Leu106→Phe) and L5F/F102A/L106F mutant proteins were identical. The L5F, V102A, L5F/V102A and V102A/L106F mutants showed distorted far-UV CD profiles; however, only the L5F and L5F/V102A mutants showed significant decreases in Tm. Alterations in the far-UV CD spectra correlated with decreased myotoxicity and protein-induced release of a liposome-entrapped marker. However, the V102A/L106F and L5F/V102A/L106F mutants, which presented high myotoxic activities, showed significantly reduced membrane-damaging activity. This demonstrates that the topology of the substrate-binding region of BthTx-I has a direct effect on the Ca2+-independent membrane damage, and implies that substrate binding retains an important role in this process.
Keywords: bothropstoxin-I, Lys49-phospholipase A2 (Lys49-PLA2), mutagenesis, myotoxicity, substrate-binding site, topology
Abbreviations: CK, creatine kinase; PLA2, phospholipase A2; Lys49-PLA2, phospholipase A2 with Lys49; BthTx-I, bothropstoxin-I; EYPC, egg yolk phosphatidylcholine
INTRODUCTION
Phospholipases A2 (PLA2s; EC 3.1.1.4) catalyse the hydrolysis of the sn-2 acyl bonds of sn-3 phospholipids [1], and are currently classified into 11 groups on the basis of amino acid sequence similarity [2]. The hydrolysis of phospholipids by group I/II PLA2s involves a His48/Asp99 pair in the catalytic site, which activates a positionally conserved water molecule, thereby initiating the nucleophilic attack on the sn-2 position of the substrate. A Ca2+ ion cofactor is bound to the carboxyl oxygen atoms of Asp49 and carbonyl main-chain oxygens of the neighbouring calcium-binding loop stabilizes the tetrahedral intermediate during catalysis [3,4].
A subfamily of class IIB PLA2s in which Asp49 is substituted by a lysine residue have been identified in several Asiatic and new world Viperid snake venoms (see [5–7] for reviews). Initial reports suggested that these Lys49-PLA2s (PLA2s with Lys49) retained low levels of catalytic activity [8–10]; however, subsequent studies with both native [11–13] and recombinant [14] proteins have failed to detect hydrolysis of phospholipid substrates. The crystal structures of Lys49-PLA2s have demonstrated that the ε-amino group of the Lys49 is located in the position occupied by Ca2+ in Asp49-PLA2s [4,14,15], and it has been suggested that the reduction or elimination of catalytic activity results from either the reorientation of the Cys29–Gly30 peptide bond [14] or the reduced binding affinity of the Ca2+ cofactor [16]. Despite the absence of detectable catalytic activity, the Lys49-PLA2s demonstrate membrane-damaging activity through a Ca2+-independent, non-hydrolytic mechanism [17–19].
Bothropstoxin-I (referred to as BthTx-I) is a Lys49-PLA2 isolated from the venom of Bothrops jararacussu, which forms homodimers both in solution [19] and in the crystalline state [20]. Flexibility at the dimer interface permits a quaternary structure transition between ‘open’ and ‘closed’ forms, and it has been proposed that, in the membrane-bound form, this transition may disrupt the packing of the bilayer lipid, thereby resulting in disruption of the membrane [20]. The model predicts that, as a result of the conformational transition, the position of the C-terminal loop region of the protein is altered, which promotes a more intimate interaction of this region with the membrane bilayer. Indeed, site-directed mutagenesis has demonstrated that residues in the C-terminal loop region play an important role in the Ca2+-independent membrane-damaging activity [21].
Comparison of the Lys49-PLA2 amino acid sequences with those of the Asp49-PLA2s has revealed that amino acid substitutions that are specific to the Lys49-PLA2s are grouped in three distinct clusters in the crystal structure [22]. In addition to residue clusters in the β-wing and active-site regions, the highly conserved Phe5/Ala102/Phe106 triad in the hydrophobic substrate-binding cleft of the Asp49-PLA2s is substituted by Leu5/Val102/Leu106 in the Lys49-PLA2s [22,23]. Previously, it has been demonstrated that the catalytic activity of the Asp49-PLA2s is highly sensitive to site-directed mutagenesis of these residues [24,25], and therefore, the changes observed in the substrate-binding region of the Lys49-PLA2s are particularly intriguing. This cluster of amino acid substitutions results in a significant alteration in the topology of the substrate-binding region; however, it is tolerated in the sense that the product is a biologically functional (although catalytically inactive) protein. The effects of substituting the highly conserved Phe5/Ala102/Phe106 triad in the Lys49-PLA2s are not known, and to understand the effects of these specific substitutions in the substrate-binding region of the Lys49-PLA2s, we have sequentially mutated the Leu5/Val102/Leu106 residues to the Phe5/Ala102/Phe106 residues encountered in the Asp49-PLA2s. The effects of these mutations were evaluated using several structural and functional assays, which have revealed that the topology of the substrate-binding region influences the calcium-independent membrane-damaging activity, and suggest a functional linkage between the substrate-binding and C-terminal loop regions.
EXPERIMENTAL
Native protein purification and sample preparation
BthTx-I was purified from crude freeze-dried B. jararacussu venom using cation-exchange chromatography as described previously [26]. Protein purity was routinely evaluated by silver staining of SDS/polyacrylamide gels, and aliquots of the purified protein were stored at −20 °C until further use.
Site-directed mutagenesis
A full-length cDNA encoding BthTx-I has been isolated previously from B. jararacussu venom gland cDNA by reverse transcriptase–PCR [27] (GenBank® accession no. X78599), and subcloned into the expression vector pET3-d [28]. Nucleotide sequencing has confirmed the construct in which Ser1 of the BthTx-I is preceded by a methionine residue, and a stop codon immediately follows Cys133 (the homology numbering scheme reported by Heinrickson [29] is used throughout the present study). After linearization of this construct with ScaI, site-directed mutagenesis of the BthTx-I was performed by PCR mutagenesis [30] to introduce the following single mutations: L5F (oligonucleotide 5′-CATTTTCCCGAACTCAAACAG-3′; base changes are underlined), V102A (5′-GCAGATTGCCGCGGCCTTGTC-3′), L106F (5′-ATTTTCTCGGAAGCAGATTGC-3′) and L106F (5′-ATTTTCTCGGGCGCAGATTGC-3′). The double mutants L5F/V102A, L5F/L106F and V102A/L106F, together with the triple mutant L5F/V102A/L106F, were obtained by mutagenesis of the respective single- and double-mutant constructs. PCRs were performed using oligonucleotides complementary to the vector sequences flanking the BthTx-I insert, which contained restriction sites for XbaI (5′-extremity) and BamHI (3′-extremity). After digestion with these enzymes, the amplified fragments were subcloned into the equivalent sites in the expression vector pET3d and fully sequenced.
Recombinant protein expression and purification
Growth medium (400 mg/l; 25 g of yeast extract, 15 g of tryptone, 10 mM MgSO4, 35 μg of chloramphenicol and 100 μg of ampicillin, pH 7.5) was inoculated with the Escherichia coli strain BL21-(DE3){pLysS} transformed with the native or mutant constructs, and grown at 37 °C to an absorbance A600 0.6. Recombinant protein expression was induced by the addition of 0.6 mM isopropyl β-D-thiogalactoside, and the culture was grown for an additional 7 h. Inclusion bodies were isolated from bacterial pellets by repeated rounds of sonication in 20 ml of lysis buffer (50 mM Tris/HCl, pH 8.0, 1 mM EDTA, 0.4 M urea and 1% Triton X-100), followed by centrifugation at 12000 g. The method for the solubilization and refolding of recombinant BthTx-I in the presence of a gel-filtration medium was performed as described previously [28]. The refolded protein was purified by cation-exchange chromatography using a 100 mm×7 mm column packed with Resource S resin (Amersham Biosciences, São Paulo, Brazil) previously equilibrated in 20 mM Tris/HCl, pH 8.0 (buffer A). After application of the sample, the column was eluted with buffer A supplemented with 1 M NaCl, and the absorbance of the eluate was monitored continuously at 280 nm. The eluted protein was dialysed against 5 mM Tris/HCl (pH 8.0) for 36 h with buffer changes every 12 h, and finally stored at 4 °C until further use.
MS analysis
Protein was diluted in 50% (v/v) acetonitrile and 0.1 M ammonium bicarbonate and reduced with 45 mM dithiothreitol for 1 h at 56 °C, and subsequently alkylated with 100 mM iodoacetamide for 1 h at room temperature (25 °C) in the dark. The protein solution was diluted 5-fold and 0.5 μg of modified trypsin (Promega, Madison, WI, U.S.A.) was added; after 24 h, trypsin hydrolysis was stopped with 5 μl of formic acid. The tryptic peptides were desalted with Poros R2 (PerSeptive Biosystems, Hertford, U.K.) and eluted in 60% (v/v) methanol and 5% (v/v) formic acid, and MS analysis of tryptic peptides was performed in an electrospray triple-quadrupole mass spectrometer Quatro II (Micromass, Manchester, U.K.). The samples were infused using a syringe pump at a flow rate of 300 nl/min under the following conditions: capillary voltage was maintained at 2.8 kV, cone voltage at 40 V and cone temperature at 100 °C. The parameters for MS scanning were optimized with a synthetic peptide for the highest signal-tonoise ratio. The spectrum was collected as the average of 14–25 scans (2–5 s/scan) over the range 300–2000 atomic mass units, and processed using MassLynx software v.3.3 (Micromass). The site-directed mutant tryptic peptides were submitted to CID (collision-induced dissociation)–MS/MS to produce fragment ions type b and y, which were used to deduce the amino acid sequence. The collision cell was set to a collision energy of 20–25 eV for double-charged ions and 38–45 eV for single-charged ions, and argon was used as the collision gas at a pressure of 0.4 mPa (3.0×10−3 mTorr). Each spectrum was collected as the average of 13–20 (3 s/scan) and processed using MassLynx software.
CD spectroscopy
Far-UV CD spectra of BthTx-I and the mutants were measured between 200 and 250 nm in 20 mM Hepes (pH 7.0) at 25 °C with a JASCO J-810 spectropolarimeter (Jasco, Tokyo, Japan) using 1 mm path-length cuvettes and a protein concentration of 70–150 μg·ml−1 BthTx-I. In each case, a total of nine spectra were collected, averaged and corrected by subtraction of a buffer blank. Thermal denaturation studies were performed using the J-810 spectropolarimeter equipped with a Peltier-type temperature controller (PTC423S; Jasco) with a heating rate of 1 °C·min−1 using 1 cm path-length cuvettes and a protein concentration of 50–150 μg·ml−1. Changes in the ellipticity signal at 222 nm were monitored over the temperature range 30–90 °C (303.16–363.16 K). The melting temperature Tm was determined from the thermal denaturation curve by assuming a two-state mechanism and using a least-squares fitting routine derived from the van't Hoff equation as described previously [31].
Release of entrapped fluorescent markers from liposomes
Membrane damaging activity was determined by measuring the release of the liposome-entrapped, self-quenching fluorescent dye calcein. Loss of liposome membrane integrity results in dilution of the fluorophore, with a consequent increase in the fluorescence signal [18,19]. Liposomes composed of EYPC (egg yolk phosphatidylcholine)/DMPA (dimyristoyl phosphatidic acid) at a molar ratio of 9:1 were prepared by reverse-phase evaporation with a buffer (25 mM Hepes and 150 mM NaCl, pH 7.0) containing 25 mM calcein (Sigma). The liposomes were passed through a 400 nm polycarbonate filter and applied to a Sephadex G50 column (1 cm×8 cm) to separate the liposomes from the free calcein. Proteins and liposomes were mixed to a final protein/lipid molar ratio of 1:400, and the kinetics of membrane damage was monitored by the increase in fluorescence with emission at 520 nm and excitation at 490 nm, and the signal was expressed as percentage of total calcein release after the addition of 5 mM Triton X-100.
Myotoxic activity
Protein (30 μg) in a total volume of 50 μl of PBS was injected into the gastrocnemius muscle of male Swiss mice (weighing 18–22 g). After 3 h, a blood sample was collected from the tail in a heparinized capillary tube and the plasma was separated by centrifugation. The plasma CK (creatine kinase) activity was determined using 4 μl of plasma using the CK-UV kinetic kit (Sigma) according to the manufacturer's instructions. CK activity was expressed in terms of unit·l−1, where 1 unit is defined as the amount of enzyme that produces 1 mmol of NADH·min−1 under the standard conditions used in the assay. Animals used as negative controls received 50 μl of PBS, and all results were expressed as the mean plasma CK activity from a minimum of five experiments.
RESULTS
As reported previously [28], BthTx-I is expressed in the form of inclusion bodies in E. coli, and all mutants were successfully expressed using this host cell (results not shown). After solubilization, chemical modification and refolding, the recombinant proteins were purified using cation-exchange chromatography, and assessment of purity by silver staining of SDS/polyacrylamide gels revealed a single protein band in all cases (results not shown). All proteins were exposed to trypsin digestion, and the resulting peptides corresponding to amino acids 1–7 (BthTx-I1–7) and 101–107 (BthTx-I101–107) were analysed using ESI (electrospray ionization)–MS. Table 1 demonstrates that these peptides present identical masses both in the native and wild-type recombinant proteins, and the observed and calculated masses of BthTx-I1–7 and BthTx-I101–107 show excellent agreement for all mutant proteins. The tryptic mutant peptides were also submitted to ESI–CID–MS/MS, and the deduced amino acid sequences shown in Table 1 unequivocally demonstrated the presence of the desired mutants.
Table 1. Results of MS analysis of the native and wild-type recombinant BthTx-I and mutant peptides.
| Calculated mass† | Observed mass | ||||
|---|---|---|---|---|---|
| Mutant | Peptide sequence* | [M+H+] | [M+2H+] | [M+H+] | [M+2H+] |
| Native | S1LFELGK7 | 793.5 | 397.3 | 793.5 | − |
| A101VAICLR107 | 802.5 | 401.7 | 802.5 | 401.8 | |
| Recombinant | S1LFELGK7 | 793.5 | 397.3 | 793.4 | − |
| A101VAICLR107 | 802.5 | 401.7 | 802.4 | 401.8 | |
| L5F | S1LFEFGK7 | 827.9 | 414.2 | 827.5 | − |
| V102A | A101AAICLR107 | 774.4 | 387.7 | − | 387.8 |
| L106A | A101VAICAR107 | 760.4 | 380.7 | 760.5 | − |
| L106F | A101VAICFR107 | 836.5 | 418.7 | 836.6 | 418.1 |
| L5F/V102A | S1LFEFGK7 | 827.9 | 414.2 | 827.5 | − |
| A101AAICLR107 | 774.4 | 387.7 | 774.5 | 387.8 | |
| L5F/L106F | S1LFEFGK7 | 827.9 | 414.2 | 827.4 | − |
| A101VAICFR107 | 836.5 | 418.7 | − | 418.9 | |
| V102A/L106F | A101AAICFR107 | 808.4 | 404.7 | − | 404.2 |
| L5F/V102A/L106F | S1LFEFGK7 | 827.9 | 414.2 | 827.5 | 414.3 |
| A101AAICFR107 | 808.4 | 404.7 | 808.5 | 404.8 | |
* Mutations from the native amino acid sequence are shown in boldface. The superscript numbers indicate the initial and final amino acid positions of the peptide according to the homology numbering scheme reported by Heinrickson [29].
† The mass of cysteine residue was calculated by modification to carboxyamidomethylcysteine.
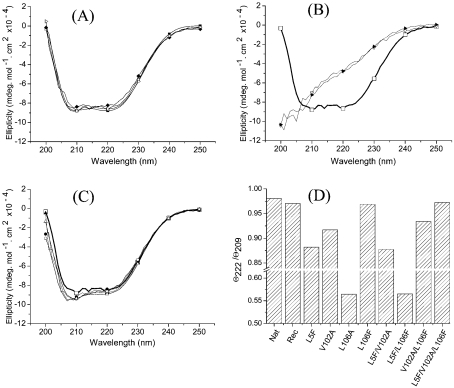
The secondary structure of the purified recombinant protein was evaluated by far-UV CD, and as presented in Figure 1(A), both the native and the recombinant BthTx-I displayed minima at 209 and 222 nm, which indicates a predominantly α-helical secondary-structure content. These results are similar to those reported previously for BthTx-I [14,21,28], and confirm that the recombinant protein retains a total secondary-structure content similar to that found in the native protein. Figure 1(A) also shows that the L106F and L5F/V102A/L106F mutants show far-UV CD spectra that are very similar to those of the native protein purified from the snake venom and native recombinant protein. As shown in Figure 1(D), the ellipticity ratio for the minima at 222 and 209 nm (θ222/θ209) is close to unity for these four proteins. Figure 1(B) presents the far-UV CD spectra of the L106A and L5F/L106F mutants, which display significantly altered profiles, demonstrating that the secondary structure is significantly perturbed in these proteins. As shown in Figure 3, both these mutants displayed a myotoxic activity significantly different from that of the negative control, demonstrating that the loss of native protein structure is accompanied by loss of biological activity. However, Figures 1(C) and 1(D) show that the L5F, V102A, L5F/V102A and V102A/L106F mutants present slightly decreased values for the minimum at 209 nm in relation to the minimum at 222 nm. Although these differences are subtle, this observation suggests that, in these cases, mutagenesis in the hydrophobic core region results in perturbations of the secondary structure of the refolding protein.
Figure 1. Far-UV CD spectra of native and recombinant BthTx-I after refolding and purification.
(A) The native BthTx-I purified from B. jararacussu venom (▪), the wild-type recombinant BthTx-I (□) and the mutants L106F (♦) and L5F/V102A/L106F (▵). (B) Wild-type recombinant BthTx-I (thick solid line, □) and the mutants L106A (▵) and L5F/L106F (▴). (C) Wild-type recombinant BthTx-I (thick solid line, □) and the mutants L5F (•), V102A (▴), L5F/V102A (○) and V102A/L106F (▵). (D) Ratio of the observed ellipticity signals at 222 and 209 nm for native BthTx-I (Nat), wild-type recombinant BthTx-I (Rec) and substrate-binding region mutants.
Figure 3. Plasma levels of CK activity 3 h after injection of 30 μg of native BthTx-I (Nat), wild-type recombinant BthTx-I (Rec) and substrate-binding site mutants into the gastrocnemius muscle of mice.
The control experiment used an identical volume of PBS in the absence of protein. Results are expressed as the means±S.D. for a minimum of five independent experiments.
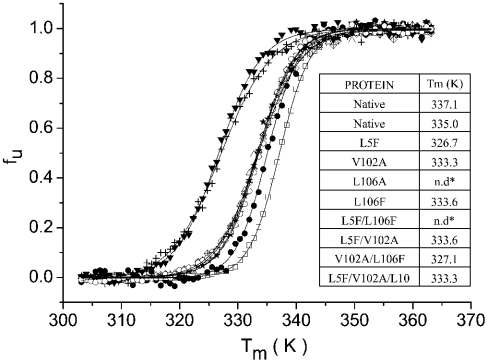
The effect of mutagenesis in the substrate-binding region on the thermal denaturation curves was determined by monitoring the changes in the far-UV CD signal at 222 nm, and the fraction of unfolded protein fu for each mutant as a function of temperature is presented in Figure 2. Estimates for the melting temperature Tm of the proteins can be derived from these denaturation curves, and as shown in Figure 2 (inset), the native, native recombinant and most of the mutant proteins show Tm values in the range 333–337 K. These results demonstrate that for most of the mutants tested, any structural change produced by site-directed mutagenesis did not significantly alter the thermal stability of the refolded proteins. However, as seen in Figure 2, the L5F and V102A/L106F mutants present reduced Tm values in the range 325–327 K. Comparison of the Tm results with the θ222/θ209 ratios (Figure 1D) shows that these two mutants present the greatest perturbations from the native far-UV CD spectra.
Figure 2. Fraction of unfolded protein fu as a function of temperature for BthTx-I and substrate-binding region mutants.
Native BthTx-I (□), wild-type recombinant BthTx-I (♦) and the mutants L5F (▾), V102A (•), L106F (○), L5F/V102A (crossed diamonds), V102A/L106F (+) and L5F/V102A/L106F (▵). The inset presents the values of melting temperature Tm of BthTx-I before and after site-directed mutagenesis of residues in the substrate-binding region; n.d., value not determined.
To evaluate the functional consequences of mutagenesis of the substrate-binding region, myotoxic activities of all these mutants were evaluated and the results are presented in Figure 3. As reported previously, the native and native recombinant proteins present identical myotoxic activities, suggesting that the recombinant protein has a completely conserved myotoxic determinant. The single-mutant L106F and the L5F/V102A/L106F mutant, which present far-UV CD spectra that are indistinguishable from the native protein, both show myotoxic activity equal to that of the native and wild-type recombinant proteins. In contrast, the single mutant V102A and the double mutants L5F/V102A and V102A/L106F show approx. 80% of the activity observed for the native protein (significant at the P<0.05 level), and the single mutant L5F shows approx. 60% of the native protein activity. Comparison of these activities with the far-UV CD data presented in Figure 1(B) shows a striking correlation: those mutants in which an altered secondary structure is observed show decreased biological activity. Furthermore, the reduction in the biological activity is proportional to the perturbation of secondary structure, which suggests that myotoxic activity is a sensitive and reliable criterion to evaluate the structural effects of site-directed mutagenesis in the substrate-binding region.
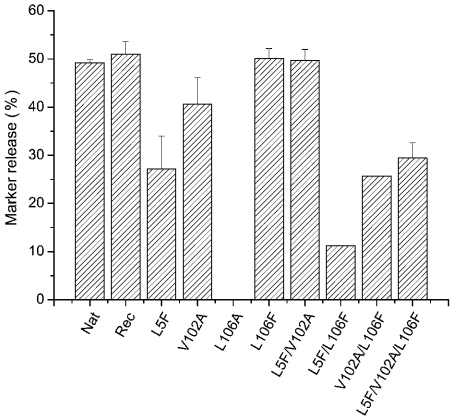
Results of the myotoxic activity tests were compared with the results from the fluorescent marker release experiments (see Figure 4), and as reported previously, the native and native recombinant proteins were found to show similar levels of membrane-damaging activity [14,21,28]. The L106A and L5F/L106F mutants, which show a little or no myotoxic activity, also present significantly reduced levels of membrane-damaging activity, which is in accord with the loss of secondary structure observed with these proteins. Reduced levels of membrane-damaging activity are observed for the L5F and V102A mutants; this correlates well with the decrease in biological activity (see Figure 3) and supports the suggestion that these mutants have suffered conformational changes that result in the loss of protein function. The mutant L106F shows a level of membrane-damaging activity that is indistinguishable from the native protein, which is consistent with the full biological activity observed for this protein. However, for the L5F/V102A, V102A/L106F and the L5F/V102A/L106F mutants, there is no correlation between the myotoxic and membrane-damaging activities. The L5P/V102A mutant shows full membrane-damaging activity and a slightly reduced myotoxic activity, whereas the other two mutants show significantly reduced membrane-damaging activities even though the myotoxic activity is fully conserved (for the L5F/V102A/L106Fe mutant) or is only slightly decreased (for the V102A/L106F mutant). It has previously been demonstrated that the homodimer is the membrane-active form of the native BthTx-I [19]; therefore, the observed decrease in membrane-damaging activities of these mutants might be the consequence of altered dimer formation. This possibility was examined by measuring the hydrodynamic radius (Rh) by dynamic light scattering, which revealed that the native and wild-type recombinant proteins and all mutant proteins had an Rh value in the range 1.8–2.1 nm. These values are consistent with previously reported Rh values for the BthTx-I homodimer in solution [32], suggesting that the observed alterations in the membrane-damaging activities are not due to changes in the quaternary structure of the mutant proteins.
Figure 4. Release of entrapped calcein from liposomes composed of EYPC/DMPA at a 9:1 molar ratio 150 s after mixing with native BthTx-I (Nat), wild-type recombinant (Rec) and substrate-binding-site mutants at a protein/lipid ratio of 1:400.
The final protein concentration in all experiments was 3 μg·ml−1, and the results are expressed as mean percentage of total calcein release after the addition of 5 mM Triton X-100.
It has previously been reported that the Lys49-PLA2s retain low levels of catalytic activity against 2-arachidonoyl-1-stearoyl-L-3-phosphatidylcholine [33,34], which raises the possibility that the altered active site of the Lys49-PLA2s supports catalysis against a specific substrate. In this case, a specific topology of the substrate-binding region in the Lys49-PLA2s may influence the orientation of the scissile ester bond of the substrate in the active-site residues, thereby contributing to the specificity of the catalytic activity. Reversion of the topology of the substrate-binding region in the Lys49-PLA2s to that found in the Asp49-PLA2s might, therefore, result in reversion of the observed catalytic inactivation of Lys49-PLA2s against mixed phospholipid substrates. However, catalytic activity tests with both the wild-type recombinant BthTx-I and the L5F/V102A/L106F mutant using a highly sensitive stopped flow Phenol Red assay [14] against either EYPC or 2-arachidonoyl-1-stearoyl-L-3-phosphatidyl-choline substrates failed to detect substrate hydrolysis (results not shown). This result is in accord with the lack of detectable activity observed previously with recombinant BthTx-I [14] and supports the suggestion that the catalytic activity reported for the native Lys49-PLA2 purified from snake venom is the result of trace contamination with catalytically active Asp49-PLA2s.
DISCUSSION
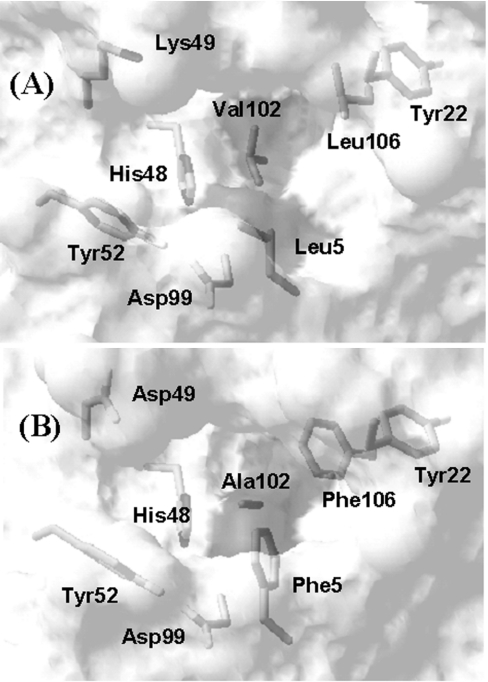
The substrate-binding cleft and active-site residues for BthTx-I and the human-secreted class II Asp49-PLA2 (hsPLA2; PDB code 1BBC; [35]) are presented in Figures 5(A) and 5(B) respectively. The three residues at positions 5, 102 and 106 in the class I/II PLA2s form a hydrophobic collar between the active-site region and the surface of the hydrophobic substrate-binding pocket. We have demonstrated previously that the Phe5/Ala102/Phe106 cluster in hsPLA2 and the Leu5/Val102/Leu106 cluster in BthTx-I have combined volumes of 468.4 and 474.4 Å3 respectively (1 Å=0.1 nm) [22], and further analysis shows that the exposed hydrophobic surface areas are 27.5 and 22.3 Å2 for the Phe5/Ala102/Phe106 and Leu5/Val102/Leu106 clusters respectively. In contrast with the conserved volumes and exposed surface areas, the atomic contacts between the three residues differ between Asp49-PLA2 and Lys49-PLA2. In the Asp49-PLA2s, all residues in the Phe5/Ala102/Phe106 cluster are in mutual contact; in contrast, in the Lys49-PLA2s, side-chain contacts are formed between Val102 and Leu5 and between Val102 and Leu106, but not between Leu5 and Leu106. Furthermore, Figure 5 demonstrates that the specific amino acid substitutions at positions 5, 102 and 106 result in an altered topology of the substrate-binding region between Asp49-PLA2 and Lys49-PLA2, and the goal of the present study is to evaluate the effects of this change on the stability and Ca2+-independent membrane-damaging activity of BthTx-I.
Figure 5. Transparent surface representation of the substrate-binding regions of (A) BthTx-I and (B) human group II secreted PLA2 (PDB code 1BBC; [35]), showing the active-site (positions 48, 49, 52 and 99) and substrate-binding residues (positions 5, 22, 102 and 106) for both proteins.
The solvent-accessible surface is shown, and the dark grey area in each Figure corresponds to the exposed surfaces of the side chains at positions 5, 102 and 106. The Figure was prepared using SwissPDB software [39].
Mutagenesis of the substrate-binding region of BthTx-I influences both the conformation and stability of the protein, a clear indication that the packing of the hydrophobic core is of prime importance in conserving the native-like secondary and tertiary structures. The systematic approach adopted in the present study using single, double and triple mutants allows a comprehensive evaluation of the roles of side-chain packing in the substrate-binding region on the function of BthTx-I. As already noted, side-chain contacts in the Lys49-PLA2s are formed between Val102 and Leu5 and between Val102 and Leu106, and although the mutation V102A reduces the side-chain volume at this position, the hydrophobic bridging contacts with Leu5 and Leu106 will be maintained. Therefore it could be expected that the structural alterations of this mutation would be minimized and, indeed, the far-UV CD spectra is only slightly perturbed and both the Tm value and the myotoxic activity are only slightly reduced with respect to the native protein.
Previous mutagenesis results with Asp49-PLA2s have demonstrated that a conserved hydrophobic volume at position 5 is essential for maintained levels of catalytic activity [25]. Superposition of the three-dimensional structures of BthTx-I and hsPLA2 reveals that the introduction of an aromatic group at position 5 by the single-mutant L5F causes side-chain clashes with Val102, and it is expected that structural reorganization of the substrate-binding region would be necessary to accommodate the introduction of the bulky aromatic ring. Although the single-mutant L5F refolds to a native-like structure with decreased myotoxic and membrane-damaging activities, the resulting protein has a low Tm value, which may reflect the structural perturbation in the hydrophobic core region. The packing conflict at position 5 is probably compounded by the additional substitution L106F and, indeed, the far-UV CD spectra demonstrate that the combination of these two mutations results in an unviable protein-refolding process. Conversely, mutations that reduce the side-chain packing problem at position 5 are more probable to favour protein refolding; thus a combination of V102A with L5F in L5F/V102A double mutant might reduce the side-chain clashes, and promote the observed gain in protein stability.
In the Asp49-PLA2s, aromatic stacking interactions are observed between the side chains of Tyr22 and Phe106, and in the Lys49-PLA2s, the side chain of the Leu106 makes contact with the aromatic ring of the conserved Tyr22. Thus, although the L106F mutant will increase the side-chain volume in the substrate-binding region, the mutant will introduce aromatic stacking interactions that may add to the stability of the contact between positions 22 and 106. Indeed, the L106F mutant retains native-like secondary structure and full myotoxic and membrane-damaging activities. These results with the Lys49-PLA2s are in broad agreement with those obtained from mutagenesis at the same positions in the Asp49-PLA2s, which demonstrated that single mutations at these positions that maintain the volume and hydrophobicity have minor effects on protein function [24]. This conclusion is supported by the loss of the ability of the L106A mutant in BthTx-I to refold to a native-like structure, emphasizing the importance of a maintained hydrophobic volume at this position. Side-chain packing also plays a role at position 106, and the reduced Tm value observed for the V102A/L106F mutant may be a consequence of the reduction in hydrophobic contacts caused by the reduction in the side-chain volume at position 102, a situation that is reverted by the completion of the hydrophobic packing by the mutation L5F in the L5F/V102A/L106F triad.
Thermal denaturation studies indicate that, in terms of stability, BthTx-I is surprisingly resilient to mutations in the substrate-binding cleft that result in the perturbation of the protein secondary structure. This may be attributed to the location of residues 5, 102 and 106 in the wall of the substrate-binding cleft, which may impart an enhanced plasticity to the packing interactions that would not be permitted for residues located deeper in the hydrophobic core of a protein. Thus alterations in the native secondary structure of BthTx-I that decrease the myotoxic and membrane-damaging activities may not decrease the thermal stability of the protein. When comparing the results of the structural studies with the changes in the Ca2+-independent membrane-damaging activity after mutagenesis in the substrate-binding region, a key question is whether the effects on the membrane-damaging activity are the consequence of specific topology changes in the substrate-binding cleft rather than unspecific structural perturbations. Five substrate-binding site mutants of BthTx-I contribute to changes in the function that are coupled with alterations in the far-UV CD spectra (see Figures 1, 3 and 4), which we suggest are the consequence of non-specific loss of the native-like protein structure. In contrast, two mutants contribute to changes in function that are not associated with changes in the far-UV CD spectra, which we suggest are probably the consequence of specific changes in the topology of the protein. Therefore, by comparing the myotoxic and membrane-damaging activities, the effects of specific and non-specific structural changes can be separated.
Both the far-UV CD spectrum and the myotoxic activity of L5F/V102A/L106F mutant are unaltered with respect to the native protein. If positions 5, 102 or 106 were direct structural determinants of the myotoxic effect, some effect of such extensive substitutions might be expected; therefore, the observed result suggests that the substrate-binding region is not a structural determinant of the myotoxic effect. This conclusion is in agreement with previous results that indicated that residues 116–122 in the C-terminal loop region directly participate in the determination of myotoxic activity [21]. Nevertheless, single and double mutations in the substrate-binding region result in a reduction of the myotoxic effect and the modification of the native-like protein structure, and the indirect effect of mutagenesis at positions 5, 102 and 106 suggest that a functional link exists between the substrate-binding and C-terminal loop regions of BthTx-I. Such a functional linkage explains the decreased myotoxic activity of Lys49-PLA2s after the modification of His48 in the active site by p-bromophenacyl bromide [36–38].
As for the case of the myotoxic effect, the results demonstrate that modification of the native-like structure resulting from mutagenesis in the substrate-binding region decreases the membrane-damaging activity. However, in contrast with the effects on myotoxicity, several mutants serve to demonstrate that structural changes in the substrate-binding region directly influence the membrane-damaging activity. This is clearly seen in the L5F/V102A/L106F mutant, which retains full myotoxicity, yet presents approx. 50% of the membrane-damaging activity observed for the native and recombinant wild-type proteins. Previous mutagenesis studies have demonstrated that the C-terminal loop of BthTx-I is not only a myotoxic determinant, but is also involved in the Ca2+-independent membrane-damaging activity [21], and the specific effects of mutagenesis in the substrate-binding region expand the known structural determinants of the calcium-independent membrane-damaging activity.
The question remains as to how topological changes in the substrate-binding region may influence the Ca2+-independent membrane-damaging activity of the protein. The involvement of the substrate-binding region implies that, although substrate hydrolysis is not important, binding of phospholipid to the protein remains an important factor in the calcium-independent membrane-damaging process. Changes in the topology of the substrate-binding region may influence either the affinity of the substrate for the protein, the type of phospholipid that binds or the orientation of the bound lipid. In the present study, we have presented evidence supporting a functional linkage between the substrate-binding and C-terminal loop regions, which suggests that occupancy of the substrate-binding site may be a crucial event in the activity of the protein.
Acknowledgments
This work was partially supported by FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) grants 98/14569-3 (to J.M.S.), 98/14686-0 (to R.R.), 01/06388-3 (to T.L.F.), 00/11165-0 (to A.H.C.O.) and 96/11165-3 (to R.J.W.) and CNPq (Conselho Nacional De Desenvolvimento Científico e Tecnológico) grant 300725/98-1 (to R.J.W.).
References
- 1.van Deenan L. L. M., de Haas G. H. The substrate specificity of phospholipase A2. Biochim. Biophys. Acta. 1963;70:538–553. doi: 10.1016/0006-3002(63)90792-3. [DOI] [PubMed] [Google Scholar]
- 2.Six D. A., Dennis E. A. The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochim. Biophys. Acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 3.Verheij H. M., Volwerk J. J., Jansen E. H. J. M., Puyk W. C., Dijkstra B. W., Drenth J., de Haas G. H. Methylation of histidine-48 in pancreatic phospholipase A2. Role of histidine and calcium ion in the catalytic mechanism. Biochemistry. 1980;19:743–750. doi: 10.1021/bi00545a021. [DOI] [PubMed] [Google Scholar]
- 4.Scott D. L., White S. P., Otwinowski Z., Yuan W., Gelb M. H., Sigler P. B. Interfacial catalysis: the mechanism of phospholipase A2. Science. 1990;250:1541–1546. doi: 10.1126/science.2274785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutierrez J. M., Lomonte B. Phospholipase A2 myotoxins from Bothrops snake venoms. Toxicon. 1995;33:1405–1424. doi: 10.1016/0041-0101(95)00085-z. [DOI] [PubMed] [Google Scholar]
- 6.Arni R. K., Ward R. J. Phospholipase A2 – a structural review. Toxicon. 1996;34:827–841. doi: 10.1016/0041-0101(96)00036-0. [DOI] [PubMed] [Google Scholar]
- 7.Ward R. J., de Azevedo W. F., Jr, Arni R. K. At the interface: crystal structures of phospholipases A2. Toxicon. 1998;36:1623–1633. doi: 10.1016/s0041-0101(98)00155-x. [DOI] [PubMed] [Google Scholar]
- 8.Maraganore J. M., Merutka G., Cho W., Welches W., Kezdy F. J., Heinrickson R. L. A new class of phospholipase A2 with lysine in place of aspartate 49. J. Biol. Chem. 1984;259:13839–13843. [PubMed] [Google Scholar]
- 9.Maraganore J. M., Heinrickson R. L. The lysine-49 phospholipase A2 from the venom of Agkistrodon piscivorus piscivorus. Relation of structure and function to other phospholipases A2. J. Biol. Chem. 1986;261:4797–4804. [PubMed] [Google Scholar]
- 10.Homsi-Brandenburgo M. I., Queiroz L. S., Santo-Neto H., Rodrigues-Simoni L., Giglio J. R. Fractionation of Bothrops jararacussu snake venom: partial chemical characterization and biological activity of bothropstoxin. Toxicon. 1988;26:615–627. doi: 10.1016/0041-0101(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 11.Condrea E. Comparison of enzymatic and pharmacological activities of lysine-49 and aspartate-49 phospholipases A2 from Agkistrodon piscivorus piscivorus snake venom. A reconsideration. Toxicon. 1989;27:705–706. doi: 10.1016/0041-0101(89)90036-6. [DOI] [PubMed] [Google Scholar]
- 12.Francis B., Gutiérrez J. M., Lomonte B., Kaiser I. I. Myotoxin II from Bothrops asper (Terciopelo) venom is a lysine-49 phospholipase A2. Arch. Biochem. Biophys. 1991;284:352–359. doi: 10.1016/0003-9861(91)90307-5. [DOI] [PubMed] [Google Scholar]
- 13.Holland D. R., Clancy L. L., Muchmore S. W., Rydel T. J., Einspahr H. M., Finzel B. C., Heinrickson R. L., Watenpaugh K. D. The crystal structure of a lysine 49 phospholipase A2 from the venom of the cottonmouth snake at 2.0 Åresolution. J. Biol. Chem. 1990;266:17649–17656. doi: 10.2210/pdb1ppa/pdb. [DOI] [PubMed] [Google Scholar]
- 14.Ward R. J., Chioato L., de Oliveira A. H. C., Ruller R., Sá J. M. Active-site mutagenesis of a Lys49-phospholipase A2: biological and membrane-disrupting activities in the absence of catalysis. Biochem. J. 2002;362:89–96. doi: 10.1042/0264-6021:3620089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arni R. K., Ward R. J., Gutiérrez J. M., Tulinsky A. Structure of a calcium-independent phospholipase-like myotoxic protein from Bothrops asper. Acta Crystallogr. D. 1995;51:311–317. doi: 10.1107/S0907444994011455. [DOI] [PubMed] [Google Scholar]
- 16.Scott D. L., Achari A., Vidal J. C., Sigler P. B. Crystallographic and biochemical studies of the (inactive) Lys49 phospholipase A2 from the venom of Agkistrodon piscivorus piscivorus. J. Biol. Chem. 1992;267:22645–22657. [PubMed] [Google Scholar]
- 17.Díaz C., Gutiérrez J. M., Lomonte B., Gene J. A. The effect of myotoxins isolated from Bothrops snake venoms on multilamellar liposomes: relationship to phospholipase A2, anticoagulant and myotoxic activities. Biochem. Biophys. Acta. 1991;1070:455–460. doi: 10.1016/0005-2736(91)90086-n. [DOI] [PubMed] [Google Scholar]
- 18.Rufini S., Cesaroni P., Desideri R. F., Gubensek F., Gutiérrez J. M., Luly P., Maassoud R., Morero R., Pedersen J. Z. Calcium ion independent membrane leakage induced by phospholipase-like myotoxins. Biochemistry. 1992;31:12424–12430. doi: 10.1021/bi00164a018. [DOI] [PubMed] [Google Scholar]
- 19.de Oliveira A. H. C., Giglio J. R., Andrião-Escarso S. H., Ito A. S., Ward R. J. A pH induced dissociation of the dimeric form of a lysine49 phospholipase A2 abolishes Ca2+-independent membrane damaging activity. Biochemistry. 2001;40:6912–6920. doi: 10.1021/bi0026728. [DOI] [PubMed] [Google Scholar]
- 20.da Silva Giotto M. T., Garratt R. C., Oliva G., Mascarenhas Y. P., Giglio J. R., Cintra A. C., de Azevedo W. F., Jr, Arni R. K., Ward R. J. Crystallographic and spectroscopic characterization of a molecular hinge: conformational changes in bothropstoxin I, a dimeric Lys49-phospholipase A2 homologue. Proteins. 1998;30:442–454. doi: 10.1002/(sici)1097-0134(19980301)30:4<442::aid-prot11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 21.Chioato L., de Oiveira A. H. C., Ruller R., Sá J. M., Ward R. J. Distinct sites for myotoxic and membrane-damaging activities in the C-terminal region of a Lys49 phospholipase A2. Biochem. J. 2002;366:971–976. doi: 10.1042/BJ20020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward R. J., Rodrigues Alves A., Rugierro Neto J., Arni R. K., Casari J. A SequenceSpace analysis of Lys49 phospholipases A2: clues towards identification of residues involved in a novel mechanism of membrane damage and in myotoxicity. Protein Eng. 1998;11:285–294. doi: 10.1093/protein/11.4.285. [DOI] [PubMed] [Google Scholar]
- 23.Maraganore J. M., Merutka G., Cho W., Welches W., Kezdy F. J., Heinrickson R. L. A new class of phospholipase A2 with lysine in place of aspartate 49. J. Biol. Chem. 1984;259:13839–13843. [PubMed] [Google Scholar]
- 24.Dupureur C. M., Yu B.-Z., Ramone A., Jain M. K., Tsai M.-D. Phospholipase A2 engineering. The structural and functional roles of aromaticity and hydrophobicity in the conserved phenylalanine-22 and phenylalanine-106 aromatic sandwich. Biochemistry. 1992;31:10576–10583. doi: 10.1021/bi00158a021. [DOI] [PubMed] [Google Scholar]
- 25.Liu X., Zhu H., Huang B., Rogers J., Yu B.-Z., Kumar A., Jain M. K., Sundaralingham M., Tsai M.-D. Phospholipase A2 engineering: probing the structural and functional roles of N-terminal residues with site-directed mutagenesis, X-ray, and NMR. Biochemistry. 1995;34:7322–7334. doi: 10.1021/bi00022a005. [DOI] [PubMed] [Google Scholar]
- 26.Homsi-Brandenburgo M. I., Queiroz L. S., Santo-Neto H., Rodrigues-Simoni L., Giglio J. R. Fractionation of Bothrops jararacussu snake venom: partial chemical characterization and biological activity of bothropstoxin. Toxicon. 1988;26:615–627. doi: 10.1016/0041-0101(88)90244-9. [DOI] [PubMed] [Google Scholar]
- 27.Ward R. J., Monesi N., Arni R. K., Larson R. E., Paço-Larson M. L. Sequence of a cDNA encoding bothropstoxin I, a myotoxin from the venom of Bothrops jararacussu. Gene. 1995;156:305–306. doi: 10.1016/0378-1119(95)00099-r. [DOI] [PubMed] [Google Scholar]
- 28.Ward R. J., de Oliveira A. H., Bortoleto R. K., Rosa J. C., Faca V. M., Greene L. J. Refolding and purification of Bothropstoxin-I, a Lys49-phospholipase A2 homologue, expressed as inclusion bodies in Escherichia coli. Protein Expr. Purif. 2001;21:134–140. doi: 10.1006/prep.2000.1353. [DOI] [PubMed] [Google Scholar]
- 29.Heinrickson R. L. Dissection and sequence analysis of phospholipase A2. Methods Enzymol. 1991;197:201–215. doi: 10.1016/0076-6879(91)97146-p. [DOI] [PubMed] [Google Scholar]
- 30.Nelson M. R., Long G. L. A general method of site-specific mutagenesis using a modification of the Thermus aquaticus polymerase chain reaction. Anal. Biochem. 1989;180:147–151. doi: 10.1016/0003-2697(89)90103-6. [DOI] [PubMed] [Google Scholar]
- 31.Yadav S., Ahmad F. A new method for the determination of stability parameters of proteins from their heat-induced denaturation curves. Anal. Biochem. 2000;283:207–213. doi: 10.1006/abio.2000.4641. [DOI] [PubMed] [Google Scholar]
- 32.Ruller R., Ferreira T. L., de Oliveira A. H. C., Ward R. J. Chemical denaturation of a homodimeric lysine-49 phospholipase A2: a stable dimer interface and a native monomeric intermediate. Arch. Biochem. Biophys. 2003;411:112–120. doi: 10.1016/s0003-9861(02)00712-9. [DOI] [PubMed] [Google Scholar]
- 33.Shimohigashi Y., Tani A., Matsumoto H., Nakashima K., Yamaguchi Y. Lysine-49-phospholipases A2 from Trimeresurus flavoviridis venom are membrane-acting enzymes. J. Biochem. (Tokyo) 1995;118:1037–1044. doi: 10.1093/jb/118.5.1037. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi Y., Shimohigashi Y., Chiwata T., Tani A., Chijiwa T., Lomonte B., Ohno M. Lys-49-phospholipases A2 as active enzyme for β-arachidonoyl phospholipid bilayer membranes. Biochem. Mol. Biol. Int. 1997;43:19–26. doi: 10.1080/15216549700203771. [DOI] [PubMed] [Google Scholar]
- 35.Wery P., Schevitz R. W., Clawson D. K., Bobbitt J. L., Dow E. R., Gamboa G., Goodson T., Hermann R. B., Kramer R. M., McClure D. B., et al. Structure of recombinant human rheumatoid arthritic synovial fluid phospholipase A2 at 2.2 Å. Nature (London) 1991;352:79–86. doi: 10.1038/352079a0. [DOI] [PubMed] [Google Scholar]
- 36.Diaz C., Gutiérrez J. M., Lomonte B., Nunez L. p-Bromophenacyl bromide modification of Bothrops asper myotoxin II, a lysine-49 phospholipase A2, affects its pharmacological activities. Toxicon. 1993;31:1202–1206. doi: 10.1016/0041-0101(93)90136-7. [DOI] [PubMed] [Google Scholar]
- 37.Andrião-Escarso S. H., Soares A. M., Rodrigues V. M., Ângulo Y., Diaz C., Lomonte B., Gutierrez J. M., Giglio J. R. Myotoxic phospholipases A(2) in Bothrops snake venoms: effect of chemical modifications on the enzymatic and pharmacological properties of bothropstoxins from Bothrops jararacussu. Biochimie. 2000;82:755–763. doi: 10.1016/s0300-9084(00)01150-0. [DOI] [PubMed] [Google Scholar]
- 38.Soares A. M., Andrião-Escarso S. H., Bortoleto R. K., Rodrigues-Simioni L., Arni R. K., Ward R. J., Gutierrez J. M., Giglio J. R. Dissociation of enzymatic and pharmacological properties of piratoxins-I and -III, two myotoxic phospholipases A2 from Bothrops pirajai snake venom. Arch. Biochem. Biophys. 2001;387:188–196. doi: 10.1006/abbi.2000.2244. [DOI] [PubMed] [Google Scholar]
- 39.Guex N., Peitsch M. C. SWISS-MODEL and the Swiss-Pdb viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]