Abstract
The X-rays emitted during CT scans can increase solid cancer risks by damaging DNA, with the risk tied to patient-specific organ doses. This study aims to establish a new method to predict patient specific abdominal organ doses from CT examinations using minimized computational resources at a fast speed. The CT data of 247 abdominal patients were selected and exported to the auto-segmentation software named DeepViewer to generate abdominal regions of interest (ROIs). Radiomics feature were extracted based on the selected CT data and ROIs. Reference organ doses were obtained by GPU-based Monte Carlo simulations. The support vector regression (SVR) model was trained based on the radiomics features and reference organ doses to predict abdominal organ doses from CT examinations. The prediction performance of the SVR model was tested and verified by changing the abdominal patients of the train and test sets randomly. For the abdominal organs, the maximal difference between the reference and the predicted dose was less than 1 mGy. For the body and bowel, the organ doses were predicted with a percentage error of less than 5.2%, and the coefficient of determination (R2) reached up to 0.9. For the left kidney, right kidney, liver, and spinal cord, the mean absolute percentage error ranged from 5.1 to 8.9%, and the R2 values were more than 0.74. The SVR model could be trained to achieve accurate prediction of personalized abdominal organ doses in less than one second using a single CPU core.
Keywords: Abdominal CT scanning, Patient-specific modeling, Radiation dosage, Radiomics, Support vector regression
Subject terms: Information theory and computation, Risk factors, Computational science, Scientific data
Introduction
Abdominal diseases are hazards to people worldwide because they cause symptoms and complications in abdominal organs and tissues. According to the WHO's Global Health Estimates liver diseases caused 1,312,480 (4.6%) of 28,444,814 deaths in the Asia-Pacific region1 Irritable bowel syndrome (IBS), gastroesophageal reflux disease (GERD), and functional dyspepsia account for 14% to 45% of the prevalence of functional gastrointestinal disorders, depending on diagnosis criteria or region2,3. About 20% of people suffer from infectious intestinal diseases in the United Kingdom each year, and serious complications (i.e., dehydration, malabsorption, sepsis, etc.) could also happen for those people4. Besides, stomach tumors have a relatively high incidence and may cause deadly complications or even deaths. In 2020, the estimated new stomach cancer cases and deaths respectively were 1.03 million and 782,6855. Thus, it is necessary to diagnose abdominal diseases as early as possible to make treatments more effective and enhance patients’ quality of life.
Computed tomography (CT) is widely applied in diagnosing abdominal diseases because of its ability to provide slice-wise anatomical images of abdominal organs and tissues. CT imaging could detect various abdominal diseases including infections, traumas, inflammations, hemorrhages, etc. For general surgery, CT imaging almost detects 95% of ordinary abdominal disease cases6. In the emergency department, 50% to 60% of patients with abdominal pain get examined by CT7. The radiation exposure from such a huge amount of abdominal CT imaging could be a risk factor that leads to secondary cancers in the long term. The data from the American Cancer Society (ACS) indicates a CT scan of the abdomen and pelvis makes a patient get as much as 10 mSv X-ray radiation8. This amount of radiation dose is almost equivalent to 3 years of natural background exposure. Australian retrospective research concluded that a 24% increase in overall cancer incidence was observed among those individuals who get exposed to CT radiation9. Thus, it is urgent to investigate the personalized organ dose from abdominal CT imaging so that the radiation-induced risk and potential complications can be accurately evaluated and controlled.
To date, size-specific dose estimate (SSDE) and Monte Carlo (MC) dose calculation, are the main methods for estimating personalized organ dose from CT diagnosis10–19. SSDE just considers size information but neglects the information regarding anatomy, tissue, and geometric shape, which may cause higher errors. MC simulation could calculate personalized organ dose directly from the patient’s CT and segmented regions of interest (ROIs) accurately, but it requires intensive computational resources of the central processing unit (CPU), graphics processing unit (GPU), or even in computing time depending on the MC software used. Until now, there were almost no previous studies that predicted personalized abdominal organ doses by training SVR models. Thus, it is necessary to explore new methods to accurately predict personalized abdominal organ doses from CT examinations within a short time using fewer computational resources.
In this study, the robust SVR model20 was trained based on the abdominal radiomics features and GPU MC-calculated organ doses to predict personalized abdominal organ dose within a short time, with relatively better robustness, using fewer abdominal patient cases and minimized computational resources. The abdominal radiomics features were extracted from auto-segmented organ contours to consider much more intensity, anatomical, and tissue properties beyond the water-equivalent diameter (Dw) or patient size used by the SSDE prediction strategy. Abdominal organ contours were generated by auto-segmentation which was approved to be effective to accurately delineate internal organ contours21.
The SVR model trained based on radiomics features sufficiently reflected the correlation between complex abdominal patient features and organ doses, thus precisely predicting personalized abdominal organ doses. The performance of the organ dose estimation method was evaluated by calculating the relative root mean squared error (RRMSE), mean absolute percentage error (MAPE), and coefficient of determination (R2) on the test sets. The robustness of the SVR model was verified by randomly allocating patient samples to the train set and test set 20 times when keeping the proportion between the two sets as 0.8 versus 0.2.
Ethics
The study was conducted following the Declaration of Helsinki. Informed consents was obtained from patients who underwent abdominal CT scans for diagnosing purpose. We used the existing CT data in our hospital to achieve retro research. The research was approved by the Ethics Committee of Shanghai Zhongye Hospital to access the CT data for research purposes. The study followed the ethical guidelines of the hospital. The CT data were anonymized, and no personal information was disclosed.
Results
Predicted organ doses
In this section, the mean values of reference and predicted abdominal organ doses and their corresponding standard deviation (SD) were computed on the test sets. As shown in Table 1, in terms of the mean organ doses on the test sets, the predicted organ doses fitted very well with the reference organ doses, the maximal difference between predicted and reference doses was smaller than 1 mGy. This indicated that the SVR model trained based on the abdominal radiomics features could achieve a relatively small deviation between the predicted and reference dose values.
Table 1.
Mean reference and predicted abdominal organ doses on the test sets.
| Organ | Mean reference dose (mGy) | SD of reference dose (mGy) | Mean predicted dose (mGy) | SD of predicted dose (mGy) |
|---|---|---|---|---|
| Body | 31.06 | 10.01 | 30.83 | 9.45 |
| Bowel | 25.52 | 11.39 | 25.16 | 10.56 |
| Kidney_L | 26.73 | 9.73 | 26.27 | 8.62 |
| Kidney_R | 25.68 | 10.29 | 26.02 | 8.34 |
| Liver | 29.54 | 13.34 | 29.69 | 9.73 |
| Spinal cord | 16.84 | 7.78 | 17.06 | 5.56 |
In Table 1, the “KidneyL” and “KidneyR” respectively denote the left kidney and right kidney.
Regression metrics for organ dose prediction
The RRMSE, MAPE, and R2 were calculated for the patient samples on the test sets. As exhibited in Table 2, the SVR prediction achieved the best performance as the RRMSE, MAPE, and R2 of 4.54%, 2.24%, and 0.96 for the body. For the abdominal organs except the body, the RRMSE values ranged from 7.34% to 12.78%, while the MAPE values were smaller than 8.89%. For all the investigated organs, the R2 values were more than 0.74 and reached up to the maximal value of 0.96 for the body.
Table 2.
Calculated regression metrics on the test sets.
| Organ | RRMSE (%) | MAPE (%) | R2 |
|---|---|---|---|
| Body | 4.54 | 2.24 | 0.96 |
| Bowel | 7.34 | 5.15 | 0.90 |
| Kidney_L | 10.12 | 5.94 | 0.79 |
| Kidney_R | 10.90 | 7.35 | 0.77 |
| Liver | 12.78 | 8.89 | 0.74 |
| Spinal cord | 9.17 | 8.63 | 0.75 |
In Table 2, the “KidneyL” and “KidneyR” respectively denote the left kidney and right kidney.
Robustness of the SVR dose prediction model
In this section, the patient samples on the training and test sets were randomly allocated 20 times keeping the proportion of the two sets as 0.8 versus 0.2. The regression metrics of the 20 patient allocation strategies were shown in Fig. 1. As seen in Fig. 1a, the interquartile range of RRMSE was from 1 to 2.5% for the abdominal organs, indicating that the performance of the SVR model remained stable when the patient samples were randomly rearranged. As illustrated in Fig. 1b, the interquartile range of MAPE was smaller than 2.2%. As shown in Fig. 1c, the interquartile range of R2 was less than 0.07%, which suggested that the performance of the SVR model remained stable against the variations in patient sample allocation strategies.
Figure 1.
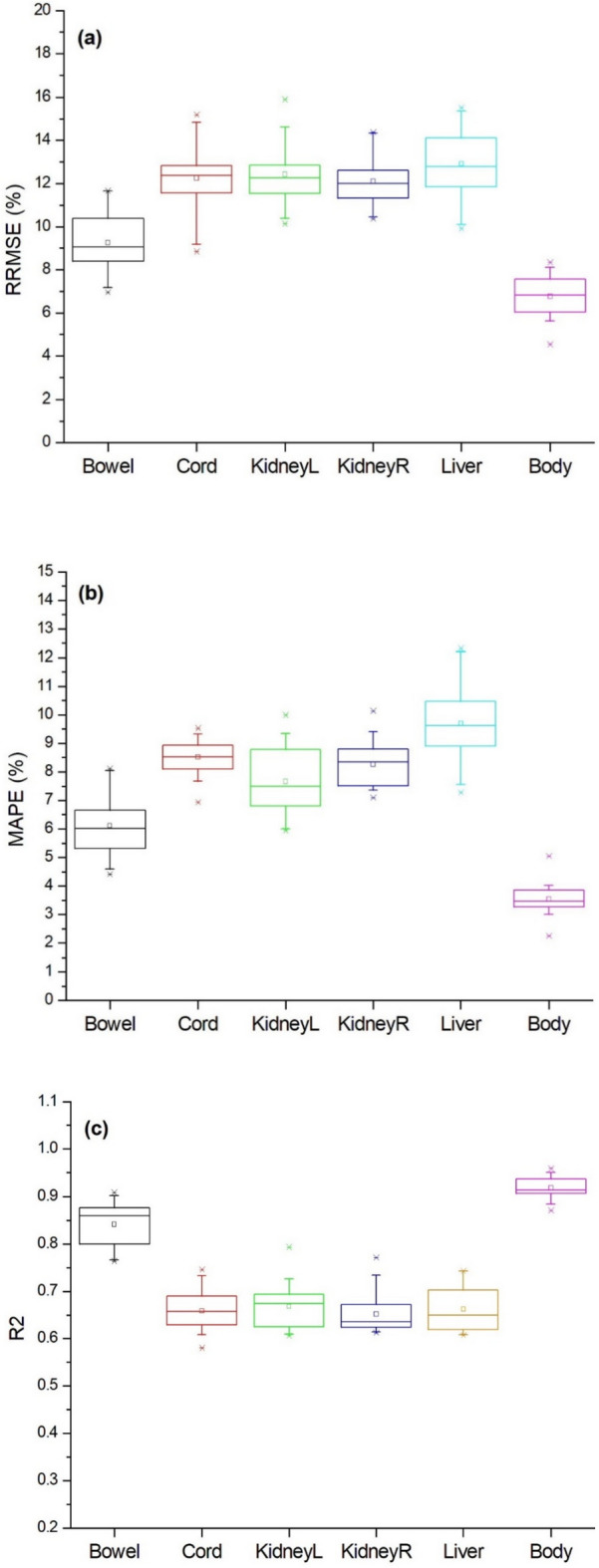
Box charts for RRMSE (a), MAPE (b), and R2 (c) for 20 random patient allocation strategies. In Fig. 2, the “KidneyL” and “KidneyR” respectively denote the left kidney and right kidney.
Discussion
Abdominal CT imaging plays an essential role in detecting abdominal diseases, monitoring abdominal disease progression, and monitoring treatment outcomes. However, the ionization radiation from abdominal CT scans could cause radiation complications or even secondary cancers. This study aimed to explore a novel method to predict accurate and robust personalized abdominal organ doses when the resources for prediction were minimized, even minimized down to a single CPU core. Previous studies merely considered the patient size and some other simple features. Unlike existing studies, this study adopted the radiomics features to consider the characteristics of patient tissue, anatomy, and geometric shape, and train the SVR model for predicting personalized abdominal organ doses. The MAPE and R2 of the SVR regression model were about 0.08 and 0.7, respectively, which suggested that radiomics features could reflect the abdominal organ features (e.g. geometric shape, size, tissue, and anatomy) as much as possible.
There were already previous studies investigating personalized organ doses for different diagnosed sites (i.e. head, chest). However, studies using the SVR prediction model dedicated to abdominal CT examinations were very rare. For the previous studies that adopted SSDE, neural networks, convolutional networks, etc., the robustness of those models had not been validated by randomly assigning patient samples to the train and test sets. For the previous MC simulation studies, a massive amount of computing resources of multiple CPUs or GPUs were required, even professional computer science was one of the prerequisites to use MC simulation to calculate personalized organ doses. The trained SVR model just needs a simple Python script to predict organ doses in one second, with minimized computing resources, even with a single CPU. In practical abdominal CT examinations, the proposed method can be applied in two main ways. First, the method can be utilized immediately after CT reconstruction through three steps: importing the CT images into segmentation software (e.g., 3D Slicer) for abdominal organ delineation, extracting radiomics features quickly, and performing fast SVR-based prediction of abdominal organ doses for clinical organ dose evaluation. Second, the method can be applied to a large amount of retrospective abdominal CT data to enable fast batch prediction of abdominal doses for research purposes. The predictive performance in MAPE for previous studies with deep learning is from 2 to 10%, which is in the similar range with the performance of SVR-based abdominal organ dose prediction models. This indicates the proposed SVR-based models could achieve accurate and fast predict using much less computation resources compared with deep learning models. This valuable advancement could significantly reduce computational costs in terms of time, hardware, and electrical energy when predicting organ doses from ionizing radiation examinations on extremely large datasets for research purposes. We also compared the performance of SVR and fully connected neural networks (FCNN) in predicting chest organ doses from CT scans. For the right lung, left lung, overall lungs, esophagus, heart, and trachea, the trained SVR model achieved a MAPE between 0.10 and 0.18, with R-squared values ranging from 0.75 to 0.89 on the test sets. In comparison, the trained FCNN model had a MAPE ranging from 0.11 to 0.15 and R-squared values from 0.74 to 0.86 on both the training and test sets.
This study has several defects as well. First, we just trained the SVR prediction model from the abdominal radiomics features of the patients diagnosed in our institution. The accuracy and robustness of the model could vary among institutions. In the future, the collaboration of multiple institutions is still necessary to verify the cross-institution effectiveness of the proposed method. Second, the GPU-calculated reference organ doses were merely validated by CTDIvol of the CT device in our institution. It is still necessary to train the SVR model for multiple CT device types. Although we investigated most of the abdominal organs, we did not include the stomach. The gastric contents varied severely among abdominal patients, causing PyRadiomics to be pretty hard to extract effective radiomic features. This variation hindered the ability to distinguish patient-specific characteristics between different patients. In the future, it is necessary to investigate the prediction model that has the potential to obtain more detailed image information inside the stomach. This study adopted resampling in Pyradiomics module to decrease the impact of voxel size discrepancy among different patients on standardized feature extraction. However, resampling also could impact feature extraction as well. Thus, the impacts of voxel size discrepancy and resampling still need to be assessed and compared in the future.
Methods
Data collection
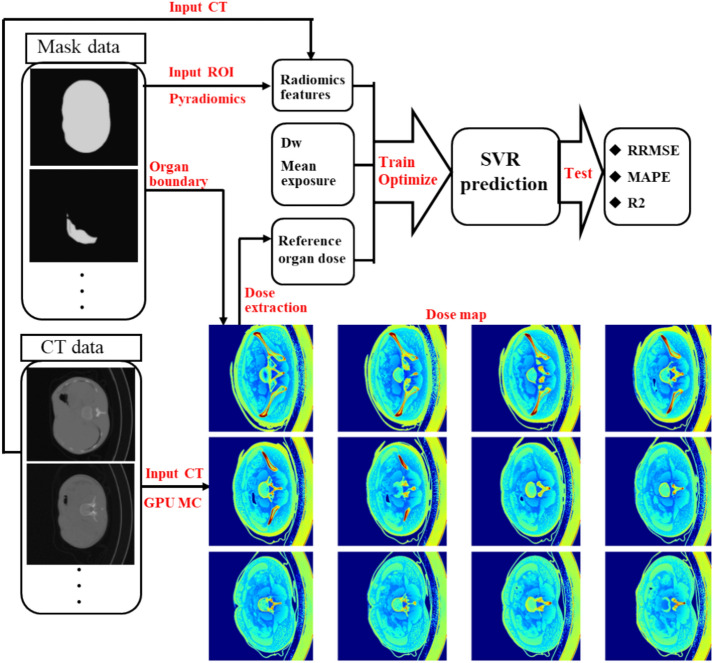
In this study, we randomly selected 247 abdominal patients who got abdominal CT examinations at Shanghai Zhongye Hospital. The CT images of the selected patients were exported to an auto-segmentation system named DeepViewer22 to segment abdominal ROIs. The segmented ROIs respectively were liver, bowel, left kidney, right kidney, and spinal cord. The abdominal CT images and ROIs of each patient were converted from DICOM to Nifti using dcmstruct2nii23 so as to generate the CT and mask data for radiomics feature extraction. The scan voltage was set as 100 kV, and tube current modulation was used.
Radiomics feature extraction
The Pyradiomics module24 was applied to extract radiomics features from each patient’s CT and ROIs. The main procedures included image preprocessing, feature calculation, and feature selection. In the first step, the spatial resolution of the CT and masks for each patient were resampled to (1, 1, 5) to standardize radiomics feature calculation via the Pyradiomics parameter “resamplePixelSpacing”. Data augmentation25–27 was performed to enhance the robustness of the SVR model. Data augmentation involved slight shifts and rotations. Zooming was excluded as it could potentially alter the patients’ dosimetric properties. In the second step, 107 radiomics features were extracted for each organ of the patient without using filters.
The radiomics features could be divided into 7 types including Gray Level Co-occurrence Matrix, First Order Statistics, Neighboring Gray Tone Difference Matrix, Gray Level Dependence Matrix, Gray Level Run Length Matrix, Shape-based, and Gray Level Size Zone Matrix. Those features reveal the ROIs’ correlation, homogeneity, contrast, intensity distribution, etc., and were used as input data for training the SVR model. In the third step, the f-regression function the scikit-learn module was used to select the relevant features to avoid overfit28 and to enhance the robustness of the SVR model. The main hypotheses of the F-test—including independence, linearity, homoscedasticity, and normality of errors—should ideally be respected for all radiomic features to ensure the validity of the test. A p-value threshold was not explicitly used to select the relevant features; instead, the top 30 features were selected based on their ranking by the F-statistic. The feature extraction and selection were performed with double AMD EPYC 7551 CPUs in the Anaconda 329 environment.
Reference organ dose calculation
Based on the CT image and masks of each patient, the reference organ doses were calculated by GGEMS, a GPU-based MC particle transport code that can address complex geometries, heterogeneous materials, and multiple radiation sources (photons and electrons)30. GGEMS could process MC simulation much faster than CPU-based MC simulation code almost without sacrificing organ dose calculation accuracy. For each patient, the GPU-calculated organ doses were used as the reference organ doses when training the SVR model to predict the organ doses of the liver, stomach, bowel, left kidney, right kidney, and spinal cord. The GPU-based MC simulations were performed with two Nvidia RTX4090 graphics cards to obtain the slice-wise dose distribution of each patient with less than 2% error in each voxel. Auto-tube current was taken into account for MC dose calculation.
SVR prediction model
This study adopted the trained SVR regression model to reflect the relationship between the input radiomics features and reference organ doses, and predict personalized abdominal organ doses accurately. SVR is a supervised learning algorithm that is used to predict continuous values31–33. SVR uses the same principle as support vector machine (SVM), which are classifiers that find the optimal hyperplane that separates two classes of data. Instead of finding a hyperplane that maximizes the margin between the classes, SVR finds a hyperplane that minimizes the error between the predicted and actual values, while allowing some tolerance for deviation.
The performance of the model was assessed by RRMSE, APE, MAPE, and R2 on the test set for the ROIs including the liver, stomach, bowel, left kidney, right kidney, and spinal cord. The robustness of the model was verified by randomly assigning patient samples to the train and test sets while keeping the proportion of the two sets as 0.8:0.2 and comparing the regression metrics of different patient sample assigning strategies. The SVR was trained and tested using two Nvidia GeForce RTX4090 GPUs and two Nvidia GeForce RTX3080ti GPUs in the Anaconda environment. Figure 1 shows the General flowchart of training and testing the SVR-based abdominal organ dose prediction model.
Figure 2.
General flowchart of preparing input data plus training and testing the SVR-based abdominal organ dose prediction model.
Regression metrics for the dose prediction
For assessing the performance of abdominal organ dose prediction, we adopted the regression metrics of RMSE, MAPE, and R2. RMSE is the root mean squared error. It measures how large the root mean squared error. A lower RMSE indicates a better fit. RMSE can be calculated as:
| 1 |
where n is the number of patients, is the reference organ dose, and is the predicted organ dose. MAPE is the average of the absolute percentage differences between the actual and predicted values. It measures how close the predictions are to the actual values in terms of percentage, without considering the direction of the error. The formula can be expressed as:
| 2 |
R2 is the proportion of the variance in the output variable that is explained by the input variables. It measures how well the regression model fits the data. A higher R2 indicates a better fit. The formula is:
| 3 |
Acknowledgements
The authors have no conflicts of interest. The authors would like to thank Professor George X.Xu and his group for their technical support regarding the auto-segmentation of ROIs using DeepViewer.
Author contributions
Wencheng Shao contributed to concept design, data calculation, data analysis, writing, and revising the manuscript; Xin Lin, Wentao Zhao, and Ying Huang to GPU-accelerated dose calculation; Wentao Zhao and Liangyong Qu to data collection, Weihai Zhuo to concept design; Haikuan Liu to concept design and revising the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (12075064), and the National Key R&D Program of China (2019YFC0117304).
Data availability
Data availability Data sets generated during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Weihai Zhuo, Email: whzhuo@fudan.edu.cn.
Haikuan Liu, Email: liuhk@fudan.edu.cn.
References
- 1.Sarin, S. K. et al. Liver diseases in the Asia-Pacific region: A lancet gastroenterology & hepatology commission. Lancet Gastroenterol. Hepatol.5, 167–228. 10.1016/S2468-1253(19)30342-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Bortoli, N. et al. Gastroesophageal reflux disease, functional dyspepsia and irritable bowel syndrome: Common overlapping gastrointestinal disorders. Ann. Gastroenterol.31, 639–648. 10.20524/aog.2018.0314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee, S. Y. et al. Prevalence and risk factors for overlaps between gastroesophageal reflux disease, dyspepsia, and irritable bowel syndrome: A population-based study. Digestion79, 196–201. 10.1159/000212077 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Tam, C. C. et al. Longitudinal study of infectious intestinal disease in the UK (IID2 study): Incidence in the community and presenting to general practice. Gut61, 69–77. 10.1136/gut.2011.238386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249. 10.3322/caac.21660 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Nan, Y. et al. Role of CT Images in the diagnosis of common acute abdominal diseases in general surgery. J. Healthc. Eng.10.1155/2022/5732357 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Cartwright, S. L. & Knudson, M. P. Evaluation of acute abdominal pain in adults. Am. Fam. Physician91, 452–459 (2015). [PubMed] [Google Scholar]
- 8.American Cancer Society. Understanding radiation risk from imaging tests. https://www.cancer.org/cancer/diagnosis-staging/tests/imaging-tests/understanding-radiation-risk-from-imaging-tests.html. Accessed 28 Jan 2024
- 9.Mathews, J. D. et al. Cancer risk in 680000 people exposed to computed tomography scans in childhood or adolescence: Data linkage study of 11 million Australians. BMJ.346, f2360. 10.1136/bmj.f2360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajaraman, V., Ponnusamy, M. & Halanaik, D. Size specific dose estimate (SSDE) for estimating patient dose from CT used in myocardial perfusion SPECT/CT. Asia Ocean J. Nucl. Med. Biol.8, 58–63. 10.22038/aojnmb.2019.40863.1276 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, J. et al. Personalized organ dose estimation for chest CT based on deep learning segmentation techniques. Phys. Med. Biol.68, 035006. 10.1088/1361-6560/ac0e7a (2023). [Google Scholar]
- 12.Cagni, E. et al. Personalized Monte Carlo-based organ dose estimates in spiral CT examinations using a portable optical scanner. Phys. Med. Biol.68, 045003. 10.1088/1361-6560/ac0f5a (2023). [Google Scholar]
- 13.Myronakis, M., Stratakis, J. & Damilakis, J. Rapid estimation of personalized organ doses using a deep learning network. Med. Phys.50(11), 7236–7244. 10.1002/mp.16356 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Salimi, Y., Akhavanallaf, A., Mansouri, Z., Shiri, I. & Zaidi, H. Real-time, acquisition parameter-free voxel-wise patient-specific Monte Carlo dose reconstruction in whole-body CT scanning using deep neural networks. Eur Radiol.33(12), 9411–9424. 10.1007/s00330-023-09839-y (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tzanis, E. & Damilakis, J. A novel methodology to train and deploy a machine learning model for personalized dose assessment in head CT. Eur Radiol32(9), 6418–6426. 10.1007/s00330-022-08756-w (2022). [DOI] [PubMed] [Google Scholar]
- 16.Maier, J., Klein, L., Eulig, E., Sawall, S. & Kachelrieß, M. Real-time estimation of patient-specific dose distributions for medical CT using the deep dose estimation. Med. Phys.49(4), 2259–2269. 10.1002/mp.15488 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Myronakis, M., Stratakis, J. & Damilakis, J. Rapid estimation of patient-specific organ doses using a deep learning network. Med. Phys.50(11), 7236–7244. 10.1002/mp.16356 (2023). [DOI] [PubMed] [Google Scholar]
- 18.Tzanis, E., Stratakis, J., Myronakis, M. & Damilakis, J. A fully automated machine learning-based methodology for personalized radiation dose assessment in thoracic and abdomen CT. Phys Med.117, 103195. 10.1016/j.ejmp.2023.103195 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Principi, S. et al. Deterministic linear Boltzmann transport equation solver for patient-specific CT dose estimation: Comparison against a Monte Carlo benchmark for realistic scanner configurations and patient models. Med. Phys.47, 6470–6483 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, T. & Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (2016). 10.1145/2939672.2939785
- 21.Maffei, N. et al. Radiomics classifier to quantify automatic segmentation quality of cardiac sub-structures for radiotherapy treatment planning. Phys. Med.83, 278–286. 10.1016/j.ejmp.2021.05.009 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Peng, Z., et al. Validation and clinical application of DL-based automatic target and OAR segmentation software, DeepViewer. In Proceedings of the American Association of Physicists in Medicine Annual Meeting (Vancouver, BC, July 12–16) 123–124 (2020).
- 23.Li, X. et al. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J. Neurosci. Methods264, 47–56. 10.1016/j.jneumeth.2016.03.001 (2016). [DOI] [PubMed] [Google Scholar]
- 24.van Griethuysen, J. J. M. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res.77, e104–e107. 10.1158/0008-5472.CAN-17-0339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rebuffi, S. A., et al. Data augmentation can improve robustness. In Advances in Neural Information Processing Systems 34 (NeurIPS 2021) (Neural Information Processing Systems Foundation).
- 26.Dwibedi, D., Misra, I. & Hebert, M. Cut, paste and learn: Surprisingly easy synthesis for instance detection. In Proceedings of the IEEE International Conference on Computer Vision 1301–1310 (2017). 10.1109/ICCV.2017.144
- 27.Zhang, Z., et al. A Robustness-oriented data augmentation method for DNN. In 2021 IEEE 21st International Conference on Software Quality, Reliability and Security Companion (QRS-C) 1–6 (2021).
- 28.Guyon, I. & Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res.3, 1157–1182 (2003). [Google Scholar]
- 29.Anaconda. The World’s Most Popular Data Science Platform. https://www.anaconda.com. Accessed 28 Jan 2024.
- 30.Bert, J., et al. GGEMS: GPU GEant4-based Monte Carlo Simulation platform. In 2016 IEEE Nuclear Science Symposium, Medical Imaging Conference and Room-Temperature Semiconductor Detector Workshop (NSS/MIC/RTSD) 1–4 (2016). 10.1109/NSSMIC.2016.8069508
- 31.Smola, A. J. & Schölkopf, B. A tutorial on support vector regression. Stat. Comput.14, 199–222 (2004). [Google Scholar]
- 32.Vapnik, V. N. The Nature of Statistical Learning Theory 2nd edn. (Springer, New York, 2000). [Google Scholar]
- 33.Drucker, H. et al. Support vector regression. In Advances in Neural Information Processing Systems 9 (NIPS 1996) (eds Mozer, M. C. et al.) 155–161 (MIT Press, Cambridge, 1997). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data availability Data sets generated during the current study are available from the corresponding author on reasonable request.