Abstract
Purpose
Application of metagenomic next-generation sequencing (mNGS) in identifying nosocomial central nervous system (CNS) infections in critical care units remains understudied.
Methods
We conducted a retrospective analysis of microbiological results through both mNGS and routine examination of cerebrospinal fluid (CSF) samples from patients with nosocomial CNS infections. The aim of this study was to assess the clinical diagnostic effect of nosocomial mNGS in this population.
Results
The study included 26 cases of nosocomial CNS infections in total. A total of 69.2% (18/26) of the samples tested positive for mNGS, which is substantially greater than the 7.7% (2/26; p<0.05) detected through conventional techniques. Administration of antibiotics before culture is most likely the cause of the low CSF culture rate. Twenty-five pathogenic strains that were missed by standard testing. Three pathogens that were consistent with the mNGS results were positive by routine tests. Eight cases were negative by mNGS due to low pathogen CSF titres. Compared to traditional testing, mNGS demonstrated 100% sensitivity and 33.3% specificity in diagnosing CNS infections. The thirty-day mortality rate was 26.9% (7/26).
Conclusion
Routine microbiologic testing frequently falls short of detecting all neuroinvasive pathogens. Our research suggests that mNGS offers an alternative means of detecting nosocomial CNS infections. By applying mNGS to CSF samples from patients with meningitis or encephalitis, we were able to improve the ability to diagnose nosocomial neurologic infections.
Keywords: metagenomic next-generation sequencing, cerebrospinal fluid, central nervous system infections, diagnosis
Central nervous system (CNS) infections affecting the brain and spinal cord are primarily caused not only by bacteria and viruses but also by fungi and parasites. CNS infections are severe and life-altering conditions, and they are the most frequent cause of hospitalizations in many developing countries, making them a significant public concern.1 Globally, CNS infectious disease has a high mortality rate.2
It is difficult to diagnose CNS infections. Due to limited diagnostic capacity and difficulties involved in performing exploratory lumbar punctures, there is a dearth of information regarding aetiology. PCR and culture are useful techniques for detecting pathogenic bacteria, but uncommon pathogenic bacteria involving CNS infections remain difficult to identify by PCR and culture.3 First, antibiotic treatment reduces the detection rate of CSF culture, which is the gold standard for diagnosing bacterial meningitis, and the majority of patients receive antibiotic treatment prior to lumbar puncture. Moreover, different media and cultivation conditions-some of which are intricate and time consuming-are necessary for microbial culture. The presence of Bartonella, for instance, which grows in culture for roughly two weeks,4 reduces diagnostic efficiency. Furthermore, accurate primers are needed for PCR amplification procedures, and if the primers do not match the target nucleic acid sequence, the results may be erroneous. Thus, the rate of positive detection for PCR amplification techniques is low.5 Overall, conventional detection techniques have several limitations,6 and it is difficult to culture organisms from patients with CNS infections,7 almost half of CNS infection patients cannot receive an aetiological diagnosis.8 CNS infection diagnosis is a very challenging task. Nevertheless, precise identification of CNS infections (mixed infections) plays a vital role in clinical therapy.
Recently, next-generation sequencing (NGS) has opened a window into the infectious diseases. Pathogen-targeted NGS (ptNGS) utilizes certain microorganism-specific primers, enriches specific genetic targets for sequencing, and facilitate target pathogen identification. ptNGS had superior performance in CSF for the common pathogen, but ptNGS has a low detection rate for atypical pathogens.9 Since metagenomic NGS (mNGS) ensures detailed sequencing of the total DNA or RNA content of the microbiome in a single test, sequencing and complete mNGS analysis can be completed within 24–72h.10 mNGS has proven to be an effective technology for identifying all pathogenic bacteria, and it is particularly useful for identifying uncommon, complex and atypical infectious diseases in clinical practice.7 mNGS of CSF demonstrates the promising potential and can improve the diagnosis of neurologic infections.9
Traditional detection methods are time-consuming, and it is difficult to culture such organisms from patients with CNS infections.8 A key detection method for CNS infections is use of mNGS of nucleic acid isolated from CSF or brain tissue.3 However, studies on application of mNGS for the diagnosis of CNS infections in critical care units are limited. Thus, we conducted a retrospective analysis of data from critical care unit patients who had undergone both mNGS and traditional testing, such as CSF culture. The efficacy and clinical diagnosis of mNGS compared with that of routine examination were retrospectively analysed.
Materials and Methods
Ethics Statement and Informed Consent
This research was conducted in accordance with the guidelines and regulations of the institutional review board of Renmin Hospital of Wuhan University in China. Human specimens were used, and written informed consent was obtained from all patients or their legal representatives, in compliance with the Declaration of Helsinki and STROBE guidelines. The retrospective review protocol was approved by the institutional review board of Renmin Hospital of Wuhan University in China (2023-9-1, Approval No. WDRY2023-K140).
Study Population
Patients older than 18 years old with CNS infections who were admitted to the Department of Critical Care Medicine, Renmin Hospital of Wuhan University of Hubei Province, were enrolled for a retrospective cohort analysis. The research period was from July 1, 2021, to August 15, 2023. We included individuals who were examined by mNGS at Renmin Hospital of Wuhan University.
The patients were adults who presented with clinical manifestations consistent with CNS infection.3 A lack of specimens to perform mNGS, insufficient clinical data, and lumbar puncture rejection were among the exclusion criteria.11,12 Every patient underwent both conventional and mNGS tests using CSF samples obtained from their initial lumbar puncture, with 1–4 days needed to analyse the mNGS results. mNGS was only performed once for each patient owing to its relatively high cost, whereas traditional tests were repeated after every other test became negative. Brain magnetic resonance imaging (MRI) and computed tomography (CT) were also performed for each patient.
This study included 26 individuals with CNS infection. Four patients had meningitis alone, 21 had encephalitis with or without meningitis, and 1 had myelitis with meningitis. The patient population were postoperative meningitis, sinusitis, intracranial trauma and secondary bloodstream infections.
Specimen Collection
In clinical practice, patients suspected of having CNS infections undergo lumbar puncture and imaging after providing informed consent. CSF samples were regularly tested, including smears of bacteria and fungi, acid-fast staining, culture of bacteria and fungi, and testing for the full complement of viruses. mNGS of CSF was performed for all patients. We collected mNGS and microorganism culture data for this investigation.
mNGS Procedure
CSF samples were collected and transported to the sequencing facility. Following the manufacturer’s instructions, DNA was extracted using a QIA Pathogen Kit from Qiagen (Germany) and quantified using a Qubit fluorometer from Thermo Fisher Scientific (United States). The NextSeq 550Dx platform was utilized for the sequencing process. To eliminate human sequences, the qualifying reads were mapped to the human reference genome. To identify pathogens, the remaining reads were aligned to the NCBI microbial genome database. The sequencing data were analysed, and the mNGS results were available quickly. The threshold criteria of mNGS are based on standardized reads, coverage, and clinical relevance. For bacteria or fungi, a positive detection result was reported if the reads per million ratio (RPM-r) of a given species or genus was ≥10, where RPM-r was defined as the RPMsample/RPMNTC.13
Statistical Analysis
Statistical analysis was performed using SPSS 26.0 software (IBM Corp, Armonk, NY, United States). The mean ± standard deviation (SD) was used to represent continuous variables with normal distributions; medians and interquartile ranges (IQRs) were used to represent continuous variables with nonnormal distributions. Categorical variables are presented as counts (no.) and percentages (%). The chi-squared test was used to compare the pathogen detection rate between mNGS and traditional approaches. The sensitivity, specificity, positive predictive value and negative predictive value were calculated using the culture method as the reference standard. p < 0.05 was considered to indicate statistical significance.
Results
Patient Characteristics
The features of the 26 individuals enrolled in this retrospective study on CNS infections are detailed in Table 1. Among the patients, 57.7% (15/26) were male, with a median age of 52.5 years (range 21–75). The laboratory test data, including white blood cell count, C-reactive protein level, procalcitonin level and CSF biochemistry results, are provided in Table 1. The median protein concentration in the CSF was 285.5 mg/L (IQR: 704.93–2676.25 mg/L), and the median CSF glucose concentration was 4.37±2.1 mmol/L. The median CSF nucleated cell count was 41.5 mm3 (range 24–220.1 mm3). The patients in the study had a mean length of stay of 22.4 days (range 3 to 84) and spent an average of 11.2 days in the ICU. The overall 30-day mortality rate was 26.9% (7/26). Meningeal and encephalar enhancement (11/26, 42.31%) was the most prevalent cerebral imaging characteristic among the patients, as illustrated in Figure 1.
Table 1.
Baseline Characteristics of the Participants
| Variables | Total(n=26) |
|---|---|
| Demographics | |
| Age (y, Mean) | 52.5 (21, 75) |
| Gender, no. (%) | |
| Male | 15(57.7) |
| Female | 11(42.3) |
| Laboratory test | |
| WBCs (109/L), Mean±SD | 12.25±6.94 |
| Procalcitonin (ng/mL), median (IQR) | 0.83(0.17, 4.14) |
| CRP (mg/L), median (IQR) | 30.6(8.11, 124.15) |
| Syndrome, no. (%) | |
| Meningitis alone | 4 (15.4) |
| Encephalitis with or without meningitis | 21 (80.8) |
| Myelitis with or without meningitis | 1 (3.8) |
| CSF (mean±SD or median, interquartile range) | |
| Protein (mg/L) | 285.5(704.93,2676.25) |
| Chlorine (mmol/L) | 124.2(113.8, 129.9) |
| Glucose (mmol/L) | 4.37±2.1 |
| ADA (U/L) | 1.46(0.58, 4.23) |
| Nucleated cells (106 /L) | 41.5(24, 220.1) |
| Immunocompromised, no. (%) | |
| Solid-organ transplant | 1(3.8) |
| Chemotherapy | 5(19.2) |
| Immunosuppression for non-neoplastic condition | 2(7.7) |
| Death within 30 days, no. (%) | 7(26.9) |
| Mean length of stay (range), days | |
| In hospital | 22.4(3–84) |
| In ICU | 11.2(3–26) |
Abbreviations: IQR, interquartile range; WBC, white blood cell; CRP, C-reactive protein; CSF, cerebrospinal fluid; CSF cerebrospinal fluid; ICU, intensive care unit;
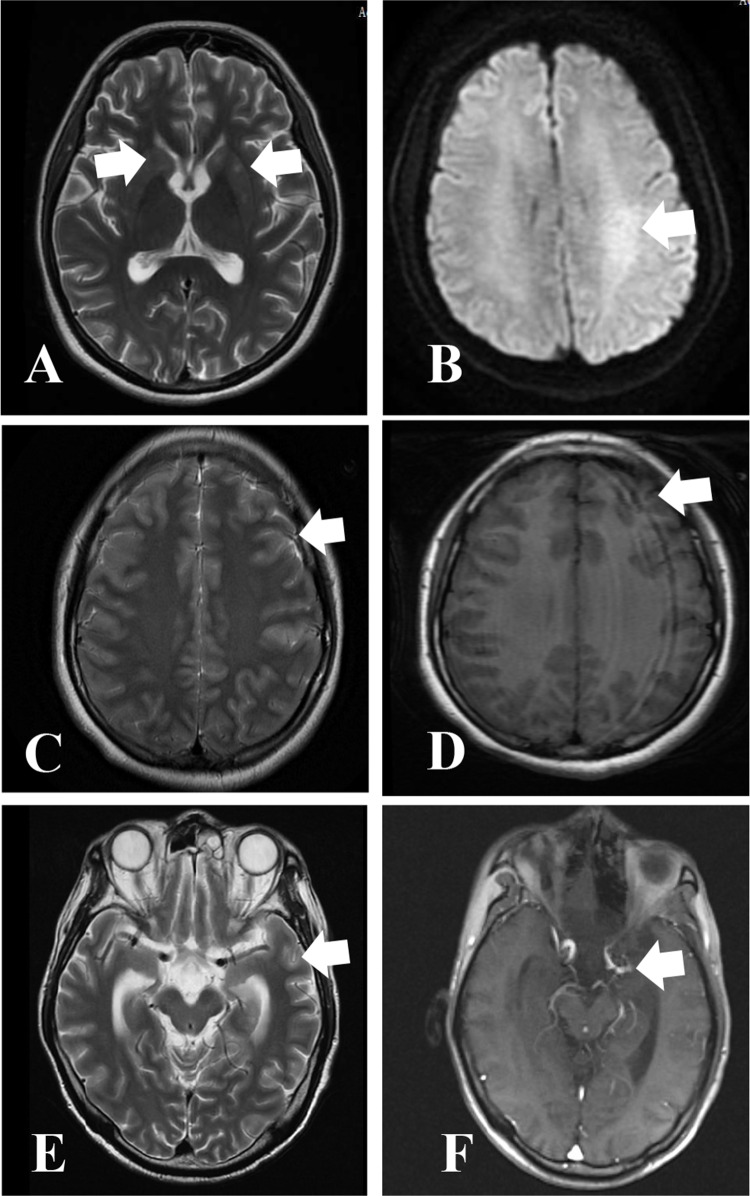
Figure 1.
Magnetic resonance imaging images of selected CNS infections cases. Abnormal signal in bilateral basal ganglia in Case 22 (A) (Arrows: bilateral basal ganglia). Abnormal signals in Left semioval center and parieto-occipital lobe, possible inflammatory lesions in case 15 (B). (Arrows: Left semioval center) Multiple streak-like high signals are seen in the cerebral sulci in case 1 (C), (Arrows: cerebral sulci) Patchy high signal intensity was seen in the left temporal lobe in case 1 (D). (Arrows: left temporal lobe) Linear enhancement on the surface of the brain gyrus in case 11 (E), (Arrows: brain gyrus) thickening and enhancement of the ependyma in the left temporal horn of the ventricle in case 11 (F).(Arrows: Left temporal horn of the ventricle).
Concordance Between mNGS and Conventional Diagnostic Testing
Two cases in our investigation were positive by mNGS and routine testing (Table 2). There were 16 cases in which pathogens tested positive according to mNGS alone, whereas no pathogens tested positive according to traditional methods alone. The two double-positive cases showed perfect consistency between mNGS and traditional techniques. Eight cases were negative for both conventional detection and mNGS.
Table 2.
Samples with Positive CSF mNGS
| Patient ID | mNGS Result | CSF Culture Result |
|---|---|---|
| C1 | K. pneumoniae, P. stutzeri, C. albicans | Negative |
| C2 | Negative | Negative |
| C3 | E. coli, E. corrodens, H. parainfluenzae, C. tropicalis |
Negative |
| C4 | A. baumannii, A.versicolor | Negative |
| C5 | A. fumigatus, S. marcescens | A. fumigatus, S. marcescens |
| C6 | K. pneumoniae | K. pneumoniae |
| C7 | Negative | Negative |
| C8 | P. aeruginosa, M. restricta | Negative |
| C9 | R. oryzae | Negative |
| C10 | E. miricola | Negative |
| C11 | P. micra, F. nucleatum | Negative |
| C12 | A. graevenitzii, C. acnes, A. viridans | Negative |
| C13 | Negative | Negative |
| C14 | P. oryzihabitans | Negative |
| C15 | Negative | Negative |
| C16 | Negative | Negative |
| C17 | P. luteola | Negative |
| C18 | E. coli | Negative |
| C19 | E.cloacae, Photorhabdus | Negative |
| C20 | E. coli, A. baumannii | Negative |
| C21 | Negative | Negative |
| C22 | E. coli | Negative |
| C23 | Negative | Negative |
| C24 | S. equorum, S. oralis, C. albicans | Negative |
| C25 | M. osloensis | Negative |
| C26 | Negative | Negative |
Abbreviations: E. coli, Escherichia coli; E. corrodens, Eikenella corrodens; K. pneumoniae, Klebsiella pneumoniae; C. albicans, Candida albicans; A. versicolor, Aspergillus versicolor; C. tropicalis, Candida tropicalis; A. fumigatus, Aspergillus fumigatus; S. marcescens, serratia marcescens; P. stutzeri, Pseudomonas stutzeri; H. parainfluenzae, Haemophilus parainfluenzae; P. aeruginosa, Pseudomonas aeruginosa; E. miricola, Elizabethkingia miricola; R. oryzae, Rhizopus oryzae; P. micra, Parvimonas micra; F. nucleatum, Fusobacterium nucleatum; C. acnes, Cutibacterium acnes; A. viridans, Aerococcus viridans; P. oryzihabitans, Pseudomonas oryzihabitans; P. luteola, Pseudomonas luteola; Enterobacter E. cloacae, cloacae; S. equorum, staphylococcus equorum; S. oralis, Streptococcus oralis; Moraxella M. osloensis, osloensis; A. graevenitzii, Actinomyces graevenitzii; M. restricta, Malassezia restricta; A. baumannii, Acinetobacter baumannii.
mNGS-Positive Results
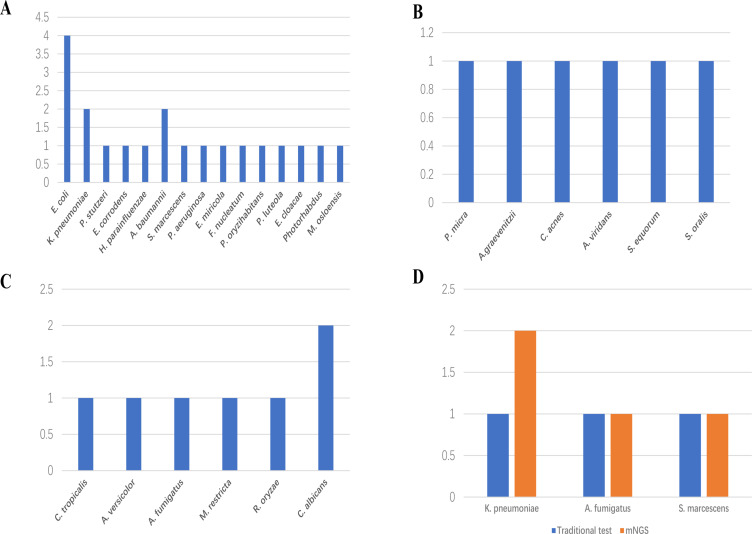
In this retrospective investigation, six gram-positive bacteria, fifteen gram-negative bacteria, and six fungal species were identified by mNGS (Figure 2). Four E. coli strains, two k pneumoniae strains and two A. baumannii strains were the gram-negative bacteria most often with positive results. The most widespread type of fungus was Candida (n=3), which included C. albicans in two patients and C. tropicalis in one patient. mNGS also detects rare fungal infections, such as R. oryzae. Three pathogens, A. fumigatus, S. marcescens and K. pneumoniae, were identified using conventional approaches and mNGS. Overall, mNGS is a better tool for detecting pathogen species and quantity than conventional approaches, especially for detecting rare pathogenic bacteria.
Figure 2.
Species distribution of (A) Gram-negative bacteria, (B) Gram-positive bacteria and (C) fungi detected by mNGS. (D) Pathogens detected by both conventional and mNGS methods. Horizontal axis indicates pathogens, vertical axis indicates pathogens number of cases.
Comparison of the Detection Rates of mNGS and Conventional Tests
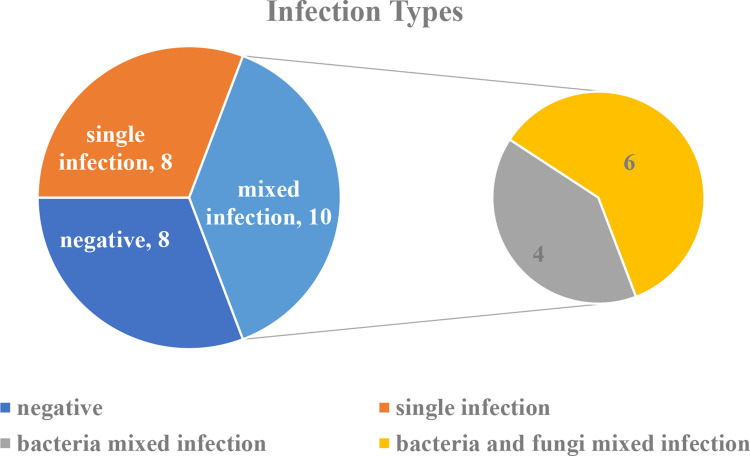
The detection rate of mixed infections with mNGS alone was 38.5% (10/26), and only one case (3.85%) of mixed infection was found by cultivation. The detection rates of mixed infection using mNGS and conventional testing were considerably different (p < 0.05). The most prevalent mixed infection was bacterial and fungal mixed infection (n=6), followed by bacterial mixed infection (n=4) (Figure 3).
Figure 3.
Mixed infections detected by mNGS.
A total of 8 (30.8%) cases with single-pathogen infections were identified by mNGS, comprising 7 bacterial infections and 1 fungal infection, but only one case (3.85%) with a single-pathogen infection was identified by traditional detection. The rates of mNGS and traditional testing for single-pathogen infection were significantly different (30.8% vs 3.85%; p < 0.05). One case of single-pathogen infection was found by mNGS and conventional detection. In this investigation, a patient infected with 2 or more species of pathogenic microbe was referred to as having a mixed infection.
Overall, this study revealed that mNGS is more effective than conventional techniques for assessing both single and mixed infections.
Diagnostic Performance of mNGS
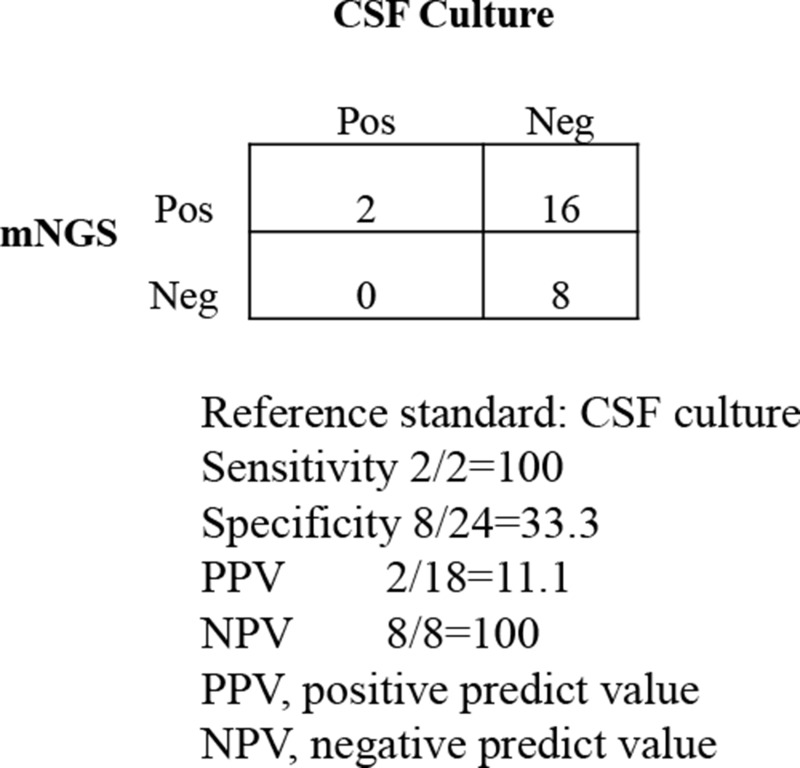
Compared to the culture method, mNGS demonstrated a sensitivity of 100% (2/2) and a specificity of 33.3% (8/24) in diagnosing CNS infections. Additionally, the positive and negative predictive values were 11.1% and 100%, respectively (without virus, Figure 4). There was a statistically significant difference (p<0.01) between the two approaches in 26 samples: the percentage of positive pathogens detected by the mNGS technique was 69.2% (18/26), while the percentage of positive pathogens detected by the conventional method was 7.69% (2/26). mNGS generally outperformed conventional techniques for identifying CNS infections in this study.
Figure 4.
Performance of different methods for the diagnosis of CNS infections.
Discussion
CNS infections impose heavy financial and psychological burdens on critical care unit patients owing to the high mortality rate and possibility of serious neurological sequelae.14 As a result, exploring application of mNGS for critically ill patients with CNS infections is important.
Twenty-six individuals with CNS infections were involved in this investigation in which CSF was detected by mNGS. We examined the diagnostic efficacy of mNGS and compared outcomes with those of CSF culture techniques. In line with previous studies, mNGS was found to be superior to traditional techniques for identifying infections involving particular pathogens as well as mixed infections. The detection rate of mNGS in our research was 69.2%, which was in line with that reported by Venkatesan et al.15 According to our statistics, mNGS outperforms conventional detection techniques in terms of the diagnostic rate. Furthermore, it offers a fresh viewpoint when diagnosing uncommon organisms causing CNS infections.16 mNGS also outperforms traditional techniques in terms of turnaround time (TAT).17 The mean TAT of mNGS in this research was 1.9 days. Conventional bacterial cultures can take 3–7 days, and fungi grow more slowly, delaying diagnosis. Although pathogen-specific testing is the mainstay of traditional testing methods, mNGS can overcome its limitations by evaluating all nucleic acids in a sample. Culture is generally considered the gold standard for detecting bacterial pathogens. Nonetheless, its utility is impeded by stringent cultivation conditions and previous antibiotic treatment. Compared to the 69.2% rate found by mNGS, the CSF culture detection rate in this study was only 7.69%. Administration of antibiotics before culture is most likely the cause of the low CSF culture rate. By comparison, the advantages of mngs can be better reflected. The detection rate of CSF mNGS in patients with CNS infections is higher than traditional methods, especially in patients who have been given antibiotics.18
The most common bacterial aetiologies are S. pneumoniae and N. meningitidis, while Listeria monocytogenes has become one of the prevalent pathogens causing bacterial meningitis.19 Nonetheless, the primary pathogens in our investigation were E. coli, K. pneumoniae, A. baumannii and C. albicans. This might be because various regions have different kinds of pathogens. Treatment of bacterial meningitis caused by K. pneumoniae and A. baumannii is challenging due to the emergence of carbapenem-resistant strains, the high mortality rate of this disease and severe neurological sequelae, such as paralysis.20 E. coli, the most common gram-negative bacillus organism, is the leading cause of neonatal meningitis, though it rarely causes meningitis in adults.21,22 E. coli meningitis can occur following penetration of the blood‒brain barrier secondary to head and spinal trauma, CSF leakage, and neurosurgical intervention and is associated complications, such as gastrointestinal perforation.23 E. coli meningitis has a high risk of morbidity and mortality.24 E. coli accounts for 0.5% to 3% of adult meningitis cases and has poor prognosis; nevertheless, there are few data, and genetic features of causative strains are frequently missing.25 Further study is needed.
There were no cases of tuberculous encephalitis or meningitis detected by either approach in this study. Diagnosis of tuberculous meningitis is challenging due to the difficulty in detecting M. tuberculosis in CSF, and delayed diagnosis and drug resistance are two of the main issues in tuberculous meningitis. Tuberculous meningitis is the most deadly type of tuberculous infection, with a mortality rate of up to 50%.26 mNGS is recommended the primary method used for diagnosing tuberculous meningitis.27 The low detection rate of tuberculous encephalitis or meningitis in our research may be due to the different kinds of pathogens in different regions. This may also be because a provincial hospital was involved, and patients from lower-level hospitals are not transferred to these hospitals.
Fungal infections of the CNS have a high mortality rate, especially in immunocompromised individuals.28 In our study, there were two cases of C. albicans infection, one case of C. tropicalis infection, one case of A. versicolor infection, one case of A. fumigatus infection, one case of M. restricta infection, and one case of R. oryzae infection. Except for the R. oryzae case, the other six cases were diagnosed as mixed infections by mNGS. Intracranial R. oryzae infection is relatively rare, mostly originating from the sinuses and orbit. The condition tends to worsen when the body’s immunity is low or the meningeal barrier is damaged.29 This patient experienced orbital cellulitis caused by trauma. He had a history of hypertension, diabetes and coronary heart disease; in addition, his blood sugar was poorly controlled, which aggravated the progression of the disease. He was hospitalized for 11 days after his condition worsened, and he died. The thick and easily broken fungal cell wall and the difficulty in obtaining DNA fragments may impact the fungal detection rate. We advise use of a combination of mNGS and conventional testing techniques for patients with complicated CNS infections. However, the complexity of mNGS technology means that there is still a long way to go before mNGS can be promoted and applied in CNS infections.
Limitations
Our research showed that mNGS is superior to conventional methods for diagnosing mixed infections and infections caused by specific pathogens in critically ill patients. However, this study has several shortcomings. First, this was a retrospective, single-centre study with a small sample size, CFS of nucleic acid detection failure caused by too little or error detection results, and bias is inevitable Prospective studies with large sample sizes and multiple centres are needed. Moreover, viral infections of the CNS have high incidence and mortality rates. Our mNGS testing method does not include testing for viruses. Further research on the detection efficiency of mNGS for viral CNS infections is needed in the future.
Conclusion
Our study indicates that mNGS is an alternative method for detecting nosocomial CNS infections. mNGS of CSF obtained from patients with meningitis or encephalitis improved diagnosis of nosocomial CNS infections who received antimicrobials.
Acknowledgment
Liying Zhan, Zhihua Lv and Yunjing Zhang are co-first authors for this study. We thank American Journal Experts for language editing.
Funding Statement
This work was supported by the National Natural Science Foundation of China (82302418) and Natural Science Foundation of Hubei Province (2022CFB728). The funders had no part in the study design, conduct or data analysis and did not have any authority over these activities.
Data Available Statement
The datasets available from the corresponding author on reasonable request.
Disclosure
The authors have no conflicts of interest to declare for this work.
References
- 1.Yu L, Zhang Z, Zou Y, et al. Next-generation sequencing in the diagnosis of neurobrucellosis: a case series of eight consecutive patients. Ann Clin Microbiol Antimicrob. 2023;22(1):44. doi: 10.1186/s12941-023-00596-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian Y, Gao R, Wang Y, et al. Economic impact of metagenomic next-generation sequencing versus traditional bacterial culture for postoperative central nervous system infections using a decision analysis mode: study protocol for a randomized controlled trial. mSystems. 2023;8(6):e0058123. doi: 10.1128/msystems.00581-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Zhu W, Jiao M, Guo W, Luo Y. Clinical application value of metagenomic next-generation sequencing in the diagnosis of central nervous system infections. Front Bioeng Biotechnol. 2023;11:885877. doi: 10.3389/fbioe.2023.885877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liesman RM, Pritt BS, Maleszewski JJ, Patel R. Laboratory diagnosis of infective endocarditis. J Clin Microbiol. 2017;55(9):2599–2608. doi: 10.1128/JCM.00635-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie L, Yang M, Liu M, Li Q, Luo C, Luo J. Integrating rapid pathogen identification and antimicrobial susceptibility testing through multiplex TAQMAN qPCR assay. J Microbiol Methods. 2024;217-218:106888. doi: 10.1016/j.mimet.2023.106888 [DOI] [PubMed] [Google Scholar]
- 6.van Zeggeren IE, Pennartz CJ, Horst LT, van de Beek D, Brouwer MC. Diagnostic Accuracy Of Clinical And Laboratory Characteristics In Suspected Nonsurgical Nosocomial Central Nervous System Infections. J Hosp Infect. 2024;145:99–105. doi: 10.1016/j.jhin.2023.12.015 [DOI] [PubMed] [Google Scholar]
- 7.Jiang X, Yan J, Huang H, et al. Development of novel parameters for pathogen identification in clinical metagenomic next-generation sequencing. Front Genet. 2023;14:1266990. doi: 10.3389/fgene.2023.1266990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramachandran PS, Wilson MR. Metagenomics for neurological infections-expanding our imagination. Nat Rev Neurol. 2020;16(10):547–556. doi: 10.1038/s41582-020-0374-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen W, Liu G, Cui L, et al. Evaluation of metagenomic and pathogen-targeted next-generation sequencing for diagnosis of meningitis and encephalitis in adults: a multicenter prospective observational cohort study in China. J Infect. 2024;88(5):106143. doi: 10.1016/j.jinf.2024.106143 [DOI] [PubMed] [Google Scholar]
- 10.Han D, Li Z, Li R, Tan P, Zhang R, Li J. mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol. 2019;45(5–6):668–685. doi: 10.1080/1040841X.2019.1681933 [DOI] [PubMed] [Google Scholar]
- 11.van de Beek D, Brouwer M, Hasbun R, Koedel U, Whitney CG, Wijdicks E. Community-acquired bacterial meningitis. Nat Rev Dis Primers. 2016;2(1):16074. doi: 10.1038/nrdp.2016.74 [DOI] [PubMed] [Google Scholar]
- 12.van de Beek D, Cabellos C, Dzupova O, et al. ESCMID guideline: diagnosis and treatment of acute bacterial meningitis. Clin Microbiol Infect. 2016;22 Suppl 3:S37–62. [DOI] [PubMed] [Google Scholar]
- 13.Miller S, Naccache SN, Samayoa E, et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019;29(5):831–842. doi: 10.1101/gr.238170.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chala TK, Lemma TD, Godana KT, Arefayine MB, Abdissa A, Gudina EK. The cost of suspected and confirmed bacterial meningitis cases treated at jimma university medical center, Ethiopia. Ethiop J Health Sci. 2022;32(4):765–772. doi: 10.4314/ejhs.v32i4.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkatesan A, Habis R, Geocadin RG. Approach to acute encephalitis in the intensive care unit. Curr Opin Crit Care. 2023;29(2):89–98. doi: 10.1097/MCC.0000000000001028 [DOI] [PubMed] [Google Scholar]
- 16.Zhan L, Huang K, Xia W, et al. The diagnosis of severe fever with thrombocytopenia syndrome using metagenomic next-generation sequencing: case report and literature review. Infect Drug Resist. 2022;15:83–89. doi: 10.2147/IDR.S345991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing F, Yang Q, Deng C, et al. Clinical impact of next-generation sequencing on laboratory diagnosis of suspected culture-negative meningitis and encephalitis. J Infect. 2022;85(5):573–607. doi: 10.1016/j.jinf.2022.08.026 [DOI] [PubMed] [Google Scholar]
- 18.Shangguan L, Xue L, Shang J, Wang H. The application value of metagenomic next-generation sequencing in community-acquired purulent meningitis after antibiotic intervention. BMC Infect Dis. 2023;23(1):683. doi: 10.1186/s12879-023-08672-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pajor MJ, Long B, Koyfman A, Liang SY. High risk and low prevalence diseases: adult bacterial meningitis. Am J Emerg Med. 2023;65:76–83. doi: 10.1016/j.ajem.2022.12.042 [DOI] [PubMed] [Google Scholar]
- 20.Hu H, Wang H, Yu M, et al. Clinical and microbiological characteristics of carbapenem-resistant Enterobacteriaceae causing post-operative central nervous system infections in China. J Glob Antimicrob Resist. 2023;35:35–43. doi: 10.1016/j.jgar.2023.08.006 [DOI] [PubMed] [Google Scholar]
- 21.Yang R, Wang X, Liu H, et al. Egr-1 is a key regulator of the blood-brain barrier damage induced by meningitic Escherichia coli. Cell Commun Signal. 2024;22(1):44. doi: 10.1186/s12964-024-01488-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasbun R. Progress and challenges in bacterial meningitis: a review. JAMA. 2022;328(21):2147–2154. doi: 10.1001/jama.2022.20521 [DOI] [PubMed] [Google Scholar]
- 23.Bijlsma MW, Brouwer MC, Kasanmoentalib ES, et al. Community-acquired bacterial meningitis in adults in the Netherlands, 2006-14: a prospective cohort study. Lancet Infect Dis. 2016;16(3):339–347. doi: 10.1016/S1473-3099(15)00430-2 [DOI] [PubMed] [Google Scholar]
- 24.Moussiegt A, Birgy A, Cointe A, Duval X, Bidet P, Bonacorsi S. Escherichia coli community-acquired meningitis in adults: a case series of 29 patients in France. Clin Microbiol Infect. 2022;28(9):1304–1305. doi: 10.1016/j.cmi.2022.04.020 [DOI] [PubMed] [Google Scholar]
- 25.Bodilsen J, Brouwer MC, Kjaergaard N, et al. Community-acquired meningitis in adults caused by Escherichia coli in Denmark and The Netherlands. J Infect. 2018;77(1):25–29. doi: 10.1016/j.jinf.2018.05.009 [DOI] [PubMed] [Google Scholar]
- 26.Soria J, Chiappe A, Gallardo J, Zunt JR, Lescano AG. Tuberculous meningitis: impact of timing of treatment initiation on mortality. Open Forum Infect Dis. 2021;8(7):ofab345. doi: 10.1093/ofid/ofab345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin A, Cheng B, Han X, Zhang H, Liu X, Liu X. Value of next-generation sequencing in early diagnosis of patients with tuberculous meningitis. J Neurol Sci. 2021;422:117310. doi: 10.1016/j.jns.2021.117310 [DOI] [PubMed] [Google Scholar]
- 28.Sharma S, Acharya J, Rijal N, et al. Cryptococcal meningitis in people living with human immunodeficiency virus in Nepal: perspectives from resource limited setting. Mycoses. 2023;66(1):47–51. doi: 10.1111/myc.13526 [DOI] [PubMed] [Google Scholar]
- 29.Acuna M, Benadof D, Yohannessen K, Leiva Y, Clement P. FilmArray(R) Meningoencephalitis panel in the diagnosis of central nervous system infections: stewardship and cost analysis in a paediatric hospital in Chile. BMC Pediatr. 2022;22(1):182. doi: 10.1186/s12887-022-03241-1 [DOI] [PMC free article] [PubMed] [Google Scholar]