Abstract
Retinoic acids and long-chain fatty acids are lipophilic agonists of nuclear receptors such as RXRs (retinoic X receptors) and PPARs (peroxisome-proliferator-activated receptors) respectively. These agonists are also ligands of intracellular lipid-binding proteins, which include FABPs (fatty acid-binding proteins). We reported previously that L (liver-type)-FABP targets fatty acids to the nucleus of hepatocytes and affects PPARα activation, which binds together with an RXR subtype to a PPRE (peroxisome-proliferator-responsive element). In the present study, we first determined the optimal combination of murine PPAR/RXR subtypes for binding to known murine FABP-PPREs and to those found by computer search and then tested their in vitro functionality. We show that all PPARs bind to L-FABP-PPRE, PPARα, PPARγ1 and PPARγ2 to A (adipocyte-type)-FABP-PPRE. All PPAR/RXR heterodimers transactivate L-FABP-PPRE, best are combinations of PPARα with RXRα or RXRγ. In contrast, PPARα heterodimers do not transactivate A-FABP-PPRE, best combinations are of PPARγ1 with RXRα and RXRγ, and of PPARγ2 with all RXR subtypes. We found that the predicted E (epidermal-type)- and H (heart-type)-FABP-PPREs are not activated by any PPAR/RXR combination without or with the PPAR pan-agonist bezafibrate. In the same way, C2C12 myoblasts transfected with promoter fragments of E-FABP and H-FABP genes containing putative PPREs are also not activated through stimulation of PPARs with bezafibrate applied to the cells. These results demonstrate that only PPREs of L- and A-FABP promoters are functional, and that binding of PPAR/RXR heterodimers to a PPRE in vitro does not necessarily predict transactivation.
Keywords: Dynabead streptavidin solid-phase method, fatty acid-binding protein, peroxisome-proliferator-activated receptor, peroxisome-proliferator-response element, reporter gene assay
Abbreviations: CRABP, cellular retinoic acid-binding protein; DR-1, direct repeat with one spacer nucleotide; FABP, fatty acid-binding protein; I-BABP, intestinal bile acid-binding protein; iLBP, intracellular lipid-binding protein; PPAR, peroxisome-proliferator-activated receptor; PPRE, peroxisome-proliferator-responsive element; RAR, retinoic acid receptor; RXR, retinoic X receptor
INTRODUCTION
Many small, biologically important lipophilic compounds regulate cellular function by modulating the rates of transcription of various target genes. Such compounds include retinoic acids and long-chain fatty acids and some of their metabolites. They bind to proteins that are members of the family of the 14–15-kDa iLBPs (intracellular lipid-binding proteins), which include the CRABPs (cellular retinoic acid-binding proteins)-I and -II, the I-BABP (intestinal bile acid-binding protein) and nine types of FABPs (fatty acid-binding proteins), which are traditionally named after the tissue of their first isolation, e.g. L (liver-type)-, A (adipocyte-type)-, E (epidermal-type)- and H (heart-type)-FABP. It is believed that FABPs solubilize and protect their ligands in aqueous spaces and facilitate their transport across the cytosol [1,2]. In recent years, it has become increasingly clear that, in addition, some CRABPs and FABPs transport their ligands to ligand-activated-transcription factors, thus being co-activators of the RARs (retinoic acid receptors), RXRs (retinoic X receptor) and PPARs (peroxisome-proliferator-activated receptors) [3–5].
PPAR subtypes α, β and γ have been described, the latter being expressed in mice as isoforms γ1 [6] and γ2 [7], which is 90 nt longer at the 5′-end as a result of alternative splicing. For transcriptional regulation of target genes, PPARs heterodimerize with the α-, β- or γ-subtypes of the 9-cis-RXR for binding to a PPRE (peroxisome-proliferator-responsive element), a DR-1 (direct repeat with one spacer nucleotide) of the DNA consensus sequence 5′-AGGTCA. Binding affinity of the PPAR/RXR heterodimer for a PPRE increases further when A is the DR-1 spacer nucleotide and when the 5′-extension has the C(A/G)(A/G)A(A/T)CT consensus sequence, the latter drawing PPAR to the 5′-half site [8,9]. Thus the DR-1 motif is not sufficient to constitute a PPRE; moreover, the 5′-extension promotes the binding of PPAR/RXR heterodimers over potential competitors, e.g. RXR homodimers [10,11]. The two consensus sequences, DR-1 and the 5′-flank, are considered to be the idealized sequence (ideal-PPRE) for PPAR/RXR binding [10].
PPREs have been found in promotors of various enzymes of peroxisomal β-oxidation (acyl-CoA oxidase, bifunctional enzyme, β-ketoacyl-CoA thiolase) [8,12,13], general lipid metabolism (lipoprotein lipase) [14] and of fatty acid transporters [15,16]. PPREs have also been found in the promoter region of L-FABP [17,18] and A-FABP [19]. Their transcriptional regulation by PPAR agonists has also been described in [20,21]. In view of the observation that FABPs bind ligands that are quite reminiscent of PPAR agonists, Issemann et al. [17] postulated a mechanism by which FABP binds fatty acid in the cytosol for transport into the nucleus, where it is transferred to PPAR to affect transcription of target genes. This hypothesis was verified by Wolfrum et al. [4], who showed that, in HepG2 cells, L-FABP functions as a mandatory vehicle for transport of PPARα and PPARγ agonists into the nucleus and thus initiates transactivation. Moreover, a PPRE in the promotor of L-FABP affects expression of this vehicle, which constitutes a signal enhancing mechanism in itself [4].
As it is known that PPAR and RXR subtypes and FABP types are expressed cell specifically, the question arises if the phenomenon observed for PPARα and PPARγ and L-FABP in HepG2 cells might be a principle applicable to other combinations of PPAR/RXR subtypes and FABP types to indicate further autoregulatory mechanism in other cell types. Therefore we determined the optimal combination of murine PPAR and murine RXR subtypes for binding to known murine FABP-PPREs and by computer-aided analysis (in silico) predicted murine FABP-PPREs and tested the functionality of these PPREs in transient activation assays.
EXPERIMENTAL
Vectors for murine PPAR and RXR subtypes
Expression plasmids for PPAR subtypes, i.e. pSG5-mPPARα, -mPPARβ and -mPPARγ1, were kindly provided by Professor W. Wahli (University of Lausanne) and plasmids for RXR subtypes, i.e. pSG5-mRXRα, -mRXRβ and -mRXRγ by Professor P. Chambon (Université Louis Pasteur de Strasbourg). For transient transfection assays, reporter constructs were obtained by cloning a single copy of the respective PPRE into the mammalian expression vector pCAT3 promoter (Promega).
Murine PPARγ2 cDNA was amplified by PCR (Taq DNA polymerase; Gibco BRL, Eggenstein, Germany) from mouse liver RNA after conversion into cDNA with reverse transcriptase (Gibco BRL). Primers were synthetic oligonucleotides comprising 20 nt of the 5′-end (5′-mPPARγ2 primer: 5′-TTATGGGTGAAACTCTGGGA-3′) and 20 nt complementary to the 3′-end (3′-mPPARγ2 primer: 5′-CTGCTAATACAAGTCCTTGT-3′) of the mPPARγ2 cDNA coding region [7]. Amplified mPPARγ2 cDNA was cloned into the pcDNA3 vector (pcDNA3-mPPARγ2) and verified by double-strand sequencing (SeqLab, Göttingen, Germany).
In vitro transcription/translation of PPARs and RXRs
Receptors were synthesized using the rabbit reticulocyte lysate Transcription/Translation kit (Promega) according to the manufacturer's instructions. For radioactive labelling, [35S]methionine (1 Ci/mol; Amersham Biosciences) was applied. To access equimolar amounts of labelled receptors for application in binding experiments, the following procedure was used: radioactively labelled proteins of the lysate were separated by SDS/PAGE (13.5% gels). Gels were dried, and labelled receptors were quantified on X-ray film by laser densitometric scanning (UltraScan XL; Amersham Biosciences); staining of the films by labelled PPAR and RXR subtypes was normalized to the number of methionine residues in the respective protein, as described by Juge-Aubry et al. [9].
Binding assay
In the Dynabead streptavidin solid-phase method, the double-stranded biotinylated oligonucleotides (Genset, San Diego, CA, U.S.A.) (Table 1) were immobilized on streptavidin-modified magnetic beads according to the manufacturer's instructions (Dynal, Oslo, Norway). Equimolar amounts of radioactively labelled pairs of PPAR and RXR subtypes (each 4 μl) were incubated with 200 ng of biotinylated oligonucleotides in binding buffer [100 mM Hepes, 5 mM (NH4)2SO4, 5 mM dithiothreitol, 1% (w/v) Tween 20 and 150 mM KCl, pH 7.6] in a final volume of 25 μl. After incubation at room temperature (21 °C) for 30 min, the Dynabead-bound DNA–protein complex was washed twice with PBS and the 96-well plate was placed for 2 min into the magnet stand furnished with the kit to separate the complex from unbound material. After removing the supernatant by aspiration, Dynabeads with bound DNA–protein complex were resuspended in 100 μl of PBS and transferred into a scintillation vial. After adding 1 ml of AQUASAFE 500 (Zinsser Analytic, Maidenhead, Berks., U.K.), radioactivity was measured by liquid-scintillation counting (Beckman, Geneva, Switzerland). With this Dynabead streptavidin solid-phase method, approx. 200 samples could be processed in 6 h.
Table 1. Murine PPAR/RXR target boxes identified by in silico analysis.
Alignment of FABP-PPREs tested for PPAR/RXR subtype binding and transactivation. The idealized consensus sequence (ideal-PPRE) served as positive control, a randomized DNA sequence as negative control (random). All PPREs consist of a DR-1 element (underlined), a 5′-flanking region of 7 nt, which is critical for binding of PPAR/RXR heterodimers and 2 nt at the 3′-end of the DR-1 element. For PPRE-binding experiments a spacer of 6 nt (GGAGGA) was added to the 3′-end to exclude sterical obstruction of DNA–protein interaction by biotin. For transactivation assays PPREs consisted of 5 extra nt at each site (GATCT at 5′-end and GAGCT at 3′-end) for cloning into the vector.
| Element | Sequence | GenBank® accession no. | Position | Reference |
|---|---|---|---|---|
| Ideal-PPRE | 5′-CAAAACTAGGTCAAAGGTCACA-3′ | [9] | ||
| L-PPRE | 5′-CAATCACTGACCTATGGCCTAT-3′ | Y14660 | –121 to –100 | [20] |
| A-PPRE* | 5′-TCTTACTGGATCAGAGTTCACT-3′ | M84651 | −5220 to −5199 | [41] |
| E-PPRE | 5′-CTGAGAAAGGTCATTCCACACA-3′ | AJ223066 | −1786 to −1765 | [22] |
| H-PPRE | 5′-TGGCACAAGCTCAGAGGTCAGT-3′ | U02884 | −792 to −771 | [23] |
| Random | 5′-TCGATGATAAGTCCCTTCAGTC-3′ |
* ARE7, see the Discussion section.
Reporter gene assay in HepG2 cells
For experiments shown in Figure 2, HepG2 cells (A.T.C.C., HB-8065 or Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany, ACC 180) were grown to 60–70% confluency in 6-well dishes (Nunc, Wiesbaden, Germany) in Dulbecco's modified Eagle's medium supplemented with 10% basal-medium-supplement artificial medium (both from Biochrom, Berlin, Germany). Cells were co-transfected with the aid of Fugene6 transfection reagent (Roche Diagnostics) with expression plasmids of respective PPAR/RXR combinations (1.5 μg/well each), the pCAT3 promoter vector (1.5 μg; Promega) containing either ideal-PPRE (positive control), L-, A-, E- or H-FABP-PPREs (Table 1) and pSV-β-galactosidase (β-Gal) (Promega) as internal reference (0.5 μg/well). For negative control (Figure 2), HepG2 cells were co-transfected with CAT reporter plasmid without PPRE and with the expression plasmids for respective PPAR and RXR subtypes. New medium was used for experiments shown in Figure 3 as the supply of old brand had ceased. Thus HepG2 cells were grown to 60–70% confluency in 6-well dishes (Nunc) in RPMI 1640 medium supplemented with 0.2% serum effective substitute both from Biochrom. For further evaluation of the transactivation of E- and H-FABP-PPRE, cells were co-transfected with the CAT reporter plasmid containing PPRE (1.5 μg) without (but with stuffer DNA) and with plasmids for PPARγ1 and RXRγ (1.5 μg each); after transfection, cells were kept in the same medium for 48 h in these assays without ligand treatment. Then cells were harvested, and expression of CAT and β-Gal proteins was measured with respective ELISAs (Roche Diagnostics) according to the manufacturer's instructions.
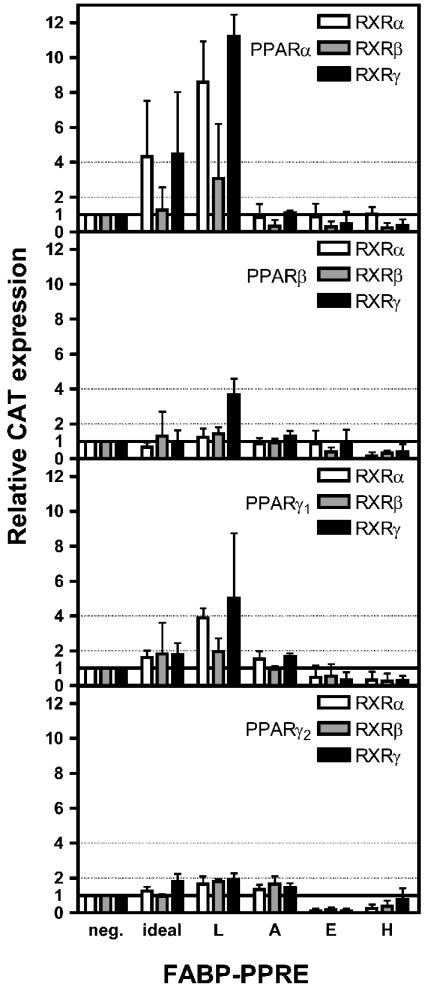
Figure 2. Dependence of transactivation potentials of PPAR/RXR heterodimers on FABP-PPREs.
HepG2 cells were transiently co-transfected with plasmids pSG5-PPARα, -PPARβ, -PPARγ1, pcDNA3-PPARγ2 and pSG5-RXR subtypes respectively together with the pCAT3 promoter plasmid containing a PPRE and pSV-β-Gal. After 42 h, cells were harvested. β-Gal and CAT concentrations were determined by ELISAs; experiments with empty pCAT3 plasmid served as control and were set to unity (neg.). Results are means±S.D. for two independent experiments, each performed in triplicate (n=6).
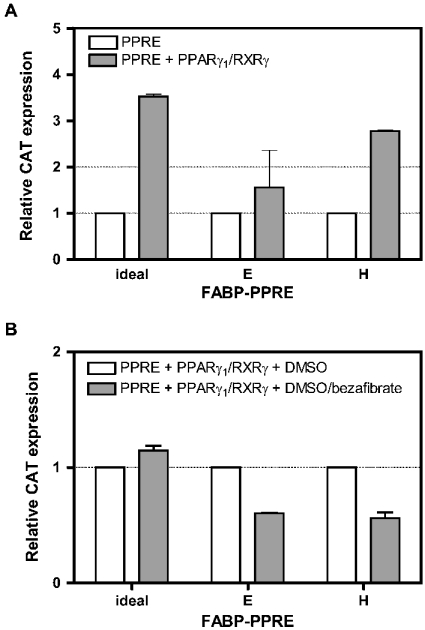
Figure 3. Transactivation of PPARγ1/RXRγ heterodimers via E- and H-FABP-PPRE without and with ligand.
(A) HepG2 cells were transiently co-transfected with pCAT3 promoter plasmid containing the PPRE, β-Gal, without and with plasmids for PPARγ1 and RXRγ. After 42 h, cells were harvested, and β-Gal and CAT concentrations were determined by ELISAs. For each PPRE, experiments were normalized to those without ectopic PPARγ1/RXRγ, set to unity. Results are means±S.D. for two independent experiments, each performed in triplicate (n=6). (B) HepG2 cells were transiently co-transfected with pCAT3 promoter plasmid containing the PPRE and plasmids for β-Gal, PPARγ1 and RXRγ. DMSO and DMSO plus bezafibrate respectively were added to the medium 4 h after transfection (final concentrations 1% DMSO and 100 μM bezafibrate) and cells were incubated for a further 38 h. After harvest, β-Gal and CAT concentrations were determined by ELISAs. For each PPRE, experiments with ligand were normalized to those with DMSO alone, set to unity. Results are means±S.D. for two independent experiments, each performed in triplicate (n=6).
For assays in the presence of solvent and ligand, co-transfections were performed as above with plasmids encoding ideal-, E- and H-FABP-PPRE respectively together with those for PPARγ1 and RXRγ. Cells were cultured 4 h after transfection in the same medium, but now containing 1% (v/v) DMSO and 1% DMSO plus 100 μM bezafibrate respectively. After 38 h, cells were harvested and expression of CAT and β-Gal proteins were measured with respective ELISAs.
Reporter gene assay in C2C12 myoblasts
Murine promoters of E-FABP gene (Fabpe [22]) and H-FABP gene (Fabph [23]) respectively served as templates to generate promoter fragments for functional analysis of putative PPREs; Fabpe2447 (nt −2447 to +35) and Fabph1514 (nt −1514 to +36) containing the putative PPREs as well as Fabpe1716 (nt −1716 to +35) and Fabph817 (nt −817 to +36) without a PPRE were synthesized by PCR and cloned each into the pCAT3 promoter vector (Promega). C2C12 myoblasts (A.T.C.C., CRL-1772) were maintained in growth medium [Dulbecco's modified Eagle's medium with 4.5 g/l glucose, 15% (v/v) fetal calf serum and 4 mM glutamine] and cells were seeded on the previous day at 50% confluency. Cells were transfected with promoter construct and pSV-β-Gal (0.5 μg/well each) on day 0 with the aid of Fugene6 transfection reagent. After transfection, cells were cultured in a medium containing 1% DMSO as control or 1% DMSO supplemented with 100 μM bezafibrate. After 48 h, total RNA was isolated using the RNA isolation kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. By a slot blot procedure, 15 μg of extracted RNA per sample was transferred on to a nylon membrane. After hybridization with DIG-labelled antisense-RNA, visualized transcripts of lacZ and cat respectively were quantified by laser-scanning densitometry (UltraScan XL; Amersham Biosciences) where each signal on the film was in the linear portion of the grey scale.
RESULTS
On the basis of the phylogenetic relationship and the structure and conformation of bound ligands, iLBPs have been classified within four subfamilies [24], i.e. (i) the retinoid-binding proteins, (ii) intestinal-type (I-) FABP, (iii) L-FABP and I-BABP, and (iv) A-, E-, H-, B (brain-type)-, M (myelin-type)- and T (testis-type)-FABP. The common denominator of subfamily (iv) is binding of one fatty acid molecule in an U-shaped conformation [25]. After in silico analysis of known promoters of all FABPs, we identified PPREs in promoters of L- and A-FABP, which have already been functionally identified [7,18] and in addition in E- and H-FABP promoters (Table 1). As a first test for functionality, we analysed PPAR/RXR binding to these repetitive DNA elements. Then we turned to transactivation experiments in HepG2 cells and finally to promoter analysis in C2C12 myoblasts to explore the physiological context.
Binding of PPAR/RXR heterodimers to PPREs
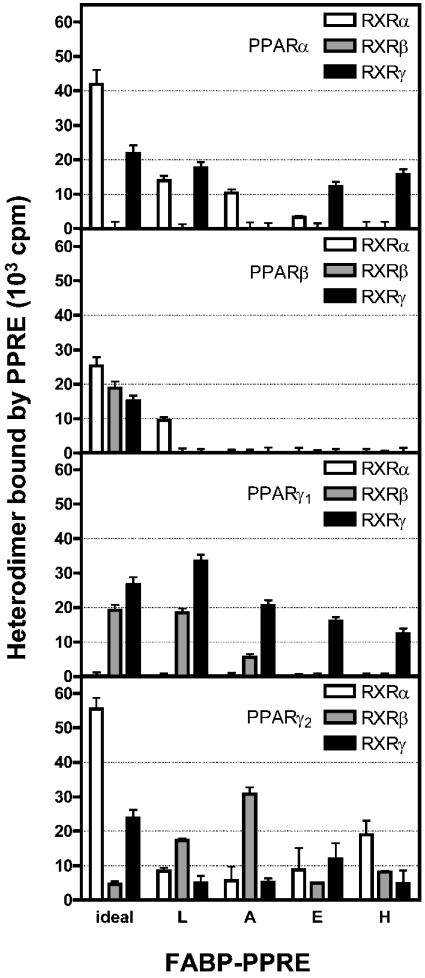
To confirm that a one-to-one PPAR/RXR complex was formed for binding to ideal-PPRE, we applied equimolar mixtures of, first radioactively labelled PPARα and unlabelled RXRα, second of unlabelled PPARα and radioactively labelled RXRα and third of both receptors radioactively labelled to immobilized ideal-PPRE, and compared the radioactivity of the complexes bound to the responsive element. The level of radioactivity of the PPARα/RXRα heterodimer with one unlabelled receptor was nearly 50% in either case of that of the all-labelled PPARα/RXRα heterodimer (results not shown). Thus in accordance with the literature [10,11], RXRα did not homodimerize in the presence of PPARα. Next, to test binding of PPAR/RXR heterodimers to a given PPRE, we added the equimolar mixture of PPAR and RXR subtypes to the four double-stranded FABP-PPREs and analysed binding. Taking ideal-PPRE as positive control and randomized DNA as negative control, we considered bound radioactivity above 40000 c.p.m. as strong binding, from 20000 to 40000 c.p.m. as moderate binding and from 10000 to 20000 c.p.m. as weak binding.
Figure 1 reveals that PPARα with RXRα or RXRγ binds to L-FABP-PPRE in a weak manner, whereas PPARβ in combination with RXRα is the only heterodimer of this PPAR subtype that may reach the threshold value for binding. Clear binding of PPARγ1 with RXRβ or RXRγ and PPARγ2 with RXRβ to L-FABP-PPRE was observed, although in the weak to moderate range. Weak interaction of the PPARα/RXRα heterodimer with A-FABP-PPRE can be deduced from the results shown in Figure 1 and no binding of PPARβ heterodimers. Yet, moderate binding to this PPRE was measured for PPARγ1/RXRγ and PPARγ2/RXRβ.
Figure 1. Binding of PPAR/RXR heterodimers to FABP-PPREs in vitro, determined with the aid of magnetobeads.
Biotinylated FABP-PPREs were immobilized on streptavidin-coated magnetic particles and incubated with equimolar amounts of 35S-labelled PPAR and RXR subtypes. After 30 min incubation at room temperature, radioactivity of bound heterodimers was measured. Results are means±S.D. for four independent experiments.
Rather restricted binding of receptor heterodimers to E- and H-FABP-PPREs was observed (Figure 1). Thus combinations of PPARβ with RXR subtypes do not bind at all, whereas PPARα and PPARγ1 subtypes in combination with RXRα and RXRγ bind to either PPRE weakly but clearly. The same is true for interaction of PPARγ2/RXRγ with E-FABP-PPRE and PPARγ2/RXRα with H-FABP-PPRE.
Taken together, heterodimers with PPARβ may not play a role at all in binding to FABP-PPREs as seen from these in vitro results and RXRγ appears to be the preferred partner for heterodimerization with PPARα or PPARγ1 for interaction with FABP-PPREs. It is noteworthy that distinct differences were observed for PPARγ isoforms in their choice of dimerization partners and in their binding to FABP-PPREs.
Impact of FABP-PPREs on the transactivation potential of PPAR/RXR heterodimers
Ideally, transactivation should correlate with binding. This was put to test by co-transfection of HepG2 cells with PPRE-responsive CAT promoter construct and with various combinations of expression vectors for PPAR and RXR subtypes. Furthermore, functionality of the putative E- and H-FABP-PPREs was additionally tested by attempting to stimulate transactivation with the PPAR pan-agonist bezafibrate [26,27]. Taking ideal-PPRE as positive control and the CAT reporter plasmid without PPRE as negative control, we considered values for CAT expression between 1 and 2 as weak, between 2 and 4 as moderate and above 4 as strong with respect to the transactivation potentials of the PPAR/RXR heterodimers tested.
Figure 2 shows that for L-FABP-PPRE, both PPARα and PPARγ1 in combination with all three RXR subtypes, have the highest transactivation potentials, i.e. in the moderate to strong range. Transactivation potentials of PPARβ and PPARγ2 with all three subtypes of RXR are in the weak range with the exception of the PPARβ/RXRγ heterodimer, which reached almost the strong range.
PPARα and PPARβ heterodimerized with all RXR subtypes, but did not trigger transactivation via A-FABP-PPRE; PPARγ1 and PPARγ2 heterodimers respectively revealed a transactivation potential, albeit in the weak range (Figure 2).
Transactivation via E- and H-FABP-PPREs of all PPAR/RXR heterodimer combinations was not detectable (Figure 2), although the binding assay still revealed a weak binding of PPARα and PPARγ isoforms in combination with some RXR subtypes to these PPREs (Figure 1). This necessitated a more detailed analysis for which we chose the PPARγ1/RXRγ heterodimer due to relatively binding best to E- and H-FABP-PPRE (Figure 1). In contrast with the transactivation assays shown in Figure 2, where values obtained were all normalized to those obtained with the empty pCAT promoter plasmid, we determined first the basal values of transactivation via individual PPREs with cells without ectopic nuclear receptors. These absolute values for relative CAT expression were 0.26±0.026 for ideal-PPRE, 0.37±0.081 for E-FABP-PPRE and 0.14±0.006 for H-FABP-PPRE (means±S.D., n=6). The absolute values were set to 1 for normalization of relative CAT expression when plasmids encoding PPARγ1 and RXRγ respectively, were co-transfected with the pCAT promoter plasmid containing the PPRE (Figure 3A). As expected, the transactivation potential via ideal-PPRE increased more than 3-fold in this positive control. Whereas the error bar in the experiment with E-FABP-PPRE did not indicate induction, the increase for transactivation via H-FABP-PPRE was surprising. A look at the absolute values presented above, however, reveals that the value for this PPRE was particularly low.
In the next experiment, we checked the possibility whether or not addition of bezafibrate would further stimulate transactivation (Figure 3B). In the present study, we prepared HepG2 cells that were transfected with PPRE, PPARγ1 and RXRγ and normalized the values obtained after administration of the ligand to that obtained with the solvent alone (set to one). Whereas a small but clear induction (1.1-fold) was observed with ideal-PPRE, clear decreases in relative CAT expression were seen for E- and H-FABP-PPRE in response to bezafibrate. This demonstrated that for these in vitro assay systems experiments were performed at the level of background noises for the latter two PPREs.
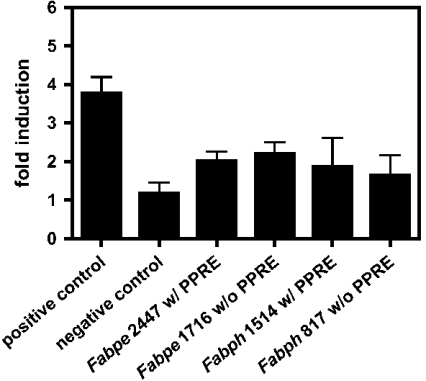
Finally, we turned to direct examination of Fabpe and Fabph promoter fragments in transfected C2C12 myoblasts and administered to these cells bezafibrate, to test whether the potential PPREs identified in silico are functional. This appeared to be a more physiological approach as we had found earlier that E- and H-FABP genes are inversely expressed in this muscle cell line during differentiation from myoblast to myotube stages [28]. Moreover, C2C12 cells express all PPAR subtypes (results not shown). According to the rationale applied, C2C12 cells transfected with Fabpe2447 and Fabph1514 containing the putative PPREs should respond to administration of bezafibrate by enhanced expression of the CAT reporter gene, in comparison with Fabpe1716 and Fabph817 without the PPREs. To verify that PPAR transactivation can be monitored in this model, we made use of ideal-PPRE as positive control and the vector alone as negative control. The basal activity of ideal-PPRE under unfed conditions of 6.6±1.2 compared with pCAT reporter gene leads to a 3.8-fold induction after incubation with bezafibrate and indicates the ability to activate a functional PPRE in this model. The vector alone was not activated by bezafibrate (Figure 4).
Figure 4. Transactivation of Fabpe and Fabph promoter fragments in C2C12 myoblasts affected by bezafibrate.
C2C12 myoblasts were transfected with pCAT promoter containing one copy of ideal-PPRE (positive control) or with pCAT promoter alone (negative control). In the tests, proper cells were transfected with pCAT promoter preceded by Fabpe2447 and Fabph1514 promotor fragments containing the putative PPRE or Fabpe1716 and Fabph817 without the proposed PPRE respectively and pSV-β-Gal. Cells were cultured for 48 h after transfection in fresh medium supplemented with 1% DMSO alone as control or with 1% DMSO and 100 μM bezafibrate respectively, and contents of lacZ and cat transcripts were evaluated by Northern blotting. Bars represent the induction of promoter activity in bezafibrate treated cells, normalized to DMSO control. Results are expressed as means±S.D. for four independent experiments.
In the further experiments shown in Figure 4, transactivation of all promoter fragments affected by bezafibrate treatment of the cells was observed, when compared with respective DMSO treatment. The basal activity of Fabpe with putative PPRE increased from 1.2±0.1 to 2.5±0.2 (with PPRE, 2.1±0.3-fold) and of Fabpe without a putative PPRE from 1.7±0.1 to 4.0±0.3 (without PPRE, 2.3±0.4-fold). The Fabph activity with putative PPRE increased from 1.2±0.3 to 2.4±0.8 (with PPRE, 2.0±1.1-fold) and of Fabph without putative PPRE from 2.8±0.1 to 4.5±0.6 (without PPRE, 1.6±0.7-fold). These differences in transactivation between PPRE-containing and non-containing promoters of both, Fabpe and Fabph are not significant.
DISCUSSION
The involvement of certain iLBPs in fatty acid signalling and subsequent gene regulation has been recognized only recently. A mechanism emerged by which the fatty acid is bound by the iLBP in the cytosol and translocated into the nucleus, where the ligand of the binding protein becomes the agonist of a nuclear receptor on protein–protein interaction of the former with the latter. This has been shown for all-trans retinoic acid as ligand/agonist for the pair CRABP-II/RAR [3,29] and for straight-chain fatty acids as ligands/agonists for the pair L-FABP/PPARα [4]. By the same mechanism specific xenobiotics served as ligands/agonists for the pairs A-FABP/PPARγ and E-FABP/PPARβ respectively [5]. As RAR and PPAR subtypes on heterodimerization with RXR subtypes bind to PPRE containing genes, gene induction is affected. The latter appears to be trivial by today's knowledge; however, with the results obtained in the present study some further predictions can be made.
First, with regard to FABPs, we know now that autoregulatory modulation of their expression applies to L- and A-FABP only, due to functional PPREs in their promoters. One example in the cellular context is the heavy induction of L-FABP expression in liver affected by increased in vivo levels of phytanic acid, an L-FABP/PPARα ligand/agonist produced after phytol feeding to mice [20]. The other example is the remarkable induction of A-FABP expression in human monocytes after administration of PPARγ agonists [21]. PPARγ isoforms are involved in the preadipocyte and in the monocyte/macrophage differentiation programmes where the A-FABP gene is a late target for these nuclear receptors. The murine A-FABP gene contains a fat-specific enhancer bearing two response elements, namely ARE6 and ARE7. ARE6 is more divergent from ideal-PPRE when compared with ARE7 and, interestingly, it was shown in electrophoretic mobility-shift assays that there was virtually no binding of PPARα and PPARβ to the ARE6 element, whereas PPARγ2 still bound significantly. All PPAR subtypes and isoforms bind to the ARE7 element, with PPARγ2 having the highest affinity for this element [30]. In contrast with the results available in the literature, the present study revealed that PPARβ heterodimers do not bind to the ARE7 element, but confirmed the binding of PPARα/RXRα, PPARγ1/RXRγ and PPARγ2/RXRβ to ARE7; although the former in the weak range, the latter two in the medium range. These binding data are borne out by our transactivation assays, where both PPARγ1 and PPARγ2 clearly transactivate the ARE7 element.
For H-FABP, an autoregulatory modulation can also be assumed. Holst et al. [31] showed that fasting animals for 24 h resulted in an increase in muscular PPARβ expression, which coincided with up-regulation of genes implicated in fatty acid catabolism, e.g. H-FABP. Furthermore, exposure of C2C12 cells to a specific PPARβ agonist promoted expression of H-FABP [31]. Our results revealed binding of PPARα and PPARγ1 and PPARγ2 subtypes to H-FABP PPRE. In the light of these findings, H-FABP has a non-functional PPRE, the concomitant up-regulation of PPARβ and H-FABP, however, is not due to a direct interaction of PPARβ with the H-FABP promoter via a PPRE.
Secondly, our study reveals the optimum heterodimer combination for interaction with FABP-PPREs. This includes the specific differences observed for PPARγ isoform heterodimerization as PPARγ1 interacted with RXRβ and RXRγ only, and PPARγ2 with all three RXR subtypes in the process of binding to ARE7. It is important to note that heterodimer binding shown in vitro does not automatically imply transactivation of the gene. This became evident by our testing of the putative PPREs in the H- and E-FABP promoters in transactivation assays and by our promoter study in C2C12 cells. A similar observation was reported in the literature for the gene encoding the membrane-bound fatty acid transport protein, whose PPRE in the promoter interacted with the PPARβ/RXRα heterodimer in a gel-shift assay, but was not active in transactivation in contrast with other PPAR/RXR heterodimer combinations [16]. As true functionality beyond binding can be demonstrated by transactivation assays, we tested RXR homodimer binding indirectly via transactivation to the functional elements ideal-PPRE and L-FABP PPRE, as they bind and transactivate PPAR/RXR heterodimers. In the present study, it is important to note that binding and transactivation of RXR homodimers to FABP-PPREs can be excluded and does not influence PPAR/RXR transactivation.
Experiments from our and other laboratories indicated that the binding proteins CRABP-II, L-, A-, H- and E-FABP and the nuclear receptor subtypes of RAR, PPAR and RXR together with ligands/agonists can form transient complexes to bind to PPREs in genes as part of the transcriptional machinery, which induces gene expression. L- and A-FABP via their respective PPREs can thus induce their own synthesis in the form of a feedforward regulation. This is an important aspect for cells that must cope with heavy intracellular fluxes of fatty acids under various physiological situations. Such cells are intestinal epithelial cells and liver parenchymal cells as well as adipocytes, where L- and A-FABP respectively are found in high amounts to transport fatty acids to the respective organelles [21,32]. At low intracellular fatty acid concentration, the signalling function to the nucleus of L- and A-FABP may be prevalent and FABPs having no functional PPRE in the gene can be active in this process as well. Indeed, such a finding was reported for fatty acid signalling via E-FABP to PPARβ in COS-7 cells [5].
Thirdly, the finding that all PPAR/RXR heterodimer combinations transactivate via L-FABP-PPRE would predict a role for all the nuclear receptor subtypes in L-FABP-expressing cells. A look into the literature reveals that in cells of adult rodents L-FABP is found in high concentrations in hepatocytes of the liver, in proximal tubular cells of the kidney, in intestine cells, decreasing from duodenum to colon and in lower amounts in stomach cells [33–35]. PPARα and PPARβ are also expressed in these L-FABP-expressing tissue cells, whereas PPARγ1 is found in cells of the large intestine and in very low levels in some stomach cells, in proximal tubular cells of the kidney and in the liver [36,37]. Restriction of expression is also found for the PPAR heterodimer partners, the RXR subtypes; however, detailed cell-specific expression data are missing. RXRα is abundantly expressed in liver and kidney and in less amounts in the intestine. RXRβ is also ubiquitously expressed, but at low levels in liver, kidney and intestine, whereas RXRγ expression is restricted to liver and kidney [36,38,39].
In contrast, analysis of the A-FABP-PPRE revealed that transactivation is obtained only with the help of the PPARγ/RXR heterodimers. In adult mouse A-FABP as well as PPARγ1 and PPARγ2 are expressed together only in adipocyte cells and in macrophages. RXR subtypes are also expressed in adipocytes, RXRγ on a very low level [40], whereas there is no detailed literature about RXR expression in macrophages.
In conclusion, the regulation of FABPs is not only affected by the presence and functionality of a PPRE in their genes, but more strikingly by the spatiotemporal occurrence of the PPAR and RXR subtypes in L- and A-FABP-expressing cells in particular.
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SP 135/10-2). C.S. and T.E. gratefully acknowledge scholarships from the Stiftungsfonds Unilever and A.S. was supported by the ERASMUS programme.
References
- 1.Coe N. R., Bernlohr D. A. Physiological properties and functions of intracellular fatty acid-binding. Biochim. Biophys. Acta. 1998;1391:287–306. doi: 10.1016/s0005-2760(97)00205-1. [DOI] [PubMed] [Google Scholar]
- 2.Storch J., Thumser A. E. The fatty acid transport function of fatty acid-binding proteins. Biochim. Biophys. Acta. 2000;1486:28–44. doi: 10.1016/s1388-1981(00)00046-9. [DOI] [PubMed] [Google Scholar]
- 3.Noy N. Retinoid-binding proteins: mediators of retinoid action. Biochem. J. 2000;348:481–495. [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfrum C., Borrmann C. M., Börchers T., Spener F. Fatty acids and hypolipidemic drugs regulate peroxisome proliferator-activated receptors α- and γ-mediated gene expression via liver fatty acid binding protein: a signaling path to the nucleus. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2323–2328. doi: 10.1073/pnas.051619898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan N. S., Shaw N. S., Vinckenbosch N., Liu P., Yasmin R., Desvergne B., Wahli W., Noy N. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol. Cell. Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Y., Alvares K., Huang Q., Rao M. S., Reddy J. K. Cloning of a new member of the peroxisome proliferator-activated receptor gene family from mouse liver. J. Biol. Chem. 1993;268:26817–26820. [PubMed] [Google Scholar]
- 7.Tontonoz P., Hu E., Graves R. A., Budavari A. I., Spiegelman B. M. mPPAR γ2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 8.Tugwood J. D., Issemann I., Anderson R. G., Bundell K. R., McPheat W. L., Green S. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 1992;11:433–439. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juge-Aubry C., Pernin A., Favez T., Burger A. G., Wahli W., Meier C. A., Desvergne B. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. Importance of the 5′-flanking region. J. Biol. Chem. 1997;272:25252–25259. doi: 10.1074/jbc.272.40.25252. [DOI] [PubMed] [Google Scholar]
- 10.Palmer C. N., Hsu M. H., Griffin H. J., Johnson E. F. Novel sequence determinants in peroxisome proliferator signaling. J. Biol. Chem. 1995;270:16114–16121. doi: 10.1074/jbc.270.27.16114. [DOI] [PubMed] [Google Scholar]
- 11.IJpenberg A., Jeannin E., Wahli W., Desvergne B. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid x receptor heterodimer binding to DNA. J. Biol. Chem. 1997;272:20108–20117. doi: 10.1074/jbc.272.32.20108. [DOI] [PubMed] [Google Scholar]
- 12.Marcus S. L., Miyata K. S., Zhang B., Subramani S., Rachubinski R. A., Capone J. P. Diverse peroxisome proliferator-activated receptors bind to the peroxisome proliferator-responsive elements of the rat hydratase/dehydrogenase and fatty acyl-CoA oxidase genes but differentially induce expression. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5723–5727. doi: 10.1073/pnas.90.12.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hijikata M., Wen J. K., Osumi T., Hashimoto T. Rat peroxisomal 3-ketoacyl-CoA thiolase gene. Occurrence of two closely related but differentially regulated genes. J. Biol. Chem. 1990;265:4600–4606. [PubMed] [Google Scholar]
- 14.Schoonjans K., Peinado-Onsurbe J., Lefebvre A. M., Heyman R. A., Briggs M., Deeb S., Staels B., Auwerx J. PPARα and PPARγ activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 15.Tontonoz P., Nagy L., Alvarez J. G., Thomazy V. A., Evans R. M. PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell (Cambridge, Mass.) 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 16.Frohnert B. I., Hui T. Y., Bernlohr D. A. Identification of a functional peroxisome proliferator-responsive element in the murine fatty acid transport protein gene. J. Biol. Chem. 1999;274:3970–3977. doi: 10.1074/jbc.274.7.3970. [DOI] [PubMed] [Google Scholar]
- 17.Issemann I., Prince R., Tugwood J., Green S. A role for fatty acids and liver fatty acid binding protein in peroxisome proliferation? Biochem. Soc. Trans. 1992;20:824–827. doi: 10.1042/bst0200824. [DOI] [PubMed] [Google Scholar]
- 18.Simon T. C., Roth K. A., Gordon J. I. Use of transgenic mice to map cis-acting elements in the liver fatty acid-binding protein gene (Fabpl) that regulate its cell lineage-specific, differentiation-dependent, and spatial patterns of expression in the gut epithelium and in the liver acinus. J. Biol. Chem. 1993;268:18345–18358. [PubMed] [Google Scholar]
- 19.Tontonoz P., Graves R. A., Budavari A. I., Erdjument-Bromage H., Lui M., Hu E., Tempst P., Spiegelman B. M. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPARγ and RXRα. Nucleic Acids Res. 1994;22:5628–5634. doi: 10.1093/nar/22.25.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolfrum C., Ellinghaus P., Fobker M., Seedorf U., Assmann G., Börchers T., Spener F. Phytanic acid is ligand and transcriptional activator of murine liver fatty acid binding protein. J. Lipid Res. 1999;40:708–714. [PubMed] [Google Scholar]
- 21.Pelton P. D., Zhou L., Demarest K. T., Burris T. P. PPARγ activation induces the expression of the adipocyte fatty acid binding protein gene in human monocytes. Biochem. Biophys. Res. Commun. 1999;261:456–458. doi: 10.1006/bbrc.1999.1071. [DOI] [PubMed] [Google Scholar]
- 22.Bleck B., Hohoff C., Binas B., Rüstow B., Dixkens C., Hameister H., Börchers T., Spener F. Cloning and chromosomal localisation of the murine epidermal-type fatty acid binding protein gene (Fabpe) Gene. 1998;215:123–130. doi: 10.1016/s0378-1119(98)00262-5. [DOI] [PubMed] [Google Scholar]
- 23.Treuner M., Kozak C. A., Gallahan D., Grosse R., Müller T. Cloning and characterization of the mouse gene encoding mammary-derived growth inhibitor/heart-fatty acid-binding protein. Gene. 1994;147:237–242. doi: 10.1016/0378-1119(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 24.Banaszak L., Winter N., Xu Z., Bernlohr D. A., Cowan S., Jones T. A. Lipid-binding proteins: a family of fatty acid and retinoid transport proteins. Adv. Protein Chem. 1994;45:89–151. doi: 10.1016/s0065-3233(08)60639-7. [DOI] [PubMed] [Google Scholar]
- 25.Hanhoff T., Lücke C., Spener F. Insights into binding of fatty acids by fatty acid binding proteins. Mol. Cell. Biochem. 2002;239:45–54. [PubMed] [Google Scholar]
- 26.Brown P. J., Winegar D. A., Plunket K. D., Moore L. B., Lewis M. C., Wilson J. G., Sundseth S. S., Koble C. S., Wu Z., Chapman J. M., et al. A ureido-thioisobutyric acid (GW9578) is a subtype-selective PPARα agonist with potent lipid-lowering activity. J. Med. Chem. 1999;42:3785–3788. doi: 10.1021/jm9903601. [DOI] [PubMed] [Google Scholar]
- 27.Peters J. M., Aoyama T., Burns A. M., Gonzalez F. J. Bezafibrate is a dual ligand for PPARα and PPARβ: studies using null mice. Biochim. Biophys. Acta. 2003;1632:80–89. doi: 10.1016/s1388-1981(03)00065-9. [DOI] [PubMed] [Google Scholar]
- 28.Bleck B., Buhlmann C., Hohoff C., Müller M., Börchers T., Spener F. Inversely related expression of epidermal- and heart-type fatty acid binding proteins during myogenic differentiation of C2C12 myoblasts. Eur. J. Lipid Sci. Technol. 2002;104:88–97. [Google Scholar]
- 29.Dong D., Ruuska S. E., Levinthal D. J., Noy N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J. Biol. Chem. 1999;274:23695–23698. doi: 10.1074/jbc.274.34.23695. [DOI] [PubMed] [Google Scholar]
- 30.Brun R. P., Tontonoz P., Forman B. M., Ellis R., Chen J., Evans R. M., Spiegelman B. M. Differential activation of adipogenesis by multiple PPAR isoforms. Genes Dev. 1996;10:974–984. doi: 10.1101/gad.10.8.974. [DOI] [PubMed] [Google Scholar]
- 31.Holst D., Luquet S., Nogueira V., Kristiansen K., Leverve X., Grimaldi P. A. Nutritional regulation and role of peroxisome proliferator-activated receptor δ in fatty acid catabolism in skeletal muscle. Biochim. Biophys. Acta. 2003;1633:43–50. doi: 10.1016/s1388-1981(03)00071-4. [DOI] [PubMed] [Google Scholar]
- 32.Gordon J. I., Alpers D. H., Ockner R. K., Strauss A. W. The nucleotide sequence of rat liver fatty acid binding protein mRNA. J. Biol. Chem. 1983;258:3356–3363. [PubMed] [Google Scholar]
- 33.Veerkamp J. H., Paulussen R. J., Peeters R. A., Maatman R. G., van Moerkerk H. T., van Kuppevelt T. H. Detection, tissue distribution and (sub)cellular localization of fatty acid-binding protein types. Mol. Cell. Biochem. 1990;98:11–18. doi: 10.1007/BF00231362. [DOI] [PubMed] [Google Scholar]
- 34.Hohoff C., Spener F. Fatty acid binding proteins and mammary-derived growth inhibitor. Fett/Lipid. 1998;100:252–263. [Google Scholar]
- 35.Iseki S., Kanda T., Hitomi M., Ono T. Ontogenic appearance of three fatty acid binding proteins in the rat stomach. Anat. Rec. 1991;229:51–60. doi: 10.1002/ar.1092290107. [DOI] [PubMed] [Google Scholar]
- 36.Braissant O., Foufelle F., Scotto C., Dauca M., Wahli W. Differential expression of peroxisome proliferator-activated receptors. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 37.Escher P., Braissant O., Basu-Modak S., Michalik L., Wahli W., Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142:4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 38.Mangelsdorf D. J., Borgmeyer U., Heyman R. A., Zhou J. Y., Ong E. S., Oro A. E., Kakizuka A., Evans R. M. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 39.Mano H., Ozawa T., Takeyama K., Yoshizawa Y., Kojima R., Kato S., Masushige S. Thyroid hormone affects the gene expression of retinoid X receptors in the adult rat. Biochem. Biophys. Res. Commun. 1993;191:943–949. doi: 10.1006/bbrc.1993.1308. [DOI] [PubMed] [Google Scholar]
- 40.Kamei Y., Kawada T., Kazuki R., Sugimoto E. Retinoic acid receptor γ2 gene expression is up-regulated by retinoic acid in 3T3-L1 preadipocytes. Biochem. J. 1993;293:807–812. doi: 10.1042/bj2930807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Graves R. A., Tontonoz P., Spiegelman B. M. Analysis of a tissue-specific enhancer: ARF6 regulates adipogenic gene expression. Mol. Cell. Biol. 1992;12:3313. doi: 10.1128/mcb.12.7.3313-a. [DOI] [PMC free article] [PubMed] [Google Scholar]