Abstract
The Mycobacterium tuberculosis oriC (the origin of chromosomal replication) region contains 13 non-perfect DnaA boxes. The M. tuberculosis initiator protein, DnaA, was overexpressed in Escherichia coli as a soluble His-tagged fusion protein. The purified protein His6MtDnaA was investigated for its binding properties to DnaA boxes from the oriC region. Gel retardation demonstrated that the DnaA from M. tuberculosis requires two DnaA boxes for efficient binding. Electron microscopy as well as DNase I footprinting showed that the His6MtDnaA protein binds to four specific regions, which correspond to the location of 11 out of 13 previously identified DnaA boxes within the M. tuberculosis oriC. Probably, in M. tuberculosis, DnaA molecules by co-operative binding of numerous ‘non-perfect’ DnaA boxes assemble along the oriC region and subsequently form a massive nucleoprotein complex.
Keywords: DnaA, initiation of chromosome replication, Mycobacterium tuberculosis, origin of chromosomal replication (oriC)
Abbreviations: IPTG, isopropyl β-D-thiogalactoside; Ni-NTA, Ni2+-nitrilotriacetate; oriC, origin of chromosomal replication
INTRODUCTION
Replication of chromosomes is strictly regulated to ensure that all copies are replicated exactly once per cell cycle [1,2]. In all branches of life, the replication is controlled at the initiation stage. Therefore the events that occur at the replication origin play a central role in the cell cycle. In bacteria, replication starts at the unique site, oriC (origin of chromosomal replication), and continues bidirectionally around the chromosome until the two replication forks meet in the terminus region opposite to the oriC or reach chromosomal ends. Initiation of bacterial chromosome replication is mediated by the initiator DnaA protein, which interacts with repetitive, non-palindromic, nonamer sequences, the DnaA boxes, which are localized within the oriC region [3,4]. Among bacteria, the initiation of replication is best understood in Escherichia coli. Binding of 10–20 DnaA protein molecules to five DnaA boxes of the E. coli oriC region promotes unwinding within the AT-rich region of the oriC [4].
TB (tuberculosis) kills 2 million people each year. This global epidemic is growing and becoming more dangerous. The World Health Organization (http://www.who.int/en/) estimates that between 2002 and 2020, approx. 1000 million people will be newly infected, over 150 million people will get sick and 36 million will die of TB, if control is not further strengthened. In man, TB is usually caused by Mycobacterium tuberculosis. The genus Mycobacterium includes numerous species ranging from rapid-growing organisms, M. smegmatis and M. fortuitum (generation time is 3–4 h), to slow-growing organisms, such as M. tuberculosis (24 h) and M. leprae (2 weeks) [5]. Assuming that the rate of chromosomal DNA synthesis in mycobacteria is approximately the same as in other bacterial organisms (such as E. coli), the mycobacterial DNA replication machinery must be completely repressed for most of the cell cycle, particularly in slow growers. Recently, the genome sequences of four different mycobacteria species including two M. tuberculosis strains [6,7] were determined. However, our knowledge of mycobacterial cell cycle and initiation of chromosome replication is scarce. In spite of the fact that the DNA fragments containing the dnaA–dnaN intergenic region from M. avium, M. leprae, M. smegmatis and M. tuberculosis were shown to promote autonomous replication [8–12], little is known about interactions between the two key elements of the initiation of mycobacterial chromosome, oriC and DnaA protein. In contrast with E. coli, the structure of mycobacterial oriC region is more complex; the oriC is longer than the one in E. coli and possesses numerous ‘non-perfect’ DnaA boxes. Each of the 13 DnaA boxes located within the M. tuberculosis oriC [12] region differs at least by one base from the perfect E. coli DnaA box (TTATCCACA). Thus it is important to understand more of the initiation of DNA replication in these slow-growing organisms. In the present study, we describe the procedure for purification of the soluble M. turbeculosis DnaA protein and we characterize binding requirements of this protein.
MATERIALS AND METHODS
Bacterial strains, media and culture conditions
Epicurian Coli™ XL1-Blue [endA1, gyrA46, hsdR17, lac, recA1, supE44, thi; F′lac:lacIq, Δ(lacZ)M15, Tn10, proA+, proB+] and DH5α [F−, Φ80dlacZΔM15, recA1, endA1, gyrA96, thi-1, hsdR17, (rk−,mk+), supE44, relA1, deoR, Δ(lacZYA-argF)U169] [13] served as host for cosmids and plasmids. E. coli BL21 [BF−dcm ompT hsdS(rB− mB)gal] was the host for overproduction of the fusion protein His6–DnaA. E. coli strains were grown in Luria–Bertani medium at 30 or 37 °C. Antibiotics were used at the following concentrations: ampicillin (100 μg/ml) for E. coli, kanamycin (30 μg/ml) for plasmids and tetracyclin (12.5 μg/ml) for Epicurian Coli™ XL1-Blue.
DNA manipulations
Cosmids, plasmids and DNA fragments were purified using kits according to the manufacturer's instructions (Qiagen, Chatsworth, CA, U.S.A.). DNA fragments for DNase I footprinting experiments and gel retardation were PCR-amplified using primers, one of which was 5′-end-labelled using [γ-32P]ATP and T4-polynucleotide kinase. Enzymes were supplied by Roche, MBI Fermentas (Vilnius, Lithuania) and Gibco BRL (Gaithersburg, MD, U.S.A.). Isotopes were obtained from MP Biomedicals (Irvine, CA, U.S.A.). The oligonucleotides used for PCR or for sequencing were from Bionovo (Bioresearch Laboratory & Biochemicals, Legnica, Poland). DNA sequencing was performed using a Thermo Sequenase cycle sequencing kit (Amersham Biosciences).
DnaA purification
The M. tuberculosis dnaA gene PCR amplified by pdnaABamHIfw and pdnaANotIrv primers (using cosmid I441 as a template) was cloned into pGEM-T Easy. The authenticity of the pGEM-T EasyMtdnaA construct was checked by sequencing of both strands. The dnaA gene was cut out from the pGEM-T EasyMtdnaA by BamHI, NotI and cloned into pET-28a(+) vector using the same restriction sites. The E. coli BL21 cells were transformed with pET-28a(+)His6MtdnaA plasmid. The His6MtDnaA protein was overproduced from a 2-litre culture of E. coli BL21 cells containing pET-28a(+)His6MtdnaA plasmid, by 3 h induction with 0.05 mM IPTG (isopropyl β-D-thiogalactoside) at 30 °C. The cells were harvested by centrifugation (4500 g, 10 min, 4 °C). The pellet was frozen at −80 °C and kept until further purification steps. The bacterial pellet was thawed and suspended in ice-cold buffer L [25 mM Hepes/KOH (pH 7.6), 100 mM potassium glutamate and 5 mM 2-mercaptoethanol] (4 ml/g of wet biomass) with addition of one tablet of Complete Mini, EDTA-free Protease Inhibitor Cocktail Tablets (Roche) [14,15]. The lysozyme was added to a final concentration of 1 mg/ml and the cell suspension was incubated on ice for 30 min. The cells were lysed by sonication and centrifuged at 34500 g for 50 min. The supernatant was precipitated with 0.34 g/ml ammonium sulphate. The precipitated proteins were centrifuged at 27000 g for 30 min, the pellet was suspended in 2.5 ml of LG buffer [45 mM Hepes/KOH (pH 7.6), 200 mM potassium glutamate, 10 mM magnesium acetate, 0.5 mM EDTA and 5 mM 2-mercaptoethanol] and desalted on a PD-10 column (Amersham Biosciences) according to the manufacturer's instructions. The sample was loaded on to the Ni-NTA (Ni2+-nitrilotriacetate)–agarose column, previously equilibrated with LG buffer. After washing with LG buffer, the His6MtDnaA protein was eluted with an imidazole gradient (1–100 mM imidazole in LG buffer). The fractions were collected and analysed by SDS/PAGE.
Gel retardation assay
For binding assays, 32P-labelled DNA was incubated with DnaA protein in the presence of non-specific competitor [poly(dI/dC); Roche] at room temperature (20 °C) for 30 min in Marians' binding buffer [20 mM Hepes/KOH (pH 7.6), 5 mM magnesium acetate, 1 mM EDTA, 4 mM dithiothreitol, 0.2% Triton X-100, 3 mM ATP and 50 μg/ml BSA] [16]. The bound complexes were separated by PAGE (4 or 6% gel) [0.25× TBE (22.25 mM Tris/borate/0.25 mM EDTA), at 4 V/cm, 4 °C]. Gels were dried and analysed by a Typhoon 8600 Variable Mode Imager and ImageQuant software.
Electron microscopy
Electron microscopy analysis of DnaA–DNA interactions was performed by the method described previously [18]. Briefly, the M. tuberculosis oriC region was PCR-amplified using pMtoriCfw and pGEM-TEasyrvbiot primers (Table 1). The PCR DNA fragment was purified by filtration through Amicon Microcon-PCR Centrifugal Filter Devices (Millipore) and then DNA was incubated with streptavidin (Promega) for 5 min at room temperature. Excess of streptavidin was removed by filtration as above. M. tuberculosis DnaA protein was incubated at room temperature with the biotin/streptavidin DNA fragment in binding buffer (as indicated above). After fixation in 0.2% glutaraldehyde (15 min, room temperature), samples were prepared for electron microscopy by adsorption on mica [19]. They were analysed with a transmission electron microscope (Philips CM100) at 60 kV. The lengths of DNA fragments and the position of complexes were measured by using an electronic digitizer and evaluated with a computer program [14].
Table 1. Cosmids, plasmids and oligonucleotides used in the present study.
Restriction sites are given in boldface.
| Relevant characteristics, reference | |
|---|---|
| Cosmids and plasmids (symbol) | |
| I441 | ampr, M. tuberculosis cosmid containing oriC region and dnaA gene [26] |
| pGEM-T Easy | ampr, T vector for cloning PCR-amplified fragments (Promega) |
| pET-28a(+) | kanr, expression vector, His-tag coding sequence (Novagen) |
| pGEX-6P-1 | ampr, used as a cloning vector (Amersham Biosciences) |
| pGEM-T Easy MtoriC | pGEM-T Easy derivative containing the M. tuberculosis oriC region (this study) |
| pGEM-T Easy MtdnaA | pGEM-T Easy derivative containing the M. tuberculosis dnaA gene (this study) |
| pET-28a(+)His6MtdnaA | pET-28a(+) derivative containing M. tuberculosis dnaA gene (this study) |
| pGEX-6P-1MtoriC214 | pGEX-6P-1 derivative, containing 214 bp of M. tuberculosis oriC region (BamHI–SalI fragment), recloned from pGEM-T Easy MtoriC (this study) |
| pGEX-6P-1MtoriC321 | pGEX-6P-1 derivative, containing 321 bp of M. tuberculosis oriC region (SalI–BamHI fragment), recloned from pGEM-T Easy MtoriC (this study) |
| Oligonucleotides (symbol) | Sequence |
| pMtoriCfw | CGGGATCCCACGGCGTGTTCTTCCGACAACG |
| pMtoriCrv | CGGGATCCTGCGCCCTTTCACCTCACGATGAG |
| pGEM-T Easyrvbiot | biot-CAGGCGGCCGCGAATTCACTAGTG |
| pdnaABamHIfw | CGGATCCTTGACCGATGACCCCGGTTCAGG |
| pdnaANotIrv | GCGGCCGCTAGCGCTTGGAGCGCTGACGG |
| fwDnaAbox-1 | GAGACACTTGTCCACACAACT |
| rvDnaAbox-1 | AGTTGTGTGGACAAGTGTCTC |
| fwDnaAbox-2 | AGACACTTGTCCACAGGCTGGGGACAACAACTT |
| rvDnaAbox-2 | AAGTTGTTGTCCCCAGCCTGTGGACAAGTGTCT |
| fwnonbox | GGTATTATTGCAACGGATTGTTG |
| rvnonbox | GTGTCAAAGATTTCAAAGCCCAC |
| pGEXfw | GGGCTGGCAAGCCACGTTTGGTG |
| pGEXrv | CCGGGAGCTGCATGTGTCAGAGG |
DNase I footprinting
The 5′-end-radiolabelled DNA fragments (approx. 10 fmol) were incubated with different amounts of DnaA protein in binding buffer (see above) at room temperature for 30 min. Then DNase I digestion was performed as described by Majka et al. [17]. The DNase I cleavage products were separated in an 8% polyacrylamide/urea sequencing gel. Gels were dried and analysed by a Typhoon 8600 Variable Mode Imager and ImageQuant software.
RESULTS
Expression and purification of a soluble M. tuberculosis DnaA protein
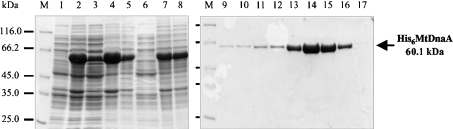
The vector pET-28a(+) was chosen to overexpress the M. tuberculosis DnaA as a His-tagged protein (see the Materials and methods section for details). To isolate the recombinant His6MtDnaA protein in its native form, the best conditions for expression of the soluble protein in E. coli were established. E. coli BL21 cells harbouring the pET-28a(+)His6MtdnaA vector were grown to an A600 0.65 at 37 °C, and then the expression of the His6MtDnaA protein was induced in the presence of a low concentration of IPTG (0.05 mM) at the reduced temperature (30 °C) (Figure 1). The resulting His6MtDnaA fusion protein (60.1 kDa, Figure 1) was purified from the soluble fraction by affinity chromatography on the Ni-NTA–agarose column as described in the Materials and methods section. The His6MtDnaA protein, similar to other DnaA proteins, exhibits a tendency to aggregate. Application of potassium glutamate in the isolation buffer (adapted from [14,15]) significantly reduced self-aggregation of the His6MtDnaA protein molecules during the purification procedures. Our procedure allowed us to isolate approx. 2.5 mg of soluble His6MtDnaA protein from 1000 ml of E. coli BL21 culture. The purified His6MtDnaA protein was more than 98% homogeneous as judged by SDS/PAGE analysis (Figure 1, fractions from tracks 14–15).
Figure 1. Expression and purification of the recombinant M. tuberculosis DnaA.
DnaA protein was isolated from E. coli BL21 cells harbouring the pET-28a(+)His6MtdnaA vector. Proteins were analysed by SDS/PAGE. Tracks: M, molecular-mass standard (protein molecular-mass standard; MBI Fermentas); 1, whole extract from uninduced cells; 2, whole extract from the cells after 3 h of induced expression; 3, soluble cell lysate from induced cells; 4, non-soluble cell debris from induced cells; 5, protein pellet after (NH4)2SO4 precipitation; 6, soluble protein fraction after (NH4)2SO4 precipitation; 7, (NH4)2SO4 protein pellet dissolved in LG buffer; 8, protein fraction desalted on PD-10 column; 9–17 fractions eluted from Ni-NTA–agarose column with LG containing an increased amount of imidazole: 9, 0 mM imidazole; 10, 1 mM imidazole; 11–12, 10 mM imidazole; 13–16, 50 mM imidazole; 17, 100 mM imidazole.
Binding requirements of the M. tuberculosis DnaA protein
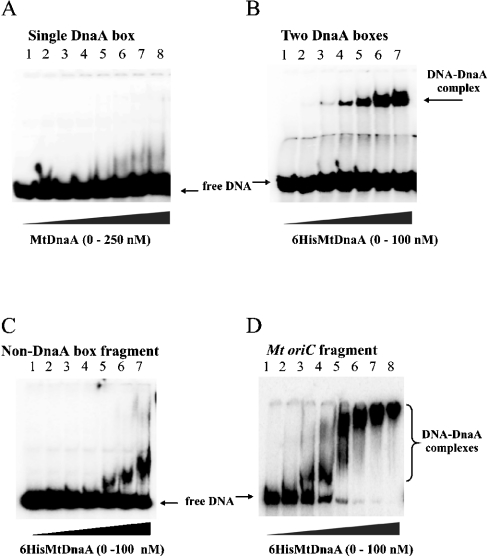
Comparison of DnaA boxes from several Mycobacteria oriC regions allowed the consensus sequence to be determined: TT(G/C)TCCACA (named a ‘perfect’ DnaA box) [8]. It corresponds to the Streptomyces consensus sequence for DnaA box (TTGTCCACA) [20]. To elucidate the binding requirements of the His6MtDnaA protein, gel retardation assays were performed. In our studies, the following four DNA fragments were chosen: (i) oligonucleotides containing a single DnaA box; (ii) oligonucleotide containing two DnaA boxes: the ‘perfect’ DnaA box and a DnaA box with one mismatch; (iii) the M. tuberculosis oriC region; and (iv) DNA fragment that does not contain any DnaA box motif (non-DnaA box fragment) (Figure 2). 32P-labelled DNA fragments were incubated with various amounts of the His6MtDnaA protein, and then nucleoprotein complexes were analysed in a native polyacrylamide gel. A single DnaA box, including the ‘perfect’ one, was not bound by the His6MtDnaA even at high protein concentration (250 nM, Figure 2A), whereas incubation of the His6MtDnaA protein with the DNA fragment containing two DnaA boxes caused the appearance of bands with decreased mobility already at 5 nM protein concentration (Figure 2B). Incubation of the His6MtDnaA protein with the oriC region led to the formation of very high-molecular-mass complexes (Figure 2D, tracks 5–8), indicating that the multiple DnaA-binding motifs and/or protein–protein interactions were engaged in the formation of the analysed complexes. Specificity of the interaction between the M. tuberculosis DnaA and the oriC region is corroborated by the inability of the His6MtDnaA protein to form high-molecular-mass complexes with the DNA fragment (348 bp) that lacks the DnaA-binding site (non-DnaA box DNA, Figure 2C). However, it has to be noted that at high protein concentration, a fast migrating complex appeared when the non-DnaA box fragment was incubated with the His6DnaA. This may suggest that a longer DNA fragment is unspecifically bound by the His6MtDnaA protein. Incubation of the oriC region with low amounts of the His6DnaA protein also led to the formation of low-molecular-mass complexes. However, these complexes may represent not fully matured nucleoprotein complexes.
Figure 2. Binding requirements of the His6MtDnaA protein.
The gel retardation assays were performed using 32P-labelled DNA fragments and increasing amounts of the His6MtDnaA. (A) The double-stranded oligonucleotide containing the single ‘perfect’ DnaA box (TTGTCCACA, 21 bp). Tracks: 1, 0; 2, 2.5; 3, 5; 4, 10; 5, 25; 6, 50; 7, 100; 8, 250 nM His6MtDnaA. (B) The double-stranded oligonucleotide containing two DnaA boxes (TTGTCCACA and TTGTCCCCA, 33 bp). Tracks: 1, 0; 2, 1; 3, 5; 4, 10; 5, 25; 6, 50; 7, 100 nM His6MtDnA. (C) Non-DnaA box DNA fragment (348 bp), tracks: 1, 0; 2, 2.5; 3, 5; 4, 10; 5, 25; 6, 50; 7, 100 nM His6MtDnaA. (D) The oriC fragment (541 bp). Tracks: 1, 0; 2, 1; 3, 2.5; 4, 5; 5, 10; 6, 25; 7, 50; 8, 100 nM His6MtDnaA.
Thus taking into account the results of gel retardation assays, it could be concluded that the M. tuberculosis DnaA protein requires at least two DnaA boxes for efficient binding.
Identification of DnaA-binding sites within the M. tuberculosis oriC region
A total of 13 putative DnaA box motifs, each with at least one mismatch from the consensus sequence, were found within the oriC region of M. tuberculosis [10] (Figure 3). Such a relatively high abundance of ‘non-perfect’ DnaA boxes raised the question of how many of them are specifically bound by DnaA protein. To answer this question, the interactions between the His6MtDnaA protein and multiple putative DnaA boxes of the oriC region were analysed by electron microscopy and DNase I footprinting.
Figure 3. Structure of the M. tuberculosis oriC region.
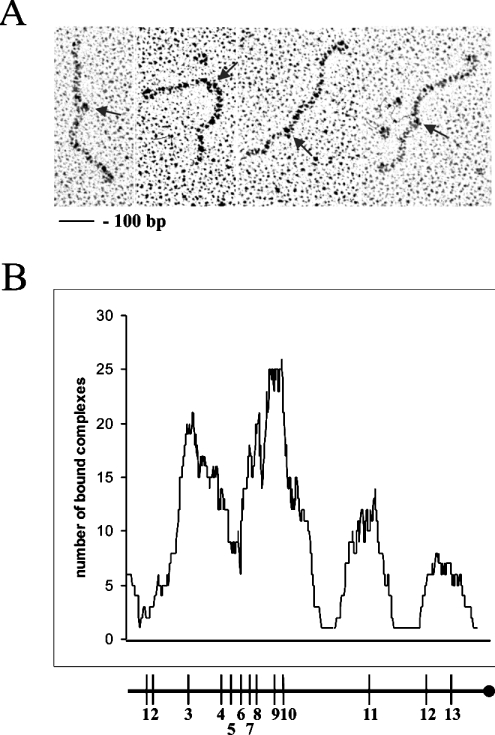
For electron microscopy analysis, the M. tuberculosis oriC region was PCR-amplified using a pair of primers pMtoriCfw and biotinylated pGEM-TEasyrvbiot. The PCR product was labelled with a streptavidin that served as reference point for measurements of the position of the nucleoproteins complex. At a ratio of 0.5:1 DnaA protein/DnaA box, the nucleoprotein complexes visualized by electron microscopy were small and their numbers did not exceed one or two per DNA molecule. The relative positions of the analysed nucleoprotein complexes corresponded to the position of most of the putative DnaA boxes along the oriC region (Figure 4). Within the analysed oriC region, the DnaA protein forms four separated complexes. The highest incidence of protein binding occurred at the two adjacent DnaA boxes (9th and 10th) and at the 3rd DnaA box, the only DnaA box with a single mismatch (Figures 3 and 4). Interestingly, the single 11th DnaA box separated from other boxes is also bound by the His6MtDnaA protein. The lowest incidence of protein binding took place at the more distal DnaA boxes (12th and 13th). According to the electron microscopy analysis, 1st and 2nd DnaA boxes were not bound at all. However, if putative-binding motifs are located close to the ends of the analysed DNA fragment, protein binding is frequently not observed (protein slips off the ends).
Figure 4. Electron microscopic visualization of M. tuberculosis nucleoprotein complexes, DnaA–oriC.
(A) Electron micrographs of 570 bp DNA fragment complexed with streptavidin (end-labelled) and DnaA protein. Arrows indicate nucleoprotein complexes. (B) Nucleoprotein complexes formed with the His6MtDnaA protein at the oriC region. Histogram of complexes of DnaA protein with the oriC region. DnaA boxes are indicated by vertical bars.
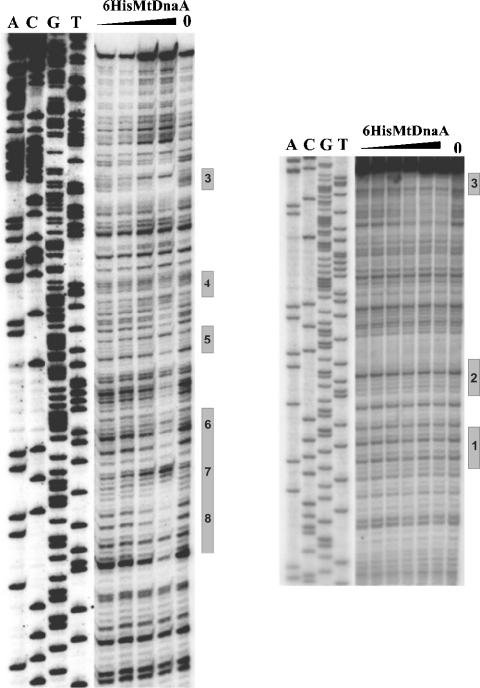
For DNase I footprinting, two DNA fragments (representing the entire M. tuberculosis oriC region) labelled at one 5′-end (lower and upper strands) were incubated with various amounts of the His6MtDnaA protein, and then subjected to limited DNase I cleavage (Figure 5). DNase I footprinting analyses corroborate electron microscopy results; the protected sites correspond to the locations of nucleoprotein complexes visualized by electron microscopy (Figure 5; results not shown). DNase I footprinting also showed that DnaA boxes 1 and 2 are not bound by the His6MtDnaA protein (Figure 5; although the boxes are located far away from the ends of the DNA fragment exposed to the DNase I), whereas the two single DnaA boxes 3 (Figure 5) and 11 (results not shown) are bound by the protein. DnaA box 3 is particularly well protected by the His6MtDnaA protein (Figure 5).
Figure 5. Interactions of the His6MtDnaA protein with the oriC region.
DNase I footprinting was performed with the 32P-labelled oriC fragments which were incubated with increasing amounts of the His6MtDnaA protein. Labelled fragments, 307 bp (left site) and 296 bp (right site) were obtained by PCR amplification using pGEX-6P-1MtoriC214 plasmid (Table 1) as a template and pairs of primers: pMtoriCfw, 32P-pGEXrv and pGEXrv, 32P-pGEXfw (Table 1) respectively. Tracks: A, C, G and T are sequencing reactions prepared with the labelled pGEXrv primer. Positions of the DnaA boxes are shown on the right side of each panel.
DISCUSSION
Studies on bacterial initiation of chromosome replication were mostly focused on a model organism, E. coli. Little is known about the corresponding process in other bacteria, particularly in slow growing organisms such as mycobacteria. Knowledge about the interactions of M. tuberculosis DnaA protein with DNA may provide fresh insights into the function of this protein, and may further help in understanding the regulation of initiation of mycobacterial chromosome replication. In the present study, we describe the purification of the M. tuberculosis DnaA protein and identify its binding sites within the M. tuberculosis oriC region.
Recently, it has been reported that two mycobacterial DnaA proteins (M. avium and M. tuberculosis) [21,22] were isolated. However, both proteins were purified from inclusion bodies as His-tagged proteins on Ni2+-affinity column under denaturing conditions. To avoid these problems, we designed the procedure for isolation of the M. tuberculosis DnaA protein in its native form from the E. coli BL21 cells harbouring pET 28a(+)His6MtdnaA expression vector. Reduction of temperature and concentration of the inducer (IPTG) during expression of the recombinant His6MtDnaA protein allowed us to obtain the protein in soluble fraction in sufficient quantity for purification on affinity chromatography. The purified His6MtDnaA protein was used for investigating its binding properties.
The oriC region of M. tuberculosis, unlike its E. coli counterpart, possesses 13 putative DnaA boxes, which differ significantly from the ‘perfect’ E. coli sequences (TTATCCACA). The consensus sequence for Mycobacterium DnaA box is TTG/CTCCACA [8]. Within the M. tuberculosis oriC, there is only one DnaA box with a single mismatch from the consensus sequence (DnaA box no. 3, Figure 3). The remaining DnaA boxes differ by two or even three bases from the Mycobacterium consensus sequence. The M. tuberculosis oriC region contains the same number of DnaA boxes as the oriC region from Thermus thermophilus [23]. However, within the latter, all but two of the 13 DnaA sequences match the E. coli ‘perfect’ DnaA box.
As demonstrated by gel retardation analysis, the His6MtDnaA does not bind a single DnaA box, not even the Mycobacterium consensus sequence DnaA box (TTGTCCACA; Figure 2A). Recently, in contrast with our results, Dziadek et al. [24] showed by surface plasmon resonance technique that the M. tuberculosis DnaA protein binds a single DnaA box with three mismatches from the consensus sequence (DnaA box no. 4, CCGTTCACA). However, our DNase I footprinting as well as electron microscopy analyses demonstrated that this DnaA box, being in the vicinity of other DnaA boxes, is not efficiently protected by DnaA protein (see Figures 4 and 5). In addition, DnaA box 4 is not essential for oriC function; mutations in this DnaA box did not abolish chromosome replication [24].
Our gel retardation assay demonstrated that if another DnaA box accompanies the ‘perfect’ one, both of them are bound with a high affinity by the M. tuberculosis DnaA (Figure 2B). Thus, in contrast with E. coli, all other DnaA proteins from Helicobacter pylori, Streptomyces lividans as well as M. tuberculosis require two DnaA boxes for efficient binding if these contain mismatches. The DnaA protein of T. thermophilus requires even more DnaA boxes for efficient binding (at least three ‘perfect’) [23]. Our results suggest that interaction of the M. tuberculosis DnaA with two DnaA boxes exhibits co-operativity.
Electron microscopy as well as DNase I footprinting demonstrated that the purified His6MtDnaA protein binds to four specific regions, which correspond to the location of 11 out of the 13 previously identified DnaA boxes within the M. tuberculosis oriC. Two DnaA boxes located at the 5th end of the oriC region are not bound by the His6MtDnaA protein. Salazar et al. [9] have identified only seven putative DnaA boxes within the M. tuberculosis oriC region. Our electron microscopy studies demonstrated that the M. tuberculosis DnaA protein binds preferentially the fragment of the oriC region that contains these seven DnaA boxes (Figure 4, DnaA boxes 3–10).
Binding of the M. tuberculosis DnaA protein to the oriC region led to the formation of high-molecular-mass nucleoprotein complexes; a discrete nucleoprotein complex was not observed (see the Gel retardation assay section; Figure 2D). In contrast, DnaA proteins from E. coli [14] and H. pylori [15] bind sequentially five DnaA boxes in their oriC regions; at least four distinct nucleoprotein complexes are formed. In S. lividans [25] and T. thermophilus [23] organisms, which also possess numerous DnaA boxes within their oriC regions (19 and 13 respectively), only high-molecular-mass complexes were observed. Probably, in M. tuberculosis, T. thermophilus and S. lividans, DnaA molecules by co-operative binding of numerous DnaA boxes assemble along the oriC region and subsequently form the initial complex. Formation of such a massive nucleoprotein complex suggests that the DnaA is involved not only in DNA–protein but also in protein–protein interactions. In S. lividans, two domains, I and III, of DnaA protein are independently involved in protein–protein interactions [25].
Thus our results suggest that in M. tuberculosis, DnaA protein binds co-operatively to multiple ‘non-perfect’ DnaA boxes in oriC. Probably, the presence of a non-perfect DnaA box within the mycobacterial oriC region is compensated by their abundance.
Acknowledgments
This work was supported by the Ministry of Scientific Research and Information Research (The State Committee for Scientific Research, grant 3 P04A 079 22). Cosmid I441 was kindly provided by R. Brosch (Institut Pasteur, Paris, France). We thank D. Kamińska (Institute of Pharmacology, Krakow, Poland) and B. Lis (Institute of Immunology and Experimental Therapy, Wroclaw, Poland) for the help in M. tuberculosis oriC cloning and isolation of the M. tuberculosis DnaA protein respectively. We also thank W. Messer for his comments on the paper.
References
- 1.Kornberg A., Baker T. Regulation of chromosomal replication and cell division. In: Kornberg A., Baker T., editors. The DNA Replication. 2nd edn. New York, NY: W. H. Freeman; 1992. pp. 731–769. [Google Scholar]
- 2.Baker T. A., Bell S. P. Polymerases and the replisome: machines within machines. Cell (Cambridge, Mass.) 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- 3.Messer W., Blaesing F., Jakimowicz D., Krause M., Majka J., Nardmann J., Schaper S., Seitz H., Speck Ch., Weigel Ch., et al. Bacterial replication initiator DnaA. Rules for DnaA binding and roles of DnaA in origin unwinding and helicase loading. Biochimie. 2001;83:5–12. doi: 10.1016/s0300-9084(00)01216-5. [DOI] [PubMed] [Google Scholar]
- 4.Messer W. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 2002;26:355–374. doi: 10.1111/j.1574-6976.2002.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 5.Hartmans S., De Bont J. A. N. The genus Mycobacterium-nonmedical. In: Balows A., editor. The Prokaryotes. 2nd edn. New York: Springer-Verlag; 1992. pp. 1214–1237. [Google Scholar]
- 6.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Fleischamnn R. D., Alland D., Eisen J. A., Carpenter L., White O., Peterson J., DeBoy R., Dodson R., Gwinn M., Haft D., et al. Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J. Bacteriol. 2002;184:5479–5490. doi: 10.1128/JB.184.19.5479-5490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajagopalan M., Qin M.-H., Nash D. R., Madiraju M. V. V. S. Mycobacterium smegmatis dnaA region and autonomous replication activity. J. Bacteriol. 1995;177:6527–6535. doi: 10.1128/jb.177.22.6527-6535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salazar L., Fsihi H., de Rossi E., Riccardi G., Rios C., Cole S. T., Takiff H. E. Organization of the origins of replication of the chromosomes of Mycobacterium smegmatis Mycobacterium leprae and Mycobacterium tuberculosis and isolation of a functional origin from M. smegmatis. Mol. Microbiol. 1996;20:283–293. doi: 10.1111/j.1365-2958.1996.tb02617.x. [DOI] [PubMed] [Google Scholar]
- 10.Qin M. H., Madiraju M. V., Rajagopalan M. Characterization of the functional replication origin of Mycobacterium tuberculosis. Gene. 1999;233:121–130. doi: 10.1016/s0378-1119(99)00148-1. [DOI] [PubMed] [Google Scholar]
- 11.Madiraju M. V., Qin M. H., Yamamoto K., Atkinson M. A., Rajagopalan M. The dnaA gene region of Mycobacterium avium and the autonomous replication activities of its 5′ and 3′ flanking regions. Microbiology. 1999;145:2913–2921. doi: 10.1099/00221287-145-10-2913. [DOI] [PubMed] [Google Scholar]
- 12.Qin M.-H., Madiraju M. V. V. S., Zachariah S., Rajagopalan M. Characterization of the oriC region of Mycobacterium smegmatis. J. Bacteriol. 1997;179:6311–6317. doi: 10.1128/jb.179.20.6311-6317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J., Russel D. W. A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 2001. Molecular Cloning. [Google Scholar]
- 14.Weigel C., Schmidt A., Ruckert B., Lurz R., Messer W. DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 1997;16:6574–6583. doi: 10.1093/emboj/16.21.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zawilak A., Durrant M. C., Jakimowicz P., Backert S., Zakrzewska-Czerwińska J. DNA binding specificity of the replication initiator protein, DnaA from Helicobacter pylori. J. Mol. Biol. 2003;334:933–947. doi: 10.1016/j.jmb.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Parada C. A., Marians K. J. Mechanism of DNA A protein-dependent pBR322 DNA replication. DNA A protein-mediated trans-strand loading of the DNA B protein at the origin of pBR322 DNA. J. Biol. Chem. 1991;266:18895–18918. [PubMed] [Google Scholar]
- 17.Majka J., Jakimowicz D., Messer W., Schrempf H., Lisowski M., Zakrzewska-Czerwińska J. Interactions of the Streptomyces lividans initiator protein DnaA with its target. Eur. J. Biochem. 1999;260:325–335. doi: 10.1046/j.1432-1327.1999.00168.x. [DOI] [PubMed] [Google Scholar]
- 18.Szalewska-Pałasz A., Weigel C., Speck C., Śrutkowska S., Konopa G., Lurz R., Marszalek J., Taylor K., Messer W., Węgrzyn G. Interaction of the Escherichia coli DnaA protein with bacteriophage λDNA. Mol. Gen. Genet. 1998;259:679–688. doi: 10.1007/s004380050863. [DOI] [PubMed] [Google Scholar]
- 19.Spiess E., Lurz R. Electron microscopic analysis of nucleic acids and nucleic acids–protein complexes. Meth. Microbiol. 1988;20:293–323. [Google Scholar]
- 20.Jakimowicz D., Majka J., Messer W., Speck Ch., Fernandez M., Cruz Martin M., Sanchez J., Schauwecker F., Keller U., Schrempf H., et al. Structural elements of the Streptomyces oriC region and their interactions with the DnaA protein. Microbiology. 1998;144:1281–1290. doi: 10.1099/00221287-144-5-1281. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto K., Muniruzzaman S., Rajagopalan M., Madiraju M. V. V. S. Modulation of Mycobacterim tuberculosis DnaA protein–adenine-nucleotide interactions by acidic phospholipids. Biochem. J. 2002;363:305–311. doi: 10.1042/0264-6021:3630305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto K., Rajagopalan M., Madiraju M. Phospholipids promote dissociation of ADP from the Mycobacterim avium DnaA protein. J. Biochem. (Tokyo) 2002;131:219–224. doi: 10.1093/oxfordjournals.jbchem.a003091. [DOI] [PubMed] [Google Scholar]
- 23.Schaper S., Nardmann J., Luder G., Lurz R., Speck C., Messer W. Identification of the chromosomal replication origin from Thermus thermophilus and its interaction with the replication initiator DnaA. J. Mol. Biol. 2000;299:655–665. doi: 10.1006/jmbi.2000.3764. [DOI] [PubMed] [Google Scholar]
- 24.Dziadek J., Rajagopalan M., Parish T., Kurepina N., Greendyke R., Kreiswirth B. N., Madiraju M. V. V. S. Mutations in the CCGTTCACA DnaA box of Mycobacterium tuberulosis oriC that abolish replication of oriC plasmids are tolerated on the chromosome. J. Bacteriol. 2002;184:3848–3855. doi: 10.1128/JB.184.14.3848-3855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakimowicz D., Majka J., Konopa G., Węgrzyn G., Messer U., Schrempf H., Zakrzewska-Czerwińska J. Architecture of the Streptomyces lividans DnaA protein-replication origin complexes. J. Mol. Biol. 2000;298:351–364. doi: 10.1006/jmbi.2000.3686. [DOI] [PubMed] [Google Scholar]
- 26.Bange F. C., Collins F. M., Jacobs W. R., Jr Survival of mice infected with Mycobacterium smegmatis containing large DNA fragments from Mycobacterium tuberculosis. Tuber. Lung Dis. 1999;79:171–180. doi: 10.1054/tuld.1998.0201. [DOI] [PubMed] [Google Scholar]