Abstract
Roberts syndrome (RBS) is an autosomal recessive disorder with profound growth deficiency and limb reduction caused by ESCO2 loss-of-function variants. Here, we elucidate the pathogenesis of limb reduction in an Esco2fl/fl;Prrx1-CreTg/0 mouse model using bulk- and single-cell-RNA-seq and gene co-expression network analyses during embryogenesis. Our results reveal morphological and vascular defects culminating in hemorrhage of mutant limbs at E12.5. Underlying this abnormal developmental progression is a pre-apoptotic, mesenchymal cell population specific to mutant limb buds enriched for p53-related signaling beginning at E9.5. We then characterize these p53-related processes of cell cycle arrest, DNA damage, cell death, and the inflammatory leukotriene signaling pathway in vivo. In utero treatment with pifithrin-α, a p53 inhibitor, rescued the hemorrhage in mutant limbs. Lastly, significant enrichments were identified among genes associated with RBS, thalidomide embryopathy, and other genetic limb reduction disorders, suggesting a common vascular etiology among these conditions.
Subject terms: Angiogenesis, Development, Gene expression, Computational biology and bioinformatics, Genetic models
Roberts syndrome (RBS) is an autosomal recessive disorder with profound growth deficiency and limb reduction caused by ESCO2 loss-of-function variants. Here, the authors show that the pathogenesis of limb reduction in an Esco2 cohesinopathy mouse model of Roberts syndrome has an underlying vascular etiology that is mediated by p53-signaling, sharing commonality with thalidomide embryopathy.
Introduction
Cohesinopathies are a group of genetic disorders caused by pathogenic variants in genes encoding the cohesin subunits or their various regulators that participate in chromosome segregation and gene transcription1–4. These disorders are heterogeneous and include Roberts syndrome (RBS) and Cornelia de Lange syndrome, as well as CHOPS, chronic atrial and intestinal dysrhythmia, holoprosencephaly 13, intellectual disability-hypotonic facies, and Warsaw breakage syndromes4–9. Among the most severe is RBS, a monogenic disorder with an autosomal recessive pattern of inheritance caused by loss-of-function (LOF) pathogenic variants in the Establishment of Sister Chromatid Cohesion N-Acetyltransferase 2 (ESCO2) gene, which encodes a regulator of the cohesin complex7,10,11.
Infants with RBS experience profound growth deficiency of prenatal onset and present with various congenital anomalies, most notably with phocomelia or severe limb reduction, and craniofacial abnormalities10,12–14. There is phenotypic variability between cases with a positive correlation of severity between the craniofacial abnormalities and the limb reduction5,8,14. The most striking clinical presentation includes complete, bilateral symmetric reduction of the limbs, while less severe cases exhibit hypomelia due to mesomelic or rhizomelic shortening12,14. Typically, the upper and lower limb reduction or absence are accompanied by digit malformations, often presenting with oligodactyly, thumb aplasia or hypoplasia, syndactyly, clinodactyly, and elbow and knee flexion contractures12,14. Given the syndromic nature of the disease, survival is poor beyond the neonatal period, although mild cases can survive into adulthood with varying degrees of physical and intellectual disability. The phenotypic similarity of RBS limb defects to thalidomide embryopathy has spurred the eponymous description of RBS as ‘pseudothalidomide’ syndrome, suggesting that the similar limb phenotypes may result from a common underlying mechanism13. Much research has been devoted to uncovering the teratogenic mechanisms of thalidomide, yet the mechanisms that dysregulate limb development in RBS have not been well studied, despite progress in understanding ESCO2’s molecular functions15–25.
ESCO2 is a highly conserved N-acetyltransferase that acetylates the SMC3 subunit of the cohesin complex8,26–28. Pathogenic variants of ESCO2 result in hypoacetylation and instability of cohesin and its downstream processes29. The establishment of cohesion is essential for tethering sister chromatids together until their separation during anaphase30,31; for the extrusion of DNA loops that modulate genome organization and gene transcription32–36, and for facilitating the repair of double-strand DNA breaks by inducing genome-wide cohesion during homologous recombination37–41. Reduction of ESCO2 in human, mice, or zebrafish tissues induces mitotic failure and increases apoptosis10,42–44. On a cellular level, lagging chromosomes, aneuploidy, and micronuclei are observed resulting from chromosome segregation defects45. In human RBS patient-derived cells, activation of the mitotic spindle checkpoint halts cell growth, which results in mitotic delay and impaired cell proliferation45,46, and exhibits hypersensitivity to DNA damaging agents47–49.
We generated an Esco2 conditional knockout (CKO) mouse line introducing a deletion that recapitulates a human mutation found in RBS patients10. Using mesenchyme-expressing Prrx1-CreTg/0, we created a model system that resembles the human limb phenotype. By applying bulk and single-cell RNA-sequencing (bulk RNA-seq; scRNA-seq), multi-scale embedded gene co-expression network analysis (MEGENA), the Ensemble of Multiple Drug Repositioning Approaches (EMUDRA), and various histological techniques, we investigated the temporal dynamics and consequences of Esco2 loss on limb development during early embryogenesis. We then leveraged this data in an enrichment analysis to identify genes commonly dysregulated among thalidomide embryopathy, genetic limb reduction disorders, and RBS.
Results
Esco2fl/fl;Prrx1-CreTg/0 mouse model resembles the human RBS limb phenotype
To study the function of ESCO2 in the context of limb development, we generated an Esco2fl/fl conditional knockout (CKO) mouse line bred with Prrx1-CreTg/0 to preferentially eliminate Esco2 from limb mesenchyme. We introduced a deletion in exon 4 resulting in a premature stop codon, replicating the effect of a documented human pathogenic variant in ESCO2 (c.879_880delAG (p.D292fsX47); ClinVar ID: 21250) that is predicted to produce a truncated Esco2 transcript with nonsense-mediate decay (Supplementary Fig. 1a–c)10. The first appearance of the limb buds and the activity of Prrx1-induced Cre recombinase are observed at E9.550. Prrx1 is expressed in a proportion of forelimb mesenchymal cells and completes recombination in all forelimb mesenchymal cells at E10.5.
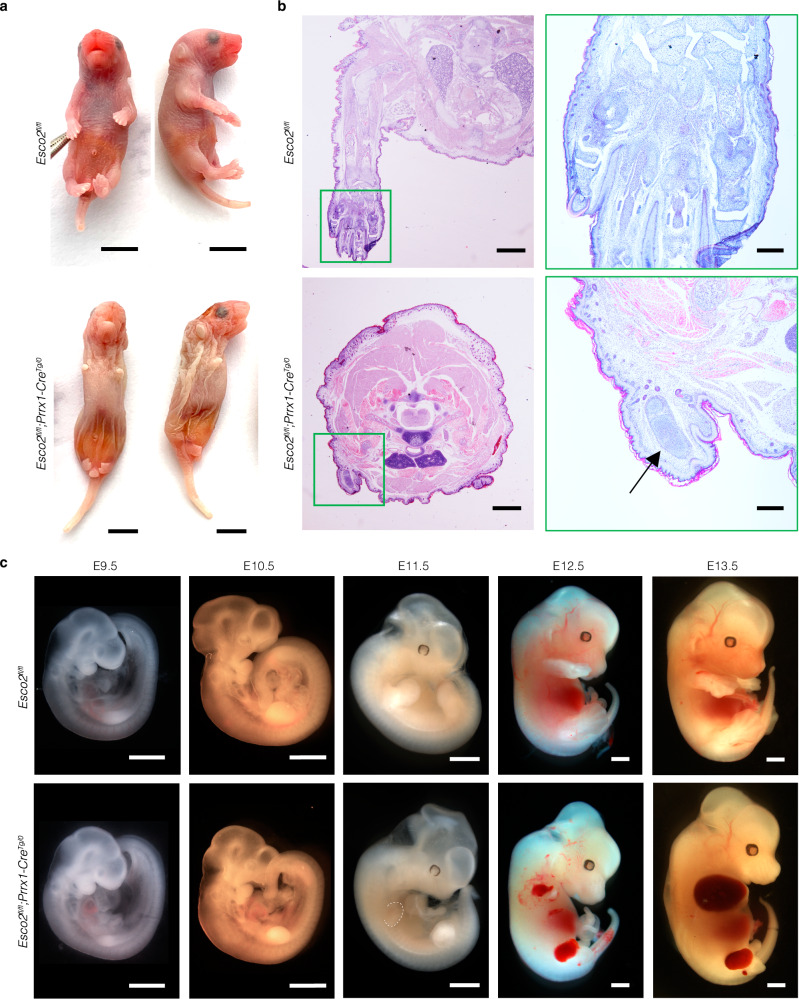
While most developing Esco2fl/fl;Prrx1-CreTg/0 embryos resulted in perinatal lethality, some were livebirths. Esco2fl/fl;Prrx1-CreTg/0 pups at P0 were born with striking limb reduction that resembled the human RBS phenotype (Fig. 1a). This deletion was highly penetrant with 100% of n = 120 Esco2fl/fl;Prrx1-CreTg/0 embryos displaying the RBS limb phenotype (Fig. 1a). Post-mortem autopsy analysis of mutants revealed islands of disorganized chondrocytes and poorly developed chondroid matrix compared to controls (Fig. 1b). The long bones of the fore- and hindlimbs were unidentifiable, with loose mesenchyme replacing skeletal muscle around the bones (Fig. 1b, Supplementary Fig. 1d, e). Internal organs were not significantly affected where Prrx1 was not expressed51,52 (Fig. 1a).
Fig. 1. Esco2fl/fl;Prrx1-CreTg/0 mouse model of Roberts syndrome.
a P0 mice in dorsal and lateral views with indicated genotypes. Mutant mice present with phocomelia and craniofacial abnormalities including skull flattening and micrognathia. Scale bar: 5000 µm. b Hematoxylin and eosin (H&E) staining of coronal sections of Esco2fl/fl (n = 3) and Esco2fl/fl;Prrx1-CreTg/0 (n = 3) forelimbs displayed in Fig. 1a. Scale bar: 1000 µm. Higher magnification of control and mutant forelimbs in outlined green boxes. Arrow points to islands of disorganized chondrocytes and palely amphophilic chondroid matrix. Humeral, radial, and ulna bones are unidentifiable in mutant compared to control pups. Scale bar: 5000 µm. c Gross morphology of E9.5–E13.5 embryos post-harvesting with indicated genotypes. Notably smaller limbs with hemorrhaging in mutant compared to control embryos. Dashed white line outlines E11.5 forelimb with edema. Scale bar: 1000 µm.
To explore morphological changes during embryogenesis, we compared Esco2fl/fl;Prrx1-CreTg/0 and Esco2fl/fl embryonic limbs from E9.5-E13.5 (Fig. 1c). These embryonic stages coincided temporally with the initiation of limb development. There was no notable difference in size or morphology of the mutant and control limbs bud at E9.5 (Fig. 1c). However, obvious morphological differences including a reduced size of the limb bud were detected in Esco2fl/fl;Prrx1-CreTg/0 embryos at E10.5, followed by the accumulation of fluid and edema at E11.5 (Fig. 1c). By E12.5, hemorrhage was detected in the proximal region of Esco2fl/fl;Prrx1-CreTg/0 limb buds, which persisted with more severe bleeding into the entire region at E13.5 (Fig. 1c). At the cellular level, we observed the presence of abnormal nuclear morphology, blebbing, and micronuclei, similar to cytogenetic profiles of human RBS patients (Supplementary Fig. 1f). Taken together, the abnormal limb morphology with hemorrhage and skeletal defects provided the first insights into potential developmental processes compromised in RBS.
Transcriptomic profiling of E9.5–E11.5 limb buds identifies a p53 driven pre-apoptotic mesenchymal cell population in mutant limbs
After identifying early and severe morphological differences in the mutant limb bud, we sought to explore expression changes at the tissue level by performing bulk RNA-seq of Esco2fl/fl;Prrx1-CreTg/0 and Esco2fl/fl limb buds at E9.5, E10.5 and E11.5 (Supplementary Fig. 2a, b). We first characterized differential gene expression between the mutant and control at each stage and found a progressive increase in the number of differentially expressed genes (DEGs) from 28 at E9.5, to 2972 at E10.5, and then to 3617 at E11.5 (Fig. 2a), consistent with the progressive severity of the mutant limb phenotype observed at the later stages. Early gene expression changes at E9.5 involved only genes upregulated in the mutant mice, while later stages showed similar numbers of both up and down-regulated genes (at E10.5 1570 and 1402, and at E11.5 1798 and 1819 up- and down-regulated genes, respectively) (Fig. 2a).
Fig. 2. Bulk RNA-seq profiling of Esco2fl/fl and Esco2fl/fl;Prrx1-CreTg/0 developing limbs from E9.5-E11.5.
a Venn diagram (top) and gene ontology (GO) enrichment (bottom) for upregulated (left) and downregulated (right) DEGs at E9.5 (blue), E10.5 (purple), and E11.5 (green). Gene Ontology enrichment analysis was performed with a p-value ranked query and multiple testing correction by the g:SCS method (p ≤ 0.05). Dashed line indicates the threshold of significance (0.05). BP, Biological Process; CC, Cellular Compartment; MF, Molecular Function. b Heatmap of the average expression changes (log2 fold change) of the 25 overlapping upregulated genes across the three ages. Gene names are to the right of the matrix and GO terms below.
At E9.5, the 28 DEGs were enriched in gene ontology (GO) processes for DNA damage response, signal transduction by p53 class mediator, and leukotriene signaling (Fig. 2a, Supplementary Data 1). Interestingly, 25 of the 28 DEGs were known members of the p53 pathway (Fig. 2b) and all 28 DEGs were implicated in additional GO cellular and biological processes at later stages (Fig. 2a, b, Supplementary Data 1). Using immunohistochemistry (IHC), we validated the most significant DEG across each age, CDKN1A, which demonstrated increased expression in mutants compared to controls (Supplementary Fig. 2c, d). The remaining 3 DEGs were members of the leukotriene signaling pathway, an inflammatory process activated in the presence of tissue damage (Fig. 2a, Supplementary Data 1). Genes involved in key patterning pathways (e.g. Shh, Fgf8, Bmp4) were not amongst the most significant DEGs in both the bulk- and scRNA-seq analyzes (Supplementary Data 1). Together, these changes highlight the complexity of limb development and implicate a role for early changes in p53 and leukotriene signaling prior to the severe morphological and vascular phenotypes observed at later stages.
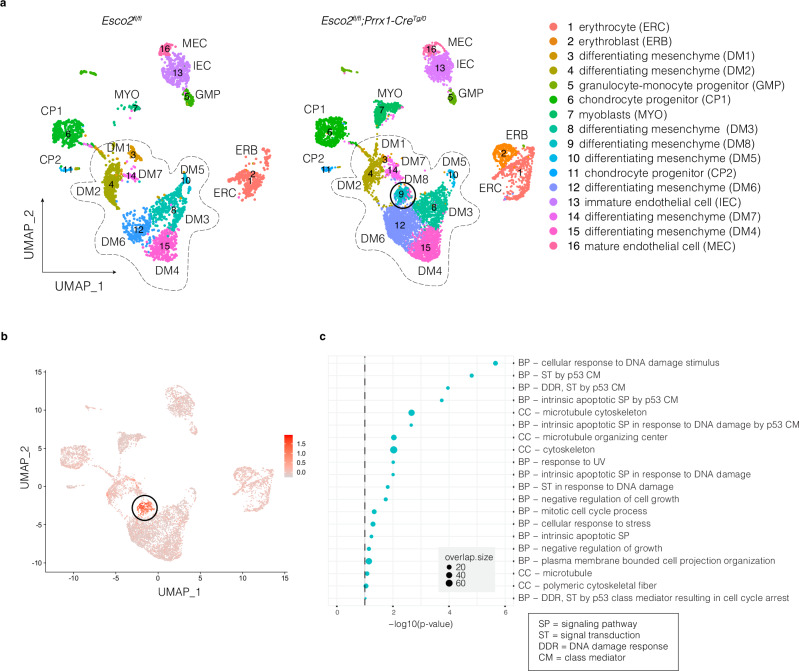
While this transcriptomic data aligned with the various RBS phenotypes at given stages, it did not necessarily allow us to dissect the specific effects of Esco2 loss on the cell populations in the developing limb. To understand how loss of Esco2 modulated cell-specific gene expression and to further resolve the transcriptional signature at E9.5, we performed scRNA-seq of E9.5 limb buds (Fig. 3). A total of 12,245 cells were analyzed from two mutant replicates and a control, each contained six pooled limb buds (Fig. 3a). UMAP analysis identified 16 distinct clusters (Fig. 3a, Supplementary Fig. 3a). Most cells were part of a larger supercluster that spanned a continuum of differentiating mesenchymal (DM) cell populations that were identified by the expression of Prrx1 (Fig. 3a, Supplementary Data 2). We also identified more differentiated populations of chondrocyte progenitors expressing Sox5, Sox6, and Sox9 (CP1), and muscle progenitors expressing Pax3 and Lbx1 (MYO) (Fig. 3a, Supplementary Data 2). Mesenchymal cells were more abundant in the mutant group, but the proportions of endothelial cells were similar in each sample, across both genotypes (Supplementary Data 2).
Fig. 3. Single-cell RNA-seq characterization of Esco2fl/fl and Esco2fl/fl;Prrx1-CreTg/0 clusters at E9.5.
a Uniform Manifold Approximation and Projection (UMAP) plot of cell clusters detected by unsupervised graph clustering of cell populations from Esco2fl/fl (left) and Esco2fl/fl;Prrx1-CreTg/0 (right) limb buds. b UMAP of the 25 marker genes expressed in DM8 that intersect with the DEGs identified in the bulk RNA-seq data at E9.5. c Gene ontology (GO) enrichment for upregulated and downregulated markers in DM8. Gene Ontology enrichment analysis was performed with a p-value ranked query and multiple testing correction by the g:SCS method (p ≤ 0.05). Dashed line indicates the threshold of significance (0.05).
Importantly, we observed a single cluster within the larger DM population, DM8, which was only present in mutant tissues and absent in control tissues (Fig. 3a). The specific marker genes that uniquely identified this cluster were highly enriched (p = 0.000) for the DEG signature identified in the bulk RNA-seq data at E9.5, accounting for 25 of the 28 DEGs (Fig. 3b, Supplementary Data 3). The top GO terms for the DM8 cluster included p53 and DNA damage response pathways (Fig. 3c), cellular responses to DNA damage stimulus and apoptotic signaling, as well as cellular component (CC) associations with microtubule organization and the cytoskeleton (Fig. 3c, Supplementary Data 2). Hierarchical clustering of the cell populations indicated that DM8 was most similar to DM3, myoblasts (MYO), and chondrocyte progenitors (CP1) (Supplementary Fig. 3a).
We next assessed whether the specific enrichment of the E9.5 DEG signature in DM8 could have been due to preferential activation of Prrx1-CreTg/0 and Cre recombinase expression in this population. We found that the average expression of Prrx1 was relatively higher in DM8 compared to most other DM populations, except DM4 (Supplementary Fig. 3c). We also detected that the Cre recombinase expression was higher on average in DM8 than other DM populations, except DM6, but was expressed in less than 5% of DM8 cells (Supplementary Fig. 3c, d). Given ESCO2’s role in establishing functional cohesion during the S phase, we further considered whether there were changes in the cell cycle distribution of the DM8 population, but found no evidence of increased cell counts in S-phase arrest in DM8 compared to other mesenchymal populations (Supplementary Fig. 3b). We also found no evidence of major copy number variants or chromosomal aberrations (Supplementary Fig. 3e), further supporting a specific activation of p53 signaling, apoptotic signaling, and DNA damage response pathways as the major driver of the distinct DM8 population in the Esco2fl/fl;Prrx1-CreTg/0 mutant.
We probed the remaining marker genes for various cell death markers not specified in the GO analysis (Supplementary Data 2), which included extrinsic apoptotic signaling genes Fadd and Fas. Thus, there were other marker genes beyond the bulk DEG signature that defined the unique DM8 population in mutant limb buds. Finally, we conducted trajectory inference and velocity analyzes to organize DM cell populations in a pseudo-temporal manner. When we included all DEGs in the analysis, there was no trajectory linking DM8 with other DM populations (Supplementary Fig. 4a). After masking the highly distinct signature of 25 DEGs identified from the bulk RNA-seq data at E9.5, the same analyzes suggested a potential trajectory linking DM8 to DM6 that then projects to DM4 (Supplementary Fig. 4b).
A vasculature related gene co-expression module is nested within an apoptosis-related module
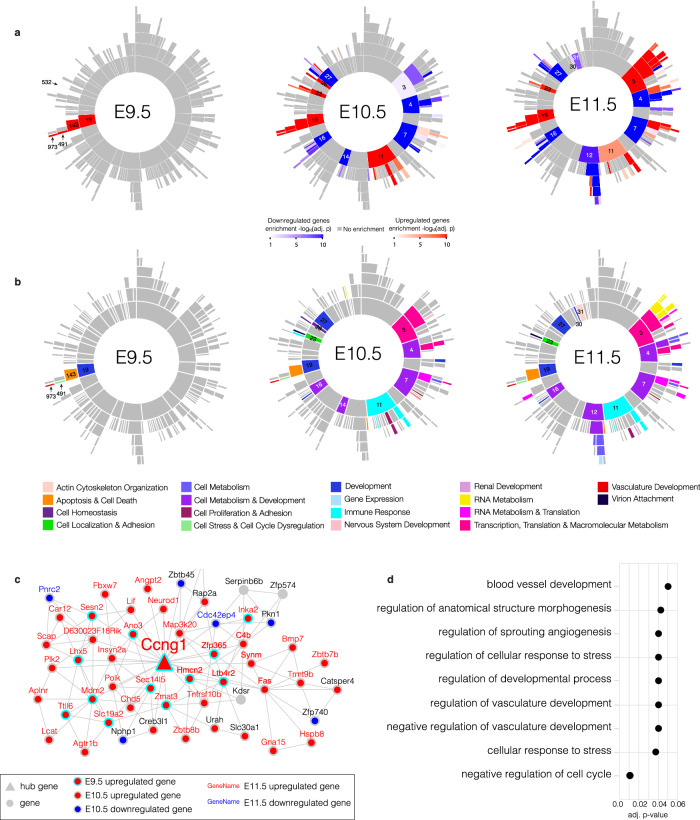
To further dissect the transcriptomic signatures during early development, we performed MEGENA of the bulk RNA-seq data across all stages. Our analysis produced a hierarchical network with a total of 187 interconnected modules (Fig. 4a, Supplementary Fig. 5a–c). We assessed each module for DEG enrichment, yielding 5 modules enriched for DEGs at E9.5, 75 modules at E10.5, and 90 modules at E11.5 (Fig. 4a, Supplementary Data 3). Each module was also annotated for its most significant GO processes to map the dominant biological pathway(s) or process(es) represented by each module (Fig. 4b, Supplementary Data 3).
Fig. 4. Multi-scale embedded gene co-expression network analysis from E9.5-E11.5.
a Sunburst plots of the MEGENA based on bulk RNA-seq data from Esco2fl/fl and Esco2fl/fl;Prrx1-CreTg/0 limb buds from E9.5-E11.5. The inner most ring represents the root and largest parent modules, which branch off in hierarchical fashion to smaller child modules. Color intensity of red (upregulated) or blue (downregulated) is proportional to −log10(corrected FET p-value). Significant correlations between gene pairs were identified using the Pearson method with FDR < 0.05. b Sunburst plots with significant modules colored according to the most significantly enriched GO BP from E9.5-E11.5. c Module 973 (M973) from E9.5-E11.5. Hub genes and genes are marked by triangles and circles, respectively. Upregulated genes at E9.5 are marked by a red circle with a cyan border. Up- and down-regulated genes at E10.5 are marked by a red or blue circle with a gray border, respectively. Up- or down-regulated genes at E11.5 are marked by red or blue gene names, respectively. Gray circles are genes that are not enriched for DEGs. Hub gene name for Ccng1 is enlarged. d GO enrichment for BPs of M973 (adj. p-values ≤ 0.05). Gene set enrichment was performed against using Fisher’s exact test and Bonferroni correction to account for multiple hypothesis testing.
Of the 28 DEGs identified at E9.5 in the bulk RNA-seq data and enriched in the DM8 cell population, 22 were contained within a single hierarchy of four interconnected modules (M19, M143, M491, M973) (Fig. 4a). The root module M19 (containing 548 genes) and M143 (containing 385 genes) were both enriched for apoptosis, cell death, and developmental related processes (Fig. 4b, Supplementary Fig. 5d, Supplementary Data 3), and the two sub-modules M491 (71 genes), and M973 (53 genes), each contained 13 E9.5 DEGs (Supplementary Data 3). M491 was enriched for processes related to cell stress, the cell cycle, and morphogenesis, while M973 showed further enrichment for cell cycle stress and vasculature-related processes (Fig. 4c, d, Supplementary Data 3). M973 contained one hub gene, Ccng1, a p53-induced cyclin kinase induced by DNA damage to promote cell proliferation (Fig. 4c). Interestingly, DM8 was the only cluster with significant upregulation of Ccng1, while it was downregulated in all other clusters (Supplementary Data 2). We validated CCNG1 using IHC at E9.5, which showed increased non-localized expression in mutant limb buds compared to controls (Supplementary Fig. 2d). The direct relationship between M973 and vasculature processes suggests that these co-expression signatures may underlie the angiogenic defects that resulted in hemorrhaging of the mutant limb buds observed at E12.5.
At later E10.5 and E11.5 stages we found three modules partitioned in one branch (M11, M89, and M342) and two independent modules (M97 and M161) enriched for immunological processes related to macrophage and TNF-α recruitment (Fig. 4b, Supplementary Data 3). Upregulation of DEGs in these immunological-related modules coincided with the inflammation observed in the proximal region of the limbs at E11.5 (Fig. 1c). Two additional modules (M27 and M173) related to mesenchymal proliferation, chondrogenesis, and skeletal development were enriched for downregulated genes (i.e. Sox9, Sox5) at E10.5 and E11.5, which was consistent with an absence of chondrogenesis at E10.5 and E11.5, and overall skeletal reduction of the RBS mutant limb postnatally (Supplementary Fig. 5e, Supplementary Fig. 6, Supplementary Data 3). In sum, this analysis captured the chronology of dysregulated processes underlying RBS limb reduction and revealed a relationship between the p53-signaling and angiogenesis at E9.5.
In vivo validation of DNA damage, apoptosis, and abnormal vasculature
We reasoned that the limited expression changes observed at E9.5 were more likely to be directly related to partial loss of ESCO2 function whereas the more extensive changes at later stages could be due to secondary effects from compromised developmental and cellular processes. Therefore, we assessed the impact of early cellular changes on the expression of genes involved in apoptosis, inflammation, and vasculature development.
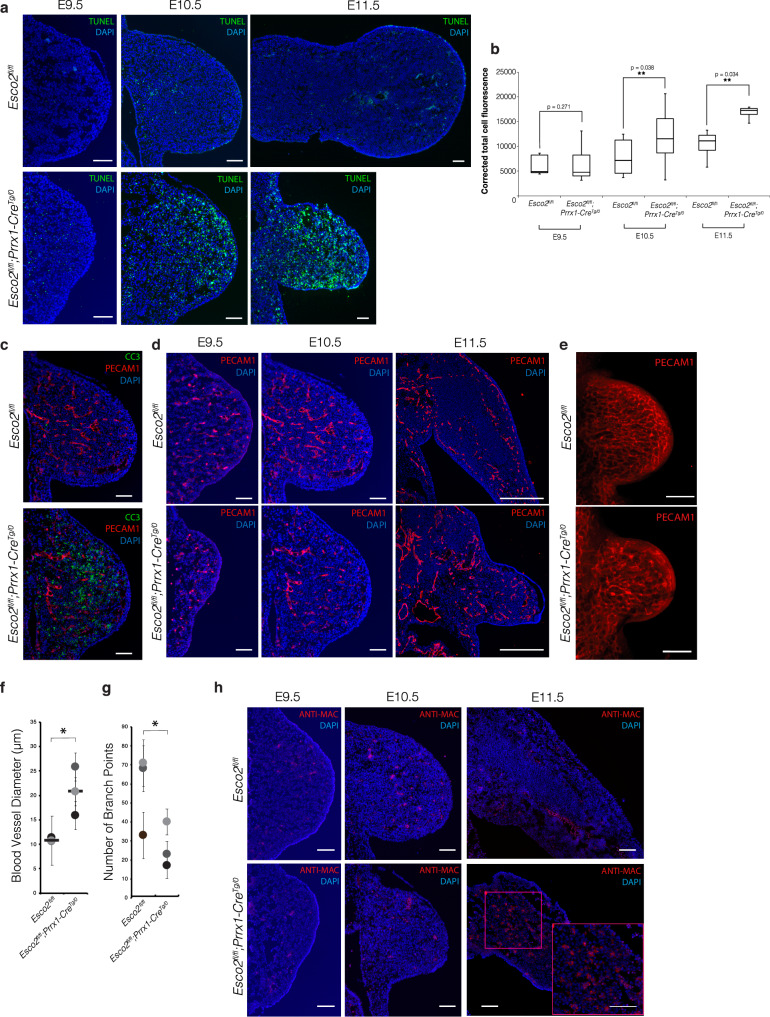
First, we determined the extent of cell death that occurred in the limb bud with TUNEL assay. At E9.5, there was no significant difference in TUNEL+ signal between mutant and control limb buds (p = 0.271) (Fig. 5a). However, a significant increase in DNA fragmentation and apoptosis was apparent in mutant limb buds at E10.5 and increased progressively by E11.5 (Fig. 5a). Quantitative analysis revealed progressive damage between control and mutant limbs at E10.5 and E11.5 (p = 0.038 and p = 0.034, respectively), with a 121% increase in TUNEL+ signal from E9.5 to E10.5 and a 22% increase in TUNEL+ signal from E10.5 to E11.5 in mutant limbs (Fig. 5b). We also performed IHC for cleaved-caspase 3 (CC3), a marker for apoptotic cell death induction, co-stained with platelet endothelial cell marker (PECAM1) (Fig. 5c) to determine whether this cell death occurred in the vasculature (Fig. 5c). Compared to controls, we observed increased CC3 expression in the surrounding mesenchyme of the vasculature, but not within the vasculature itself (Fig. 5c). To further explore the vasculature patterning of the developing limb, we stained for PECAM1 from E9.5-E11.5. Per normal limb development, the vasculature of control limb buds remained unpatterned at E9.5 and E10.5 with evidence of vascular regression at E11.5 (Fig. 5d). Mutant limb buds at both E9.5 and E10.5 displayed an obvious reduction in the size of the bud with sparser distribution of blood vessels; however, by E11.5, there was no evidence of vascular regression or patterning (Fig. 5d).
Fig. 5. In vivo validation of differential gene expression from bulk and single-cell RNA-seq analysis.
a TUNEL assay of Esco2fl/fl limb buds (n = 3) and Esco2fl/fl;Prrx1-CreTg/0 limb buds (n = 3) from E9.5-E11.5 in the coronal plane. Scale bar: 100 µm. b Corrected total cell fluorescence (CTCF) of TUNEL+ signal between Esco2fl/fl and Esco2fl/fl;Prrx1-CreTg/0 limbs at E9.5, E10.5 and E11.5. n = 4 for Esco2fl/fl and Esco2fl/fl;Prrx1-CreTg/0 limb buds at E9.5, n = 5 for each genotype at E10.5, and n = 2 for each genotype at E11.5. In each box plot, the center line indicates the median, the edges of the box represent the first and third quartiles, and the whiskers extend to span a 1.5 interquartile range from the edges. Asterisk indicates a significant difference using a one-tailed, Student’s t-test (p ≤ 0.05; p = 0.271, p = 0.038, p = 0.034 for E9.5, E10.5, and E11.5 respectively). c Immunofluorescent staining of Esco2fl/fl (n = 3) and Esco2fl/fl;Prrx1-CreTg/0 (n = 3) limb buds at E10.5 (coronal plane) with cleaved-caspase 3 (CC3) and PECAM1. All samples were co-stained with DAPI. Scale: 100 µm. d Immunofluorescent staining of Esco2fl/fl (n = 3) and Esco2fl/fl;Prrx1-CreTg/0 (n = 3) limb buds at E9.5 (coronal plane), E10.5 (coronal plane) and E11.5 (transverse plane) with PECAM1. All samples were co-stained with DAPI. Scale: 200 µm. e Whole mount immunofluorescent staining of Esco2fl/fl and Esco2fl/fl;Prrx1-CreTg/0 limb buds at E10.5. Scale: 20 mm. f Average blood vessel diameter (µm) of Esco2fl/fl (n = 3) and Esco2fl/fl;Prrx1-CreTg/0 (n = 3) limb buds at E10.5. Littermate pairs are color matched by gray-scale. Data shown are the average measurements from each sample with mean ± SD. Black bar indicates median. Asterisk indicates a significant difference using a one-tailed, Student’s t-test (p ≤ 0.05; p = 0.013). g Number of branch points per sample of Esco2fl/fl (n = 3) and Esco2fl/fl;Prrx1-CreTg/0 (n = 3) limb buds at E10.5. Littermate pairs are color matched by gray-scale. Data shown are the total measurements from each sample with mean ± SD. Asterisk indicates a significant difference using a one-tailed, Student’s t-test (p ≤ 0.05; p = 0.047). h Immunofluorescent staining of Esco2fl/fl (n = 3) limb buds and Esco2fl/fl;Prrx1-CreTg/0 (n = 3) limb buds from E9.5–E11.5 with Anti-macrophage in the coronal plane. Scale: 100 µm. All samples were co-stained with DAPI. Magenta box delineates region with high density of macrophages, magnified in bottom right corner. Source data for 5b, 5f, and 5 g are provided with this paper.
Our findings in the mutant were consistent with previous reports that indicated that deficient vascular regression prevents the mesenchymal condensation, a prerequisite for chondrogenesis and skeletal development53,54. We questioned whether this lack of regression contributed to defective chondrogenesis and long bone hypoplasia or aplasia of RBS mutant limbs (Fig. 1a). To test this hypothesis, we stained for SOX9, the master regulator of chondrogenesis. We observed reduction of SOX9 expression in the mid-region of the limb bud at E10.5 and E11.5 (Supplementary Fig. 6), further aligning with the vascular defects observed. However, there was some expression of SOX9 in the distal tip of the mutant limb bud, suggesting this might have given rise to some long bone growth and digit formation (Supplementary Fig. 6), which further supports the P0 limb phenotype observed (Fig. 1a).
To gain a deeper understanding of how these vascular anomalies affected the architecture of vasculature networks, we performed whole-mount IHC on E10.5 limb buds for PECAM1 and imaged them with confocal microscopy to obtain 3D structural details (Fig. 5e). In control limb buds, we observed a highly branched, patterned, and robust vascular network compared to the disorganized and dilated vasculature of mutant limb buds (Fig. 5e). To quantify these differences, we measured the average blood vessel diameter of littermate mutants (n = 3) and controls (n = 3), which indicated a significant increase in vessel diameter of mutants (p = 0.013) (Fig. 5f). We next counted the number of branch points and observed a significant reduction in the mutants (p = 0.047) compared to controls (Fig. 5g). Taken together, these results suggested that DNA damage and apoptosis beginning as early as E9.5 precede abnormal vascular patterning in Esco2fl/fl;Prrx1-CreTg/0 limb buds.
To determine whether these abnormal processes coincided with the inflammatory response detected in the transcriptomic signature at E9.5, we performed IHC for Anti-Macrophage (Fig. 5h). Although the RNA-seq data indicated differential expression of leukotriene specific signaling genes at E9.5 and E10.5, we did not detect expression of Anti-Macrophage until E11.5 (Fig. 5h). At E11.5, there was protein expression localized to the mesenchymal core (Fig. 5h), which suggested that the inflammatory response was a potential consequence of earlier DNA damage, fragmentation, apoptosis and vascular defects.
p53 inhibition suppresses hemorrhage in mutant limbs
The upregulation of p53 signaling genes at E9.5 led us to reason that p53 inhibition might ameliorate the RBS limb phenotype by reducing the downstream effects of p53 signaling, particularly on cell death in tissues necessary for limb outgrowth. Pifithrin-α is a p53-inhibitor frequently used to investigate p53-dependent cellular responses by reducing p53-dependent apoptosis and protecting cells from genotoxic stress, such as double-stranded breaks (DSBs) caused by irradiation55. It was previously applied in a rescue experiment for Treacher Collins syndrome, which shares a similar transcriptomic profile to that of our RBS mouse model at E9.556. We reasoned that the p53 signature in combination with the hypersensitivity of RBS cells to DNA damaging agents supported use of this drug for potential rescue.
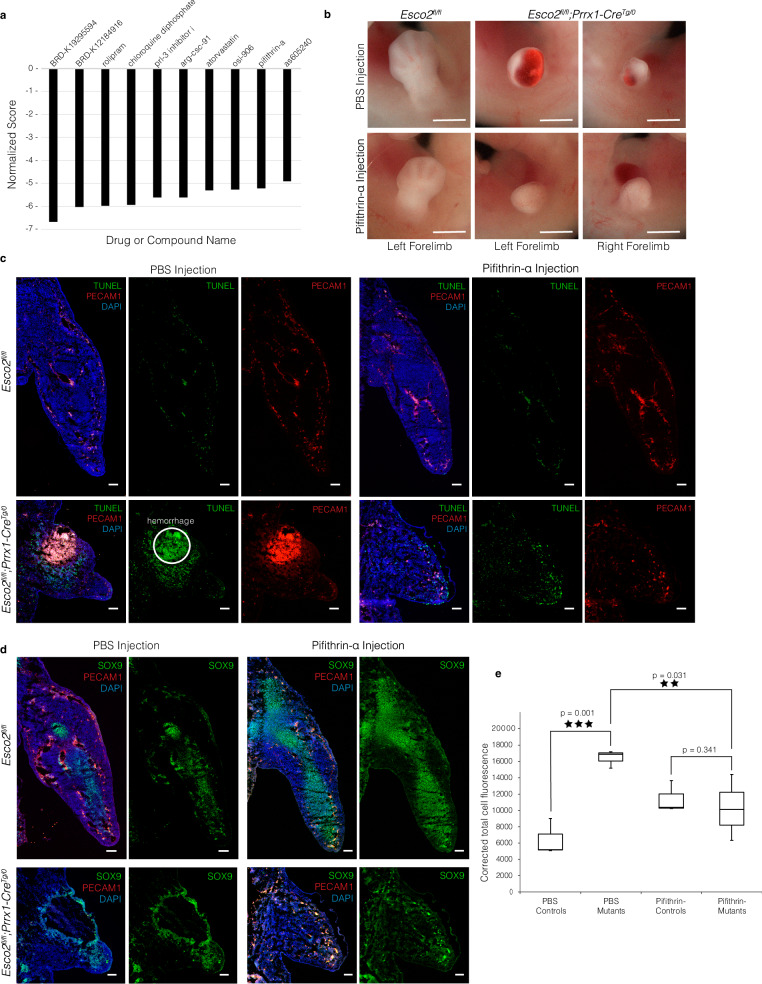
We confirmed the suitability of pifithrin-α using the Ensemble of Multiple Drug Repositioning Approaches (EMUDRA) to match transcriptomic DEG signatures with drug-induced signatures to identify candidate therapeutic agents57. We performed the analysis with input data from the DEG profile at E10.5 as there were too few DEGs at E9.5 to be accurately matched with drug-induced signatures (Supplementary Data 4). EMUDRA analysis produced 354 drug candidates (Fig. 6a and Supplementary Data 4). Pifithrin-α was ranked ninth among the top ten candidate compounds by normalized score (Fig. 6a and Supplementary Data 4), which provided further support that the overall RBS transcriptional signature was associated with the p53 pathway. The other nine compounds were either under development, lacked mechanistic information, or were unavailable for purchase.
Fig. 6. Rescue of hemorrhage in Esco2fl/fl;Prrx1-CreTg/0 limbs by pifithrin-α treatment.
a Bar chart of normalized scores for top 10 drug candidates identified by EMUDRA. Drugs are ranked by score with more negative values indicating stronger association. b Gross morphology of Esco2fl/fl;Prrx1-CreTg/0 and Esco2fl/fl embryonic limb buds at E12.5 treated with either PBS (top) or pifithrin-α (bottom). Scale bar: 1 mm. c TUNEL assay co-stained with PECAM1 of Esco2fl/fl (n = 3) and Esco2fl/fl;Prrx1-CreTg/0 (n = 3) limbs treated with PBS (left) or pifithrin-α (right) at E12.5 in the transverse plane. Scale bar: 100 µm. d Immunofluorescent staining of Esco2fl/fl (n = 3) and Esco2fl/fl;Prrx1-CreTg/0 (n = 3) limbs treated with PBS (left) or pifithrin-α (right) for SOX9 and PECAM1 at E12.5 in the transverse plane. Scale: 100 µm. e Box and whisker plot showing the corrected total cell fluorescence (CTCF) of TUNEL+ signal between Esco2fl/fl (n = 3) and Esco2fl/fl;Prrx1-CreTg/0 (n = 3) limbs treated with PBS or pifithrin-α. In each box plot, the center line indicates the median, the edges of the box represent the first and third quartiles, and the whiskers extend to span a 1.5 interquartile range from the edges. Asterisk indicates a significant difference using a one-tailed, Student’s t-test (p ≤ 0.05; p = 0.001 for PBS comparisons, p = 0.341 for pifithrin-α comparisons, and p = 0.031 for Esco2fl/fl;Prrx1-CreTg/0 comparisons between PBS and pifithrin-α groups). Source data are provided with this paper.
We injected pifithrin-α into pregnant females once daily from E8.5 to E11.5 and harvested the embryos at E12.5 to determine whether inhibiting p53 would reduce apoptosis, improve the morphology of the limb bud, eliminate the hemorrhage, and restore chondrogenesis. Control matings were also generated in which pregnant females were either untreated or injected with phosphate buffered saline (PBS) vehicle. This regimen was established to coincide with the initiation of limb development at E9.5 and aligned with the earliest presentation of p53 signaling in our data. Upon recovery of the embryos at E12.5, we observed a successful rescue of the hemorrhage in pifithrin-α treated mutant embryos compared to PBS-treated mutants (Fig. 6b). While the size and morphology of the limb bud at E12.5 were not fully recovered to normal, the elimination of hemorrhage supported our hypothesis that the vascular defects were related to p53-induced apoptosis.
To determine whether pifithrin-α treatment reduced the extent of apoptosis, we evaluated the tissues using TUNEL assay co-stained with PECAM1. Pifithrin-α-treated mutant embryos displayed a significant reduction in the number of TUNEL+ cells in the mesenchymal region of the limb (p = 0.050) (Fig. 6c, e) compared to PBS-treated mutants with significant TUNEL+ signals surrounding the region of hemorrhage (Fig. 6c). We also observed increased expression of PECAM1 in endothelial cells of pifithrin-α treated mutants compared to PBS-treated mutants, indicating improvement in angiogenesis (Fig. 6c). Lastly, to determine whether p53 inhibition could restore chondrogenesis signaling and subsequent limb outgrowth, we co-stained for SOX9 and PECAM1. However, we observed a decrease in the number of SOX9 positive cells in pifithrin-α-treated mutant embryos (n = 3) compared to PBS-treated mutants (n = 3), particularly in the distal region of the limb (Fig. 6d), showing a lack of improvement with p53 inhibition. This observation suggested that the chondrogenesis defects are not a direct or specific result of increased cell death in chondrogenic or surrounding tissues and are potentially influenced by ESCO2 via another mechanism.
In sum, our results identified p53 inhibition as a direct mediator of vascular hemorrhage, but not chondrogenesis in mutant limbs. The morphology of the limb was not fully recovered, which suggests a more primary role of p53 in supporting the vascular phenotype.
Mesenchymal clusters are enriched for downregulated genes in human genetic limb reduction disorders and thalidomide embryopathy
Human limb and digit reduction have various causes, including genetic disorders and teratogenic drug exposure to developing fetuses in utero. We identified 103 downregulated genes spanning 50 genetic limb reduction disorders and 54 up- and 169 down-regulated genes associated with thalidomide embryopathy by performing a literature search and OMIM search for key terms (Supplementary Data 5). We then determined the degree of enrichment between these gene sets and our cluster markers identified by scRNA-seq at E9.5 (Supplementary Data 5 and Fig. 7).
Fig. 7. Cluster marker enrichment for human genetic limb reduction disorders and thalidomide embryopathy genes.
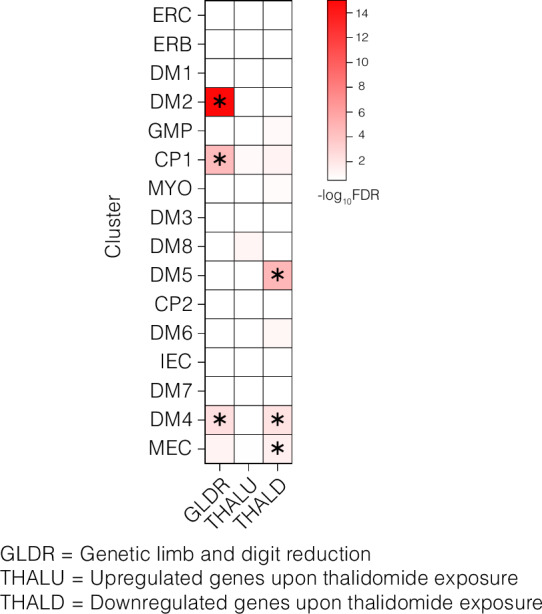
Heatmap of -log10FDR. Each row represents a single cell cluster, and each column represents a gene list associated with downregulated genes that cause genetic limb reduction disorders (GLRD), and upregulated (THALU) or downregulated (THALD) genes associated with thalidomide exposure. Significantly enriched clusters are asterisked (p ≤ 0.05). P-values for enrichment were determined with a one-sided Fisher’s exact test, with Bonferroni correction for multiple testing.
Mesenchymal-related clusters DM2, DM4, and CP1 were significantly enriched for genes associated with unique genetic limb reduction disorders (Fig. 7 and Supplementary Data 5). DM2 was enriched for several ribosomal genes associated with Diamond Blackfan anemia (p = 5.49E-16); DM4 was enriched for genes associated with omodysplasia 1 (GPC6), acromesomelic dysplasia 3 (BMPR1), Langer mesomelic dysplasia (ROR2), Holt-Oram syndrome (TBX5) and Fanconi anemia (FANCD2) (p = 0.004); lastly, CP1 was enriched for genes associated with split-hand/foot malformation 4 (TRP63), achondrogenesis type 2 (COL2A1), split-hand/foot malformation with long bone deficiency (BHLHA9), tetraamelia syndrome 1 (WNT3), Robinow syndrome (WNT5A), hypophosphatasia, infantile (ALPL), ulna and fibula, absence of, with severe limb deficiency (WNT7A), split-hand/foot malformation 1; and split-hand/foot malformation 1 with or without deafness (DLX5) (p = 2.97E-05) (Supplementary Data 5).
Significant enrichments also were identified among human thalidomide-induced downregulated genes with clusters DM5 (p = 1.60E-05), DM4 (p = 0.007) and MEC (p = 0.031) (Fig. 7). The genes associated with these enrichments were related to blood vessel morphogenesis, vascular repair, chondrocyte differentiation, macrophage recruitment, and inflammation (Supplementary Data 5). We observed no significant enrichments in any cluster for thalidomide-induced upregulated genes (Fig. 7 and Supplementary Data 5).
To further understand potential similarities between RBS and thalidomide, we analyzed the scRNA-seq data to determine if genes comprising subunits of the CRL4CRBN complex, to which thalidomide binds and exerts its teratogenic effect, were differentially expressed in our RBS mouse model. In humans, this complex is composed of four subunits: cullin4 protein (CUL4A/B), DNA damage binding protein 1 (DDB1), ROC1 protein with a RING domain, ring box 1 (RBX1), and the substrate receptor cereblon (CRBN)21,22. We observed that Cul4b, Rbx1, Ddb1, and Crbn1 were significantly downregulated in many clusters: Cul4a and Cul4b in 8/16 and 14/16 clusters, respectively; Ddb1 in 15/16 clusters, including all DM clusters; Rbx1 in 11/16 clusters; and Crbn1 in 13/16 clusters (Supplementary Data 2). In DM8 specifically, Cul4b, Rbx1, Ddb1, and Crbn1 were significantly downregulated. DM1 was the only cluster in which all genes were downregulated (Supplementary Data 2).
In sum, the significant enrichments identified among cluster markers at E9.5 with other human genetic limb reduction disorders and thalidomide embryopathy indicated potential mechanistic commonality between these various phenocopies, and further suggested that some of these genes or proteins might be potential targets of ESCO2.
Discussion
Major contributions of our work are the identification of a pre-apoptotic and p53-dependent mesenchymal cell population, only present in the RBS limb buds, and the further elucidation of the chronology of cellular perturbations underlying RBS limb reduction. We were able to capture some of the earliest gene expression and signaling perturbations to demonstrate a primary p53-induced apoptotic etiology and secondary vascular and inflammatory etiology of limb reduction in RBS. The application of various transcriptomic approaches to the limb buds of our Esco2fl/fl mouse model with conditional expression of Cre recombinase in only a subset of Prxx1-expressing mesenchymal cells at E9.5 allowed us to analyze transcriptomic and cellular events prior to extensive cell death at E10.5 and E11.5.
The observed upregulation of p53-related genes in DM8 at E9.5 could be explained by DNA damage induced by reactive oxygen species (ROS) production or increased cleavage rates at a mutated site during replication58–61 as evidenced by increased expression of several genes involved in the NFkβ, MAPK and peroxiredoxin pathways, as well as increased expression of genes associated with Hsp40, cytochrome P450, and chaperonin containing TCP1 (Supplementary Datasets 1 and 2). It has also been shown that eco1 mutations in yeast and human RBS cell lines display increased ROS production with elevated apoptosis rates61–65. DNA damage-induced ROS also may induce cell death via oxidation of cohesin subunits through oxidation of effector proteins such as p53, the c-Jun N-terminal kinase signaling kinase, tumor necrosis factor (TNF)-α cytokine, mitochondrial cytochrome c-release regulators, or DNA-damage induced apoptosis that directly triggers caspase activation66–73. At E9.5, we also show evidence of extrinsic in addition to the intrinsic apoptosis, and DNA-damage induced cellular senescence as indicated by the upregulation of genes such as Fadd, Fas, Cdkn1a, and Ccng1, and the presence of micronuclei. The extrinsic apoptosis indicates that additional external factors in the tissue environment are contributing to cell death independent of intrinsic cell signaling. These external signals may be derived from necrotic tissue, or perturbations secondary to ESCO2. For example, apoptotic cells can secrete chemotactic and immunogenic signals that result in the infiltration of macrophages to remove cellular debris74,75. The Anti-Macrophage protein expression observed at E11.5 supports this process.
We also observed no evidence of the orchestrated vascular regression and mesenchymal condensation in our RBS mouse53. We had presumed that the extensive cell death in the mesenchyme was the underlying cause of the limb deficiency, but pifithrin-α treatment only recovered the hemorrhage in mutant limbs and not the chondrogenesis. The implication of this result is that the role of p53 is only supported in the mutant vasculature phenotype. Although it is possible that more time was needed to allow repopulation of the chondrocytes, the vasculature was still not entirely recovered to normal and therefore it is unlikely that chondrogenesis would be fully recovered as well. Explanation for the lack of recovery might be p53-independent cell death of chondrocyte progenitors or vascular-dependent chondrogenesis. Esco2 knockdown in zebrafish also show transcriptional perturbations of developmental genes and fin regeneration defects independent of apoptosis23. This role for ESCO2 is further supported in various studies suggesting it either acts as a transcription factor itself or mediates cohesin’s distant interactions with other transcriptional regulators23,29,42,76–81.
Another major contribution of our study is the identification of a p53-driven vascular etiology in the pathogenesis of a cohesinopathy and potentially other limb reduction defects. More specifically, several studies support an emerging relationship between RBS and thalidomide embryopathy. Human pluripotent stem cell-derived neural constructs exposed to thalidomide display significant downregulation of ESCO2, suggesting that ESCO2 may be a target of thalidomide or that thalidomide exposure affects a pathway to which ESCO2 belongs18. It remains unknown whether ESCO2 acts via the CRL4 complex in the affected limb bud tissues as does thalidomide, but the CRL4 complex does target and promote ESCO2 degradation during mitosis to prevent excess cohesion formation25. Esco2 and cohesin have been shown to co-regulate ddb1 transcription in yeast, thus identifying a common target between thalidomide and RBS15. Our results further support a unifying mechanism, such that significant enrichments of genes associated with ROS, and vascular and chondrogenic differentiation were identified.
Vascular etiologies have been reported in teratogenically induced limb reduction, such as thalidomide embryopathy, but not in most genetic limb reduction disorders82,83. Our data show how loss of ESCO2 causes limb reduction phenotypes, through p53 induction, cell death, and vascular disruption. We also demonstrate that loss of ESCO2 causes the downregulation of several genes linked to other limb reduction syndromes, including genes targeted by thalidomide, demonstrating commonality between these limb reduction conditions. In our Esco2 fl/fl;Prrx1-Cre mouse model, we observed significant enrichments for genes previously established to regulate thalidomide’s angiogenic response (e.g. Tgfbr3, Hand1, and Efnb2)84, suggesting RBS and other cohesinopathies potentially share similar downstream targets with thalidomide. Thalidomide also targets blood vessels, causes cell death, localized gene expression changes, and tissue damage. This limb damage results either through loss of chondrogenic precursors or the resulting vascular changes that induce the mispositioning of vessels such that the bony elements cannot form normally82,83. There are also other studies suggesting loss or mispositioning of vessels during development results in limb reduction phenotypes via a variety of injury situations, as reviewed in Vargesson & Hootnick83. The combination of these studies and our analyzes provide evidence for a shared pathogenic etiology between RBS, thalidomide embryopathy, and potentially other limb reduction conditions.
Methods
Ethics statement
This research complies with all relevant ethical regulations. Studies using murine samples were approved by the Institutional Animal Care and Use Committee (IACUC) at the Icahn School of Medicine at Mount Sinai (IACUC #07-0757) and Mayo Clinic (IACUC # A00007293-23). No human subjects were used in this study.
Mice
All mice were bred and maintained in animal facilities certified by Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) under IACUC-approved protocols. Mice from both genders were maintained on the C57BL/6J genetic background and were housed under a 12-h light/12-h dark cycle, at temperatures of 20–24 °C with 45–65% humidity. The animals for experiments were euthanized by cervical dislocation with anesthesia. The Body Condition Score (BCS) of all mice was monitored and those with BCS < 2 (underconditioned) were euthanized. The Esco2 conditional knockout mouse line was generated by gene targeting. The Esco2 targeting vector, which contained two homology arms, two loxP sites flanking exon 4, an FRT-flanked neomycin cassette, and a diphtheria toxin selection marker, was electroporated into 129/SvPas ES cells. Neomycin-resistant ES cell clones were screened by Southern blotting and polymerase chain reaction (PCR). Verified ES cell clones were injected into C57BL/6 J blastocysts (Mus musculus). Resulting chimeric animals were mated with mice ubiquitously expressing FLP recombinase to generate floxed mice (Esco2fl/+), which were bred to homozygosity. Female Esco2fl/fl mice were bred with male mice heterozygous for Prrx1-Cre (from Jackson Laboratory) to obtain Esco2fl/+;Prrx1-CreTg/0 mice. Male Esco2fl/+;Prrx1-CreTg/0 mice were bred with female Esco2fl/fl mice to generate the desired Esco2fl/fl;Prrx1-CreTg/0 genotype. The resulting embryos were genotyped by PCR of genomic DNA obtained from embryonic yolk sacs using custom primers manufactured by Integrated DNA Technologies. All primers used for genotyping purposes in this study are listed in Supplementary Data 6. The genotypes of the mutants were Esco2fl/fl;Prrx1-CreTg/0 and the control mice were Esco2fl/fl. Sex genotypes were identified by PCR of genomic DNA obtained from embryonic yolk sacs using primers for the X- and Y-chromosomes as described previously85,86 and listed in Supplementary Data 6. Timed matings were made to obtain embryos through ages E9.5 – P0. Dissected embryos were fixed in 4% paraformaldehyde and stored at 4 °C. Only forelimb buds were used for all experimentation (use of the term “limb bud” refers to forelimbs throughout the text). Limb buds were embedded in the desired plane in Optimal Cutting Temperature (OCT) compound (Sakura, item number: 4583) in labeled square plastic molds (Thermo Fisher Scientific, Disposable Base Molds) and stored at −80 °C until cryosectioning.
Immunohistochemistry
Embryos were fixed in 4% paraformaldehyde and cryopreserved in 30% sucrose prior to sectioning in the coronal, sagittal or transverse planes along the dorsal-ventral, anterior-posterior or proximal-distal axes, respectively, at 12 μm thickness. Samples were then washed with phosphate buffered saline (PBS) and circled with a hydrophobic barrier pen to confine the staining reagents. Samples were permeabilized with 0.2% Triton-X100 (Sigma), blocked with 1% bovine serum albumin in PBS for 1 hour at room temperature, and incubated for 1 hour at room temperature in primary antibody (1:100–1:1000). After washing, embryos were incubated with secondary antibody (1:1000) and co-stained with 1 μg/mL DAPI (1:50,000). Samples were covered with fluorescent mounting medium and imaged using a Nikon E600 microscope. Primary antibodies include: Anti-CD31/PECAM-1 Antibody [MEC 7.46] (Abcam: ab7388; 1:100); Anti-Macrophage Antibody (Abcam: ab56297); Anti-SOX9 Antibody (Abcam: ab76997; 1:500); Cleaved-caspase 3 (Cell Signaling Technology, 9661S); Cyclin G1 Polyclonal Antibody (Invitrogen: PA5-92375; 1:200); Recombinant Anti-p21 antibody [EPR18021] (Abcam: ab188224; 1:500). Secondary antibodies include: Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488) (ab150077); Goat Anti-Rabbit IgG H&L (Alexa Fluor® 594) (ab150080); Goat Anti-Rat IgG H&L (Alexa Fluor® 594) (ab150160). Note: Heat mediated antigen retrieval was performed for three minutes using 1X Citrate Buffer, pH 6.0, (Sigma: C9999-100ML, diluted from 10×) for Anti-p21 and Cyclin G1 antibodies.
TUNEL assay
TUNEL assay was performed with the In Situ Cell Death Detection Kit, POD (Roche Applied Science: #11684817910) following the manufacturer’s instructions. After the reaction, samples were stained with DAPI (1:50,000). TUNEL signal was detected by fluorescence microscopy.
Whole mount immunofluorescence and confocal imaging
All steps were carried out in 1.5 mL Eppendorf tubes until imaging. Embryos were fixed in 4% paraformaldehyde in PBS overnight at 4 °C, followed by permeabilization in 1% TritonX-100 (Sigma-Aldrich) in PBS for 30 min three times. Embryos were then blocked for 1 h at room temperature in blocking buffer (1% TritonX-100 in PBS + 10% Fetal Calf Serum) twice and incubated with Anti-CD31/PECAM-1 (1:1000 dilution, [MEC 7.46] (Abcam: ab7388)) at 4 °C for 2 days. After three successive washes in blocking buffer for 1 hour each, embryos were incubated in secondary antibody, BD anti-rat antibody (1:500 dilution) for 1 day at room temperature and counterstained with 1 µg/mL DAPI in PBS for 1 h, followed by two washes with PBS. Samples were then dehydrated with glycerol (20%, 40% 80%, 100%) in PBS and stored and imaged in 100% glycerol. All images were taken using the Leica SP5 DMI at the Microscopy Core of the Icahn School of Medicine at Mount Sinai.
Corrected total cell fluorescence (CTCF) calculation
Post-immunohistochemistry staining, images were processed in FIJI imaging software87. We applied the following formula to calculate the corrected total cell fluorescence (CTCF):
To count the number of cells, a median filter was applied to 32-bit images, followed by a Bernsen-based auto local-thresholding method and watershed segmentation to identify cell boundaries88. To count the number of fluorescent cells, the ‘Analyze particles’ function was used.
RNA isolation, library preparation, and bulk RNA-sequencing
Embryos were harvested at three stages according to the Theiler Staging Criteria for Mouse Embryo Development (E9.5, E10.5, and E11.5). To exclude confounding sex differences, only male embryos were selected for RNA-seq. We exclusively selected right forelimbs, to further eliminate potential differences between right and left forelimbs. Individual limb buds were isolated and purified using Qiagen’s RNeasy Micro Kit (Qiagen: 74004). High quality RNA with RIN scores >9.5 were subjected to library preparation and whole transcriptome RNA-sequencing at the Genetics Resources Core Facility at Johns Hopkins University, School of Medicine. Sequencing library preparation was performed by the Core Facility using the NEBNext® UltraIITM DNA Library Prep Kit for Illumina (NEB #E7103) by following the manufacturer’s instructions. A total of 18 libraries were generated based on triplicate samples of each genotype at each stage (three replicates of Esco2fl/fl;Prrx1-CreTg/0 and Esco2fl/fl at E9.5, E10.5 and E11.5). The input was normalized to 78 ng, the lowest amount of sample, and sequenced on Illumina NovaSeqTM 6000 using standard protocols for 300 cycle paired-end sequencing with a read length of 150 bp. Average yield was ~200–250 Gb of raw sequencing data per lane, or ~1.3–1.6 billion paired-end reads. To limit potential batch effects, the samples were arranged by alternating genotypes for each stage consecutively and sequenced in a single HiSeq run.
RNA-seq analysis
Illumina data were processed through Illumina’s Real-Time Analysis (RTA) software generating base calls and corresponding base call quality scores. CIDRSeqSuite 7.1.0 was used to convert compressed bcl files into compressed fastq files. High-throughput RNA-sequencing data was subjected to adapter removal with cutadapt v1.889 and base quality trimming to remove 3′ read sequences if more than 20 bases with Q < 20 were present. Paired-end reads were then mapped to the mouse (mm10) reference genome using STAR90 v2.7.3a and gene count summaries were generated using featureCounts91 v2.0.0 and GENCODE vM23 (Ensembl 98) annotations. Raw fragment (i.e. paired-end read) counts were then combined into a numerical matrix, with genes in rows and experiments in columns, and used as input for differential gene expression analysis with the Bioconductor Limma v3.42.0 package92 after multiple filtering steps to remove low-expressed genes. First, gene counts were converted to fragments per kb per million reads (FPKM) using the RSEM package93 with default settings in strand specific mode, and only genes with expression levels above 1 FPKM in at least 50% of samples were retained for further analysis. Additional filtering removed genes with less than 50 total reads across all samples or less than 200 nucleotides in length. Finally, normalization factors were computed on the filtered data matrix using the weighted trimmed mean of M-values method, followed by voom94 mean-variance transformation in preparation for Limma linear modeling. Data were fitted to a design matrix that contained all sample groups and pairwise comparisons were performed between sample groups (littermate mutants versus controls). eBayes adjusted p-values were corrected for multiple testing using the Benjamin-Hochberg method and were used to select genes with significant expression differences (q ≤ 0.05).
Gene ontology enrichment analyzes
Gene ontology (GO) enrichment for biological process (BP), molecular function (MF), and cellular component (CC) were computed using the R gProfileR R v0.6.4 package95. The background gene set included all detected genes. An ordered query was used, ranking genes by log2 fold-change for differential gene expression analyzes. To account for multiple comparisons, p-values were corrected using the g:SCS algorithm.
scRNA-seq library preparation
A total of 3 libraries were generated based on three replicates of each genotype (Esco2fl/fl;Prrx1-CreTg/0 and Esco2fl/fl). Each replicate consisted of three pooled samples (2 limb buds per sample, comprising a total of 6 limb buds per replicate). Samples were genotyped by PCR and each replicate was sex balanced with two male samples and one female sample.
Embryos were harvested at E9.5 according to the Theiler Staging Criteria for Mouse Embryo Development. Right and left forelimbs (a pair) from a single embryo were gently cut off the body using #5SF (11252-00), #5 (11254-20) and/or #55 (11255-20) forceps from Fine Science Tools. Additional tissues flanking the limb buds were removed, and each limb bud pair was transferred into a single Eppendorf tube for further processing. Dissections were performed under a stereomicroscope in multiple changes of cold phosphate-buffered saline (PBS). Limb buds were digested at 37 °C in 0.5 mL of 0.25% trypsin/EDTA (Invitrogen, 25200-072) for 5 min at 37 °C with trituration by pipetting at 2.5 and 5 min. Trypsinization was stopped by adding 0.5 mL of cell culture medium DMEM with 10% FBS. Cell suspensions were centrifuged at 300 g and 4 °C for 3 min. Supernatant was removed, and cells were resuspended in 0.5 mL of pre-chilled (4 °C) 10% DMSO in FBS. Cell suspensions were then transferred into cryovials, placed into a pre-chilled freezing container at −80 °C overnight, and transferred to −150 °C for long term storage the following day. Once sufficient samples were generated, cryovials were thawed at 37 °C and transferred into a 15 mL centrifuge tube. 1 mL of pre-warmed (37 °C) cell culture medium was added dropwise with gentle tube shaking, followed by an additional 2 mL of medium. Three samples were then pooled into one 15 mL centrifuge tube and mixed well prior pelleting at 300 g for 3 min. The media was then aspirated slowly leaving approximately 50 µl of supernatant and cells were resuspended in 1 mL of freshly made 0.04% BSA in PBS. Cell suspensions were then strained through a 40 μm filter (Falcon, 352340), pelleted at 300 g for 3 min, aspirated to approximately 50 µl of supernatant, and resuspended in 50 uL of 0.04% BSA in PBS. scRNA-seq 3’ expression libraries were prepared on a Chromium instrument (10X Genomics, model GCG-SR-1) using the Chromium Single Cell Gene Expression kit (version 3) by the New York Genome Center (NYGC). Sequencing was conducted by on an Illumina NovaSeq using standard protocols for paired-end sequencing with a read length of 50 bp. Sequencing used standard Illumina primers, where read 1 was the UMI, read 2 was the library index, and read 3 was the transcript. Esco2fl/fl;Prrx1-CreTg/0 and Esco2fl/fl libraries consisted of a final number of cells 10,094 and 6527 cells, respectively.
scRNA-seq analysis
Preprocessing of 10X Genomics scRNA-seq data, clustering, and cell type identification was performed using CellRanger software version 7.0.0. Filtered gene and barcode count matrices were used for scRNA-seq analysis with a custom R pipeline centered around the Seurat v4.1.0 package96,97, in addition to other publicly available R toolkits. To remove potential artifacts and outliers, only cells with >1000 genes detected and genes detected in >5 cells were included. Cells with outlier numbers of detected genes (>11,000) and/or high expression of mitochondrial genes (>20%) were removed. The cells from both replicates and both embryonic days were normalized using regularized binomial negative regression98 and integrated based on common transcripts or ‘anchors’97. The cleaned and integrated data was then used for initial clustering. The top eighteen principal components (PCs) were used to perform unsupervised shared nearest neighbor graph-based clustering (k = 100) as implemented in the package Seurat v4.0.096,97. Differential gene expression analysis among cell clusters and stages was tested by non-parametric Wilcoxon rank sum test, with an FDR ≤ 0.05 and ln2(FC) expression threshold ≤0.25.
To determine if any clusters were enriched for proliferation, cells were scored based on their expression of cell cycle associated genes with the Seurat function CellCycleScoring and visualized according to the assigned G2/M or S1 classification.
Copy number variation analysis
The R package inferCNV of the Trinity CTAT Project (https://github.com/broadinstitute/inferCNV) was used to determine copy number variation (CNV). First, genes accounting for greater than 5% of total chromosome expression were removed to avoid bias. A total of 36 genes were removed: Ptma, Rpl39, Gpc3, Rps4x, Rps20, Rps8, Rpl32, Hbb-bh1, Hbb-y, Rps12, Rpl41, Rps26, Rpl18a, Rpl13, Rpl15, Rps24, Tpt1, Gpc6, Rplp1, Eef1a1, Rpsa, Hba-x, Rps23, Rps7, Rps29, Rpl37, Rpl30, Rpl8, Rpl35a, Rpl24, Gm42418, Hsp90ab1, Rps14, Rpl17, Malat1, Fau. The filtered count matrix then was used as input for inferCNV v1.15.0. All cells in control clusters were defined as references, and the CNV analysis was applied to cells of mutant clusters. We applied the “median filtering” function of inferCNV to further remove noise in the analyzed dataset. Lastly, the heatmaps were generated using the plot_cnv function.
Trajectory inference and velocity analysis
Trajectory inference analysis was carried out using Monocle 399. The raw count matrix, along with the annotation of cells generated for Fig. 3 of our work were used as input for the new_cell_data_set function. The data was pre-processed with preprocess_cds and align_cds, using 100 dimensions in both functions, and the sample information in the latter, to correct for inter-sample differences. The reduce_dimensions, cluster_cells and learn_graph functions were called sequentially, and the generated trajectories were visualized with plot_cells. Monocle objects generated at this step were saved for used in the next procedure. Estimation of the fraction of spliced and unspliced molecules per single cell RNA-seq sample was carried out using the Python implementation of Velocyto v0.17.17100. The Velocyto run10x module was run with default options, using as input the CellRanger single cell processed outputs, along with the Mus musculus annotation in GTF format. The resulting Loom files were then processed in the R statistical computing environment101 with the use of the libraries velocyto.R, SeuratWrappers and Seurat. Briefly, the run10x Loom outputs were loaded using the ReadVelocity function and converted to a Seurat object using the as.Seurat function. The UMAP dimensional reduction embedding from the previously generated Monocle3 objects was then added to the Velocyto-Seurat converted object. This accounted for Velocyto’s lack of built-in inter-sample correction method, resulting in clusters that were independent to each sample. Finally, we applied the RunVelocity function, and the shown_velo function was called to generated the UMAP plots with the overlaid velocity arrows.
Multi-scale embedded gene co-expression network analysis (MEGENA)
The high-throughput RNA-sequencing data used for the differential gene expression analysis was also used as the input data for gene co-expression network analysis with the R package MEGENA v1.3.4102. MEGENA first identifies significant correlations between gene pairs using the Pearson method with FDR < 0.05 to identify significant correlations. The significant gene pairs are then sorted by absolute Pearson’s correlation coefficient and sequentially embedded onto a three-dimensional topological sphere following a planarity test to construct a “planar filtered network (PFN).” PFN then undergoes unsupervised clustering at multiple compactness scales by multi-clustering analysis (MCA) to identify gene modules. MCA splits the parent modules into smaller child modules, resulting in a multi-scale hierarchy. Candidate (hub) genes are identified as genes with significantly higher (p < 0.05) network connectivity than the randomly permuted planar networks102. Each module was then tested for significant gene set enrichment (FDR ≤ 0.05) against the Molecular Signatures Database (MSigDB)103. Module diagrams showing the co-expression relationships between modules and genes within each module were plotted with Cytoscape104.
Ensemble of multiple drug repositioning approaches (EMUDRA)
E9.5, E10.5, and E11.5 DEGs generated by RNA-seq were mapped to human orthologs. The mapped DEGs and their corresponding log fold changes were then used to predict compounds with the R package EMUDRA57. Normalized LINCS level 3 data from the drug treatment experiments on the ovarian cancer cell line RMUGS was downloaded from the LINCS data portal to perform the analysis. Drug-induced changes were identified by comparing drug-treated gene expression profiles to vehicle control (DMSO-treated) using the R package Limma92 at an FDR cut-off 0.05. A total of 355 drugs or compounds were tested and ranked based on their normalized score. We used the results from the E10.5 signature as there were too few DEGs in the E9.5 signature to perform the EMUDRA analysis accurately.
Pharmacological inhibition using pifithrin-α
Injections of pifithrin-α were administered according to Jones et al.56. Pifithrin-α (Enzo, BML-GR325-0005) was injected intraperitoneally at 2.2 mg per kg (body weight) in PBS. Female mice were weighed daily to determine the necessary dosage. Treatment was daily from E8.5 until the embryos were harvested at E11.5 or E12.5, and subsequently analyzed.
Gene enrichment analysis
A literature search was performed using PubMed and OMIM to identify genes implicated in genetic disorders that cause with limb reduction and thalidomide embryopathy. A total of 326 genes were collectively identified and used for the enrichment analysis (designated gene_list). Genes were converted to their mouse orthologs using babelgene package in R. Differentially expressed genes from E9.5 determined by scRNA-seq analysis were tested for significant (p ≤ 0.05) gene set enrichment against gene_list. Fisher’s exact tests with a Bonferroni correction applied for multiple comparisons testing was applied.
Other statistical analyzes
Analyzes determining statistical significance between Esco2fl/fl and Esco2fl/fl;Prrx1-CreTg/0 samples for TUNEL assays, number of branch points, and blood vessel diameter were performed using a Student’s t-test in Microsoft Excel.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We would like to thank our colleagues at Mount Sinai’s Microscopy CoRE and CCMS’s Necroscopy core. This work was supported, in part, through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai. Research reported in this paper was supported by the Office of Research Infrastructure of the National Institutes of Health (NIH) under award number S10OD026880 and NIH grants R01AG068030 (B.Z.), R03DE026814 (E.J and B.Z.), R01DE029832 (E.J. and H.B.), and R01DE029322 (E.J. and H.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
X.Y. developed and implemented LoxP-Cre strategies for creating the Esco2 CKO mouse model. M.W. and A.S.S. characterized the mouse model. A.S.G.R. and A.S.S. conducted the bulk and single-cell RNA-seq analyzes. B.V-M. conducted the InferCNV analysis, trajectory and RNA velocity analyzes and assisted with single-cell RNA-seq analyzes. X.Z., B.Z., and A.S.S. conducted the gene co-expression network and drug repositioning analyzes. A.S.S., M.W. completed all wet-lab related experimentation. A.S.S. generated all the figures and tables for the manuscript with input from H.v.B. and E.W.J. A.S.S. wrote the initial manuscript draft with input and suggestions from H.v.B. and E.W.J. H.v.B and E.W.J provided advice and guidance throughout. All the authors contributed to editing and revising the paper.
Peer review
Peer review information
Nature Communications thanks Katta Girisha, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The data generated in this study have been deposited in the Gene Expression Omnibus under the GEO accession number GSE230493 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101099]. The processed data are available in the supplementary datasets within the article. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files or from the corresponding author upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Meng Wu, Email: wu.meng@mayo.edu.
Harm van Bakel, Email: harm.vanbakel@mssm.edu.
Ethylin Wang Jabs, Email: jabs.ethylin@mayo.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-51328-3.
References
- 1.Skibbens, R. V. et al. Cohesinopathies of a feather flock together. PLoS Genet9, e1004036 (2013). 10.1371/journal.pgen.1004036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerji, R., Skibbens, R. V. & Iovine, M. K. How many roads lead to cohesinopathies? Dev. Dyn.246, 881–888 (2017). 10.1002/dvdy.24510 [DOI] [PubMed] [Google Scholar]
- 3.McNairn, A. J. & Gerton, J. L. Cohesinopathies: one ring, many obligations. Mutat. Res–Fund. Mol. M.647, 103–111 (2008). 10.1016/j.mrfmmm.2008.08.010 [DOI] [PubMed] [Google Scholar]
- 4.Piché, J., Van Vliet, P. P., Pucéat, M. & Andelfinger, G. The expanding phenotypes of cohesinopathies: one ring to rule them all! Cell Cycle18, 2828–2848 (2019). 10.1080/15384101.2019.1658476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantaputra, P. N. et al. Juberg-Hayward syndrome is a cohesinopathy, caused by mutation in ESCO2. Eur. J. Orthod.43, 45–50 (2021). 10.1093/ejo/cjaa023 [DOI] [PubMed] [Google Scholar]
- 6.Liu, J. & Krantz, I. D. Cornelia de Lange syndrome, cohesin, and beyond. Clin. Genet.76, 303–314 (2009). 10.1111/j.1399-0004.2009.01271.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vega, H. et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet.37, 468–470 (2005). 10.1038/ng1548 [DOI] [PubMed] [Google Scholar]
- 8.Schüle, B., Oviedo, A., Johnston, K., Pai, S. & Francke, U. Inactivating mutations in ESCO2 cause SC phocomelia and Roberts syndrome: no phenotype-genotype correlation. Am. J. Hum. Genet.77, 1117–1128 (2005). 10.1086/498695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kantaputra, P. N. et al. Juberg-Hayward syndrome and Roberts syndrome are allelic, caused by mutations in ESCO2. Arch. Oral Biol.119, 104918 (2020). 10.1016/j.archoralbio.2020.104918 [DOI] [PubMed] [Google Scholar]
- 10.Gordillo, M. et al. The molecular mechanism underlying Roberts syndrome involves loss of ESCO2 acetyltransferase activity. Hum. Mol. Genet.17, 2172–2180 (2008). 10.1093/hmg/ddn116 [DOI] [PubMed] [Google Scholar]
- 11.Robert, J. B. A child with double cleft of lip and palate, protrusion of the intermaxillary portion of the upper jaw and imperfect development of the bones of the four extremities. Ann. Surg.70, 252–253 (1919). [Google Scholar]
- 12.Goh, E. S. Y. et al. The Roberts syndrome/SC phocomelia spectrum—a case report of an adult with review of the literature. Am. J. Med. Genet. A.152, 472–478 (2010). 10.1002/ajmg.a.33261 [DOI] [PubMed] [Google Scholar]
- 13.Lenz, W. D., Marquardt, E. & Weicker, H. Pseudothalidomide syndrome. Birth Defects Orig. Art. Ser.11, 97–107 (1974). [Google Scholar]
- 14.Vega, H. et al. Phenotypic variability in 49 cases of ESCO2 mutations, including novel missense and codon deletion in the acetyltransferase domain, correlates with ESCO2 expression and establishes the clinical criteria for Roberts syndrome. J. Med. Genet.47, 30–37 (2010). 10.1136/jmg.2009.068395 [DOI] [PubMed] [Google Scholar]
- 15.Sanchez, A. C., Thren, E. D., Iovine, M. K. & Skibbens, R. V. Esco2 and cohesin regulate CRL4 ubiquitin ligase ddb1 expression and thalidomide teratogenicity. Cell Cycle21, 501–513 (2022). 10.1080/15384101.2021.2023304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McBride, W. G. Thalidomide and congenital abnormalities. Lancet2, 90927–90928 (1961). [Google Scholar]
- 17.Bodera, P. & Stankiewicz, W. Immunomodulatory properties of thalidomide analogs: pomalidomide and lenalidomide, experimental and therapeutic applications. Recent Pat. Endocr. Metab. Immune Drug Discov.5, 192–196 (2011). 10.2174/187221411797265890 [DOI] [PubMed] [Google Scholar]
- 18.Schwartz, M. P. et al. Human pluripotent stem cell-derived neural constructs for predicting neural toxicity. Proc. Natl. Acad. Sci.112, 12516–12521 (2015). 10.1073/pnas.1516645112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulmke, P. A. et al. Molecular profiling reveals involvement of ESCO2 in intermediate progenitor cell maintenance in the developing mouse cortex. Stem Cell Rep16, 968–984 (2021). 10.1016/j.stemcr.2021.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, X. B., Huang, B., Pan, Y. H., Su, S. G. & Li, Y. ESCO2 inhibits tumor metastasis via transcriptionally repressing MMP2 in colorectal cancer. Cancer Manag. Res.10, 6157–6166 (2018). 10.2147/CMAR.S181265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, T. et al. Identification of a primary target of thalidomide teratogenicity. Science327, 1345–1350 (2010). 10.1126/science.1177319 [DOI] [PubMed] [Google Scholar]
- 22.Ito, T., Ando, H. & Handa, H. Teratogenic effects of thalidomide: molecular mechanisms. Cell. Mol. Life Sci.68, 1569–1579 (2011). 10.1007/s00018-010-0619-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banerji, R., Skibbens, R. V. & Iovine, M. K. Cohesin mediates Esco2-dependent transcriptional regulation in a zebrafish regenerating fin model of Roberts Syndrome. Biol. Open6, 1802–1813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun, H. et al. Cul4-Ddb1 ubiquitin ligases facilitate DNA replication-coupled sister chromatid cohesion through regulation of cohesin acetyltransferase Esco2. PLoS Genet. 15, e1007685 (2019). 10.1371/journal.pgen.1007685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minamino, M. et al. Temporal regulation of ESCO2 degradation by the MCM complex, the CUL4-DDB1-VPRBP complex, and the anaphase-promoting complex. Curr. Biol.28, 2665–2672 (2018). 10.1016/j.cub.2018.06.037 [DOI] [PubMed] [Google Scholar]
- 26.Skibbens, R. V., Corson, L. B., Koshland, D. & Hieter, P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev13, 307–319 (1999). 10.1101/gad.13.3.307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tóth, A. et al. Yeast cohesin complex requires a conserved protein, Eco1p (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev13, 320–333 (1999). 10.1101/gad.13.3.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bellows, A. M., Kenna, M. A., Cassimeris, L. & Skibbens, R. V. Human EFO1p exhibits acetyltransferase activity and is a unique combination of linker histone and Ctf7p/Eco1p chromatid cohesion establishment domains. Nucleic Acids Res.31, 6334–6343 (2003). 10.1093/nar/gkg811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters, J. M., Tedeschi, A. & Schmitz, J. The cohesin complex and its roles in chromosome biology. Genes Dev22, 3089–3114 (2008). 10.1101/gad.1724308 [DOI] [PubMed] [Google Scholar]
- 30.Michaelis, C., Ciosk, R. & Nasmyth, K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell91, 35–45 (1997). 10.1016/S0092-8674(01)80007-6 [DOI] [PubMed] [Google Scholar]
- 31.DiNardo, S., Voelkel, K. & Sternglanz, R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl Acad. Sci. USA81, 2616–2620 (1984). 10.1073/pnas.81.9.2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gassler, J. et al. A mechanism of cohesin‐dependent loop extrusion organizes zygotic genome architecture. EMBO J36, 3600–3618 (2017). 10.15252/embj.201798083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson, I. F. et al. DNA loop extrusion by human cohesin. Science366, 1338–1345 (2019). 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- 34.Kim, Y., Shi, Z., Zhang, H., Finkelstein, I. J. & Yu, H. Human cohesin compacts DNA by loop extrusion. Science366, 1345–1349 (2019). 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golfier, S., Quail, T., Kimura, H. & Brugués, J. Cohesin and condensin extrude DNA loops in a cell cycle-dependent manner. eLife9, e53885 (2020). 10.7554/eLife.53885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wutz, G. et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J36, 3573–3599 (2017). 10.15252/embj.201798004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ball, A. R. Jr & Yokomori, K. Damage‐induced reactivation of cohesin in postreplicative DNA repair. Bioessays30, 5–9 (2008). 10.1002/bies.20691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Covo, S., Westmoreland, J. W., Gordenin, D. A. & Resnick, M. A. Cohesin is limiting for the suppression of DNA damage–induced recombination between homologous chromosomes. PLoS Genet6, e1001006 (2010). 10.1371/journal.pgen.1001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watrin, E. & Peters, J. M. The cohesin complex is required for the DNA damage‐induced G2/M checkpoint in mammalian cells. EMBO J28, 2625–2635 (2009). 10.1038/emboj.2009.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim, S. T., Xu, B. & Kastan, M. B. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev16, 560–570 (2002). 10.1101/gad.970602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ünal, E. et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell.16, 991–1002 (2004). 10.1016/j.molcel.2004.11.027 [DOI] [PubMed] [Google Scholar]
- 42.Mönnich, M., Kuriger, Z., Print, C. G. & Horsfield, J. A. A zebrafish model of Roberts syndrome reveals that Esco2 depletion interferes with development by disrupting the cell cycle. PloS One6, e20051 (2011). 10.1371/journal.pone.0020051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whelan, G. et al. Cohesin acetyltransferase Esco2 is a cell viability factor and is required for cohesion in pericentric heterochromatin. EMBO J31, 71–82 (2012). 10.1038/emboj.2011.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Percival, S. M. et al. Variations in dysfunction of sister chromatid cohesion in esco2 mutant zebrafish reflect the phenotypic diversity of Roberts syndrome. Dis. Model Mech.8, 941–955 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jabs, E. W., Tuck-Muller, C. M., Cusano, R. & Rattner, J. B. Centromere separation and aneuploidy in human mitotic mutants: Roberts syndrome. Prog. Clin. Biol. Res.318, 111–118 (1989). [PubMed] [Google Scholar]
- 46.Tomkins, D. J. & Sisken, J. E. Abnormalities in the cell-division cycle in Roberts syndrome fibroblasts: a cellular basis for the phenotypic characteristics? Am. J. Hum. Genet.36, 1332 (1984). [PMC free article] [PubMed] [Google Scholar]
- 47.Burns, M. A. & Tomkins, D. J. Hypersensitivity to mitomycin C cell-killing in Roberts syndrome fibroblasts with, but not without, the heterochromatin abnormality. Mutat. Res., Sect. Environ. Mutagen. Relat. Subj216, 243–249 (1989). [DOI] [PubMed] [Google Scholar]
- 48.van der Lelij, P. et al. The cellular phenotype of Roberts syndrome fibroblasts as revealed by ectopic expression of ESCO2. PloS One4, e6936 (2009). 10.1371/journal.pone.0006936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Den Berg, D. J. & Francke, U. Sensitivity of Roberts syndrome cells to gamma radiation, mitomycin C, and protein synthesis inhibitors. Somat. Cell Mol. Genet.19, 377–392 (1993). 10.1007/BF01232749 [DOI] [PubMed] [Google Scholar]
- 50.Logan, M. et al. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis33, 77–80 (2002). 10.1002/gene.10092 [DOI] [PubMed] [Google Scholar]
- 51.Martin, J. F. & Olson, E. N. Identification of a prx1 limb enhancer. Genesis26, 225–229 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Durland, J. L., Sferlazzo, M., Logan, M. & Burke, A. C. Visualizing the lateral somitic frontier in the Prx1Cre transgenic mouse. J. Anat.212, 590–602 (2008). 10.1111/j.1469-7580.2008.00879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin, M. & Pacifici, M. Vascular regression is required for mesenchymal condensation and chondrogenesis in the developing limb. Dev. Dyn.222, 522–533 (2001). 10.1002/dvdy.1212 [DOI] [PubMed] [Google Scholar]
- 54.Eshkar-Oren, I. et al. The forming limb skeleton serves as a signaling center for limb vasculature patterning via regulation of Vegf. Development136, 1263–1272 (2009). 10.1242/dev.034199 [DOI] [PubMed] [Google Scholar]
- 55.Komarov, P. G. et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science285, 1733–1737 (1999). 10.1126/science.285.5434.1733 [DOI] [PubMed] [Google Scholar]
- 56.Jones, N. C. et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med.14, 125–133 (2008). 10.1038/nm1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou, X., Wang, M., Katsyv, I., Irie, H. & Zhang, B. EMUDRA: ensemble of multiple drug repositioning approaches to improve prediction accuracy. Bioinformatics34, 3151–3159 (2018). 10.1093/bioinformatics/bty325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mfarej, M. G. & Skibbens, R. V. An ever-changing landscape in Roberts syndrome biology: implications for macromolecular damage. PLoS Genet16, e1009219 (2020). 10.1371/journal.pgen.1009219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salmon, T. B., Evert, B. A., Song, B. & Doetsch, P. W. Biological consequences of oxidative stress-induced DNA damage in Saccharomyces cerevisiae. Nucleic Acids Res.32, 3712–3723 (2004). 10.1093/nar/gkh696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowe, L. A., Degtyareva, N. & Doetsch, P. W. DNA damage-induced reactive oxygen species (ROS) stress response in Saccharomyces cerevisiae. Free Radic. Biol. Med.45, 1167–1177 (2008). 10.1016/j.freeradbiomed.2008.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang, M. et al. DNA damage induces reactive oxygen species generation through the H2AX-Nox1/Rac1 pathway. Cell Death Dis.3, e249 (2012). 10.1038/cddis.2011.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, B., Lee, K. K., Zhang, L. & Gerton, J. L. Stimulation of mTORC1 with L-leucine rescues defects associated with Roberts syndrome. PLoS Genet.9, e1003857 (2013). 10.1371/journal.pgen.1003857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu, B., Gogol, M., Gaudenz, K. & Gerton, J. L. Improved transcription and translation with L-leucine stimulation of mTORC1 in Roberts syndrome. BMC Genom.17, 1–18 (2016). 10.1186/s12864-015-2354-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McKay, M. J. et al. A Roberts syndrome individual with differential genotoxin sensitivity and a DNA damage response defect. Int. J. Radiat. Oncol. Biol. Phys.103, 1194–1202 (2019). 10.1016/j.ijrobp.2018.11.047 [DOI] [PubMed] [Google Scholar]
- 65.Ren, Q., Yang, H., Gao, B. & Zhang, Z. Global transcriptional analysis of yeast cell death induced by mutation of sister chromatid cohesin. Compar. Funct. Genom.2008, e634283 (2008). 10.1155/2008/634283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perkins, A. T., Das, T. M., Panzera, L. C. & Bickel, S. E. Oxidative stress in oocytes during midprophase induces premature loss of cohesion and chromosome segregation errors. Proc. Natl Acad. Sci. USA113, 6823–6830 (2016). 10.1073/pnas.1612047113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Polyak, K., Xia, Y., Zweier, J. L., Kinzler, K. W. & Vogelstein, B. A model for p53-induced apoptosis. Nature389, 300–305 (1997). 10.1038/38525 [DOI] [PubMed] [Google Scholar]
- 68.Norbury, C. J. & Zhivotovsky, B. DNA damage-induced apoptosis. Oncogene23, 2797–2808 (2004). 10.1038/sj.onc.1207532 [DOI] [PubMed] [Google Scholar]
- 69.Shen, H. M. & Liu, Z. G. JNK signaling pathway is a key modulator in cell death mediated by reactive oxygen and nitrogen species. Free Radic. Biol. Med.40, 928–939 (2006). 10.1016/j.freeradbiomed.2005.10.056 [DOI] [PubMed] [Google Scholar]
- 70.Madesh, M. & Hajnóczky, G. VDAC-dependent permeabilization of the outer mitochondrial membrane by superoxide induces rapid and massive cytochrome c release. J. Cell Biol.155, 1003–1016 (2001). 10.1083/jcb.200105057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simon, H. U., Haj-Yehia, A. & Levi-Schaffer, F. Role of reactive oxygen species (ROS) in apoptosis induction. Apoptosis5, 415–418 (2000). 10.1023/A:1009616228304 [DOI] [PubMed] [Google Scholar]
- 72.Gotoh, Y. & Cooper, J. A. Reactive oxygen species-and dimerization-induced activation of apoptosis signal-regulating kinase 1 in tumor necrosis factor-α signal transduction. J. Biol. Chem.273, 17477–17482 (1998). 10.1074/jbc.273.28.17477 [DOI] [PubMed] [Google Scholar]
- 73.Lopez, K. E. & Bouchier-Hayes, L. Lethal and non-lethal functions of caspases in the DNA damage response. Cells11, e1887 (2022). 10.3390/cells11121887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schaub, F. J. et al. Fas/FADD-mediated activation of a specific program of inflammatory gene expression in vascular smooth muscle cells. Nat. Med.6, 790–796 (2000). 10.1038/77521 [DOI] [PubMed] [Google Scholar]
- 75.Lauber, K. et al. Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell113, 717–730 (2003). 10.1016/S0092-8674(03)00422-7 [DOI] [PubMed] [Google Scholar]
- 76.Horsfield, J. A. et al. Cohesin-dependent regulation of Runx genes. Development134, 2639–2649 (2007). 10.1242/dev.002485 [DOI] [PubMed] [Google Scholar]
- 77.Kim, B. J. et al. Esco2 is a novel corepressor that associates with various chromatin modifying enzymes. Biochem. Biophys. Res. Commun.372, 298–304 (2008). 10.1016/j.bbrc.2008.05.056 [DOI] [PubMed] [Google Scholar]
- 78.Rhodes, J. M., McEwan, M. & Horsfield, J. A. Gene regulation by cohesin in cancer: is the ring an unexpected party to proliferation? Mol. Cancer Res.9, 1587–1607 (2011). 10.1158/1541-7786.MCR-11-0382 [DOI] [PubMed] [Google Scholar]
- 79.Mehta, G. D., Rizvi, S. M. A. & Ghosh, S. K. Cohesin: a guardian of genome integrity. Biochim. Biophys. Acta. Mol. Cell. Res.1823, 1324–1342 (2012). 10.1016/j.bbamcr.2012.05.027 [DOI] [PubMed] [Google Scholar]
- 80.Skibbens, R. V., Marzillier, J. & Eastman, L. Cohesins coordinate gene transcriptions of related function within Saccharomyces cerevisiae. Cell Cycle9, 1601–1606 (2010). 10.4161/cc.9.8.11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waldman, T. Emerging themes in cohesin cancer biology. Nat. Rev. Cancer20, 504–515 (2020). 10.1038/s41568-020-0270-1 [DOI] [PubMed] [Google Scholar]
- 82.Vargesson, N. The teratogenic effects of thalidomide on limbs. J Hand Surg Eur Vol.44, 88–95 (2019). 10.1177/1753193418805249 [DOI] [PubMed] [Google Scholar]
- 83.Vargesson, N. & Hootnick, D. Arterial dysgenesis and limb defects: clinical and experimental examples. Reprod. Toxicol.70, 21–29 (2017). 10.1016/j.reprotox.2016.10.005 [DOI] [PubMed] [Google Scholar]
- 84.Meganathan, K. et al. Identification of thalidomide-specific transcriptomics and proteomics signatures during differentiation of human embryonic stem cells. PloS One7, e44228 (2012). 10.1371/journal.pone.0044228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bean, C. J., Hunt, P. A., Millie, E. A. & Hassold, T. J. Analysis of a malsegregating mouse Y chromosome: evidence that the earliest cleavage divisions of the mammalian embryo are non-disjunction-prone. Hum. Mol. Genet.10, 963–972 (2001). 10.1093/hmg/10.9.963 [DOI] [PubMed] [Google Scholar]
- 86.Wang, Y. et al. Activation of p38 MAPK pathway in the skull abnormalities of Apert syndrome Fgfr2+ P253R mice. BMC Dev. Bio.10, 1–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods9, 676–682 (2012). 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bernsen, J. Dynamic thresholding of grey-level images fcV. Proceeding of the 8 International Conference O11 Pattern Recognition, 1251–1255 (1986).
- 89.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J17, 10–12 (2011). 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 90.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics30, 923–930 (2014). 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- 92.Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.43, e47 (2015). 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform12, 1–16 (2011). 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol.15, 1–17 (2014). 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reimand, J., Kull, M., Peterson, H., Hansen, J. & Vilo, J. g: Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res.35, W193–W200 (2007). 10.1093/nar/gkm226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol.36, 411–420 (2018). 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–1902 (2019). 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol.20, 296 (2019). 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol.32, 381–386 (2014). 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.La Manno, G. et al. RNA velocity of single cells. Nature560, 494–498 (2018). 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.RDevelopment, C.O.R.E. TEAM 2009: R: a language and environment for statistical computing. Internet: http://www.R-project.org (2012).
- 102.Song, W. M. & Zhang, B. Multiscale embedded gene co-expression network analysis. PLoS Comput. Biol.11, e1004574 (2015). 10.1371/journal.pcbi.1004574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA102, 15545–15550 (2005). 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res.13, 2498–2504 (2003). 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study have been deposited in the Gene Expression Omnibus under the GEO accession number GSE230493 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE101099]. The processed data are available in the supplementary datasets within the article. All other relevant data supporting the key findings of this study are available within the article and its Supplementary Information files or from the corresponding author upon request. Source data are provided with this paper.