Abstract
The first IMPI (inhibitor of metalloproteinases from insects) was identified in the greater wax moth, Galleria mellonella [Wedde, Weise, Kopacek, Franke and Vilcinskas (1998) Eur. J. Biochem. 255, 535–543]. Here we report cloning and expression of a cDNA coding for this IMPI. The IMPI mRNA was identified among the induced transcripts from a subtractive and suppressive PCR analysis after bacterial challenge of G. mellonella larvae. Induced expression of the IMPI during a humoral immune response was confirmed by real-time PCR, which documented up to 500 times higher amounts of IMPI mRNA in immunized larvae in comparison with untreated ones. The IMPI sequence shares no similarity with those of tissue inhibitors of metalloproteinases or other natural inhibitors of metalloproteinases, and the recombinant IMPI specifically inhibits thermolysin-like metalloproteinases, but not matrix metalloproteinases. These results support the hypothesis that the IMPI represents a novel type of immune-related protein which is induced and processed during the G. mellonella humoral immune response to inactivate pathogen-associated thermolysin-like metalloproteinases.
Keywords: innate immunity, insect metalloproteinase inhibitor (IMPI), microbial proteinase, pathogen, thermolysin
Abbreviations: IMPI, insect metalloproteinase inhibitor; ISPI, inducible serine proteinase inhibitor; LPS, lipopolysaccharide; MALDI–TOF-MS, matrix-assisted laser-desorption ionization–time-of-flight MS; MMP, matrix metalloproteinase; RACE, rapid amplification of cDNA ends; rIMPI, recombinant insect metalloproteinase inhibitor; rIMPI-1, recombinant protein corresponding to native IMPI; rIMPI-2, recombinant protein coded by the full-length IMPI cDNA; rIMPI-1*, recombinant insect metalloproteinase inhibitor after thermolysin–Sepharose purification; SPC, subtilisin-like proprotein convertase; TIL domain, trypsin inhibitor-like cysteine-rich domain; TIMP, tissue inhibitor of metalloproteinases
INTRODUCTION
The IMPI (inhibitor of metalloproteinases from insects) isolated from the haemolymph of immunized greater wax moth (Galleria mellonella) larvae [1] represented the first peptidic inhibitor of metalloproteinases identified in invertebrates. The IMPI is a heat-stable, glycosylated, low-molecular-mass protein (8364 Da), and its partially determined amino acid sequence exhibits no similarity to those of vertebrate TIMPs (tissue inhibitors of metalloproteinases) or other natural inhibitors of metalloproteinases [1]. Vertebrate TIMPs represent a family of four related proteins that regulate MMPs (matrix metalloproteinases). TIMPs and MMPs both play a critical role in the controlled remodelling of the extracellular matrix during development, morphogenesis and wound healing [2–4]. The first TIMP homologue in insects was reported from Drosophila in 1999 [5]. Cloning and characterization of this TIMP homologue strongly suggested that it inhibits the Drosophila MMPs Dm1-MMP and Dm2-MMP [6], which participate in tissue remodelling [7].
In contrast with the TIMPs, the G. mellonella IMPI is induced in response to injected microbial elicitors of the innate immune response, such as cell wall components from bacteria or fungi. Immunized G. mellonella larvae with enhanced levels of IMPI activity within the haemolymph survive after injection of normally lethal concentrations of the bacterial metalloproteinase thermolysin [1]. In addition to the IMPI, a number of novel ISPIs (inducible serine proteinase inhibitors; i.e. ISPI-1, -2 and -3) have been discovered within the haemolymph of immunized G. mellonella larvae. On the basis of the determined amino acid sequences, G. mellonella ISPI-2 represents a novel member of the Kunitz-type inhibitor family, whereas ISPI-1 and ISPI-3 share no similarity with other known proteins. All three identified ISPIs were determined to inhibit toxic serine proteinases produced by the entomopathogenic fungus Metarhizium anisopliae in vitro [8]. Parasitic fungi utilize a spectrum of secreted proteinases to directly penetrate the integument of the infected insect host, to colonize and digest its haemolymph and tissues, and to incapacitate its immune system [9]. Interestingly, among the proteolytic enzymes produced by pathogenic bacteria and fungi, metalloproteinases in particular represent potent activators of innate immunity in insects [10]. Degradation of haemolymph proteins by bacterial thermolysin results in the formation of particularly small peptides with molecular masses below 3 kDa, which in turn induce the expression of genes encoding the IMPI and other immune proteins in G. mellonella, suggesting a feedback loop regulation of released metalloproteinases in insects [11].
In the present study we have identified the IMPI among other inducible immune-related genes in G. mellonella larvae using an equalizing subtractive hybridization technique [12–14] and obtained the full-length cDNA by RACE (rapid amplification of cDNA ends) PCR. Interestingly, the isolated cDNA encodes a peptide of 170 residues, whereas the native IMPI is less than half this size in mass terms.
To study the generation of the shorter, naturally occurring IMPI and to localize the inhibitory active domain(s) of the IMPI, two recombinant proteins were constructed: rIMPI-1, corresponding to native IMPI, and rIMPI-2, coded by the full-length IMPI cDNA. The rIMPIs were expressed in Drosophila Schneider 2 cells, as this insect expression system is reportedly able to subject recombinant proteins to glycosylation and post-translational processing [15]. The rIMPIs were tested against three bacterial metalloproteinases and six human MMPs to elucidate the potential of the IMPI as a novel structure for the development of synthetic inhibitors used in modern strategies for the targeted therapeutic regulation of metalloproteinases, associated either with pathogens or with malignant cells.
MATERIALS AND METHODS
Immunization of G. mellonella larvae and RNA isolation
Rearing, immunization and haemolymph collection was done according to Wedde et al. [1]. Briefly, 1 mg/ml LPS (lipopolysaccharide; Sigma) was dissolved in distilled water, and 10 μl per animal was injected dorsolaterally into final-instar larvae. Haemolymph was collected, transferred into CEB (citrate/EDTA buffer) and centrifuged for 5 min at 300 g to pellet haemocytes. In addition, whole larvae ground in liquid nitrogen were used for mRNA isolation. RNA isolation was done with Rneasy kits (QIAGEN). For real-time PCR, additional DNAse treatment was performed (RNase-Free DNase Set; QIAGEN).
Subtractive hybridization and suppressive PCR
Subtractive hybridization and suppressive PCR were carried out with the SMART PCR cDNA Synthesis Kit (Clontech) and the PCR Select cDNA Subtraction Kit (Clontech), followed by a screening with non-radioactive digoxygenin dot blots (as previously described in [14]). Among the clones obtained, we identified a fragment of the IMPI cDNA. 3′- and 5′-RACE PCR was performed with the SMART RACE cDNA Amplification Kit (Clontech) using the IMPI-specific internal primers 5′-ATC CCA ATG GAA CGA CGC GAA CG-3′ (5′-IMPI-RACE) and 5′-TAA CTT CAC CAT GCA TAC CAA TTT GC-3′ (3′-IMPI-RACE).
Real-time PCR
Real-time PCR was performed using the LightCycler system and software (Roche). An aliquot of 1 μg of total RNA from whole larvae was used to synthesize cDNA (GeneAmpRNA-PCR Core Kit; Roche). PCR was done with the LightCycler-DNA Master SYBR Green I Mix (Roche) using the following PCR primers: Impi-a-up, 5′-AGC ATA GTC CTA ATT TGT ACC-3′; Impi-a-low, 5′-GCG AAC GTA TTT TAG GAC AG-3′. For relative quantification of IMPI cDNA, expression of a housekeeping gene (β-actin; PCR primers: forward, 5′-GGG ACG ATA TGG AGA AGA TCT G-3′; reverse, 5′-CAC GCT CTG TGA GGA TCT TC-3′) was measured.
Cloning and expression of IMPI proteins
Reverse transcription–PCR was performed with total RNA from immunized larvae using random hexamers and MuLV reverse transcriptase (Perkin Elmer by Roche). For subsequent amplification of the target sequences, AmpliTaq DNA polymerase and PCR primers with BglII and EcoRI restriction sites were used: 5′-ACT CGA AGA TCT AGC ATA GTC CTA ATT TGT AAC-3′ (IMPI-clone-a-up) and 5′-TCG AGT CGA ATT CGC GAA CGT ATT TTA GGA CAG-3′ (IMPI-clone-a-down) for rIMPI-1, and IMPI-clone-a-up and 5′-TCG AGT CGA ATT CCT TGA CAA TCA GGG GAA TGT-3′ (IMPI-clone-b-down) for rIMPI-2. The following PCR steps were performed on a Perkin Elmer 7600 Cycler: 94 °C for 3 min; five cycles of 94 °C for 30 s, 58 °C for 40 s and 75 °C for 40 s; 35 cycles of 95 °C for 30 s, 56 °C for 30 s and 75 °C for 35 s; and a final step at 72 °C for 10 min. The resulting amplification products were purified (QIAEX-2 DNA Isolation-Kit; QIAGEN), cloned into the eukaryotic vector pMT/BiP/V5-His C (Invitrogen) and transformed in Escherichia coli with the TOPO TA Cloning Kit (Invitrogen). After plasmid preparation (QIAfilter Plasmid Maxi Kit; QIAGEN), the proteins were expressed in Schneider 2 cells using CuSO4 for induction (Drosophila Expressions System, DES; Invitrogen).
The recombinant proteins were purified from the supernatant by affinity chromatography according to the manufacturer's recommendations using chelating Sepharose as matrix and Ni2+ as ligand (Pharmacia Biotech). After elution of proteins with 50 mM sodium phosphate, 500 mM NaCl and 500 mM imidazole, pH 8, the proteins were dialysed against distilled water and dried using a Speedvac.
Purification of native IMPI from haemolymph
Cell-free immune haemolymph was precipitated with 3% (v/v) trichloroacetic acid and desalted [1]. The resulting supernatant was used for further enrichment of IMPI by preparative gel electrophoresis (Prep Cell 491; Bio-Rad) under native conditions according to the manufacturer's recommendations. The IMPI-containing fractions (which were determined by Azocoll assay; see below) were dialysed against distilled water and freeze-dried. Finally, the native IMPI was isolated with an affinity-purification step on a CNBr-activated thermolysin–Sepharose column as described previously [1].
Visualization, quantification and MS of IMPI proteins
The recombinant proteins were detected after PAGE and Western blotting using primary antibodies targeted to the V5 epitope (anti-V5 antibody, dilution 1:5000; Invitrogen), and goat anti-mouse Ig conjugated to horseradish peroxidase (dilution 1:10000, Dako) as secondary antibody. Chemiluminescent detection was done using ECL® Western blotting detection reagents (Amersham Pharmacia Biotech). The concentration of isolated proteins was determined using a MicroBC assay protein quantification kit (Uptima). MALDI-TOF (matrix-assisted laser-desorption ionization–time-of-flight) MS of purified proteins was done by AnagnosTec. A 1–4 μl aliquot of the sample was prepared on to a stainless steel template, and immediately 1 μl of matrix [10 mg/ml 2,5-dihydroxybenzoic acid in water/acetonitrile (1:1, v/v) with 0.03% trifluoroacetic acid] was added. The sample/matrix mixture was air dried at room temperature. Positive-ion mass spectra were recorded using a MALDI-TOF mass spectrometer (Voyager DE-PRO; Applied Biosystems) equipped with a reflectron, post-source decay and collision-induced dissociation options. Mass spectra were obtained from 400 to 20000 Da. All analyses were carried out in the linear and delayed extraction mode. BSA products were used for external calibration.
Enzymic assays
Nomenclature and classification of proteolytic enzymes accords with [16,17]. Inhibition of the following metalloproteinases by IMPI was assayed: thermolysin of family M4 (EC 3.4.24.27; Boehringer), bacillolysin (=dispase, protease type IX; bacterial) of family M4 (EC 3.4.24.28; Sigma), pseudolysin of family M4 (EC 3.4.24.26; Calbiochem) and human MMP-1, -2, -3, -7, -8, and -9 of family M10A (EC 3.4.24.7, -24, -17, -23, -34 and -35 respectively; Calbiochem). The MMPs except for MMP-7 were activated with 2 mM 4-aminophenylmercuric acetate (Sigma) for 2 h at 37 °C. Furthermore, we used the metalloproteinase inhibitors TIMP-2 (Calbiochem) and phosphoramidon (Sigma) as references.
MMP-1/MMP-9 substrate assays (assays I/II) were performed according to [18] with some modifications. Reaction mixtures contained 10 μM of quenched fluorescent MMP-1/MMP-9 substrate (Calbiochem), 50 mM Tris/HCl (pH 7.6), 200 mM NaCl and 5 mM CaCl2, completed with 20 μM ZnSO4 and 0.05% Brij 35 (assay I) or 0.1 μM ZnCl and 0.2% Brij 35 (assay II). Proteolytic activities were measured by monitoring fluorimetric changes after incubation for 17 h (assay I) or 19 h (assay II) at room temperature using a TECAN fluorescence microplate reader (excitation/emission 355/460 nm).
Azocoll assays were performed according to [1] with some modifications. Reaction mixtures contained 15 mg/ml Azocoll (Calbiochem), 50 mM Tris/HCl (pH 7.6), 150 mM NaCl and 5 mM CaCl2. Proteolytic activities were measured by absorbance changes at 490 nm after incubation for 18 h at 37 °C using a microplate photometer.
RESULTS
Detection and characterization of IMPI mRNA in G. mellonella haemocytes
From the subtractive hybridization and suppressive PCR analysis of G. mellonella haemocytes, one clone was identified as the IMPI mRNA by comparing the deduced amino acid sequence with the known partial sequence of the native IMPI protein [1]. The full-length sequence of the IMPI-encoding mRNA was determined by employing 3′- and 5′-RACE PCR. The cDNA has an open reading frame of 510 nucleotides, corresponding to 170 amino acids (Figure 1). Searching with BLAST in NCBI databases revealed no significant similarities to known mRNAs.
Figure 1. cDNA sequence and deduced amino acid sequence of G. mellonella IMPI.
Nucleotides are numbered from the first base at the 5′-end. Amino acids are numbered from the initiating methionine. The putative signal peptide is shown in italics, and the putative IMPI protein is underlined. The amino acid sequence in parentheses was determined by peptide sequencing. The SPC cleavage site (R-R) is given in pale grey lettering.
Inducibility of IMPI mRNA
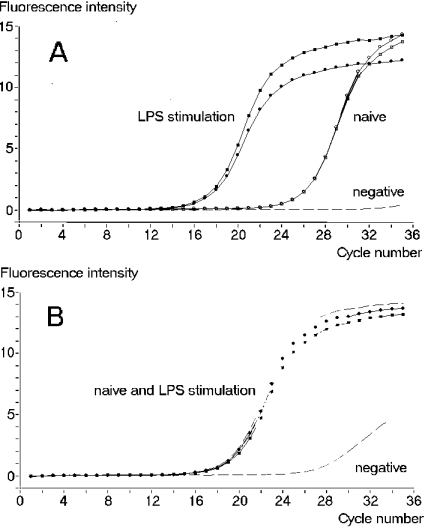
To confirm up-regulation of the IMPI gene in LPS-challenged larvae, real-time PCR (Light Cycler) was performed with IMPI-specific primers. As a control, primers specific for the housekeeping gene β-actin were used. The PCR kinetics of actin in untreated and LPS-stimulated larvae disclosed no significant differences in expression (Figure 2B), whereas IMPI mRNA exhibited far higher levels after LPS stimulation (Figure 2A). The difference of nine PCR cycles corresponds to a 500-fold increased amount of mRNA molecules in immunized animals compared with the control animals. The melting temperature plots affirmed the presence of single specific PCR products (results not shown). To distinguish between the effects of LPS injection and the effects of the injury caused by injection, control larvae were treated with distilled water without LPS. A slight up-regulation (15-fold) was observed after injection of water (results not shown). This limited immune response is typical for wounding caused by injection.
Figure 2. Relative quantification of IMPI cDNA in comparison with actin cDNA by real-time PCR.
(A) IMPI mRNA is up-regulated upon LPS stimulation, as demonstrated by the lower number of cycles needed to amplify IMPI. (B) PCR with β-actin-specific primers using the same cDNA preparations as in (A) indicates no difference in the total cDNA amount between the different probes.
IMPI protein
The amino acid sequence deduced from the cDNA contains a putative signal peptide of 18 amino acids [19,20] and a putative mature protein consisting of 152 amino acids with a calculated molecular mass of 16636 Da. The mature protein has two cysteine-rich stretches separated by the motif of a putative Arg-Arg cleavage site for SPCs (subtilisin-like proprotein convertases) (Figure 1).
In contrast with the putative mature IMPI protein deduced from the isolated cDNA, the native IMPI isolated from larval haemolymph is less than half the size, with a mass of 8364 Da [1]. Considering the previously determined sequence of 26 amino acids at the C-terminal part of the native protein, the mass of the protein without the glycosylation part (7679 Da), the presence of five disulphide bridges, the putative signal sequence (amino acids 1–18) and the novel identified SPC motif, the native IMPI protein represents the N-terminal domain of the mature protein (amino acids 19–88) and has a predicted length of 70 amino acids (Figure 1).
Searching with Pfam (http://www.sanger.ac.uk/Software/Pfam/) [21] for protein domains revealed significant identity with a TIL domain (trypsin inhibitor-like cysteine-rich domain). The similarity extends over 59 bp, starting at position 24, and is thus located in the first part of the protein between the N-terminus and the SPC cleavage site motif. A BLAST search displayed similarities with a score of more than 39 bits to mucin and to a hypothetical protein of Caenorhabditis elegans. In both cases, similarities to these proteins involve putative TIL domains.
Recombinant proteins
Two recombinant proteins were constructed based on the cDNA obtained: rIMPI-1 (amino acids 19–88), corresponding to the native IMPI, and rIMPI-2 (amino acids 19–170), which comprises the entire mature protein according to the IMPI mRNA (Figure 1). Both recombinant proteins were expressed in Drosophila Schneider 2 cells and purified by making use of the His-tag located at the C-terminus. The recombinant proteins were detected by Western blot analysis with antibodies targeted to the V5 epitope. Furthermore, we analysed the proteins by MS and examined their patterns of inhibition. To compare the inhibition kinetics of rIMPI-1 with those of native IMPI, both proteins were finally purified by affinity chromatography on a thermolysin–Sepharose column.
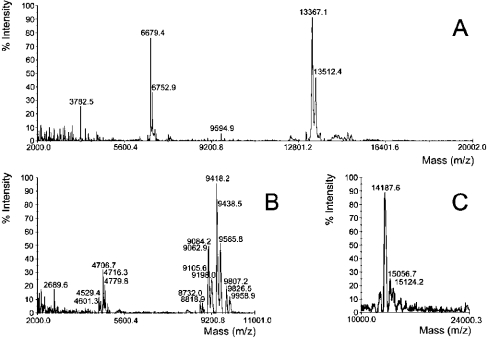
rIMPI-1 exhibited a molecular mass of 13367 Da (Figure 3A), which corresponds to the mass of the IMPI-derived sequence (7755 Da) plus the mass of the vector-derived sequences (4737 Da), notably the V5 epitope and the His-tag. The difference of 893 Da between the predicted and measured masses (Table 1) may be explained by glycosylation of the IMPI in Drosophila Schneider 2 cells in comparison with glycosylation of native IMPI in G. mellonella (685 Da). After a second purification step on a thermolysin–Sepharose column, the rIMPI-1 protein exhibited a main mass of 9418 Da and was not detectable by Western blot analysis with antibodies targeted to the V5 epitope. To discriminate between the recombinant proteins, we termed the latter purification product rIMPI-1*.
Figure 3. MALDI MS of rIMPI-1 (A), rIMPI-1* (B) and rIMPI-2 (C).
The rIMPI* was purified by affinity chromatography on a thermolysin–Sepharose column to remove the vector part from rIMPI-1 (see Figure 5). rIMPI-1 and rIMPI-2 were isolated from the supernatant of the Drosophila Schneider 2 cell culture medium by affinity chromatography using a chelating Sepharose matrix. For details, see the Materials and methods section.
Table 1. Comparison of obtained and expected molecular masses of native IMPI and rIMPIs.
The differences between the obtained and expected molecular masses are hypothetically due to distinct glycosylation in the Drosophila expression system. Therefore we include possible N- and O-glycosylation sites along the IMPI coding and vector-derived amino acid sequences
| Expected IMPI part | Expected vector part | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Molecular mass (Da) | Glycosylation sites | Glycosylation sites | |||||||
| Protein | Obtained | Expected | Difference | Mass (Da) | N | O | Mass (Da) | N | O |
| Native IMPI | 8364 | ||||||||
| rIMPI-1 | 13367 | 12474 | 893 | 7755 | 1 | 3 | 4737 | − | 7 |
| rIMPI-1* | 9418 | 8509 | 909 | 7755 | 1 | 3 | 772 | − | 1 |
| rIMPI-2 | 14188 | 21256 | −7068 | 16636 | 2 | 16 | 4638 | 1 | 7 |
| rIMPI-2 after SPC | 14188 | 12964 | 1224 | 8587 | 1 | 13 | 4395 | 1 | 6 |
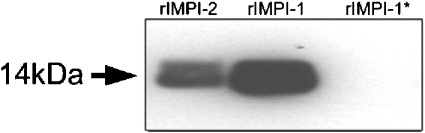
MS analysis of rIMPI-1* confirmed the removal of the V5 epitope on the thermolysin–Separose column (Figures 3B and 4). Among the possible sites for cleavage by thermolysin, the determined mass of rIMPI-1* matches best with the cleavage site illustrated in Figure 5.
Figure 4. Western blot analysis of rIMPIs.
Samples of 25 ng of rIMPI proteins were loaded on a 12% (w/v) polyacrylamide Bis-Tris gel in reducing loading buffer. After incubation with anti-V5 as primary antibody and goat anti-mouse Ig conjugated to horseradish peroxidase as secondary antibody, rIMPI-1* revealed no signal, in contrast with rIMPI-1 and rIMPI-2.
Figure 5. Possible cleavage sites for thermolysin along the vector part of rIMPI-1*.
The part of the sequence contributed by IMPI is in bold. Possible sites of cleavage by thermolysin are indicated by | (N-terminal sides of Ile, Leu, Val, Ala, Phe and Met). MS analysis of rIMPI-1* matches best with the cleavage indicated by ↓.
In contrast with our expectation, rIMPI-2 exhibited a main mass of 14188 Da (Figure 3C), which corresponds to approximately half the size of the predicted entire protein of 21256 Da plus a vector-derived sequence of 4638 Da (Table 1). Assuming post-translational processing of the expressed protein at the SPC cleavage site, the expected mass of rIMPI-2, hypothetically representing the C-terminal part of the full-length transcript, would be 12964 Da. Thus the relatively small difference between calculated and determined masses is rather consistent with SPC cleavage of the entire protein. As a hypothesis, we attribute the differences between predicted and measured masses of recombinant rIMPI-1, rIMPI-1* and rIMPI-2 to modified glycosylation of the recombinant proteins in the expression system used (Table 1).
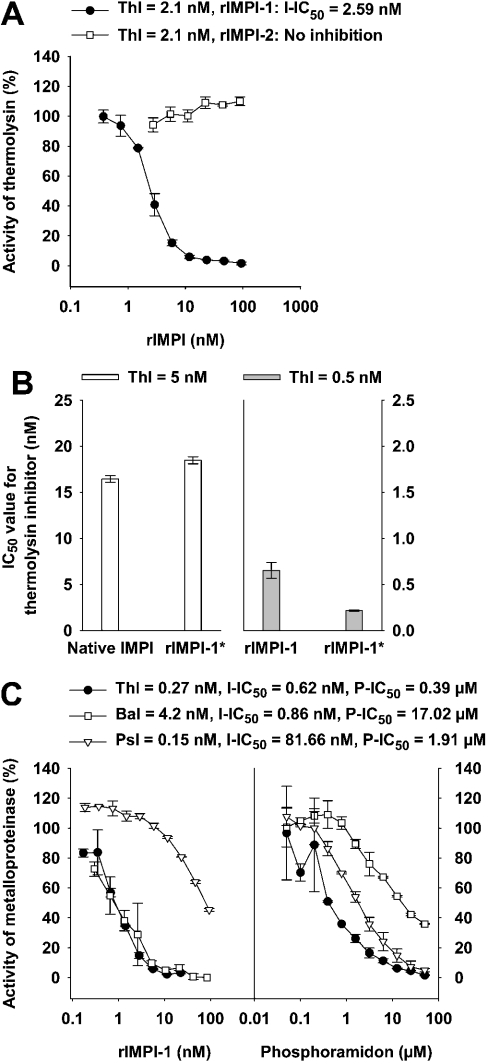
rIMPI-1/-1*, rIMPI-2 and native IMPI were analysed for their inhibitory activity against thermolysin using the MMP-1/MMP-9 substrate assay (assays I/II). rIMPI-1 was shown to inhibit bacterial thermolysin, whereas rIMPI-2 demonstrated no inhibitory activity (Figure 6A). Comparison of the inhibitory activities of native IMPI versus rIMPI-1* with truncated vector sequences revealed comparable IC50 values, whereas rIMPI-1, with a complete vector, demonstrated an approximately three times lower inhibitory activity (Figure 6B).
Figure 6. Inhibitory activity of native IMPI, rIMPI-1, rIMPI-1*, rIMPI-2 and phosphoramidon against bacterial metalloproteinases belonging to the gluzincin family.
For each assay, positive controls were performed and the proteinases were added at concentrations that provided similar proteolytic activity (Thl, thermolysin; Bal, bacillolysin; Psl, pseudolysin). The resulting proteinase activity was defined as 100%. The IC50 values for tested inhibitors (I-IC50 for IMPI and P-IC50 for phosphoramidon) were determined in the presence of inhibitors, which were serially diluted 3-fold and preincubated for 10 min with the proteinase before initiating the reaction by substrate addition. (A) Thermolysin activity was monitored in the presence of rIMPI-1 or rIMPI-2 using the MMP-1/MMP-9 substrate assay (assay I). (B) Comparison of the IC50 values for native IMPI and rIMPI-1* (both isolated on thermolysin–Sepharose columns), and comparison of IC50 values for rIMPI-1 (isolated on Ni2+-chelating Sepharose column) and rIMPI-1* (purified additionally on a thermolysin–Sepharose column) for thermolysin activity using the MMP-1/MMP-9 substrate assay (assay II). (C) Thermolysin, bacillolysin and pseudolysin activities were analysed in the presence of rIMPI-1 and phosphoramidon using the Azocoll assay.
Screening of inhibitory activity of rIMPI-1 against bacterial and human metalloproteinases
Using the Azocoll assay, we screened for inhibitory activity of rIMPI-1 against several bacterial metalloproteinases of the gluzincins, i.e. thermolysin, bacillolysin and pseudolysin. As shown in Figure 6(C), the IC50 values for rIMPI-1 were determined to be 0.62, 0.86 and 81.66 nM for thermolysin, bacillolysin and pseudolysin respectively. rIMPI-1 is therefore a potent inhibitor of all three bacterial metalloproteinases tested, with a preference for thermolysin and bacillolysin. Phosphoramidon, a highly specific inhibitor of thermolysin, exhibited the highest inhibitory activity against thermolysin, with an IC50 value of 0.39 μM at a given pH [22,23]; however, unlike rIMPI-1, it is a more potent inhibitor of pseudolysin than of bacillolysin (Figure 6C), suggesting different metalloproteinase specificities of the two inhibitors.
Furthermore, we screened for inhibitory activity of rIMPI-1 against MMP-1, MMP-2, MMP-3, MMP-7, MMP-8 and MMP-9 using the MMP-1/MMP-9 substrate assay (assay I). MMP-1 and MMP-3 in particular were inhibited by rIMPI-1 at test concentrations, but the IC50 values were more than 100 times higher than those of TIMP-2 (Table 2), which inhibits MMP-2 and MMP-9 in an equimolar manner with a preference for MMP-2 [24].
Table 2. IC50 values for rIMPI-1 and TIMP-2 on the activity of thermolysin and human MMPs.
Values are given as means±S.D. (n=3). The IC50 values for tested inhibitors were determined using either the Azocoll assay or the MMP-1/MMP-9 substrate assay. Details are described in the Materials and methods section.
| rIMPI-1 | TIMP-2 | ||||
|---|---|---|---|---|---|
| Proteinase | Proteinase concn (nM) | Concentration range (nM) | IC50 value (nM) | Concentration range (nM) | IC50 value (nM) |
| Thermolysin | 2.1 | 0–95 | 2.59±0.21 | 0–17.5 | − |
| MMP-1 | 1.1 | 0–750 | 461.65±2.01 | 0–17.5 | <1 |
| MMP-2 | 0.28 | 0–750 | − | 0–17.5 | <1 |
| MMP-3 | 4.55 | 0–750 | 664.92±13.91 | 0–17.5 | 4.01±0.21 |
| MMP-7 | 1 | 0–750 | − | 0–17.5 | 2.23±0.19 |
| MMP-8 | 1.7 | 0–750 | − | 0–17.5 | 3.81±0.34 |
| MMP-9 | 0.11 | 0–750 | − | 0–17.5 | 1.9±0.07 |
DISCUSSION
The past decade has witnessed a tremendous increase in literature about members of vertebrate MMP and TIMP families [2–4,25]. The first MMP and TIMP genes from insects were identified in Drosophila [5]; furthermore, in 1998 we identified an unusual inhibitor of metalloproteinases (IMPI) in larvae of the greater wax moth G. mellonella that has no similarity to TIMPs [1]. The present work focuses on the cloning and expression of the IMPI. Its cDNA was generated by suppression subtractive hybridization of LPS-challenged G. mellonella larvae. Using real-time PCR, marked up-regulation of IMPI mRNA was observed LPS-treated larvae; this corresponded to the increase in IMPI protein from G. mellonella in response to injected LPS [1].
Comparison of the IMPI protein based on the full-length cDNA with the structure and partially known amino acid sequence of the native IMPI protein showed that the native protein purified from haemolymph is less than half the size of the predicted full-length IMPI. Therefore we postulate that post-translational processing of the full-length protein occurs. Indeed, analysis of the deduced amino acids revealed a cleavage site for SPCs located approximately in the middle of the full-length protein. To examine both the properties of the shorter IMPI protein and the features of the full-length protein, we expressed two different IMPI proteins in Drosophila Schneider 2 cells: rIMPI-1 corresponding to native IMPI, and rIMPI-2 expected to represent the full-length protein.
MS analysis of rIMPI-2 resulted in the detection of a protein with a molecular mass of 14188 Da. This unexpected result implies that rIMPI-2 does not represent the predicted full-length protein with a calculated mass of 21256 Da, but rather a post-translationally modified protein. As a working hypothesis, we postulate post-translational processing of rIMPI-1 and rIMPI-2 in Drosophila Schneider 2 cells by SPC-mediated cleavage. In fact, insects are able to synthesize endogenous SPCs and to process synthesized precursor proteins at the corresponding cleavage sites. Homologues of mammalian furin [26,27] and amontillado, which corresponds to the mammalian PC2 protein [28,29], have been identified in Drosophila. In particular, Drosophila Schneider 2 cells are reportedly capable of proprotein processing by SPC-like enzymes [15]. The SPCs are an evolutionarily conserved family of serine proteinases that catalyse the maturation of a strikingly diverse group of propeptide substrates [30,31]. Proteolytic processing of biologically active proteins by convertases can occur intracellularly, at the cell surface or extracellularly, and allows tight post-transcriptional regulation of their temporal and spatial activities [31]. However, the postulated SPC-mediated processing of the IMPI in G. mellonella in vivo and by Drosophila Schneider 2 cells in vitro remains speculative until cleavage at this site has been proven.
MS analysis of rIMPI-1 and rIMPI-1*, which represent the native IMPI and thus the N-terminal domain of the full-length protein, revealed a mass difference of approx. 4 kDa. The determined mass difference between rIMPI-1 and rIMPI-1* obtained after thermolysin–Sepharose column preparation, and the observation that rIMPI-1* was not detectable by Western blot analysis, indicates a nearly complete removal of vector-derived sequences by thermolysin-mediated cleavage. The three-dimensional structure of the IMPI may account for its resistance to thermolysin-mediated cleavage, whereas the vector part, without an inhibitory function, is obviously sensitive. Among the possible thermolysin cleavage sites along the vector part, the detected mass of IMPI-1* matches best with the removal of the vector-derived sequence at the position illustrated in Figure 5.
In fact, rIMPI-1* with truncated vector residues, and thus best matched in size and structure with native IMPI, exhibits inhibitory activity against thermolysin similar to that of the purified native protein. Comparison of rIMPI-1 (with the complete vector sequences) and rIMPI-1* (with a truncated vector part) shows a slightly higher IC50 value for rIMPI-1. This might be explained by a sub-optimal steric conformation due to the appended vector sequences (V5 epitope and His-tag).
Among the metalloproteinases tested, the IMPI was active against several bacterial metalloproteinases of the M4 family of clan MA(E), the gluzincins. The low IC50 values against members of the M4 family indicate that the IMPI is a good inhibitor of these metalloproteinases. This family of metalloproteinases is known only from bacteria, including human pathogens such as Legionella, Listeria, Clostridium, Helciobacter, Pseudomonas and Vibrio [32]. Metalloproteinases produced by human pathogenic bacteria act as synergistic virulence factors and promote pathogenesis in a multifaceted manner. Targeted inhibition of microbial metalloproteinases has therefore become a novel strategy in the therapy of microbial diseases [33]. The IMPI inhibits pseudolysin from Pseundomonas aeruginosa, which has been shown to cause tissue damage by degrading collagens, elastin and fibronectin [34].
In contrast with the specific inhibition of the gluzincin enzymes, native and rIMPIs exhibited no, or negligible, inhibitory activity against MMPs of clan MA(M) (metzincins). In agreement with the non-TIMP-like sequence and low molecular mass of the IMPI, it does not represent a specific MMP inhibitor, in the way that TIMPs do.
Using the full-length sequence of the IMPI, a BLAST search in the NCBI databases and a search with the domain search algorithm Pfam showed no sequence similarity with other natural metalloproteinase inhibitors. The IMPI-encoding sequence does not share similarity either with those of vertebrate TIMPs or with that of their putative homologue identified in Drosophila melanogaster [5]. The IMPI also possesses no similarities to SMPI, a small peptidic inhibitor of the gluzincins from Streptomyces nigrescens [35]. To date, no such inhibitor with specificity against microbial metalloproteinases has been reported for both vertebrates and invertebrates.
Interestingly, searching for conserved protein domains with the Pfam algorithm revealed similarity of the IMPI to the TIL domain. This domain is present in trypsin inhibitors, as well as in many extracellular proteins. Like the IMPI, the trypsin inhibitors are small proteins, and have a single active domain of 10 cysteine residues building five disulphide bridges [36]. Both inhibitors are highly resistant to proteolysis and heat denaturation [37]. However, the IMPI inhibits not the serine proteinase trypsin [1], but bacterial metalloproteinases. The demonstrated similarities of the IMPI and the trypsin inhibitors indicate that they may have a common ancestor. Whether the IMPI has evolved from a trypsin inhibitor by changes in the reactive site or from other extracellular proteins remains speculative.
Regarding the putative role of the IMPI in G. mellonella, we postulate a contribution to the humoral immune response by inhibition of metalloproteinases associated with entomopathogens. Viruses, bacteria and fungi that infect and kill insects have been reported to produce thermolysin-like metalloproteinases that play an essential role as virulence factors promoting pathogen development within the infected host. There are several well demonstrated examples, which support our assumption. Enhancin is a metalloproteinase which has been shown to facilitate Nucleopolyhedrovirus infection in insects [38,39]. Bacillus thuringiensis has a M4 peptidase member that enables the pathogen to penetrate the gut and thereby enter the insect body. The entomopathogenic fungus Metarhizium anisopliae produces a thermolysin-like metalloproteinase which functions as a back-up system in case its other proteolytic enzymes are inactivated by endogenous inhibitors of the infected host [40]. Proteinases released by this pathogenic fungus have been reported to impair the cellular immune response in G. mellonella [41]. However, this insect is not defenceless against toxic microbial proteinases. Inducible inhibitors against fungal proteinases within the haemolymph of G. mellonella can delay fungal development in vivo and in vitro [42]. Since entomopathogenic bacteria and fungi utilize a number of different proteinases that promote infection and virulence, the IMPI alone cannot provide immunity for G. mellonella against pathogens. Immune-related genomics and proteomics studies in this insect have resulted in the discovery of a number of antimicrobial proteins and proteinase inhibitors that contribute synergistically to its innate immunity [8–10,14,41].
In conclusion, the IMPI represents a new natural inhibitor of the gluzincin metalloproteinases. It is part of the humoral immune response of G. mellonella, and is up-regulated at the transcriptional level after bacterial challenge. As the IMPI inhibits several microbial metalloproteinases, it has a potential role as a template for therapeutics against human pathogenic bacteria.
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft, Collaborative Research Center 506, Recombinant Proteins and Nucleic Acids for tumour therapy to A.V.
References
- 1.Wedde M., Weise C., Kopacek P., Franke P., Vilcinskas A. Purification and characterization of an inducible metalloprotease inhibitor from the hemolymph of greater wax moth larvae, Galleria mellonella. Eur. J. Biochem. 1998;255:535–543. doi: 10.1046/j.1432-1327.1998.2550535.x. [DOI] [PubMed] [Google Scholar]
- 2.Nagase H., Woessner J. F., Jr Matrix metalloproteinases. J. Biol. Chem. 1999;274:21491–21494. doi: 10.1074/jbc.274.31.21491. [DOI] [PubMed] [Google Scholar]
- 3.Brew K., Dinakarpandian D., Nagase H. Tissue inhibitors of metalloproteinases: evolution, structure and function. Biochim. Biophys. Acta. 2000;1477:267–283. doi: 10.1016/s0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- 4.Vu T. H., Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev. 2000;14:2123–2133. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- 5.Pohar N., Godenschwege T. A., Buchner E. Invertebrate tissue inhibitor of metalloproteinase: structure and nested gene organization within the synapsin locus is conserved from Drosophila to human. Genomics. 1999;57:293–296. doi: 10.1006/geno.1999.5776. [DOI] [PubMed] [Google Scholar]
- 6.Wei S., Xie Z., Filenova E., Brew K. Drosophila TIMP is a potent inhibitor of MMPs and TACE: Similarities in structure and function to TIMP-3. Biochemistry. 2003;42:12200–12207. doi: 10.1021/bi035358x. [DOI] [PubMed] [Google Scholar]
- 7.Page-McCaw A., Serano J., Sante J. M., Rubin G. M. Drosophila matrix metalloproteinases are required for tissue remodeling, but not embryonic development. Dev. Cell. 2003;4:95–106. doi: 10.1016/s1534-5807(02)00400-8. [DOI] [PubMed] [Google Scholar]
- 8.Frobius A. C., Kanost M. R., Gotz P., Vilcinskas A. Isolation and characterization of novel inducible serine protease inhibitors from larval hemolymph of the greater wax moth Galleria mellonella. Eur. J. Biochem. 2000;267:2046–2053. doi: 10.1046/j.1432-1327.2000.01207.x. [DOI] [PubMed] [Google Scholar]
- 9.Vilcinskas A., Götz P. Entomopathogenic fungi and the insect immune system. Adv. Parasitol. 2003;43:267–313. [Google Scholar]
- 10.Gillespie J. P., Bailey A. M., Cobb B., Vilcinskas A. Fungi as elicitors of insect immune responses. Arch. Insect Biochem. Physiol. 2000;44:49–68. doi: 10.1002/1520-6327(200006)44:2<49::AID-ARCH1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 11.Griesch J., Wedde M., Vilcinskas A. Recognition and regulation of metalloproteinase activity in the haemolymph of Galleria mellonella: a new pathway mediating induction of humoral immune responses. Insect Biochem. Mol. Biol. 2000;30:461–472. doi: 10.1016/s0965-1748(00)00010-2. [DOI] [PubMed] [Google Scholar]
- 12.Diatchenko L., Lau Y. F., Campbell A. P., Chenchik A., Moqadam F., Huang B., Lukyanov S., Lukyanov K., Gurskaya N., Sverdlov E. D., Siebert P. D. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. U.S.A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurskaya N. G., Diatchenko L., Chenchik A., Siebert P. D., Khaspekov G. L., Lukyanov K. A., Vagner L. L., Ermolaeva O. D., Lukyanov S. A., Sverdlov E. D. Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemaglutinin and phorbol 12-myristate 13-acetate. Anal. Biochem. 1996;240:90–97. doi: 10.1006/abio.1996.0334. [DOI] [PubMed] [Google Scholar]
- 14.Seitz V., Clermont A., Wedde M., Hummel M., Vilcinskas A., Schlatterer K., Podsiadlowski L. Identification of immunorelevant genes from greater wax moth (Galleria mellonella) by a subtractive hybridization approach. Dev. Comp. Immunol. 2003;27:207–215. doi: 10.1016/s0145-305x(02)00097-6. [DOI] [PubMed] [Google Scholar]
- 15.Denault J. B., Lazure C., Day R., Leduc R. Comparative characterization of two forms of recombinant human SPC1 secreted from Schneider 2 cells. Protein Expression Purif. 2000;19:113–124. doi: 10.1006/prep.2000.1215. [DOI] [PubMed] [Google Scholar]
- 16.White J. S., White D. C. Boca Raton, FL: CRC Press; 1997. Source Book of Enzymes. [Google Scholar]
- 17.Rawlings N. D., O'Brien E. A., Barrett A. J. MEROPS: the protease database. Nucleic Acids Res. 2002;30:343–346. doi: 10.1093/nar/30.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bickett D. M., Green M. D., Berman J., Dezube M., Howe A. S., Brown P. J., Roth J. T., McGeehan G. M. A high throughput fluorogenic substrate for interstitial collagenase (MMP-1) and gelatinase (MMP-9) Anal. Biochem. 1993;212:58–64. doi: 10.1006/abio.1993.1291. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen H., Krogh A. Prediction of signal peptides and signal anchors by a hidden Markov model. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998;6:122–130. [PubMed] [Google Scholar]
- 21.Bateman A., Birney E., Cerruti L., Durbin R., Etwiller L., Eddy S. R., Griffiths-Jones S., Howe K. L., Marshall M., Sonnhammer E. L. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitagishi K., Hiromi K. Binding between thermolysin and its specific inhibitor, phosphoramidon. J. Biochem. (Tokyo) 1984;95:529–534. doi: 10.1093/oxfordjournals.jbchem.a134635. [DOI] [PubMed] [Google Scholar]
- 23.Indig F. E., Ben Meir D., Spungin A., Blumberg S. Investigation of neutral endopeptidases (EC 3.4.24.11) and of neutral proteinases (EC 3.4.24.4) using a new sensitive two-stage enzymatic reaction. FEBS Lett. 1989;255:237–240. doi: 10.1016/0014-5793(89)81098-1. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y. E., Wang M., Greene J., Su J., Ullrich S., Li H., Sheng S., Alexander P., Sang Q. A., Shi Y. E. Preparation and characterization of recombinant tissue inhibitor of metalloproteinase 4 (TIMP-4) J. Biol. Chem. 1997;272:20479–20483. doi: 10.1074/jbc.272.33.20479. [DOI] [PubMed] [Google Scholar]
- 25.MaCawley L., Matrisian L. Matrix metalloproteinases: they're not just for matrix anymore. Curr. Opin. Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 26.De Bie I., Savaria D., Roebroek A. J., Day R., Lazure C., Van de Ven W. J., Seidah N. G. Processing specificity and biosynthesis of the Drosophila melanogaster convertases dfurin1, dfurin1-CRR, dfurin1-X, and dfurin2. J. Biol. Chem. 1995;270:1020–1028. doi: 10.1074/jbc.270.3.1020. [DOI] [PubMed] [Google Scholar]
- 27.Roebroek A. J., Creemers J. W., Pauli I. G., Kurzik-Dumke U., Rentrop M., Gateff E. A., Leunissen J. A., Van de Ven W. J. Cloning and functional expression of Dfurin2, a subtilisin-like proprotein processing enzyme of Drosophila melanogaster with multiple repeats of a cysteine motif. J. Biol. Chem. 1992;267:17208–17215. [PubMed] [Google Scholar]
- 28.Rayburn L. Y., Gooding H. C., Choksi S. P., Maloney D., Kidd A. R., III, Siekhaus D. E., Bender M. Amontillado, the Drosophila homolog of the prohormone processing protease PC2, is required during embryogenesis and early larval development. Genetics. 2003;163:227–237. doi: 10.1093/genetics/163.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siekhaus D. E., Fuller R. S. A role for amontillado, the Drosophila homolog of the neuropeptide precursor processing protease PC2, in triggering hatching behavior. J. Neurosci. 1999;19:6942–6954. doi: 10.1523/JNEUROSCI.19-16-06942.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergeron F., Leduc R., Day R. Subtilase-like pro-protein convertases: from molecular specificity to therapeutic applications. J. Mol. Endocrinol. 2000;24:1–22. doi: 10.1677/jme.0.0240001. [DOI] [PubMed] [Google Scholar]
- 31.Seidah N. G., Chretien M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45–62. doi: 10.1016/s0006-8993(99)01909-5. [DOI] [PubMed] [Google Scholar]
- 32.Barrett A. J., Rawlings N. D., Woessner J. F., Jr . San Diego, CA: Academic Press; 1998. Handbook of Proteolytic Enzymes. [Google Scholar]
- 33.Mock M., Roques B. P. Progress in rapid screening of Bacillus anthracis lethal factor activity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6527–6529. doi: 10.1073/pnas.112220599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyoshi S., Shinoda S. Microbial metalloproteases and pathogenesis. Microbes Infect. 2000;2:91–98. doi: 10.1016/s1286-4579(00)00280-x. [DOI] [PubMed] [Google Scholar]
- 35.Ohno A., Tate S., Seeram S. S., Hiraga K., Swindells M. B., Oda K., Kainosho M. NMR structure of the Streptomyces metalloproteinase inhibitor, SMPI, isolated from Streptomyces nigrescens TK-23: another example of an ancestral beta gamma-crystallin precursor structure. J. Mol. Biol. 1998;282:421–433. doi: 10.1006/jmbi.1998.2022. [DOI] [PubMed] [Google Scholar]
- 36.Huang K., Strynadka N. C., Bernard V. D., Peanasky R. J., James M. N. The molecular structure of the complex of Ascaris chymotrypsin/elastase inhibitor with porcine elastase. Structure. 1994;2:679–689. doi: 10.1016/s0969-2126(00)00068-x. [DOI] [PubMed] [Google Scholar]
- 37.Grasberger B. L., Clore G. M., Gronenborn A. M. High-resolution structure of Ascaris trypsin inhibitor in solution: direct evidence for a pH-induced conformational transition in the reactive site. Structure. 1994;2:669–678. doi: 10.1016/s0969-2126(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 38.Lepore L. S., Roelvink P. R., Granados R. R. Enhancin, the granulosis virus protein that facilitates nucleopolyhedrovirus (NPV) infections, is a metalloprotease. J. Invertebrate Pathol. 1996;68:131–140. doi: 10.1006/jipa.1996.0070. [DOI] [PubMed] [Google Scholar]
- 39.Wang P., Granados R. R. An intestinal mucin is the target substrate for a baculovirus enhancin. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6977–6982. doi: 10.1073/pnas.94.13.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.St Leger R. J., Bidochka M. J., Roberts D. W. Characterization of a novel carboxypeptidase produced by the entomopathogenic fungus Metarhizium anisopliae. Arch. Biochem. Biophys. 1994;314:392–398. doi: 10.1006/abbi.1994.1458. [DOI] [PubMed] [Google Scholar]
- 41.Griesch J., Vilcinskas A. Proteases released by entomopathogenic fungi impair phagocytic activity, attachment and spreading of plasmatocytes isolated from haemolymph of the greater wax moth Galleria mellonella. Biocontrol Sci. Technol. 1998;8:517–531. [Google Scholar]
- 42.Vilcinskas A., Wedde M. Inhibition of Beauveria bassiana proteases and fungal development by inducible protease inhibitors in the haemolymph of Galleria mellonella larvae. Biocontrol Sci. Technol. 1997;7:591–601. [Google Scholar]