Abstract
Leigh syndrome French Canadian (LSFC) is a variant of cytochrome oxidase deficiency found in Québec and caused by mutations in the LRPPRC (leucine-rich pentatricopeptide repeat cassette) gene. Northern blots showed that the LRPPRC mRNA levels seen in skeletal muscle>heart>placenta>kidney>liver>lung=brain were proportionally almost opposite in strength to the severity of the enzymic cytochrome oxidase defect. The levels of COX (cytochrome c oxidase) I and COX III mRNA visible on Northern blots were reduced in LSFC patients due to the common (A354V, Ala354→Val) founder mutation. The amount of LRPPRC protein found in both fibroblast and liver mitochondria from LSFC patients was consistently reduced to <30% of control levels. Import of [35S]methionine LRPPRC into rat liver mitochondria was slower for the mutant (A354V) protein. A titre of LRPPRC protein was also found in nuclear fractions that could not be easily accounted for by mitochondrial contamination. [35S]Methionine labelling of mitochondrial translation products showed that the translation of COX I, and perhaps COX III, was specifically reduced in the presence of the mutation. These results suggest that the gene product of LRPPRC, like PET 309p, has a role in the translation or stability of the mRNA for mitochondrially encoded COX subunits. A more diffuse distribution of LRPPRC in LSFC cells compared with controls was evident when viewed by immunofluorescence microscopy, with less LRPPRC present in peripheral mitochondria.
Keywords: cytochrome c oxidase I (COX I) and COX III mRNA, cytochrome oxidase, Leigh syndrome, leucine-rich pentatricopeptide repeat cassette (LRPPRC) protein, mitochondria
Abbreviations: COX, cytochrome c oxidase; DAPI, 4′,6′-diamidino-2-phenylindole; DTT, dithiothreitol; LRPPRC, leucine-rich pentatricopeptide repeat cassette; LSFC, Leigh syndrome French Canadian; NP40, Nonidet P40
INTRODUCTION
The LRPPRC (leucine-rich pentatricopeptide repeat cassette) motif containing LRPPRC protein encoded on chromosome 2 is a distant homologue of the yeast protein PET 309p, a protein required for efficient expression of the COX (cytochrome c oxidase) complex [1–3]. The LRPPRC gene was identified as causative for the LSFC (Leigh syndrome French Canadian variant, MIM220111) due to a founder mutation (C1119→T transition) in exon 9 predicting a missense A354V (Ala354→Val) change at a residue conserved in the mouse and human [3].
The LRPPRC protein exhibits the characteristics of a mitochondrial protein, possessing a cleavable leader sequence and a series of pentatricopeptide tandem repeats [4,5]. The function of this PET 309p homologue together with the presence of PPR repeats strongly suggests that this protein is an mRNA-binding protein [4–6]. Although an SEC domain in the C-terminal half of the protein has been found to be capable of interacting with a number of proteins, including fibronectin, a microtubule-associated protein and three proteins of unknown function, none of these appear to be mitochondrial [5]. Nuclear, cytoplasmic and mitochondrial localizations have been reported for LRPPRC protein with a preference for binding single-stranded polypyrimidine RNA [7,8]. Homologues of the LRPPRC protein in other organisms, besides yeast, are the bicoid stability factor of Drosophila melanogaster and a salt-induced protein in Arabidopsis thaliana [6]. Homologues of the PET 309 gene in lower organisms have been described, notably cya5 in Neurospora crassa and crpI in Zea mays [9,10]. The latter is necessary for the translation of petA mRNA in chloroplasts so that a failure of cytochrome f synthesis results. A majority of CRP1 protein was found to be soluble in chloroplasts and not highly associated with either chloroplast membranes or ribosomes [11]. The bicoid stability factor was isolated as a protein that contributed to the stability of the maternally derived bicoid mRNA, which encodes a protein responsible for the organization of the anterior body pattern in the embryo of Drosophila [11]. Whereas this protein exhibited a punctate immunostaining reminiscent of mitochondrial distribution, it was shown to bind elements of the bicoid mRNA in vitro, which is cytosolic in distribution. This bicoid stability factor protein (1355 amino acids) is of a similar size to LRPPRC (1361 amino acids) but both are larger than PET 309p (966 amino acids). Since much of the bicoid stability factor and LRPPRC are repeat structures, the length comparison suggests that no vital conserved motif is absent from the shorter PET 309p.
The LSFC variant of COX deficiency is unusual in that it has a phenotype that is characterized by a less severe neurological picture, which is slower in progression and less severe than is seen in other LSFC patients [12,13]. To our present knowledge, it occurs only in the Saguenay and Charlevoix regions of Québec, and the biochemical profile of the COX defect is such that liver and brain are affected more severely than heart, kidney and skeletal muscles [12,13].
In the present study, we show that skin fibroblast cell lines bearing the (A354V) mutation in LRPPC have a very reduced level of LRPPRC protein expression in mitochondria. This in turn is associated with lower detectable levels of the COX mRNA transcripts derived from mitochondrial DNA.
MATERIALS AND METHODS
Tissue culture
All fibroblast cell lines were grown in Dulbecco's modified Eagle's medium with glucose (4.5 g/l), glutamine (2.5 mM), penicillin (100 μg/ml) and streptomycin (100 μg/ml) at 37 °C with 5% CO2.
Isolation of mitochondria
Fibroblast cells were harvested by trypsin and washed twice with ice-cold PBS, and the mitochondria were prepared as described previously [14]. Human liver mitochondria were prepared from autopsy samples obtained with informed consent. Briefly, a 0.5 g of liver sample was minced and homogenized with a polytron homogenizer in mitochondrial preparation buffer [0.34 M sucrose, 100 mM KCl, 10 mM Tris (pH 7.4) and 1 mM EDTA]. After the removal of debris by centrifugation at 1023 g for 10 min, the mitochondrial fraction was prepared as described above [15].
Nuclear protein preparation
Fibroblast cells (10 millions) were suspended in a low-salt NP40 (Nonidet P40) buffer [10 mM Hepes (pH 7.9), 10 mM KCl, 1.5 mM MgCl2, 0.1% NP40 and 0.5 mM DTT (dithiothreitol) plus a cocktail of proteinase inhibitors] and incubated on ice for 5 min to break the cell membrane. After 1 min of vortex-mixing to strip off the cytoplasmic and organellar fractions, the nuclei in the insoluble fraction were pelleted by centrifuging and washed twice with the same buffer and then suspended in 50 μl of high-salt nuclear extraction buffer [20 mM Hepes (pH 7.9), 420 mM NaCl, 1.5 mM MgCl2, 25% glycerol and 0.5 mM DTT plus a cocktail of proteinase inhibitors]. After rocking vigorously at 40 °C for 15 min on a shaking platform, the nuclear protein in the insoluble fraction was collected by centrifugation at 23426 g for 10 min.
Northern-blot analysis
A filter containing RNA samples from different tissues was purchased directly from Clontech (Palo Alto, CA, U.S.A.), whereas the filter containing RNA samples from both the normal and the patient was prepared as follows. Briefly, total RNA was isolated from fibroblast cell lines by using an RNA Easy kit from Qiagen (Mississauga, ON, Canada). RNA samples (25 μg) were separated on 2% (v/v) formaldehyde–agarose gels, UV cross-linked and probed with cDNAs of LRPPRC, COX I, COX II, COX III, ND1 and β-actin [10,11]. Equal amounts of DNA (250 ng) were labelled as probes using Rediprime II Random labelling kits with 32P-α-dCTP (Amersham Biosciences, Little Chalfont, Bucks., U.K.). Hybridization was performed by using ExpressHyb hybridization solution from Clontech in a rotary oven. All hybridization and washing procedures were performed at 68 °C. The washed filter was used to expose an X-ray film at −80 °C. For reprobing, the same RNA filters were stripped in diethyl pyrocarbonate-water containing 1% SDS at 95 °C for 20 min. Stripping efficiency was confirmed by exposing to an X-ray film overnight. Reblotting was performed as described above.
Antibodies and Western blot
Anti-human complex I (49 kDa subunit) and LRPPRC antibodies were prepared by immunizing rabbits with peptides corresponding to the C-terminal 14 amino acids of the protein. Anti-human histone H1 antibody was purchased from Santa Cruz Biotechnologies (Santa Cruz, CA, U.S.A.). Protein samples were prepared either by lysis of isolated mitochondria or by diluting the nuclear extract in 1% NP40 lysis buffer [50 mM Tris (pH 8.0), 100 mM NaCl, 10 mM NaF, 0.5 mM DTT, 0.1 mM EDTA, 1 mM Na3VO4 and a cocktail of proteinase inhibitors]: 15 μg of protein for each sample was used and SDS/PAGE was performed as described previously, followed by transfer to nitrocellulose membranes [13–15]. Immunoblotting was performed as described previously with the secondary antibody goat anti-rabbit IgG conjugated with horseradish peroxidase, followed by development with ECL® solution (Amersham Biosciences) [16].
35S-labelling of mitochondrial translation products
Labelling was carried out by the method of Chomyn [17]. Cultured fibroblasts were passaged for 48 h before pulse labelling in the presence of emetine.
Mitochondria were then isolated as described above, and mitochondrial protein were separated by electrophoresis on SDS/polyacrylamide gels (10–20% linear gradient). After electrophoresis, the separated samples were transferred on to a nitrocellulose membrane and exposed to an X-ray film at room temperature (22 °C) for 2 days [17].
Import of 35S-labelled LRPPRC into rat liver mitochondria
Full-length LRPPRC cDNA was amplified by reverse transcriptase–PCR, subcloned into TA cloning vector (Invitrogen) and sequenced. The vectors were linearized, purified and quantified. The linearized vector (10 μg) was used to generate [35S]methionine-labelled protein via transcription and translation in vitro by using TNT T7 coupled reticulocyte kit (Promega) according to the manufacturer's instructions. Label efficiency was determined on an SDS/10% polyacrylamide gel. Import reaction was initiated by adding 50 μl of the translated products in equal volume of freshly isolated rat liver mitochondria (100 μg) in 2× import buffer [20 mM Hepes/KOH(pH 7.5), 1 mM EGTA, 140 mM sucrose, 440 mM mannitol, 66 mM glutamate and 4 mg/ml fatty acid-free BSA] at 300 °C. The reaction was terminated at various times as required (10–60 min) by adding a 10-fold volume of ice-cold import buffer and quickly centrifuging at 23000 g for 10 min at 40 °C to remove all un-imported protein. After another washing, the pellet was heated in 50 μl of SDS loading buffer at 95 °C for 8 min and 25 μl of each sample was loaded and separated on an SDS/10% polyacrylamide gel. After fixing and drying, the gel was exposed to an X-ray film.
Immunofluorescence staining of cells
Mito-tracker red fluorescent protein (Mito-RFP) vector was purchased from Clontech and used to transfect cells from both the normal individual and the patient using SuperFect Transfection reagent from Qiagen. After 2 weeks of selection in G418 medium, the cells were seeded in lab-tek chamber slides (Nalge Nunc International, Rochester, NY, U.S.A.) and cultured overnight. The cells were then fixed, permeabilized and immunostained by using Cytofix/Cytoperm kit from PharMingen (Mississauga, ON, Canada). Briefly, the cells were washed twice with ice-cold PBS and fixed in Cytofix/Cytoperm solution at 4 °C for 20 min. The fixed cells were washed twice with Perm/Wash solution and then incubated in 1:1000 diluted goat serum for 30 min with constant rocking to block non-specific binding sites. After removal of the blocking solution, 1:1000 diluted anti-LRPPRC antibody was added and incubated for 1 h. The cells were intensively washed three times with Perm/Wash buffer and then incubated with FITC-conjugated goat ant-rabbit antibody for another hour. Unbound secondary antibody was removed by repeated washing. After draining away any water remaining on the slide at room temperature in the dark, the wet slide was mounted in SlowFade Antifade with DAPI (4,6-diamidino-2-phenylindole) solution (Molecular Probes). DAPI-stained nuclei, mito-RFP-labelled mitochondria and immunostained LRPPRC were imaged using a confocal fluorescence microscope with appropriate light filters.
RESULTS
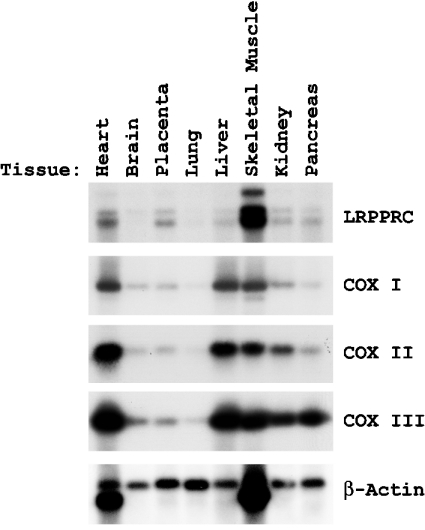
The presence of a homozygous mutation (C1119→T transition) in the LRPPRC cDNA in the majority of the LSFC patients produces a situation where the level of measurable cytochrome oxidase activity in fibroblasts is 50%, the level in liver and brain is 20% or less, but the level in kidney, skeletal muscles and heart is close to normal [12,13]. We performed Northern blotting for LRPPRC mRNA using a variety of human tissue RNA preparations (Figure 1). It is clear that the LRPPRC mRNA level is highest in skeletal muscles>heart>placenta>kidney>liver>lung=brain, reciprocal/opposite almost to the location of the enzymic COX defect in LSFC-COX deficiency and the organs showing significant pathology, i.e. brain and liver. It is also perhaps of interest that patients with LSFC-COX deficiency who die early often enter a phase of decompensation with metabolic acidosis and pulmonary oedema, a situation which may implicate lung as an organ with a metabolic problem, i.e. correlating with low expression of LRPPRC mRNA [12]. There appears to be three transcripts in all tissues, one of 7 kb and the others 5.0 and 4.7 kb respectively (Figure 1). The significance of these three transcripts at present is not clear. Separate Northern blots with probes to the 5′- and 3′-end respectively of the LRPPRC cDNA also displayed three bands (results not shown).
Figure 1. Northern blots to analyse transcript levels of LRPPRC and COX subunits in different tissues.
Tissue-specific transcription analysis of LRPPRC by Northern blotting. The filter containing different tissue mRNAs was purchased from Clontech and hybridized with 32P-labelled cDNA probes for LRPPRC, COX I, COX II, COX III and β-actin. The filter was exposed to an X-ray film to reveal the comparative labelling of the RNA species.
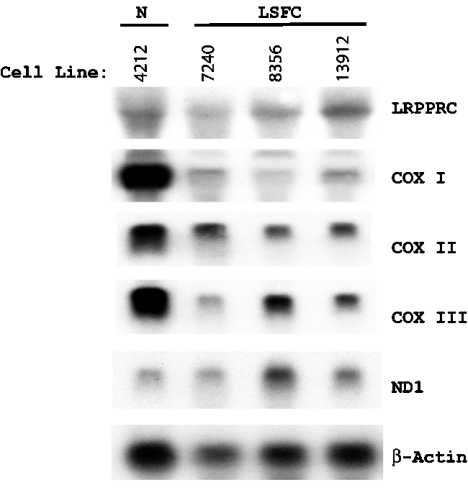
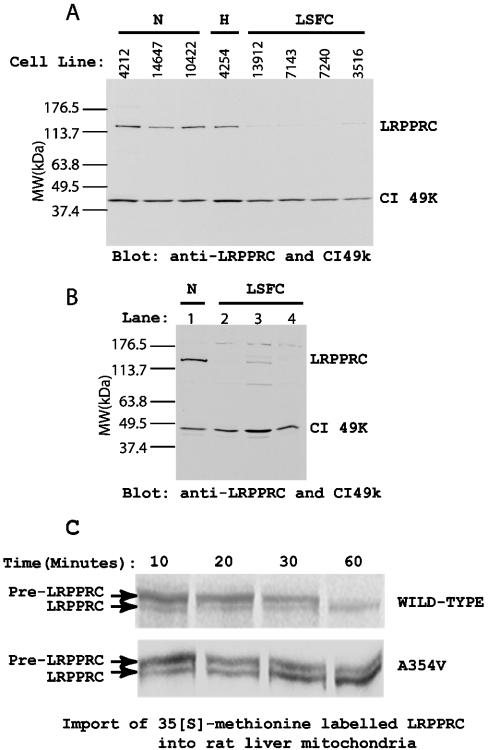
The mRNA expression pattern of mitochondrially encoded COX subunits I, II and III investigated by Northern blotting showed a pattern with the expression order being heart>liver>skeletal muscle>kidney>brain=placenta>lung (Figure 1). The effects of the LSFC mutation on the levels of LRPPRC and COX subunit mRNAs were then examined in fibroblasts from patients by Northern blotting to compare with control samples (Figure 2). Expressions of COX I and COX III were decreased in comparison with the level of β-actin mRNA, a nuclear-encoded cytoskeletal component, and NDI, an mtDNA-encoded transcript for complex I. Ratios of the expressed transcript by densitometry were 21.8, 47.0 and 30.1% for COX I, COX II and COX III transcripts with respect to control when actin transcript density was used as a baseline. Cardiac and skeletal muscles show extra bands at 1.6 kb for actin due to cross-reactivity of the β-actin (ACTB) probe with the myofibrillar form of actin, which is ignored in the calculations [18]. Comparison of transcript density showed that COX I and COX III mRNAs, in particular, were decreased in cells from patients. Immunoblotting showed that LRPPRC protein was greatly decreased in mitochondria by the presence of the mutation to <25% of the wild-type level in both fibroblasts and liver (Figures 3A and 3B). Compared with the 49 kDa subunit of complex I, the level was undetectable in two out of three samples of liver obtained at autopsy. To investigate import and processing, [35S]methionine-labelled LRPPRC protein was prepared by translation in a reticulocyte lysate and incubated with freshly isolated rat liver mitochondria. After gel separation, the labelled pre-LRPPRC precursor protein decreased and an LRPPRC band of lesser molecular mass became dominant over time (Figure 3C). When an A354V mutant transcript was translated, the processing to this lower molecular mass product was slower, indicating decreased import and cleavage of the mitochondrial targeting sequence.
Figure 2. Northern blots to analyse the transcript level of LRPPRC and COX subunits in cultured skin fibroblasts.
The same filter was probed in succession with 32P-labelled cDNA probes for LRPPRC, COX I, COX II, COX III, ND1 and β-actin. Lane 1 is the control cell line 4212. Lanes 2, 3 and 4 are cell lines 7240, 8356 and 13912 from LSFC patients respectively. The experimental procedure is outlined in the Materials and methods section.
Figure 3. Western-blot analysis of LRPPRC contents of skin fibroblasts and liver mitochondria from LSFC patients and controls.
The mitochondrial fraction was prepared by differential centrifugation and separated on an SDS/8% polyacrylamide gel. The separated samples were transferred on to a nitrocellulose membrane and blotted with antibodies to LRPPRC and the 49 kDa subunit of complex I (CI49k). After washing to remove unbound primary antibodies, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG and developed with ECL® solution. (A) LRPPRC content in fibroblast cell mitochondria. N, normal cells; H, heterozygous cells; LSFC, fibroblast cells from LSFC patients. (B) LRPPRC content of liver mitochondria. Lane 1, normal liver mitochondria; lanes 2–4, liver mitochondria from LSFC patients. Locations of LRPPRC and CI49K are indicated on the right side. Migration of standard proteins of designated molecular mass (MW) are marked on the left side. CI49K is the mitochondrial 49 kDa complex I protein, which was used as an equal loading indicator. (C) Import of [35S]methionine-labelled LRPPRC into rat liver mitochondria. A full-length LRPPRC cDNA was translated in a reticulocyte lysate using [35S]methionine incorporation as described in the Materials and methods section. Samples were taken for wild-type and A354V mutant LRPPRC incubated with rat liver mitochondria for 10, 20, 30 and 60 min and protein bands visualized after separation on an SDS/10% polyacrylamide gel. Bands are designated pre-LRPPRC and LRPPRC for the precursor and mature proteins respectively.
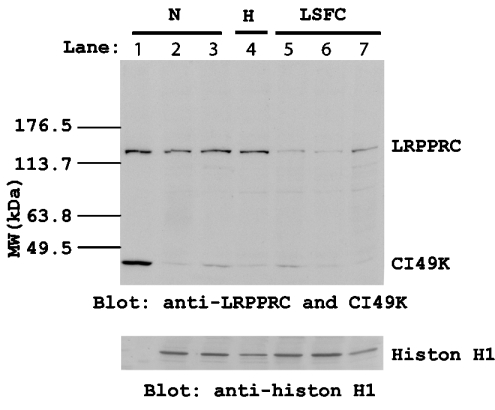
A nuclear fraction of the fibroblasts was prepared by detergent extraction of cells and differential centrifugation, and the level of LRPPRC protein was visualized by immunoblotting (Figure 4). Histone H1 immunoblots show that the level of this protein is the same in all nuclear fractions, whereas there appeared to be LRPPRC protein in the nucleus or at least in a fraction of the cells that is resistant to NP40 extraction. Surprisingly, there was detectable LRPPRC in the nuclear fractions of controls but less in LSFC fibroblasts. Antibody titres for the 49 kDa subunit of complex I were very low, but histone H1 was equivalent in controls and patients.
Figure 4. Western-blot analysis of LRPPRC content in nuclei.
Nuclear fractions were prepared by removing cell membranes, organelles and cytoplasm in a low-salt 0.1% NP40 lysis buffer followed by extraction in a high-salt extraction buffer. Samples from either normal or LSFC fibroblast cells were separated on an SDS/10% polyacrylamide gel and transferred on to a nitrocellulose membrane. The membrane was probed with antibodies to LRPPRC, CI49K and histone H1 respectively. Upper panel: anti-LRPPRC and anti-CI49k blots. Lower panel: anti-histone H1 blot. Lane 1, normal mitochondrial fraction from cell line 4212; lanes 2 and 3, normal nuclear fraction from cell lines 4212 and 3839; lane 4, nuclear fraction from 4254 heterozygous cells; lanes 5–7, nuclear fraction from cell lines 3516, 7143 and 7240 from LSFC patients. CI49K is a mitochondrial-specific protein. Histone H1 is a nuclear-specific protein.
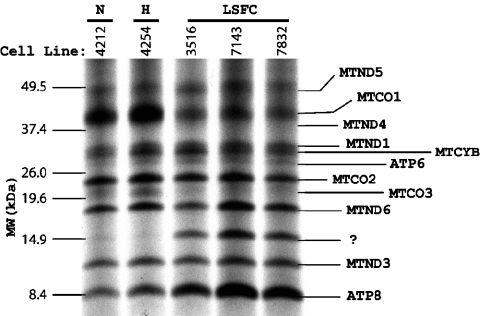
To ascertain whether the lower amount of LRPPRC protein influenced translation of COX mRNAs in mitochondria, the appearance of [35S]methionine-labelled protein in mitochondria was assessed by incubating cultured fibroblasts with [35S]methionine in the presence of emetine. When this was done, a very specific reduction in the amount of labelled COX I and COX III proteins was seen compared with all other proteins (Figure 5). In addition, a prominent extra protein band was seen below the complex I subunit, MTND6. Since this was not visible in control skin fibroblasts, we presumed that this was probably a degradation product of labelled but unassembled COX protein.
Figure 5. [35S]Methionine pulse-labelled human mitochondrial translation products.
Normal or patient fibroblast cells were pulse-labelled with [35S]methionine in the presence of 100 μg/ml emetine for 90 min. The mitochondrial fraction was prepared by differential centrifugation. Radiolabelled protein was determined by scintillation counter. Total labelled protein (10000 c.p.m.) was loaded on each lane and separated on an SDS/polyacrylamide gel (10–20% linear gradient). The separated samples were transferred on to a nitrocellulose membrane by electrophoresis and the membrane was exposed to an X-ray film. MTCO1, MTCO2 and MTCO3 denote subunits 1, 2 and 3 of COX respectively. MTND1, MTND2, MTND3, MTND4, MTND4L, MTND5 and MTND6 refer to subunits of the rotenone-sensitive NADH dehydrogenase. ATP6 and ATP8 are subunits 6 and 8 of H+-ATPase. Migration of standard proteins is indicated on the left side. N, normal cells; H, heterozygous cells; LSFC, patient cells.
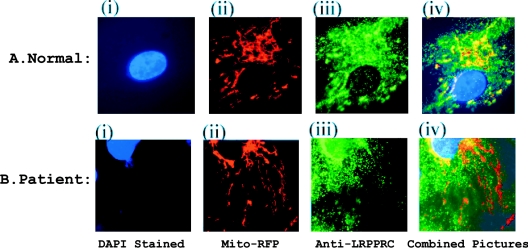
Differential intracellular localization of LRPPRC protein was investigated in LSFC fibroblasts by comparing the image obtained with mitochondrially targeted red fluorescent protein with the immunofluorescence (green) image obtained with the LRPPRC antibody (Figure 6). The control cells showed almost perfect symmetry of mitochondrial localization for both green and red fluorescence in control cells. In LSFC cells, some mitochondrial areas appeared to have little or no LRPPRC. This coincided with an increase in the speculated staining for LRPPRC in the cytoplasm.
Figure 6. Immunostaining of LRPPRC in fibroblast cells.
Both normal and LSFC fibroblast cells transfected with mitochondrially targeted red fluorescent protein were fixed, permeabilized and blocked under the same conditions. The treated cells were incubated with rabbit anti-LRPPRC antibody for 1 h and, after washing, incubated again with FITC-conjugated anti-rabbit antibody for a further 1 h. After removal of unbound secondary antibody, the cells were mounted in anti-fade plus DAPI solution. Fluorescence signals in the cells were visualized under the fluorescence microscope. Upper panel, fluorescence image in normal fibroblast cells; lower panel, fluorescence image in LSFC fibroblast cells. (i) Blue signal, DAPI stain of nucleus; (ii) mitochondrial red fluorescent protein; (iii) green signal, FITC-stained LRPPRC; and (iv) combined image to show overlap.
DISCUSSION
A deficiency of cytochrome oxidase in LSFC patients underlies the development of a phenotype in affected patients, which includes lactic acidaemia, psychomotor retardation and damage to the basal ganglia of the central nervous system [12,13]. The synthesis of a viable COX complex in mitochondria requires the co-ordinated expression of three mtDNA-encoded and ten nuclear-encoded cytochrome oxidase subunits, together with the incorporation of two haems and two copper centres with all the modifications required for their installation [19–21]. Experience with prokaryotic organisms has shown that the basic function of the COX enzyme relies heavily on a core complex within cytochrome oxidase formed by subunits COX I, COX II and COX III. The system for expression of these three subunits in yeast has been shown to be a co-ordinated process involving several key proteins. The expression of COX I protein requires the protein products of the genes PET 309, MSS1 and MTO1, COX II requires PET 111 and COX III requires PET 54, PET 494 and PET 122 [2,21–25]. Some functions of the PET genes, required in yeast and necessary for the splicing out of introns in the mRNAs encoded by yeast mitochondrial DNA, are by definition not required in mammalian systems. However, PET 309 was supposed to be needed for the stabilization and translation of the COX I mRNA, so it was not clear whether or not a homologue of PET 309p would be needed for COX I translation in humans.
The finding of a missense mutation (A354V) in the LRPPRC gene as a founder mutation in the Saguenay region of Québec associated with cytochrome oxidase deficiency implies that LRPPRC as a PET 309 homologue is necessary for the normal expression of this enzyme in certain tissues [3]. The present study shows novel effects of this mutation on the observed biochemical phenotype. Northern-blot analysis suggests that mRNA levels encoding mutant LRPPRC protein are the same as controls, ruling out an effect on RNA stability. We showed, by immunofluorescence labelling, a set of images that suggest strongly that LRPPRC is in the mitochondria in control cells. However, in LSFC cells, LRPPRC seems to be diffusely situated in the cytosol and in most of the cells almost absent from the mitochondria. Moreover, the observed marked decrease in titre of LRPPRC in LSFC mitochondria, as observed by Western blotting, suggests that the protein is either not being produced in sufficient quantity, is not being efficiently imported or is being degraded after import. Experimental examination of import suggests that the precursor of LRPPRC is processed to the mature protein faster than the mutant A354V protein. A fraction of the LRPPRC protein seems to be associated disproportionately with the nucleus. Comparison with the level of the 49 kDa complex I subunit present in mitochondrial membranes suggests only a minor contamination of the nuclear fraction with mitochondria. However, LRPPRC is also reduced in the nuclei of LSFC patients. Since LRPPRC is a leucine-rich protein and a presumptive RNA-binding protein, it is possible that a proportion of the protein synthesized is directed towards the nucleus [9]. A further clue to the mechanism of action of the LRPPRC mutation is found in the specific decrease of the [35S]methionine signal for the synthesis of COX I and COX III subunits in an experiment to view specifically the synthesis of mitochondrially encoded proteins.
The lower levels of COX I and COX III mRNAs seen in LSFC patients suggest that LRPPRC has an mRNA-stabilizing function. Since LRPPRC is present in large amounts in heart and skeletal muscles, it is possible that the LSFC mutation does not completely remove the protective effect of the RNA-binding protein in these tissues. However, in tissues where expression is lower, the mutation may have a more significant effect since the titre may be too low to give protection.
How do we reconcile the effect of the mutation (A354V) on the final COX activity with the postulated mechanism of action of LRPPRC? The LRPPRC gene has an mRNA expression profile typical of a mitochondrial protein, high in heart, skeletal muscles and kidney and low in brain and liver. On the other hand, when mutated, its impact on COX expression seems to be greatest in liver and brain, intermediate in skin fibroblasts and lowest in heart and kidney. This perhaps suggests that, wherever the expression of COX I is lowest, the assistance of LRPPRC is needed for stability. The alternative explanation for the differential effect of the mutation would be a scenario involving differences in the machinery or mechanism of COX translation or involving differences in the control of mitochondrial mRNA degradation in different tissues. At this time, there is no precedent for such differences, but such an explanation remains a possibility.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (MOP15077), the Canadian Genetic Diseases Network and L'Association D'Acidose Lactique du Saguenay Lac St Jean.
References
- 1.Manthey G. M., McEwen J. E. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manthey G. M., Przybyla-Zawislak B. D., McEwen J. E. The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur. J. Biochem. 1998;255:156–161. doi: 10.1046/j.1432-1327.1998.2550156.x. [DOI] [PubMed] [Google Scholar]
- 3.Mootha V. K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U.S.A. 2003;100:605–610. doi: 10.1073/pnas.242716699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou J., Wang F., McKeehan W. L. Molecular cloning and expression of the gene for a major leucine-rich protein from human hepatoblastoma cells (HepG2) In Vitro Cell. Dev. Biol. Anim. 1994;30A:111–114. doi: 10.1007/BF02631402. [DOI] [PubMed] [Google Scholar]
- 5.Liu L., McKeehan W. L. Sequence analysis of LRPPRC and its SEC1 domain interaction partners suggests roles in cytoskeletal organization, vesicular trafficking, nucleocytosolic shuttling, and chromosome activity. Genomics. 2002;79:124–136. doi: 10.1006/geno.2001.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacDonald P. M., Kerr K. Mutational analysis of an RNA recognition element that mediates localization of bicoid mRNA. Mol. Cell. Biol. 1998;18:3788–3795. doi: 10.1128/mcb.18.7.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milli S., Pinol-Roma S. LRP130, a pentatricopeptide motif protein with a noncanonical RNA binding domain, is bound in vivo to mitochondrial and nuclear RNAs. Mol. Cell. Biol. 2003;14:4972–4982. doi: 10.1128/MCB.23.14.4972-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuchiya N., Fukuda H., Sugimura T., Nagao M., Nakagama H. LRP130, a protein containing nine pentatricopeptide repeat motifs, interacts with a single-stranded cytosine-rich sequence of mouse hypervariable minisatellite Pc-1. Eur. J. Biochem. 2002;269:2927–2933. doi: 10.1046/j.1432-1033.2002.02966.x. [DOI] [PubMed] [Google Scholar]
- 9.Coffin J. W., Dhillon R., Ritzel R. G., Nargang F. E. The Neurospora crassa cya-5 nuclear gene encodes a protein with a region of homology to the Saccharomyces cerevisiae PET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein. Curr. Genet. 1997;32:273–280. doi: 10.1007/s002940050277. [DOI] [PubMed] [Google Scholar]
- 10.Fisk D. G., Walker M. B., Barkan A. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. EMBO J. 1999;18:2621–2625. doi: 10.1093/emboj/18.9.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mancebo R., Zhou X., Shillinglaw W., Henzel W., MacDonald P. M. BSF binds specifically to the bicoid mRNA 3′ untranslated region and contributes to stabilization of bicoid mRNA. Mol. Cell. Biol. 2001;21:3462–3471. doi: 10.1128/MCB.21.10.3462-3471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morin C., Mitchell G., Larochelle J., Lambert M., Ogier H., Robinson B. H., DeBraekeleer M. Clinical, metabolic and genetic aspects of cytochrome c oxidase deficiency in Saguenay-Lac-Saint-Jean. Am. J. Hum. Genet. 1993;53:488–496. [PMC free article] [PubMed] [Google Scholar]
- 13.Merante F., Petrova-Benedict R., MacKay N., Mitchell G., Morin C., DeBraekeleer M., Laframboise R., Gagne R., Robinson B. H. A biochemically distinct form of cytochrome oxidase (COX) deficiency in the Saguenay-Lac-Saint-Jean region of Québec. Am. J. Hum. Genet. 1993;53:481–487. [PMC free article] [PubMed] [Google Scholar]
- 14.Pitkänen S., Raha S., Robinson B. H. Diagnosis of complex 1 deficiency in patients with lactic acidemia using skin fibroblast cultures. Biochem. Mol. Med. 1996;59:134–137. doi: 10.1006/bmme.1996.0078. [DOI] [PubMed] [Google Scholar]
- 15.Rickwood D., Wilson M. T., Darley-Usmar V. M., editors. Mitochondria: A Practical Approach. Oxford: IRL Press; 1987. Isolation and characteristics of intact mitochondria; pp. 1–16. [Google Scholar]
- 16.McEachern G., Kassovska-Bratinova S., Raha S., Tarnopolsky M. A., Turnbull J., Bourgeois J., Robinson B. H. Manganese superoxide dismutase levels are elevated in a proportion of amyotrophic lateral sclerosis patient cell lines. Biochem. Biophys. Res. Commun. 2000;273:359–363. doi: 10.1006/bbrc.2000.2933. [DOI] [PubMed] [Google Scholar]
- 17.Chomyn A. In vivo labelling and analysis of human mitochondrial translation products. Methods Enzymol. 1996;264:4197–4211. doi: 10.1016/s0076-6879(96)64020-8. [DOI] [PubMed] [Google Scholar]
- 18.Pari G., Jardine K., McBurney M. W. Multiple CArG boxes in the human cardiac actin gene promoter required for expression in embryonic cardiac muscle cells developing in vitro from embryonal carcinoma cells. Mol. Cell. Biol. 1991;11:4796–4803. doi: 10.1128/mcb.11.9.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoubridge E. A. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 2001;106:46–52. doi: 10.1002/ajmg.1378. [DOI] [PubMed] [Google Scholar]
- 20.Schon E. A. Mitochondrial genetics and disease. Trends Biochem. Sci. 2000;25:555–556. doi: 10.1016/s0968-0004(00)01688-1. [DOI] [PubMed] [Google Scholar]
- 21.Robinson B. H. Human cytochrome oxidase deficiency. Pediatr. Res. 2000;48:581–585. doi: 10.1203/00006450-200011000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Li X., Li R., Lin X., Guan M.-X. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12 S rRNA A1555G mutation. J. Biol. Chem. 2002;277:27256–27264. doi: 10.1074/jbc.M203267200. [DOI] [PubMed] [Google Scholar]
- 23.Green-Willms N. S., Butler C. A., Dunstan H. M., Fox T. D. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J. Biol. Chem. 2001;276:6392–6397. doi: 10.1074/jbc.M009856200. [DOI] [PubMed] [Google Scholar]
- 24.Costanzo M. C., Fox T. D. A point mutation in the 5′-untranslated leader that affects translational activation of the mitochondrial COX3 mRNA. Curr. Genet. 1995;28:60–66. doi: 10.1007/BF00311882. [DOI] [PubMed] [Google Scholar]
- 25.Costanzo M. C., Bonnefoy N., Williams E. H., Clark-Walker G. D., Fox T. D. Highly diverged homologues of Saccharomyces cerevisiae mitochondrial mRNA-specific translational activators have orthologous functions in other budding yeasts. Genetics. 2000;154:999–1012. doi: 10.1093/genetics/154.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagai K., Oubridge C., Ito N., Avis J., Evans P. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem. Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]