Abstract
With the widespread use of yeast two-hybrid systems, many heterodimeric forms of S100 proteins have been found, although their biological significance is unknown. In the present study, S100A1 was found to interact with another S100 protein, S100P, by using the yeast two-hybrid system. The binding parameters of the interaction were obtained using an optical biosensor and show that S100P has a slightly higher affinity for S100A1 (Kd=10–20 nM) when compared with that for self-association (Kd=40–120 nM). The physical interaction of S100A1 and S100P was also demonstrated in living mammalian cells using a fluorescence resonance energy transfer technique. Preincubation of recombinant S100P with S100A1, before the biosensor assay, reduced by up to 50% the binding of S100P to a recombinant C-terminal fragment of non-muscle myosin A, one of its target molecules. Site-specific mutations of S100P and S100A1, combined with homology modelling of an S100P/S100A1 heterodimer using known S100P and S100A1 structures, allowed the hydrophobic interactions at the dimeric interface of the heterodimer to be defined and provide an explanation for the heterodimerization of S100P and S100A1 at the molecular level. These results have revealed the similarities and the differences between the S100P homodimer and the S100A1/S100P heterodimer.
Keywords: dimeric interface, heterodimerization, non-muscle myosin A, S100A1, S100P, yeast two-hybrid
Abbreviations: ECFP, enhanced cyan fluorescent protein; EYFP, enhanced yellow–green fluorescent protein; MHC IIA, non-muscle myosin A heavy chain; rh, recombinant human; wt, wild-type; for brevity the one-letter system has been used for amino acids, e.g. H1069 means His-1069
INTRODUCTION
Members of the S100 family of over 20 EF-hand type calcium-binding proteins occur exclusively in vertebrates and are distributed in a tissue-specific manner [1]. They are supposed to act by interacting with a variety of protein targets, both inside and outside the cells [1], and their increased levels have been associated with a number of disease states [2]. Although often isolated as homodimers [3,4], the diversity of their potential biological functions may be further expanded because of the formation of non-covalent heterodimers with subunits from other family members, e.g. S100A1/S100B [5], S100A8/S100A9 [6], S100B/S100A6 [7], S100A1/S100A4 [8,9] and S100B/S100A11 [7]. The S100A8/S100A9 heterodimer, rather than S100A8 or S100A9 homodimers, is the preferred functional form within cells [10]. Gribenko et al. [10a] report on the S1000P/S100Z heterodimer formation: they note that S100Z has remarkable homology with S100A1. Using the yeast-two hybrid system, we now report that S100A1 can form a heterodimer with another S100 protein, S100P. Human S100P and S100A1 are proteins of 95 and 93 amino acid residues respectively, and share approx. 50% amino acid sequence identity. From the available results, the two proteins appear to be involved in different human diseases and have very different functions, despite the similarities in their sequences and overall structures. S100A1 has been reported to be important in regulating cytoskeletal fibres, including microtubules, type III intermediate filaments and actin [11]. S100A1 is highly expressed in human heart and, as such, is an early marker for acute myocardial ischaemia [12]. S100A1 is also up-regulated in chronic pulmonary hypertension [13] and down-regulated in cardiomyopathy [14]. Overexpression of S100A1 in cardiomyocytes enhances their contractility [15], whereas impaired contractility of the heart in response to haemodynamic stress was observed in S100A1-deficient mice [16].
S100P cDNA was isolated and cloned originally from human placenta [17]. Increased levels of S100P mRNA have been reported not only in immortalized and chemically transformed epithelial cell lines [18], but also in doxorubicin-resistant colonic carcinoma cell lines relative to their parental counterparts [19], and in clinical specimens of invasive ductal breast carcinomas and ductal hyperplasia in situ relative to the adjacent normal breast tissues [18]. S100P is down-regulated following androgen deprivation in an androgen-responsive, but not in androgen-independent, prostatic cancer cell lines [20]. S100P is one of the most overexpressed gene products in a microarray screen of hormone-refractory prostatic cancer xenografts [21] and pancreatic carcinomas relative to normal tissues [22,23] and its levels are significantly associated with disease progression of prostatic and pancreatic carcinomas [24].
It is not known whether S100P is expressed in cardiomyocytes, but S100A1 is not up-regulated in most tumours investigated [2]. Unlike the S100A1/S100B heterodimer, where both subunits have very similar functions [2], the S100P/S100A1 heterodimer is composed of two subunits of potentially different functions and thus presents an interesting combination of subunits. In the present study, using an optical biosensor, it is now shown that S100P has a slightly higher affinity for S100A1 than for self-association, and with fluorescence resonance energy transfer technique, close contact between S100A1 and S100P in living mammalian cells is demonstrated, indicating that the heterodimeric form can occur in vivo. More importantly, we found that S100A1 can reduce the binding of S100P to MHC-IIA (non-muscle myosin A heavy chain) in a biosensor assay. This result suggests that the S100P/S100A1 heterodimer and the S100P homodimer may behave differently. Using site-directed mutations of S100A1 and S100P combined with simulation of the dimeric interface of S100A1/S100P, the structural similarity and difference between the heterodimer and the S100P homodimer are now revealed at the molecular level. The structural difference adjacent to the C-terminal region may account for the difference in the target binding properties between S100P homodimer and S100P/S100A1 heterodimer. Thus the formation of heterodimers in vivo may point to a regulatory mechanism among the S100 proteins.
EXPERIMENTAL
DNA constructs for the yeast two-hybrid system
To screen a human breast carcinoma-derived yeast two-hybrid library constructed with pYESTrp2 vector (Invitrogen, Groningen, The Netherlands), the Lex A bait vector with a LEU2-selectable marker was used as described previously [8]. A 282 bp cDNA corresponding to the coding region of human S100A1 mRNA was obtained by PCR of pYESTrp2 S100A1 [8] and subcloned into the LexA vector to produce the LexA-S100A1 bait construct. Nucleotide sequencing ensured correct insertion of the constructs and the correct sequence of the cDNA. Selection for this bait plasmid in yeast cells was on leucine-free plates or medium. Screening of a breast carcinoma-derived two-hybrid library was performed exactly as described previously [8]. The β-galactosidase activity of each positive transformant yeast strain was determined using a liquid β-galactosidase assay [25]. The number of Miller Units of β-galactosidase activity was calculated as described previously [8].
Fluorescence lifetime measurements
The pECFP-S100A1 (where ECFP stands for enhanced cyan fluorescent protein), pS100A1-EYFP (where EYFP stands for enhanced yellow–green fluorescent protein) and pS100P-EYFP were constructed by inserting the PCR-amplified DNA fragments containing the coding sequences of S100A1 and S100P into pECFP-C1 and pEYFP-N1 vectors (ClonTech) at the HindIII and BamHI sites. The primers for amplifying S100A1 were as follows: forward primer, 5′-GCTAAGCTTCGATGGGCTCTGAGCTGGAG (for constructing both pECFP-S100A1 and pS100A1-EYFP); and reverse primers 1, 5′-TCCGGATCCTCAACTGTTCTCCCAGAAG (for constructing pECFP-S100A1) and 2, 5′-TCCGGATCCAGACTGTTCTCCCAGAAG (for constructing pS100A1-EYFP). The primers for amplifying S100P were as follows: forward primer, 5′-TCGAAGCTTATGACGGAACTAGAGACAG; and reverse primer, 5′-TCCGGATCCGCTTTGAGTCCTGCCTTC. HeLa cells were co-transfected with pECFP-S100A1 to express donor protein, and with pS100P-EYFP, pEYFP-S100A1 or pEYFP-N1 vector to express acceptor protein. The fluorescence lifetime of ECFP-S100A1 was collected using a purpose-built scanning confocal microscope [26] from HeLa cells that co-expressed (i) ECFP-S100A1 and S100P-EYFP, (ii) ECFP-S100A1 and EYFP vectors (negative control), (iii) ECFP-S100A1 (negative control) or (iv) ECFP-S100A1 and S100A1-EYFP (positive control). The data were recorded at room temperature (20 °C), and FLIM (fluorescence lifetime imaging microscopy) images in the time domain from single cells were collected using a time-correlated single-photon counting [27] fluorescence lifetime imaging module (SPC-730; Becker & Hickl, Berlin, Germany) and a photomultiplier tube (PMH-100-1; Hamamatsu Photonics, Bridgewater, NJ, U.S.A.). FLIM images were analysed using SPCImage 2.0 (Becker & Hickl). The methods for calculation of fluorescence lifetimes and statistical analysis are described elsewhere (see [27a]).
Mutagenesis of S100A1 and S100P
The mutant constructs of S100A1 were generated using the PCR overlap-extension method of site-directed mutagenesis [28]. The two external primers used were the following: 5′-EcoRI primer 5′-CTGGAATTCATGGGCTCTGAGCTGGAG, and 3′-XhoI primer 5′-CGTCTCGAGAGGAAGGGCGCTGCCCAATG. The inner primers for the F15A mutation were as follows: forward primer, 5′-GTGGCACATGCACACTCGGGCAAAGAGG; and reverse primer, 5′-GCATGTGCCACGTTGATGAGGGTCTC. A multiple mutation (L11H+F15A+F71L) came by chance from random mutations during PCRs. All the cDNAs with the point mutations were digested with EcoRI and XhoI and inserted into the LexA vector. DNA sequencing confirmed the nucleotide changes and the coding frames for expression of fusion protein.
cDNAs encoding mutant S100P proteins, I11A+I12A, F15A, F71A, F74G/V, I75G, V76G, I75G+V76G, were constructed using the primers described previously [29].
Recombinant proteins
cDNAs encoding the 95 amino acids of human S100P protein or the 93 amino acids of human S100A1 were obtained using PCR from clones selected in the two-hybrid screen and were subcloned into the expression vector pET11a (Novagen, Madison, WI, U.S.A.). The recombinant human S100P (rhS100P) and S100A1 (rhS100A1) proteins were purified as described previously for S100A4 [30]. For biosensor experiments, rhS100P and rhS100A1 were additionally purified by gel filtration on Superdex 75 (Amersham Biosciences) to eliminate possible aggregations. Purified recombinant proteins were immediately frozen at −80 °C for storage. The rhC-MHC-IIA (recombinant C-terminal 149 amino acid fragment of human non-muscle myosin A heavy chain) was produced as described previously [31].
Binding assays
Binding reactions were performed in an IAsys two-channel resonant mirror biosensor (Thermo Electron, Basingstoke, U.K.), as described previously [32]. rhS100P and rhS100A1 were immobilized at low densities (between 150 and 250 arc s, where 600 arc s corresponds to 1 ng of protein/mm2 of sensor surface) on aminosilane surfaces using bis(sulphosuccinimidyl) suberate (Perbio, Chester, U.K.) according to the manufacturer's instructions. For the calculation of kon, low concentrations of ligate rhS100P (10 nM–3 μM) were used, whereas for the measurement of koff, concentrations of ligate in the range 3–9 μM were used. The binding parameters kon and koff were calculated from the association and dissociation phases of the binding reactions respectively using the non-linear curve-fitting FastFit software provided with the instrument (Thermo Electron). The equilibrium dissociation constant (Kd) was calculated both from the association and dissociation rate constants and from the extents of binding at equilibrium, as described previously [32]. To detect effects of rhS100A1 on the interaction of S100P with immobilized rhC-MHC-IIA, rhS100P (1 μM) was preincubated with various concentrations of rhS100A1 (from 0.1 to 20 μM) for 1 h or in equimolar ratio (0.75 μM S100A1 and 0.75 μM S100P) for various times (from 0 min to 48 h), and the mixtures were then added to the rhC-MHC-IIA biosensor surface to detect changes in the binding response.
Cell culture and Western blotting
The human malignant breast carcinoma-derived cell lines, MCF-7 and MDA-MB-231, were cultured as described previously [33]. The human neuroblastoma cell lines, SH-SY 5Y, N-type (ECACC no. 94030304), SH-EP1 S-type (kindly provided by Professor M. Schwab, Deutches Krebsforschung-zentrum, Heidelberg, Germany) and the SK-N-AS cell line (ECACC no. 94092302) were routinely cultured as for the breast cell lines to 80% confluency. The human normal B-lymphocyte cell line, Colo 720 L (ECACC no. 93052623), was cultured in suspension and harvested by centrifugation. For Western blotting, the cells were lysed using SDS-sample buffer and the resultant cell lysates were subjected to SDS/PAGE (15% gel) and electroblotted on to the Immobilon P membrane. S100A1 and S100P proteins were detected using rabbit anti-S100A1 antibody (Dako, Copenhagen, Denmark) and mouse anti-S100P antibody (BD Science, San Jose, CA, U.S.A.). Bands of S100A1 and S100P positivity from cell lines on Western blots were quantified relative to the intensities of the signals of bands of recombinant S100A1 and S100P subjected to SDS/PAGE, using IMAGE Software analysis of video images.
Homology modelling of an S100P/S100A1 heterodimer
The NMR structure of human apo-S100A1 (Protein Databank code 1K2H) [35] and the X-ray structure of human S100P (Protein Databank code 1J55) [29] at 2.0 Å (1 Å=0.1 nm) resolution were used for homology modelling. The heterodimeric structural template of S100A1 and S100P was constructed by graphically superimposing the structure of the apo-S100A1 monomer [35] on to the X-ray structure of one of the monomers within the calcium-bound S100P dimer. Molecular dynamic simulation of the heterodimeric structure was processed using the CHARMM (Chemistry at HARvard Macromolecular Mechanics) protein structure modelling software [36,37]. First, all proton positions were mapped on to the heterodimeric structural template of the S100P/S100A1 heterodimer at pH 7.0. Secondly, this initial structure was optimized by 500 steps of energy minimization with a dielectric constant 1.0 (conditions of no water molecules). Thirdly, the minimized structure was theoretically heated from 0 to 300 K in 10 ps by 10000 steps. After every 100 steps, the system temperature was increased by 3 K and the system translations and rotations were checked and adjusted. Finally, the temperature of the heated structure was kept at 300 K for 30 ps with 30000 steps, to study the structure and energy fluctuations. During molecular dynamics, the atomic velocities were distributed in a Gaussian manner and the fluctuations in the system temperature were controlled to be within 10 K.
RESULTS
Interactions of S100A1 with target proteins in yeast cells
More than 2×106 yeast cell transformants from a yeast twohybrid cDNA library, constructed from mRNA isolated from a human breast cancer specimen (see the Experimental section), were co-transfected into L40 yeast cells with an S100A1 cDNA bait vector. Target plasmids were recovered from six independently isolated blue colonies, and the purified plasmid DNAs from each colony were co-transfected into strain L40 yeast cells. Re-isolation of plasmid DNA and co-transfection were repeated once more, whereupon four colonies still showed a blue colour in lift assays with 5-bromo-4-chloroindol-3-yl β-D-galactopyranoside. Prey plasmid DNA was isolated from these four transformants and the nucleotide sequences of two target plasmids were identical with that of human S100A1 cDNA and the other two were identical with that of human S100P cDNA. The two colonies containing both the S100A1 bait and S100P prey plasmids exhibited β-galactosidase activities that were 20–40-fold higher than colonies containing only bait plasmid or only prey plasmid (Figure 1A). The above experiments show that, at least in yeast cells, S100A1 can interact in a heterologous manner with S100P.
Figure 1. Measurement of interactions between S100A1 bait and/or target vectors isolated from the breast cancer two-hybrid library using the yeast two-hybrid system.
Clones of yeast strain L40 were transformed with the DNA constructs indicated below, grown up, and β-galactosidase activity determined in Miller Units, as described in the Experimental section. (A) LexA vector; YEStrp2-S100P-1; YEStrp2-S100P-2; LexA-S100A1; LexA-S100A1 (bait)+YES-S100P-1/2: recovered target plasmids 1 or 2 containing S100P cDNA; LexA-S100A1 (bait)+YES A1-1/YES A1-2: recovered prey plasmids 1 or 2 with S100A1 cDNA; LexA-S100A1 (bait)+YES-S100P-coding sequence only and pYESTrp2-S100P-coding sequence only. (B) wtS100P, wtS100A1, wtS100A2 and wtS100A4 as a bait or a prey.
The yeast two-hybrid system was also used to test for self-association of S100P and for association between S100P and other S100 proteins, S100A1, S100A2 and S100A4 (Figure 1B). A much higher level of β-galactosidase activity was obtained for the interaction of S100P with S100A1 when compared with that for S100P with S100A2 (P=0.001, Student's t test) or S100P with S100A4 (P=0.01). Furthermore, the level of β-galactosidase activity for the interaction between S100P and S100A1 was more than twice that for the self-association of S100P (P=0.012) (Figure 1B) and was similar to that of the homodimer of S100A1 (P=0.5).
Characterization of S100P interactions in vitro using an optical biosensor
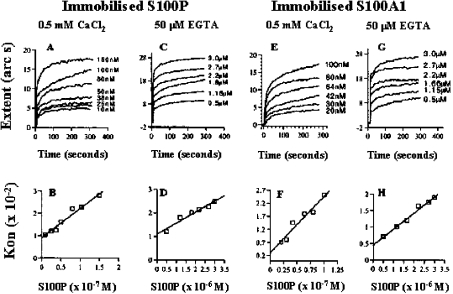
The levels of β-galactosidase activities in the yeast two-hybrid system indicate only approximately the relative binding preferences of S100A1 and S100P; thus the binding kinetics were studied using an optical biosensor. The association of rhS100P with rhS100A1 was characterized by faster association kinetics in calcium-containing (0.5 mM) than in calcium-free (50 μM EGTA) buffers. In all conditions, the association and the dissociation binding reactions were monophasic. For example, a one-site model described the binding reactions at least as closely as a two-site model (result not shown), and plots of kon against the concentration of ligate were always linear (Figure 2). Consequently, a one-site model was used to calculate all the binding parameters. In the presence of Ca2+, the association rate constant for the interaction between rhS100P and rhS100A1 [kass=(2.2±0.3)×105 M−1·s−1] (Table 1) was similar to that previously determined for the self-association of rhS100P [kass=(1.5±0.1)×105 M−1·s−1] [29]. In the absence of Ca2+, the association rate constant for the interaction between rhS100P and rhS100A1 was 30–50-fold slower [kass=(4.7±0.1)×103 M−1·s−1; Table 1] than that observed in the presence of Ca2+. Similar dependence on Ca2+ has been observed for the self-association of S100P [29]. The dissociation rate constant (kdiss) of rhS100A1 from immobilized rhS100P was slightly slower than that of rhS100P from immobilized rhS100P (Table 1), although there was no effect of Ca2+ on either of the dissociation rate constants. In the presence of calcium ions, the interaction between rhS100P and rhS100A1 had a slightly higher affinity (Kd=0–20 nM), compared with that found previously for the self-association of rhS100P (Kd=40–120 nM) [29]. In the absence of Ca2+, the two interactions had similar affinities, 20–50-fold lower than those detected in the presence of 0.5 mM Ca2+. The Kd values for the interaction of rhS100A1 with rhS100P calculated from the extents of binding observed at equilibrium were very similar to those calculated from the kinetic constants (Table 1), indicating that the binding parameters probably reflect the intrinsic values.
Figure 2. Self-association of S100P and association of S100P with S100A1 in vitro using an optical biosensor.
Recombinant S100P (A–D) and S100A1 (E–H) were immobilized on an aminosilane surface as described in the Experimental section. Various concentrations of recombinant S100P were added in the presence (A, B, E, F) or in the absence (C, D, G, H) of calcium ions, and the extents of binding were observed for 300–400 s (A, C, E, G). The concentrations of added S100P were plotted against kon using FastPlot software (B, D, F, H) and yielded a straight line (regression coefficient, r=0.95–0.98) in each case.
Table 1. Kinetics of binding of soluble S100P to immobilized S100P or S100A1 in the presence of 0.5 mM Ca2+ or in the absence of Ca2+ (50 μM EGTA).
| Immobilized protein | Ca2+or EGTA | kass (M−1·s−1)* | r† | kdiss (s−1)‡ | Kinetic Kd (nM)§ | Equilibrium Kd (nM)∥ |
|---|---|---|---|---|---|---|
| S100A1 | Ca2+ | 2.2±0.3×105 | 0.96 | 3±1×10−3 | 14±5 | 15±2.9 |
| S100A1 | EGTA | 4.7±0.1×103 | 0.98 | 5±0.7×10−3 | 1100±300 | 1800±600 |
| S100P¶ | Ca2+ | 1.5±0.1×105 | 0.98 | 9±1×10−3 | 64±24 | 94±32 |
| S100P¶ | EGTA | 4.5±0.5×103 | 0.95 | 11±1×10−3 | 2500±800 | 1900±500 |
* S.E.M. of each determination of kass is derived from the deviation of the data sets from a one-site binding model, calculated by matrix inversion using the FastFit software provided with the instrument (see the Experimental section). No evidence was found for a two-site model of association. Two independent sets of kon were determined and their errors combined.
† Correlation coefficient of the linear regression through the kon values used for obtaining kass.
‡ kdiss is the means±S.E.M. for at least six values, obtained at different concentrations of S100P. No evidence was found for a two-site model of dissociation.
§ Kinetic Kd was calculated from the ratio of kdiss/kass, and the S.E.M. is the combined S.E.M. of the two kinetic parameters.
∥ Equilibrium Kd was calculated from the extent of binding observed at or near equilibrium at six or more different concentrations of ligate in two independent experiments. S.E.M. is the combined error for the two experiments.
¶ From Zhang et al. [29].
S100A1 and S100P are co-expressed in breast carcinoma cells
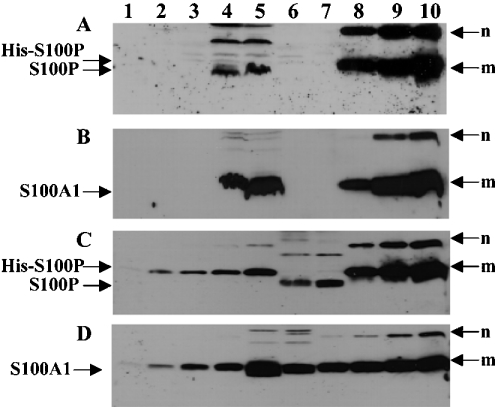
To explore the possibility of the interaction of S100A1 and S100P in mammalian cells, the co-expression of S100A1 and S100P proteins was sought in extracts from a number of benign and malignant tumour cell lines using Western-blotting procedures. Both S100A1 and S100P were co-expressed in cell lines MCF-7 and MDA-MB-231 (Figure 3). Semi-quantification of the Western blots using co-electrophoresed recombinant proteins as standards (see the Experimental section) showed that the S100A1/S100P ratio was 14–21:10 and 8–12:1 for the MDA-MB-231 and MCF-7 cells respectively.
Figure 3. Western blotting for S100A1 and S100P proteins in extracts of cell lines.
(A, B) Protein (60 μg) of each cell lysate of neuroblastoma cell lines SH-EP (lane 1), SH-SY-5Y (lane 2), SK-N-AS (lane 3), breast carcinoma cell lines, MDA MB-231 (lane 4), MCF-7 (lane 5) and the human normal B lymphocyte cell line, Colo 720 L (lane 6) and recombinant proteins, S100A1 (1 μg in lane 7 of A; 0.3, 1.0 and 3 μg in lanes 8, 9 and 10 of B), His-tagged S100P (1.0 μg in lane 7 of B; 0.3, 1.0 and 3 μg in lanes 8, 9 and 10 of A) were separated by SDS/PAGE (15% gel) and transferred on to Immobilon membranes. For semi-quantification, His-tagged S100P (C), 10, 20, 40, 80, 160, 320, 640 and 1280 ng were loaded in lanes 1, 2, 3, 4, 5, 8, 9 and 10 respectively; S100A1 (D), 10, 20, 40, 80, 160, 320, 640 and 1280 ng were loaded in lanes 1, 2, 3, 4, 7, 8, 9 and 10 respectively. MCF7 cell lysate (60 μg) was loaded in lane 6 (C) and lane 5 (D). MDA MB-231 cell lysate (60 μg) was loaded in lane 7 (C) and lane 6 (D). The membranes were incubated with either mouse anti-S100P (A, C) or rabbit anti-S100A1 (B, D) and bound antibody detected as described in the Experimental section. m, monomer; n, SDS-resistant multimers of the recombinant proteins, His-S100P (A, C) and S100A1 (B, D). The higher-molecular-mass bands in lanes 4 and 5 of (A), lanes 6 and 7 of (C) are the multimers of endogenous non-His-tagged S100P.
MCF-7 cells were co-transfected with vectors encoding ECFP-S100A1 and S100P-EYFP fusion proteins, and the transfected cells were examined by confocal microscopy. Although there was some variability in the nuclear/cytoplasmic location of the fluorescent fusion proteins, the EGFP and EYFP fluorescence is distributed in the same regions in the transfected cells observed (Figure 4).
Figure 4. Co-localization of pECFP-S100P and pEYFP-S100A1.
MCF-7 cells were routinely cultured and co-transfected with expression plasmids encoding ECFP-S100A1 and S100P-EYFP. After 24 h, the images were taken under a confocal microscope. (A–D) Show the same field. S100P-EYFP, shown in pseudocolour green (A) is present in both nucleus and cytoplasm, but in some cells S100P-EYFP is present either in the nucleus or in the cytoplasm. In one of the cells shown, S100P-EYFP is concentrated around the peri-nuclear region (arrow). ECFP-S100A1, shown in pseudocolour red (B), distributes in an almost identical pattern with that of S100P-EYFP, irrespective of the overall pattern of localization in the cell. The images in (A, B) are superimposed (D), the yellow colour confirming the co-localization of S100P-EYFP and ECFP-S100A1. (C) Phase-contrast image of the same field.
Interaction of S100A1 with S100P in mammalian cells revealed by fluorescent lifetime imaging
The cells expressing both ECFP-S100A1 and S100P-EYFP showed significantly reduced fluorescence lifetimes when compared with the cells that expressed ECFP-S100A1 only (P=0.001, Student's t test) or that co-expressed ECFP-S100A1 and EYFP (P=0.001) (Table 2). The reductions in fluorescence lifetimes observed in cells with ECFP-S100A1 and S100P-EYFP were similar to that for cells co-expressing both ECFP-S100A1 and S100A1-EYFP (P=0.37), which in turn was significantly different from cells expressing ECFP-S100A1 only (P=0.002) or ECFP-S100A1 and EYFP (P=0.003) (Table 2). The reductions of lifetime resulted from energy transfer from donor to acceptor, and thus indicated a close association between S100A1 and S100P or S100A1 and S100A1 in vivo.
Table 2. Measurement of fluorescence lifetime by fluorescent lifetime imaging.
| Donor* | Acceptor* | Fluorescence lifetime (ns) (means±S.D.)† |
|---|---|---|
| ECFP-S100A1 | S100P-EYFP | 1.55±0.33 |
| ECFP-S100A1 | EYFP | 2.63±0.25 |
| ECFP-S100A1 | None | 2.47±0.11 |
| ECFP-S100A1 | S100A1-EYFP | 1.44±0.36 |
* HeLa cells that co-express donor and acceptor proteins were used for the measurement of fluorescence lifetime.
† Means±S.D. from 7 to 12 cells for two independent transfections. The fluorescent lifetimes measured from the cells co-expressing ECFP-S100A1 and S100P-EYFP and from the cells co-expressing ECFP-S100A1 and S100A1-EYFP are similar (P=0.374, Student's t test) but significantly shorter than that of the cells co-expressing ECFP-S100A1 and EYFP (P=0.001 and P=0.003 respectively) or the cells expressing ECFP-S100A1 only (P=0.001 and P=0.002 respectively). The fluorescent lifetimes measured of the cells co-expressing ECFP-S100A1 and EYFP and the cells expressing ECFP-S100A1 only are similar (P=0.231).
Influence of S100A1 on the binding of S100P to immobilized rhC-MHC-IIA in an optical biosensor
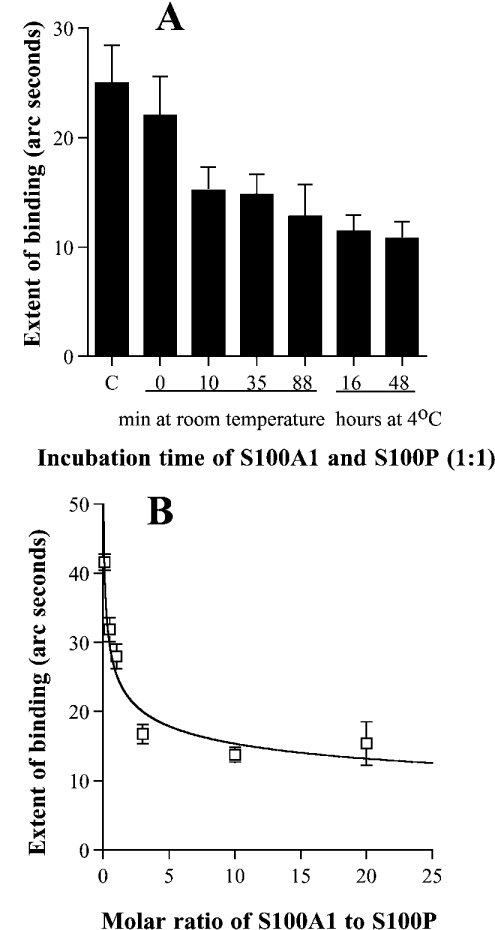
In the biosensor experiments, rhS100A1 showed a very low response of binding to rhC-MHC-IIA in the presence of 0.5 mM CaCl2, whereas, under the same conditions, rhS100P showed a strong response (G. Wang, D. G. Fernig, P. S. Rudland and R. Barraclough, unpublished work), which was 10-fold greater than that for S100A1 (P=0.001, Student's t test) at the concentrations used (results not shown). To determine whether rhS100A1 affected the interaction between rhS100P and rhC-MHC-IIA, rhS100A1 and rhS100P were preincubated at an equimolar ratio for various times before measuring the interactions of the mixture with immobilized rhC-MHC-IIA (Figure 5A). When rhS100A1 and rhS100P were mixed and immediately added to a rhC-MHC-IIA surface only a small reduction in binding to rhC-MHC-IIA was observed relative to that with S100P alone (P=0.11). However, there was a significant reduction in the binding following a 10 min preincubation at room temperature of S100P with rhS100A1 (P=0.02) (Figure 5A). Longer incubation periods, at room temperature or at 4 °C, did not significantly reduce binding any further. The incubation of rhS100P or rhS100A1, separately, for up to 3 h at room temperature and for up to 48 h at 4 °C had no obvious effect on their binding to rhC-MHC-IIA surface (results not shown). When rhS100P (1 μM) was preincubated for 1 h at room temperature with various concentrations of rhS100A1 (from 0.1 to 20 μM) to obtain molar ratios (S100A1/S100P) ranging from 1:10 to 20:1, the binding response to rhC-MHC-IIA was reduced in a dose-dependent manner (Figure 5B). When the molar ratio of S100A1/S100P was increased to or above 3:1, the binding response to rhC-MHC-IIA was significantly reduced to approx. 50% of that observed in the absence of rhS100A1 (P=0.001). These results suggest that rhS100A1 has an inhibitory effect on the binding of rhS100P to the immobilized rhC-MHC-IIA, particularly at low molar excesses of S100A1 over S100P.
Figure 5. Effect of S100A1 on the binding of S100P to immobilized rhC-MHC-IIA.
Purified rhC-MHC-IIA was immobilized on an aminosilane surface as described in the Experimental section. S100P and S100A1 were preincubated for various times at an equimolar ratio at room temperature or at 4 °C (A) or at various ratios for 60 min at room temperature (B). To detect the binding response, 2 μl of each mixture was added to rhC-MHC-IIA surface in a total volume of 30 μl [the final concentration of S100P protein in (A) was 25 nM and in (B) was 33 nM]. The extent of binding of each binding reaction was calculated using the Fastfit software. The means±S.E.M. was determined from at least four individual binding experiments. C (in A), 25 mM S100P only as a positive control.
Analysis of the binding interface of S100A1 and S100P using site-directed mutagenesis
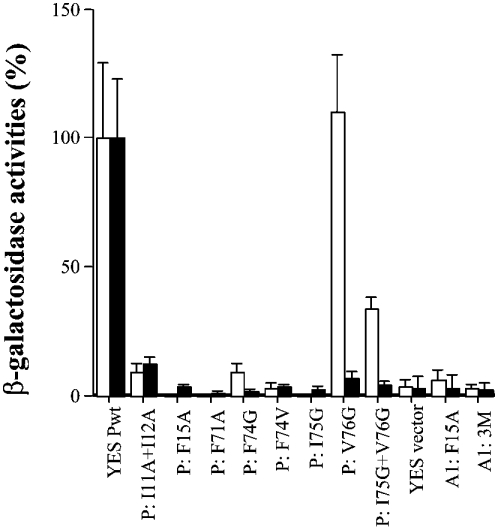
To examine in more detail the regions of S100P that can interact with another S100P or S100A1 molecule, mutations were made to residues in the dimer interface of S100A1 or S100P and these mutated molecules were tested for interaction with wtS100A1 (wild-type S100A1) or wtS100P, using the yeast two-hybrid system. A mutation, which resulted in the conversion of F15 of S100A1 into an alanine residue (termed F15A) (Figure 6), and multiple mutations, S100A1L11H+F15A+F71L (named 3M), which disrupted the interaction between the mutant and wild-type S100A1 in the yeast two-hybrid system, also disrupted the interaction between S100A1 and wtS100P (Figure 6). S100P mutants, I11A+I12A, F15A, F71A, F74G, F74V, I75G, abolished almost completely the interaction of S100P with wtS100P, and also almost completely abolished the interaction with wtS100A1 (Figure 6). These results broadly suggest that the same amino acid residues associated with homodimerization of S100A1 or S100P are involved in the S100P/S100A1 interaction and further suggest that this interaction is heterodimerization. However, the S100P mutant V76G abolished its interaction almost completely with wtS100P molecules, but did not interrupt its interaction with S100A1, an effect that was evident even when combined with the I75G mutant that on its own almost completely abolished interaction with S100A1 (Figure 6). These results suggest that, although the interface for the S100P/S100A1 interaction is similar to that of the S100P homodimer, it is not identical.
Figure 6. Effect of mutations on S100P homodimerization and heterodimerization in the yeast two-hybrid system.
The yeast strain L40 was co-transformed with the DNA constructs in pairs (bait+prey) and the transformants were grown in selected agar plates. A total of six clones were randomly chosen from each of two independent transformations of each pair of DNA constructs and their β-galactosidase activities were determined as described in the Experimental section. The relative binding affinities of bait, S100A1wt (Lex A1wt) (white) or S100Pwt (Lex Pwt) (black) to preys, S100Pwt (YES Pwt) or S100P mutants (P:=S100P) I11A+I12A, F15A, F71A, F74G, F74V, I75G, V76G, I75G+V76G or S100A1 mutants (A1:=S100A1), F15A and 3M: L11H+F15A+F71L triple mutations are expressed as the percentages of the β-galactosidase activities of Lex A1wt+YES Pwt for heterodimerization or Lex Pwt+YES Pwt for homodimerization. S100P self-association data are taken from Zhang et al. [29].
Modelling of the S100P/S100A1 heterodimer
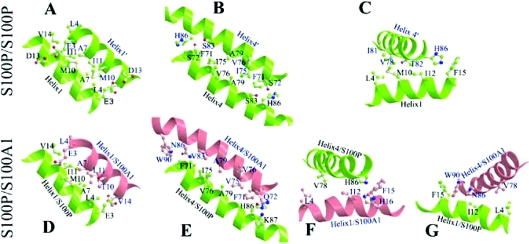
To reveal the similarities and differences between S100P/S100A1 heterodimer and S100P homodimer, a model of the structure of holo-S100P/S100A1 heterodimer was generated by homology with known S100 protein structures [29,35] using CHARMM software. The interfaces of the S100P/S100A1 heterodimer from the modelling as well as the interfaces of the S100P homodimer from X-ray crystallography are shown in Figure 7. The modelling results suggest that the basic structure and dimer interface of the S100P/S100A1 heterodimer are very similar on the whole to that of the S100P homodimer (Figures 7A and 7D, 7B and 7E, 7C and 7F); however, there are some important differences. Compared with the S100P homodimer, the heterodimer interface consists of three additional hydrophobic residues provided by the S100A1 molecule, namely W90, V83 (cf. Figures 7B and 7E) and H16 (Figure 7F) (since histidine is uncharged at pH 7.0, its imidazole ring contributes to the hydrophobic interaction). These residues contribute to the interface between helices 4 and 4′ and the interface between helix 1 of S100A1 and helix 4 of S100P, and between helix 1 of S100P and helix 4 of S100A1 (Figure 7G), thus increasing the binding energy between the heterodimeric subunits. In the interfaces between helices 4 and 4′ or 1 and 4′ or 1′ and 4, the twisting of the two subunits of the heterodimer is more than that in the homodimer, due to closer interactions of residues W90, N86 and V83 in helix 4′ of S100A1 with F71 in helix 4 of S100P (Figure 7E) and F15 in helix 1 of S100P (Figure 7G). There are also closer interactions of H86 of S100P with F71 (cf. Figures 7E and 7B), H16 and F15 of S100A1 (Figure 7F). The distances between W90 of S100A1 and F71 or F15 of S100P are below 3.5 Å, and as a result, the total contact area in these interfaces in the heterodimer becomes 8% larger than that in the S100P homodimer (Table 3).
Figure 7. Comparison of the S100P homodimeric interface and modelled heterodimeric interface with S100A1.
Ball-and-stick ribbon models of interacting helices were generated by MOLSCRIPT software for the interaction between self-associated S100P (A–C) and for dimerization with S100A1 (D–G) modelled using CHARMM software, as described in the Experimental section. Interacting amino acid residues are shown as ball and stick molecular representations. Interactions are shown between helices 1 of the interacting partners (A, D), between helices 4 of the interacting partners (B, E), and between helix 4 and helix 1 of S100P for the self-association (C), and between helix 4 of S100P and helix 1 of S100A1 (F) and between helix 1 of S100P and helix 4 of S100A1 (G) for the heterodimeric interaction. The indicated amino acids are supposed to contribute to the stability of the dimer interface.
Table 3. Areas of interacting helical surfaces for the self-association of S100P and its interaction with S100A1.
| Area (Å2)* | ||
|---|---|---|
| Helical interfaces | S100P/S100A1 | S100P/S100P |
| Helix1/helix1 | 797 | 835 |
| Helix4/helix4 | 791 | 707 |
| Helix1/helix4 | 1016 | 905 |
| Other contacts | 519 | 455 |
| Total contact area | 3123† | 2902† |
* Areas were calculated using the computer program GRASP.
† The total contact area of the heterodimer is slightly larger (nearly 8%) than that of the homodimer, suggesting that the heterodimer may be more stable than the homodimer.
DISCUSSION
Using the yeast two-hybrid system in vivo and an optical biosensor in vitro, a novel heterodimer, S100P/S100A1, has been identified, which is of interest because its two subunits are from two S100 members that are associated with different human diseases [13,23] and potentially have different functions [15,16,38,39]. It is shown that the average extent of binding of S100P to rhC-MHC-IIA, measured in the biosensor experiments, was reduced by 50% when S100P was preincubated with an S100A1/S100P molar ratio of approx. 3, suggesting that the dimerization may have potential biological effects.
On the basis of the β-galactosidase liquid assay of binding activity in the yeast two-hybrid system, it was found that the interaction between S100P and S100A1 in the yeast cells was similar at stimulating reporter gene activity to self-association of S100A1, but more effective than self-association of S100P in the same system. The interaction between S100P and S100A1 in vitro using an optical biosensor exhibited a slightly higher affinity when compared with that for the S100P self-association (Table 1), consistent with the results obtained in the yeast two-hybrid system. Using the fluorescence resonance energy transfer technique, evidence for physical interaction of S100A1 and S100P in HeLa cells has been observed. These results suggest that the heterodimerization may occur in living cells, such as certain breast cancer cells, which contain both S100A1 and S100P (Table 2). Thus these results suggest that heterodimerization of S100 proteins may be a simple regulatory mechanism and such heterodimerization may have potential biological consequences.
Although calcium binding has been shown to affect the conformation of S100 proteins and their binding properties to their targets [2], Ca2+ ions are neither essential for the S100P self-association nor for its interaction with S100A1. In a calcium-free buffer, the interaction of S100P with immobilized S100P or S100A1 can be detected in an optical biosensor, and the Kd for the S100P self-association and for the S100P–S100A1 interaction is in a range 0.8–3.3 μM (Table 1), which is within the biological range. Therefore it is not surprising that S100P can interact with itself or S100A1 in yeast cells. Although Ca2+ ions are not essential for either self-association of S100P or interaction with S100A1, the binding affinities are increased 20–130-fold by adding Ca2+ ions to 0.5 mM in the biosensor assay. This increase is solely due to Ca2+ ions increasing the association rate constant of these interactions. The present results suggest that the conformational changes caused by Ca2+ favour both homodimerization and heterodimerization of S100 proteins.
The heterodimeric interface of S100A1/S100P has been studied by site-directed mutagenesis and structural homology modelling. S100P with a single mutation, F15A, does not interact with wtS100P [40], nor with wtS100A1 (Figure 6). The corresponding F15A mutation in S100A1 also prevents both S100A1 homodimerization and S100A1–S100P interaction. These results show that F15 (which is in the dimeric interface of S100P [29]) is a key residue for both homodimerization and heterodimerization. Single mutation of I11A or I12A of S100P was shown to be not sufficient to abolish its interaction with wtS100P [40]. In the present study, the double mutation of S100P, I11A+I12A in helix 1, only partially interrupted its association with wtS100P and wtS100A1 (Figure 6), indicating that I11 and I12 contribute much less than F15 to the dimer interface of S100P. The single mutations of S100P in helix 4, F71A, F74G/V, I75G and V76G, almost completely abolished its association with wtS100P. These mutants, except V76G, also interrupted the S100P–S100A1 interaction (Figure 6). Since the residues, I11, F15, F71 and I75, are in the dimeric interface of the S100P homodimer [29], these results indicate that the interface of the S100P/S100A1 heterodimer is similar to that of the S100P homodimer. In the modelling of the three-dimensional holo-S100P/S100A1 heterodimer, the overall structure of the heterodimer has been shown to be very similar to that of the S100P homodimer (results not shown). This result is consistent with the results from mutagenesis.
However, the self-association of wtS100P was interrupted by V76G and I75G+V76G mutants, whereas these mutations had little effect on the interaction of S100P with wtS100A1 (Figure 6). The side chain of V76 is not involved in the dimeric interface of S100P or in the heterodimer. The V76G mutation may affect the stability of helix 4 of S100P and is sufficient to abolish the homodimerization but not heterodimerization. This result illustrates a structural difference between the S100P homodimer and the heterodimer with S100A1. In the model of holo-S100A1/S100P, some differences can be seen mainly in helix 4. First, more hydrophobic residues from helix 4 of both subunits are involved in the interface of the heterodimer (F71, V75, V83, A79 and W90 from S100A1, and F71, I75, and A79, H86 from S100P) (Figure 7E) compared with that in the S100P homodimer (F71, I75 and A79) (Figure 7B). Thus the heterodimer should be more stable when compared with the S100P homodimer in this region. The total contact area in the heterodimer is larger than that in the S100P homodimer (Table 3). This provides an explanation for the stronger interaction of S100P with S100A1 than S100P self-association, as shown in the yeast two-hybrid system and in the biosensor assays. This point is supported by mutation S83I of S100P, which enhanced the homodimeric interaction by adding the hydrophobic residue, isoleucine, to the interface [40]. Secondly, the stronger hydrophobic interactions at both ends of the helix 4 interfaces in the heterodimer tend to twist a helix 4 close to the partner helix 4′. The mutation V76G may enable the helix 4 of the S100P subunit to come into closer contact with the helix 4 of the S100A1 subunit in the heterodimer. Therefore the overall effect of V76G mutation of S100P on the heterodimer was not significant, although it was sufficient to interrupt the S100P homodimer. This could also be the explanation for the lower effect of the double mutation of I75G+V76G of S100P on heterodimerization when compared with that of the single mutation of I75G.
It is not clear how the homodimer or heterodimer of S100 proteins interacts with other target proteins. The differences in the dimeric interface, particularly in helix 4/4 (4′), between the S100P homodimer and the S100P/S100A1 heterodimer might be sufficient to alter the conformation of the adjacent C-terminal regions. These adjacent regions have been shown to be important both for target protein binding [41] and for sarcoplasmic Ca2+ release [42] by S100A1, and for target binding and metastasis induction by S100A4 (S. Zhang, G. Wang, D. Liu, D. G. Fernig, P. S. Rudland and R. Barraclough, unpublished work). Thus heterodimerization might be a mechanism that modulates target binding and the function of S100 proteins when more than one such protein is made by an individual cell.
Acknowledgments
This work was supported by grants from the North West Cancer Research Fund, Biotechnology and Biological Sciences Research Council, and the Cancer and Polio Research Fund.
References
- 1.Donato R. S100: a multigenic family of calcium-modulated proteins of the EF-hand type with intracellular and extracellular functional roles. Int. J. Biochem. Cell Biol. 2001;33:637–668. doi: 10.1016/s1357-2725(01)00046-2. [DOI] [PubMed] [Google Scholar]
- 2.Donato R. Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim. Biophys. Acta. 1999;1450:191–231. doi: 10.1016/s0167-4889(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 3.Maler L., Potts B. C., Chazin W. J. High resolution solution structure of apo calcyclin and structural variations in the S100 family of calcium-binding proteins. J. Biomol. NMR. 1999;13:233–247. doi: 10.1023/a:1008315517955. [DOI] [PubMed] [Google Scholar]
- 4.Vallely K. M., Rustandi R. R., Ellis K. C., Varlamova O., Bresnick A. R., Weber D. J. Solution structure of human Mts1 (S100A4) as determined by NMR spectroscopy. Biochemistry. 2002;41:12670–12680. doi: 10.1021/bi020365r. [DOI] [PubMed] [Google Scholar]
- 5.Isobe T., Ishioka N., Masuda T., Takahashi Y., Ganno S., Okuyama T. A rapid separation of S100 subunits by high performance liquid chromatography: the subunit compositions of S100 proteins. Biochem. Int. 1983;6:419–426. [PubMed] [Google Scholar]
- 6.Bhardwaj R. S., Zotz C., Zwadlo-Klarwasser G., Roth J., Goebeler M., Mahnke K., Falk M., Meinardus-Hager G., Sorg C. The calcium-binding proteins MRP8 and MRP14 form a membrane-associated heterodimer in a subset of monocytes/macrophages present in acute but absent in chronic inflammatory lesions. Eur. J. Immunol. 1992;22:1891–1897. doi: 10.1002/eji.1830220732. [DOI] [PubMed] [Google Scholar]
- 7.Deloulme J. C., Assard N., Mbele G. O., Mangin C., Kuwano R., Baudier J. S100A6 and S100A11 are specific targets of the calcium- and zinc-binding S100B protein in vivo. J. Biol. Chem. 2000;275:35302–35310. doi: 10.1074/jbc.M003943200. [DOI] [PubMed] [Google Scholar]
- 8.Wang G., Rudland P. S., White M. R., Barraclough R. Interaction in vivo and in vitro of the metastasis-inducing S100 protein, S100A4 (p9Ka) with S100A1. J. Biol. Chem. 2000;275:11141–11146. doi: 10.1074/jbc.275.15.11141. [DOI] [PubMed] [Google Scholar]
- 9.Tarabykina S., Kriajevska M., Scott D. J., Hill T. J., Lafitte D., Derrick P. J., Dodson G. G., Lukanidin E., Bronstein I. Heterocomplex formation between metastasis-related protein S100A4 (Mts1) and S100A1 as revealed by the yeast two-hybrid system. FEBS Lett. 2000;475:187–191. doi: 10.1016/s0014-5793(00)01652-5. [DOI] [PubMed] [Google Scholar]
- 10.Kerkhoff C., Klempt M., Sorg C. Novel insights into structure and function of MRP8 (S100A8) and MRP14 (S100A9) Biochim. Biophys. Acta. 1998;1448:200–211. doi: 10.1016/s0167-4889(98)00144-x. [DOI] [PubMed] [Google Scholar]
- 10a.Gribenko A. V., Hopper J. E., Makhatadze G. I. Molecular characterization and tissue distribution of a novel member of the S100 family of EF-hand proteins. Biochemistry. 2001;40:15538–15548. doi: 10.1021/bi0114731. [DOI] [PubMed] [Google Scholar]
- 11.Garbuglia M., Verzini M., Sorci G., Bianchi R., Giambanco I., Agneletti A. L., Donato R. The calcium-modulated proteins, S100A1 and S100B, as potential regulators of the dynamics of type III intermediate filaments. Braz. J. Med. Biol. Res. 1999;32:1177–1185. doi: 10.1590/s0100-879x1999001000001. [DOI] [PubMed] [Google Scholar]
- 12.Kiewitz R., Acklin C., Minder E., Huber P. R., Schäfer B. W., Heizmann C. W. S100A1, a new marker for acute myocardial ischemia. Biochem. Biophys. Res. Commun. 2000;274:865–871. doi: 10.1006/bbrc.2000.3229. [DOI] [PubMed] [Google Scholar]
- 13.Ehlermann P., Remppis A., Guddat O., Weimann J., Schnabel P. A., Motsch J., Heizmann C. W., Katus H. A. Right ventricular upregulation of the Ca2+ binding protein S100A1 in chronic pulmonary hypertension. Biochim. Biophys. Acta. 2000;1500:249–255. doi: 10.1016/s0925-4439(99)00106-4. [DOI] [PubMed] [Google Scholar]
- 14.Remppis A., Greten T., Schafer B. W., Hunziker P., Erne P., Katus H. A., Heizmann C. W. Altered expression of the Ca(2+)-binding protein S100A1 in human cardiomyopathy. Biochim. Biophys. Acta. 1996;1313:253–257. doi: 10.1016/0167-4889(96)00097-3. [DOI] [PubMed] [Google Scholar]
- 15.Most P., Remppis A., Pleger S. T., Loffler E., Ehlermann P., Bernotat J., Kleuss C., Heierhorst J., Ruiz P., Witt H., et al. Transgenic overexpression of the Ca2+-binding protein S100A1 in the heart leads to increased in vivo myocardial contractile performance. J. Biol. Chem. 2003;278:33809–33817. doi: 10.1074/jbc.M301788200. [DOI] [PubMed] [Google Scholar]
- 16.Du X. J., Cole T. J., Tenis N., Gao X. M., Kontgen F., Kemp B. E., Heierhorst J. Impaired cardiac contractility response to hemodynamic stress in S100A1-deficient mice. Mol. Cell. Biol. 2002;22:2821–2829. doi: 10.1128/MCB.22.8.2821-2829.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becker T., Gerke V., Kube E., Weber K. S100P, a novel Ca2+-binding protein from human placenta. cDNA cloning, recombinant protein expression and Ca2+ binding properties. Eur. J. Biochem. 1992;207:541–547. doi: 10.1111/j.1432-1033.1992.tb17080.x. [DOI] [PubMed] [Google Scholar]
- 18.Guerreiro Da Silva I. D., Hu Y. F., Russo I. H., Ao X., Salicioni A. M., Yang X., Russo J. S100P calcium-binding protein overexpression is associated with immortalization of human breast epithelial cells in vitro and early stages of breast cancer development in vivo. Int. J. Oncol. 2000;16:231–240. [PubMed] [Google Scholar]
- 19.Bertram J., Palfner K., Hiddemann W., Kneba M. Elevated expression of S100P, CAPL and MAGE 3 in doxorubicin-resistant cell lines: comparison of mRNA differential display reverse transcription–polymerase chain reaction and subtractive suppressive hybridization for the analysis of differential gene expression. Anticancer Drugs. 1998;9:311–317. doi: 10.1097/00001813-199804000-00004. [DOI] [PubMed] [Google Scholar]
- 20.Amler L. C., Agus D. B., LeDuc C., Sapinoso M. L., Fox W. D., Kern S., Lee D., Wang V., Leysens M., Higgins B., et al. Dysregulated expression of androgen-responsive and nonresponsive genes in the androgen-independent prostate cancer xenograft model CWR22-R1. Cancer Res. 2000;60:6134–6141. [PubMed] [Google Scholar]
- 21.Averboukh L., Liang P., Kantoff P. W., Pardee A. B. Regulation of S100P expression by androgen. Prostate. 1996;29:350–355. doi: 10.1002/(SICI)1097-0045(199612)29:6<350::AID-PROS2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 22.Logsdon C. D., Simeone D. M., Binkley C., Arumugam T., Greenson J. K., Giordano T. J., Misek D. E., Kuick R., Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 23.Crnogorac-Jurcevic T., Missiaglia E., Blaveri E., Gangeswaran R., Jones M., Terris B., Costello E., Neoptolemos J. P., Lemoine N. R. Molecular alterations in pancreatic carcinoma: expression profiling shows that dysregulated expression of S100 genes is highly prevalent. J. Pathol. 2003;201:63–74. doi: 10.1002/path.1418. [DOI] [PubMed] [Google Scholar]
- 24.Mousses S., Bubendorf L., Wagner U., Hostetter G., Kononen J., Cornelison R., Goldberger N., Elkahloun A. G., Willi N., Koivisto P., et al. Clinical validation of candidate genes associated with prostate cancer progression in the CWR22 model system using tissue microarrays. Cancer Res. 2002;62:1256–1260. [PubMed] [Google Scholar]
- 25.Becker D., Fikes J. Cloning and functional analysis of heterologous eukaryotic transcription factors in yeast. In: Hames B., Higgins S., editors. Gene Transcription: A Practical Approach. Oxford: IRL Press; 1993. pp. 295–319. [Google Scholar]
- 26.Van der Oord C., Jones G., Shaw D., Munro I., Levine Y., Gerritsen H. High-resolution confocal microscopy using synchrotron radiation. J. Microsc. 1996;182:217–224. doi: 10.1046/j.1365-2818.1996.66432.x. [DOI] [PubMed] [Google Scholar]
- 27.Pepperkok R., Squire A., Geley S., Bastiaens P. I. H. Simultaneous detection of multiple green fluorescent proteins in live cells by fluorescence lifetime imaging microscopy. Curr. Biol. 1999;9:269–272. doi: 10.1016/s0960-9822(99)80117-1. [DOI] [PubMed] [Google Scholar]
- 27a.Zhang S., Wang G., Fernig D. G., Rudland P. S., Webb S. E. D., Barraclough R., Martin-Fernandez M. Interaction of metastasis-inducing S100A4 in vivo by fluorescence lifetime imaging microscopy. Eur. Biophys. J. 2004 doi: 10.1007/s00249-004-0428-x. in the press. [DOI] [PubMed] [Google Scholar]
- 28.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Please L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H., Wang G., Ding Y., Wang Z., Barraclough R., Rudland P. S., Fernig D. G., Rao Z. The crystal structure at 2 Å resolution of Ca2+-binding protein S100P. J. Mol. Biol. 2003;325:785–794. doi: 10.1016/s0022-2836(02)01278-0. [DOI] [PubMed] [Google Scholar]
- 30.Gibbs F. E., Wilkinson M. C., Rudland P. S., Barraclough R. Interactions in vitro of p9Ka, the rat S-100-related, metastasis-inducing, calcium-binding protein. J. Biol. Chem. 1994;269:18992–18999. [PubMed] [Google Scholar]
- 31.Chen H., Fernig D. G., Rudland P. S., Sparks A., Wilkinson M. C., Barraclough R. Binding to intracellular targets of the metastasis-inducing protein, S100A4 (p9Ka) Biochem. Biophys. Res. Commun. 2001;286:1212–1217. doi: 10.1006/bbrc.2001.5517. [DOI] [PubMed] [Google Scholar]
- 32.Fernig D. G. Optical biosensor techniques to analyse protein–polysaccharide interactions. Methods Mol. Biol. 2001;171:505–518. doi: 10.1385/1-59259-209-0:505. [DOI] [PubMed] [Google Scholar]
- 33.Liu D., Rudland P. S., Sibson D. R., Barraclough R. Identification of mRNAs differentially-expressed between benign and malignant breast tumour cells. Br. J. Cancer. 2002;87:423–431. doi: 10.1038/sj.bjc.6600456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reference deleted.
- 35.Rustandi R. R., Baldisseri D. M., Inman K. G., Nizner P., Hamilton S. M., Landar A., Zimmer D. B., Weber D. J. Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR. Biochemistry. 2002;41:788–796. doi: 10.1021/bi0118308. [DOI] [PubMed] [Google Scholar]
- 36.MacKerell A. D. Molecular modeling and dynamics of neuropeptide Y. J. Comput. Aided Mol. Des. 1988;2:55–63. doi: 10.1007/BF01532053. [DOI] [PubMed] [Google Scholar]
- 37.Avram S., Movileanu L., Mihailescu D., Flonta M. L. Comparative study of some energetic and steric parameters of the wild type and mutants HIV-1 protease: a way to explain the viral resistance. J. Cell. Mol. Med. 2002;6:251–260. doi: 10.1111/j.1582-4934.2002.tb00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arumugam T., Simeone D. M., Schmidt A. M., Logsdon C. D. S100P stimulates cell proliferation and survival via RAGE. J. Biol. Chem. 2004;279:5059–5065. doi: 10.1074/jbc.M310124200. [DOI] [PubMed] [Google Scholar]
- 39.Okada M., Hatakeyama T., Itoh H., Tokuta N., Tokumitsu H., Kobayashi R. S100A1 is a novel molecular chaperone and a member of the Hsp70/Hsp90 multichaperone complex. J. Biol. Chem. 2004;279:4221–4233. doi: 10.1074/jbc.M309014200. [DOI] [PubMed] [Google Scholar]
- 40.Koltzscher M., Gerke V. Identification of hydrophobic amino acid residues involved in the formation of S100P homodimers in vivo. Biochemistry. 2000;39:9533–9539. doi: 10.1021/bi000257+. [DOI] [PubMed] [Google Scholar]
- 41.Osterloh D., Ivanenkov V. V., Gerke V. Hydrophobic residues in the C-terminal region of S100A1 are essential for target protein binding but not for dimerization. Cell Calcium. 1998;24:137–151. doi: 10.1016/s0143-4160(98)90081-1. [DOI] [PubMed] [Google Scholar]
- 42.Most P., Remppis A., Weber C., Bernotat J., Ehlermann P., Pleger S. T., Kirsch W., Weber M., Uttenweiler D., Smith G. L., et al. The C terminus (amino acids 75–94) and the linker region (amino acids 42–54) of the Ca2+-binding protein S100A1 differentially enhance sarcoplasmic Ca2+ release in murine skinned skeletal muscle fibers. J. Biol. Chem. 2003;278:26356–26364. doi: 10.1074/jbc.M303338200. [DOI] [PubMed] [Google Scholar]