Abstract
The dual signal approach, i.e. a mitochondrial signal at the N-terminus and an ER (endoplasmic reticulum) or a peroxisomal signal at the C-terminus of EGFP (enhanced green fluorescent protein), was employed in transfected HeLa cells to test for a co-translational import model. The signal peptide from OTC (ornithine transcarbamylase) or arginase II was fused to the N-terminus of EGFP, and an ER or peroxisomal signal was fused to its C-terminus. The rationale was that if the free preprotein remained in the cytosol, it could be distributed between the two organelles by using a post-translational pathway. The resulting fusion proteins were imported exclusively into mitochondria, suggesting that co-translational import occurred. Native preALDH (precursor of rat liver mitochondrial aldehyde dehydrogenase), preOTC and rhodanese, each with the addition of a C-terminal ER or peroxisomal signal, were also translocated only to the mitochondria, again showing that a co-translational import pathway exists for these native proteins. Import of preALDHsp–DHFR, a fusion protein consisting of the leader sequence (signal peptide) of preALDH fused to DHFR (dihydrofolate reductase), was studied in the presence of methotrexate, a substrate analogue for DHFR. It was found that 70% of the preALDHsp–DHFR was imported into mitochondria in the presence of methotrexate, implying that 70% of the protein utilized the co-translational import pathway and 30% used the post-translational import pathway. Thus it appears that co-translational import is a major pathway for mitochondrial protein import. A model is proposed to explain how competition between binding factors could influence whether or not a cytosolic carrier protein, such as DHFR, uses the co- or post-translational import pathway.
Keywords: co-translational import, dual signals, green fluorescent protein import, HeLa cell protein import, methotrexate, mitochondrial protein import
Abbreviations: AII, arginase II; ALDH, aldehyde dehydrogenase; DHFR, dihydrofolate reductase; DMEM, Dulbecco's modified Eagle's medium; ER, endoplasmic reticulum; the suffix-ER denotes the addition of a 40-amino-acid peptide that targets the protein to the endoplasmic reticulum; (E)GFP, (enhanced) green fluorescence protein; HA, haemagglutinin; Hsp, heat shock protein; MPP, mitochondrial processing peptidase; OTC, ornithine transcarbamylase; preOTCsp, preAIIsp and preALDHsp, signal peptides of preOTC, preAII and preALDH respectively; PTS, peroxisomal targeting signal; rhodanese-(Δ2–30), rhodanese variant lacking residues 2–30 that is not imported to mitochondria; Tom, translocase of the outer membrane of mitochondria
INTRODUCTION
Protein trafficking has been investigated in both prokaryotes and eukaryotes for many years. As might be expected, the mechanism of trafficking into each subcellular organelle differs. In almost all cases, however, an amino acid sequence on the protein is involved in the recognition process. The location and nature of the signals differ among the various proteins. For example, proteins destined for the peroxisomes possess either an N-terminal signal called PTS2 (peroxisomal targeting signal 2) or a C-terminal signal called PTS1 [1,2]. PTS1 is also used for targeting of proteins to glycosomes and glyoxysomes [3]. In contrast, proteins destined for the mitochondria typically have an extension of amino acids only at their N-terminal end [4,5]. There appear to be at least two different ways that a protein can be trafficked to the ER (endoplasmic reticulum): one uses a signal located at the C-terminal end of the protein, and the other uses a signal that is located at the N-terminal end [6]. The latter requires a complex mechanism that involves a docking protein referred to as the signal recognition particle receptor [7–9].
The point during protein translation at which trafficking occurs has not been fully established. That is, does trafficking occur while the protein is still being synthesized, or does it occur only after the entire protein has been made? Obviously, if the signal is located at the C-terminal end of the protein, it is not possible for the protein to associate with the organelle's import apparatus until the entire protein has been synthesized and leaves the ribosome. If, though, the signal is located at the N-terminal, as one finds with most mitochondrial matrix space proteins [4,5,10–12], then it is possible for trafficking to occur as the protein emerges from the ribosome.
Soon after it was realized that proteins destined for the mitochondria possess signals that are located in a processable leader sequence found at its N-terminus, it became possible to perform in vitro transport studies with newly synthesized proteins [13]. The finding that import occurred under these conditions showed that the event was truly post-translational, in that the entire protein was synthesized prior to the addition of isolated mitochondria. However, during the past decade, experimental evidence has been presented showing that in vivo import could occur while the protein was being synthesized [14,15]. Our studies have also shown that mitochondrial import in HeLa cells could be a co-translational event. We used a carrier protein possessing a signal for one organelle at its N-terminus (mitochondria) and a different signal for another organelle at its C-terminus (ER), and found the protein only in mitochondria. The interpretation of this observation was that import in transfected HeLa cells could be a co-translational event [16].
The carrier protein employed in the previous HeLa cell experiment was GFP (green fluorescent protein), and only one mitochondrial leader sequence, from rat liver ALDH (aldehyde dehydrogenase), was used. Here we expand the study to use various mitochondrial proteins, as well as different leader sequences fused to EGFP (enhanced GFP). A peroxisomal C-terminal signal was also used, in addition to the ER one used in the original experiments. The purpose was to establish how general the cotranslational model that we proposed is. Lastly, DHFR (dihydrofolate reductase) was used as a carrier protein, so that import could be studied in the presence of methotrexate, a drug that binds irreversibly to folded DHFR [17–19]. This system was employed to eliminate the possibility that differential rates of import into respective organelles could have contributed to the finding of only mitochondrial import in the dual signal construct. The data presented show that a co-translational import model can best explain the results obtained with these various precursor proteins.
MATERIALS AND METHODS
Reagents
The pEGFP-N1 vector was purchased from Clontech; primers were from IDT, Inc.; restriction enzymes and T4 DNA ligase were from New England Biolabs; DMEM (Dulbecco's modified Eagle's medium), calf serum, trypsin-EDTA and antibiotics were from Gibco BRL; PolyFect Transfection Reagent was obtained from QIAGEN; methotrexate was purchased from Sigma; anti-HA (haemagglutinin) monoclonal antibody was from Covance; horseradish peroxidase-conjugated goat anti-mouse antibody was purchased from Amersham Pharmacia Biotech; anti-porin and anti-Erp57 were from Molecular Probes and Stressgen respectively; FITC-conjugated goat anti-(mouse IgG) was from Southern Biotechnology Associates, Inc.; chemiluminescence detection reagents and X-ray film were obtained from Perkin Elmer Life Sciences, Inc.; nitrocellulose membranes were from Schleicher and Schuell Inc.; Complete Protease Inhibitor Mixture was from Roche, Inc.
Construction of plasmids
The presequence of preOTC (precursor of human liver ornithine transcarbamylase) plus the first 10 amino acid residues from its mature region, or the presequence of preAII (precursor of human liver arginase II) plus the first five amino acids from its mature region, was fused to the N-terminus of EGFP (a variant of native GFP) by the splicing by overlap extension PCR method [20,21]. PCR primers were designed to clone the products into pEGFP-N1. The PCR products were digested with BglII/NotI and subcloned into the pEGFP-N1 vector to make pOTC-EGFP-N1 and pAII-EGFP-N1. The PCR products preOTCsp–EGFP-ER, preOTCsp–EGFP–SKL, preAIIsp–EGFP-ER and EGFP–SKL were also constructed by the same method and cloned into the pEGFP-N1 vector (the sp subscript denotes the signal peptide of the preprotein indicated, -ER denotes the addition of a 40-amino-acid peptide that targets the protein to the endoplasmic reticulum, and −SKL denotes addition of the three amino acids Ser-Lys-Leu, which target the protein to the peroxisome). The pEGFP-N1 and pEGFP-ER vectors were described in a previous study [16].
Two primers encoding the HA-tag amino acid sequence with a stop codon and SpeI, EcoRV, MluI and ScaI sites were annealed and cloned into the pEGFP-N1 vector already digested with AgeI/NotI, to make the plasmid pHAC. To make the pHAER construct, a PCR product containing the ER targeting signal from rat microsomal ALDH3 was digested with ScaI/NotI and cloned into ScaI/NotI-digested pHAC vector. For ALDH, the PCR product encoding either preALDH or mature ALDH was digested by KpnI/BamHI and inserted into KpnI/BamHI-digested pHAC or pHAER. For rhodanese and OTC, the PCR products were digested with SalI/XmaI and inserted into SalI/XmaI-digested pHAC or pHAER. Using the same restriction enzymes, rhodanese-(Δ2–30) (a rhodanese variant lacking residues 2–30 that is not imported to mitochondria [22]) was also inserted into pHAC or pHAER. For DHFR, the PCR products encoding either DHFR or rat preALDH leader sequence fused to DHFR were digested with KpnI/BamHI and inserted into KpnI/BamHI-digested pHAC or pHAER.
To make the constructs containing a C-terminal peroxisome targeting signal, a PCR product encoding the amino acid sequence of the HA tag and an additional serine-lysine-leucine sequence followed by a stop codon was digested with AgeI/NotI and exchanged into all constructs with the C-terminal ER targeting signal. All DNA fragments obtained from PCR were confirmed by DNA sequencing.
Cell line and culture conditions
HeLa cells were provided by Dr Steven Broyles (Department of Biochemistry, Purdue University). The HeLa cells were cultured in DMEM supplemented with 10% (v/v) calf serum and 10 μg/ml gentamycin at 37 °C and 5% CO2. Transfection was performed as described previously [16].
Direct observation of EGFP fluorescence in cultured cells
This work was performed as described in [16].
Immunofluorescence assay
Transfected and non-transfected HeLa cells were plated on to 10-well slides. The slides were fixed in acetone/methanol (50:50, v/v) at −20 °C for 7 min and then incubated in 20% (v/v) goat serum/PBS at 4 °C overnight. The primary antibody for HA was used at 1:1000 dilution, and the slides were incubated at room temperature for 1 h. The unbound primary antibody was removed by washing the slides three times with PBS. The slides were then incubated with FITC-conjugated goat anti-mouse antibody at room temperature for 30 min; the secondary antibody was used at a 1:200 dilution. Excess secondary antibody was removed by washing the slides three times in PBS. Fluorescence microscopy was performed using an Olympus BX60 Fluorescence Microscope.
Immunoblot analysis
Whole-cell extracts were separated by SDS/PAGE and electrotransferred on to a nitrocellulose membrane. Monoclonal antibodies against GFP and the HA-tag were used. Horseradish peroxidase-conjugated goat anti-mouse antibody or alkaline phosphatase-conjugated anti-mouse antibody was employed as the secondary antibody by using standard methods.
Subcellular fractionation of HeLa cells
Culture and transfection of HeLa cells were performed as described above. Between 16 and 24 h after transfection, the cells were washed twice with ice-cold PBS, harvested, and then resuspended in ice-cold 5 mM Hepes, pH 7.4, containing 0.32 M sucrose. Subcellular fractionation was performed as described [16,23–26]. The different fractions were analysed by Western blot using anti-EGFP antibody. Antibodies against porin and Erp57 were used as mitochondrial and ER markers respectively.
Effect of methotrexate on the import of preALDHsp–DHFR in HeLa cells
Approx. 1.6×106 HeLa cells were placed on 10 cm plates in 8 ml of growth medium (DMEM plus 10% calf serum) the day before transfection. The cells, which were 40–80% confluent on the day of transfection, were transfected with 6 μg of plasmid DNA at 37 °C using PolyFect Transfection Reagent. At 4 h after transfection, methotrexate was added to a final concentration of 0, 0.2, 0.5 or 1 mM. Between 16 and 24 h after the addition of methotrexate, the cells were harvested for immunofluorescence microscopy using an Olympus BX60 Fluorescence Microscope and Western blotting.
RESULTS
Subcellular localization of chimaeras containing different signal peptides fused to EGFP detected by fluorescence microscopy and subcellular analysis in transfected HeLa cells
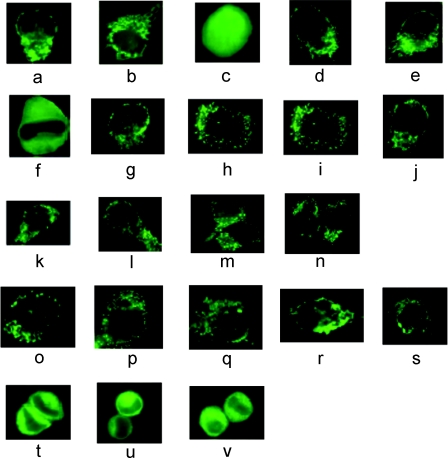
Previously, we showed that when both a mitochondrial and an ER signal were fused to EGFP, the resulting protein was translocated only to mitochondria, with no ER localization being observed [16]. To explain this, it was proposed that this construct was imported by a co-translational import pathway. We were interested to determine if other preproteins also follow the co-translational pathway. PreOTC and preAII were used for this purpose. These preproteins were selected because both are inducible enzymes, in contrast with ALDH, which is a constitutive enzyme. The mammalian expression vectors pOTC-EGFP and pAII-EGFP were used to transfect HeLa cells. After 16–24 h of transfection, GFP was localized to what appeared to be mitochondria (Figures 1a and 1b). As a control, HeLa cells were transfected with EGFP; after expression, green fluorescence appeared throughout the cell, indicative of non-specific localization of the protein (Figure 1c). To determine if the import mechanism was co-translational or post-translational, we took the advantage of our previously published method that utilized two different signals fused to EGFP [16]. The C-terminal signal peptide of microsomal ALDH, necessary for ER targeting, was fused to both preOTCsp–EGFP and preAIIsp–EGFP to make preOTCsp–EGFP-ER and preAIIsp–EGFP-ER respectively. When these two proteins were expressed in HeLa cells, only mitochondrial fluorescence was observed (Figures 1d and 1e). To verify that the ER signal could be active in vivo, EGFP possessing only the ER signal was expressed in HeLa cells. Now the fluorescence appeared to be associated with the ER (Figure 1f), confirming that the ER targeting signal could function independently. Thus it appeared that, like preALDHsp–EGFP-ER [16], both preOTCsp–EGFP-ER and preAIIsp–EGFP-ER followed the co-translational import pathway.
Figure 1. Fluorescence microscopy of HeLa cells transiently expressing EGFP fusion or HA-tagged proteins.
HeLa cells were cultured on coverslips and transfected with 2.5 μg of plasmid DNA. Fluorescence microscopy was used to localize the EGFP constructs. HA-tagged proteins were detected using immunofluorescence. The expressed proteins that lacked a mitochondrial signal were distributed throughout the cell, including the nucleus. The proteins with mitochondrial signals were localized to mitochondria, and are seen as either small dots or long cylinders. The absence of fluorescence in the cytosol and nucleus is indicative of efficient mitochondrial import. The proteins that had only an ER or peroxisomal signal were localized to the ER, appearing as a lacy network of membranes, or the peroxisome, appearing as bright dots. (a) PreOTCsp–EGFP, (b) preAIIsp–EGFP, (c) EGFP, (d) preOTCsp–EGFP-ER, (e) preAIIsp–EGFP-ER, (f) EGFP-ER, (g) preOTCsp–EGFP–SKL, (h) EGFP–SKL, (i) preALDHsp–HA, (j) rhodanese–HA, (k) preOTCsp–HA, (l) ALDH–HA–SKL, (m) OTC–HA–SKL, (n) (Δ2–30)-rhodanese–SKL, (o) preALDHsp–HA–SKL, (p) preALDHsp–HA-ER, (q) preOTCsp–HA-ER, (r) preOTCsp–HA–SKL, (s) rhodanese–HA–SKL, (t) ALDH-ER, (u) OTC-ER, (v) (Δ2–30)-rhodanese-ER.
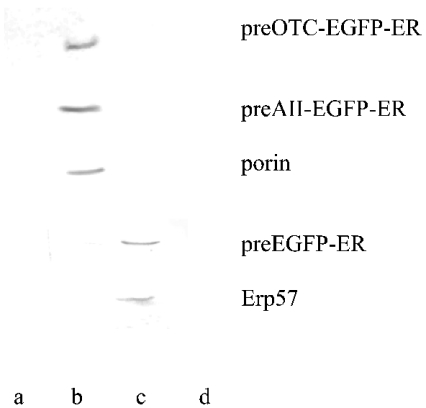
To confirm that preOTCsp–EGFP-ER and preAIIsp–EGFP-ER were imported into mitochondria only, the intracellular distributions also were analysed by subcellular fractionation. The separation of a mitochondria-rich fraction from the ER was confirmed by using Western blots of marker proteins. For the mitochondrial fraction, a monoclonal antibody against human mitochondrial outer-membrane porin was used, and anti-Erp57 was used as a marker for the ER. In Figure 2, it is shown that a monoclonal antibody against EGFP recognized the protein fraction obtained only from mitochondria, and this fraction was also recognized by the antibody against porin. The ER fraction obtained from this experiment was verified by blotting with anti-Erp57. For the EGFP-ER construct, the antibody against EGFP recognized the proteins obtained from the ER fraction. This fraction was also recognized by the antibody against Erp57 (Figure 2). Here also the mitochondrial fraction was verified by blotting with anti-porin (results not shown). Therefore the subcellular localization analysis indicated that EGFPs possessing the dual signals were transported primarily to mitochondria. These findings support the immunofluorescence data, indicating that a co-translational import pathway could exist for these two constructs.
Figure 2. Western blot analysis of preOTCsp–EGFP-ER, preAIIsp–EGFP-ER and EGFP-ER expressed in HeLa cells and after subcellular fractionation.
Subcellular fractionation was described in the Materials and methods section. Different organelles were subjected to Western blot analysis. PreOTCsp–EGFP-ER, preAIIsp–EGFP-ER and EGFP-ER were each transfected into HeLa cells and detected by the anti-EGFP antibody. Human porin was detected using its monoclonal antibody, and served as a mitochondrial marker. Anti-Erp57 antibody was used as an ER-specific marker. Lanes a, b, c and d represent nucleus-, mitochondria-, ER- and cytosol-rich fractions respectively.
A C-terminal peroxisomal signal was then used in the dual leader study to determine if a different C-terminal signal would affect subcellular localization. SKL (Ser-Lys-Leu), the tripeptide C-terminal signal involved in peroxisomal protein import [2], was fused to the C-terminus of preOTCsp–EGFP to produce preOTCsp–EGFP–SKL. What appeared to be a mitochondrial localization was observed in HeLa cells expressing this chimaeric protein that contained both signals (Figure 1g). When EGFP–SKL, a chimaeric protein containing only the peroxisomal signal, was expressed, green fluorescence was localized in what appeared to be the peroxisome (Figure 1h). Since the appearance of mitochondrial and peroxisomal localization did not differ much, immunoblot assays were performed to distinguish between mitochondrial and peroxisomal localizations (see below). Independent of the leader sequence or the C-terminal targeting signal, all of the dual-leader-containing proteins were found exclusively in mitochondria, consistent with what we demonstrated previously [16].
Subcellular localization of native preproteins in transfected HeLa cells detected by fluorescence microscopy
In the previous section, EGFP was used as the passenger protein. To rule out any effect of EGFP, a rapidly folding protein [27], on the import of different chimaeric proteins, we used native precursor proteins. The HA-tag was fused to the C-terminus of each preprotein to allow us to detect them by immunofluorescence using a FITC-conjugated secondary antibody against the anti-HA monoclonal antibody. Native mature mitochondrial proteins without a leader sequence were distributed throughout the whole cell, as detected by fluorescence microscopy (results not shown), showing that no specific targeting to any subcellular organelle was observed for constructs lacking a leader sequence. In contrast, mitochondria-associated fluorescence was observed for cells expressing the native precursor mitochondrial proteins preALDH (Figure 1i), rhodanese (Figure 1j) and preOTC (Figure 1k). ER-specific fluorescence (a lacy network of membranes surrounding the nucleus [16]) was observed when the ER targeting signal was fused to the mature part of preALDH, preOTC or rhodanese-(Δ2–30) [22] (Figures 1t, 1u and 1v). Similarly, when the peroxisome targeting signal was fused to the mature part of preALDH and preOTC or to rhodanese-(Δ2–30), the fluorescence was localized in what appeared to be peroxisomes (bright dots surrounding the nucleus) (Figures 1l, 1m and 1n). The cells expressing the constructs with dual leaders (the mitochondrial leader at the N-terminus and either the ER or the peroxisome signal at the C-terminus) showed fluorescence only in the mitochondria, with no detectable fluorescence associated with the ER or peroxisome (Figures 1o–1s). Since mitochondria and peroxisomes are very similar in shape and size, we performed immunoblotting (see below) to differentiate between them.
Subcellular localization in transfected HeLa cells of the chimaeras containing different signal peptides fused to EGFP and of native preproteins detected on the basis of molecular mass using immunoblot analysis
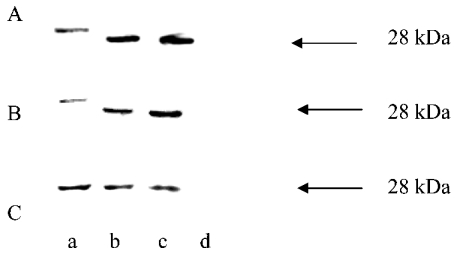
The leader sequences of most precursor proteins are removed by the MPP (mitochondrial processing peptidase) after their import into mitochondria. The difference in molecular mass between the precursor and the mature form was used to determine whether or not the expressed chimaeras were imported into mitochondria, similarly to what we showed previously [16]. EGFP, a 28 kDa protein, was detected using the monoclonal antibody in HeLa cell extracts after the cells were transfected with the pEGFP vector. This protein was used as a molecular mass marker protein in subsequent Western blots (Figure 3). PreOTCsp–EGFP, preOTCsp–EGFP-ER, preAIIsp–EGFP and preAIIsp–EGFP-ER were expressed separately in HeLa cells. The data show that the leader sequence was removed from each of them (Figures 3A and 3B). This implies that these proteins were localized to the mitochondrial matrix space. Similarly, preOTCsp–EGFP–SKL after expression had a molecular mass identical to what would be expected after removal of the leader sequence (Figure 3C). Western blot analysis showed that these constructs were all processed, implying that they must have been imported into the mitochondria independent of their C-terminal sequence.
Figure 3. Western blot analysis of EGFP-containing constructs expressed in HeLa cells.
Whole-cell extracts of HeLa cells expressing each EGFP-containing construct were separately resolved by SDS/PAGE and identified by Western blots with anti-EGFP antibody. The non-transfected HeLa cells were used as a negative control. Expressed EGFP of molecular mass 28 kDa was used as the marker indicated by the arrow. (A) Lanes: a, preOTCsp–EGFP-ER; b, preOTCsp–EGFP; c, EGFP; d, untransfected cells. (B) Lanes: a, preAIIsp–EGFP-ER; b, preAIIsp–EGFP; c, EGFP; d, untransfected cells. (C) Lanes: a, preOTCsp–EGFP–SKL; b, EGFP–SKL; c, EGFP; d, untransfected cells.
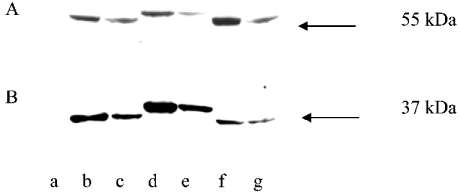
A similar set of experiments using native mammalian proteins was performed. The molecular mass of preALDH or preALDHsp–SKL is approx. 57 kDa; after import into the mitochondria, the leader sequence would be removed by MPP so that the molecular mass of the protein would be decreased to 55 kDa. The molecular mass of preALDHsp-ER is approx. 61 kDa, and that of mature ALDH-ER protein is 59 kDa. HeLa cells expressing preALDH (Figures 4A, lane b) or preALDHsp–SKL (Figure 4A, lane f) each produced a 55 kDa protein band, indicating that these proteins must have been imported into mitochondria and the leader sequences removed by MPP. The molecular mass found for ALDH-ER expressed in HeLa cells was 59 kDa, as expected for the construct associating with the ER (Figure 4A, lane e). The mass of ALDH–SKL expressed in HeLa cells was 55 kDa, as expected for this construct (Figure 4A, lane g). A processed protein of 59 kDa was observed from cells expressing preALDHsp-ER, indicative of leader sequence removal by MPP and hence mitochondrial localization (Figure 4A, lane d). These results suggest that, since the molecular masses of the products were those of the expected imported and processed proteins, preALDHsp-ER and preALDHsp–SKL were localized exclusively in the HeLa cell mitochondria. No precursor-size proteins were observed by Western blotting of HeLa cells expressing preALDH, preALDHsp-ER or preALDHsp–SKL, suggesting that the precursor proteins did not remain in the cytosol, nor were they translocated to the ER or peroxisomes.
Figure 4. Western blot analysis of precursor proteins expressed in HeLa cells.
Whole-cell extracts of HeLa cells expressing the native proteins were resolved by SDS/PAGE and identified by immunofluorescence using the monoclonal anti-HA antibody. (A) HeLa cells expressing preALDH constructs. Lane a, untransfected HeLa cell extract; lane b, preALDH; lane c, mature ALDH; lane d, preALDHsp-ER; lane e, ALDH-ER; lane f, preALDHsp–SKL; lane g, ALDH–SKL. The arrow indicates the position of a 55 kDa protein band. (B) HeLa cells expressing OTC constructs. Lane a, untransfected HeLa cells; lane b, mature OTC; lane c, preOTC; lane d, OTC-ER; lane e, preOTCsp-ER; lane f, OTC–SKL; lane g, preOTCsp–SKL. The arrow indicates the position of a 37 kDa protein band.
Again, cells expressing preOTC, preOTCsp-ER or preOTCsp–SKL produced protein bands after SDS/PAGE corresponding to the sizes of the processed products, indicating that each protein was localized to mitochondria. No protein bands corresponding to the sizes expected for ER or peroxisome localization were detected (Figure 4B).
Import of preALDHsp–DHFR into HeLa cell mitochondria in the presence of methotrexate
The results obtained led us to conclude that import was primarily a co-translational event. However, we cannot rule out the possibility that the rate of import into mitochondria was much higher than were the rates of ER or peroxisomal import. If this were case, almost all of the dual leader proteins would be found in mitochondria even though they followed the post-translational pathway. To rule out this possibility, we changed the carrier to DHFR and studied import in the presence of methotrexate. This drug is a substrate analogue and binds essentially irreversibly to DHFR. If DHFR were free in the cytosol, an event necessary for the post-translational import pathway, then it would bind the drug and would be in an import-incompatible conformation. It has already been shown that methotrexate prevented the in vitro import of a chimaera consisting of the signal peptide of cytochrome oxidase subunit IV fused to DHFR [17–19]. The most likely reason is that the drug can stabilize the folded state, a conformation that is not compatible with import. However, it was reported that methotrexate did not inhibit the import of this fusion protein in yeast cells, strongly suggesting that co-translational import occurred [14,15].
HeLa cells were transfected with preALDHsp–DHFR- or DHFR-expressing plasmids. Methotrexate was added to the cells after 4 h of transfection and the cells were then incubated for 16–24 h. The methotrexate-treated cells were harvested and immunoblot analysis was performed to determine if methotrexate affected the import of preALDHsp–DHFR. Untreated cells were harvested to serve as a control.
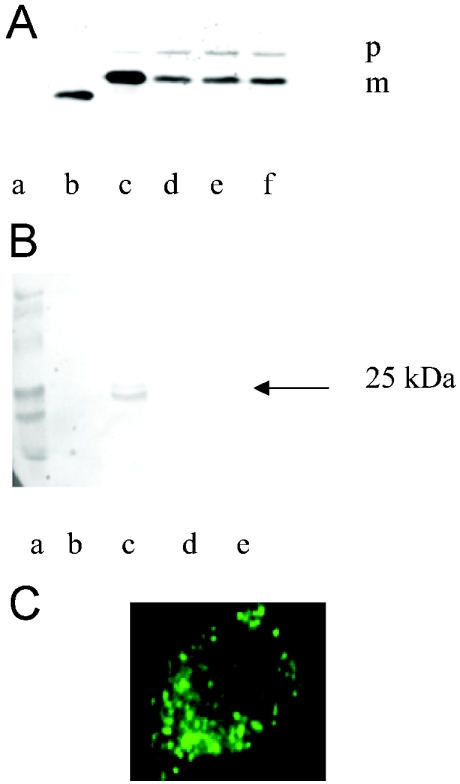
Two protein bands were observed from methotrexate-treated cells after Western blot (Figure 5A). One had a molecular mass of approx. 27 kDa, corresponding to the size of preALDHsp–DHFR; the other was 25 kDa, corresponding to the size of mature protein (DHFR plus 20 amino acids from mature ALDH). The presence of preALDHsp–DHFR implied that some of the precursor protein was not imported into mitochondria; if it were, it would have been processed. The non-processed protein was probably bound to methotrexate and hence remained in the cytosol. The ratio between the precursor form and the imported mature form was approx. 3:7; this ratio was found to be independent of the three different methotrexate concentrations employed. In the absence of methotrexate, >95% of the precursor protein was imported and processed (Figure 5A, lane c).
Figure 5. Effects of methotrexate on preALDHsp–DHFR import in HeLa cells.
(A) Effect analysed by Western blot. Whole-cell extracts of HeLa cells expressing preALDHsp–DHFR were resolved by SDS/PAGE and identified by immunoblot using the monoclonal anti-HA antibody. Lane a, untransfected cells; lane b, DHFR plus methotrexate (1 mM); lane c, preALDHsp–DHFR without methotrexate; lanes d–f, preALDHsp–DHFR with increasing concentrations (0.2, 0.5 and 1 mM respectively) of methotrexate. The ratio of precursor (indicated by p) to mature (indicated by m) protein was approx. 3:7 in the presence of methotrexate and 0.05:1 in its absence. (B) Effect analysed by Western blot after subcellular fractionation. PreALDHsp–DHFR was transfected into HeLa cells in the presence of methotrexate and, after expression of the proteins, subcellular fractionation was performed. The different fractions, i.e. nucleus (lane b), mitochondria (lane c), ER (lane d) and cytosol (lane e), were analysed by immunoblotting using the anti-HA antibody. Lane a contains molecular mass markers (Bio-Rad). (C) Fluorescence microscopy of HeLa cells transiently expressing HA-tagged preALDHsp–DHFR in the presence of methotrexate. HeLa cells were cultured on coverslips and transfected with 2.5 μg of plasmid DNA. After 4 h of transfection, 1 mM methotrexate was added to the medium. After 16–24 h of transfection, cells were isolated and used in immunofluorescence assays using anti-HA monoclonal antibody and FITC-labelled secondary antibody. The fluorescence pattern was similar to those in Figure 1 when mitochondrial import occurred.
A possible explanation for the presence of the precursor form of preALDHsp–DHFR in the cell extract is that methotrexate affected the general mitochondrial import machinery. In order to rule out this possibility, the experiment was performed with a native preALDH construct in the presence of the drug. Only processed ALDH was detected by Western blot analysis (results not shown). The absence of preALDH showed that methotrexate did not inhibit the general import machinery. This means that the existence of some precursor proteins in the presence of the drug was the result of import not being exclusively co-translational. We cannot, however, from these data exclude the possibility of a scenario where the drug binds to folded DHFR outside the mitochondria (a post-translational pathway) and only the leader portion of the DHFR construct enters the mitochondrial matrix, where it is processed by the MPP. If this were occurring, immunoblotting of the whole-cell extract would not allow us to conclude that processed DHFR would be only in the mitochondrial matrix. To exclude this possibility, after transfection of HeLa cells with preALDHsp–DHFR in the presence of 1 mM methotrexate, subcellular fractionation and immunofluorescence microscopy was performed. Immunobloting was carried out with nucleus-, mitochondria-, ER- and cytosol-rich fractions. Only the mitochondrial fraction showed positive interactions with the anti-HA antibody (Figure 5B), suggesting that preALDHsp–DHFR was transported into mitochondria even in the presence of the drug. Immunofluorescence microscopy with the anti-HA antibody and FITC-linked secondary antibody showed that the green fluorescence was localized only in the mitochondria (Figure 5C). These data support the notion that preALDHsp–DHFR was imported into the mitochondria in the presence of methotrexate. Hence a co-translational import model is most suitable to explain how the proteins entered the mitochondria. The molecules remaining in the cytosol in the presence of methotrexate must have followed the post-translational pathway.
DISCUSSION
Pioneering work from the Schatz [28] and Neupert [13] laboratories demonstrated that proteins produced in an in vitro translation system could be imported into isolated mitochondria, and hence the post-translational import pathway was established. The necessity of heat shock proteins strengthened the idea of the post-translational pathway [29–32]. During the past decade we, among others, have occasionally found a precursor protein that was not imported under the in vitro assay conditions. For example, the leader from preALDH would not allow EGFP to be imported [33], and we found that isopropylmalate synthase 4 could not be imported into isolated mitochondria [34]. It is possible that the leader interacted with the mature portion of the protein so that it was not available to bind to the import apparatus [35]. Alternatively, it is possible that the mature portion of the protein affected the conformation of the leader, such that a stable helix, a presumed prerequisite for import, could not form. A still different explanation for the finding that not all precursor proteins could be imported in vitro is that, for some, folding is faster than import. This implies that the only way these proteins could be imported in vivo would be by a co-translational import mechanism. Co-translational import implies that the precursor is not free in the cytosol, and might be imported as it is coming off the ribosome.
Among the first experimental evidence showing that mitochondrial import could be co-translational came from Verner's laboratory [15]. They showed that methotrexate did not inhibit the in vivo import of a chimaera consisting of the signal peptide of cytochrome oxidase subunit IV fused to DHFR in yeast cells, whereas it inhibited in vitro import into isolated yeast mitochondria. They concluded that the in vivo import commences before complete synthesis of the precursor protein, i.e. that it followed a co-translational pathway [15]. Crowley and Payne [36] came to a similar conclusion when they observed that ribosomes were bound to mitochondria. More recently, MacKenzie and Payne [37], using a different approach, again proposed that co-translational import occurs, as did Funfschilling and Rospert [38]. A genomic approach was used by Marc et al. [39] to conclude that a co-translational import pathway should exist.
Our laboratory employed a two-signal system to investigate whether co-translational import could occur in vivo in HeLa cells. EGFP was used as a carrier protein; a mitochondrial leader sequence was placed at the N-terminus, and an ER signal was placed at the C-terminus, of EGFP. The rationale was that if the proteins were free in solution after synthesis, then a portion would be found in both organelles. The green fluorescence was found only associated with mitochondria [16], inconsistent with a post-translational pathway.
In the above-mentioned in vivo import experiments with HeLa cells, the preALDH mitochondrial leader fused to EGFP-ER was used. In the present study, we used the leader peptides from preOTC and preAII fused to EGFP-ER. After expression, both were found only in mitochondria, with no ER-associated fluorescence being detected. This showed that, like preALDHsp–EGFP-ER, these two precursor proteins also followed the co-translational import pathway. We next used a C-terminal peroxisomal signal to examine if it altered the import pathway of the dual-leader-containing proteins. PreOTCsp–EGFP–SKL was found only in mitochondria, showing that it too followed the co-translational import pathway. These data indicated that a co-translational import mechanism could occur independent of both the leader sequence and the signal attached to the C-terminus of EGFP.
EGFP was used as a carrier protein in the in vivo import into HeLa cell mitochondria, to facilitate detection using fluorescence microscopy. EGFP, however, is a very rapidly folding protein [27], and its folding may affect the import pathway of the different proteins employed. To exclude that possibility, native preALDH, preOTC and rhodanese were used in the present study. The three preproteins contained their native mitochondrial N-terminal signal, and either an ER or an SKL signal was placed at their C-terminus. The data obtained from microscopy and immunoblotting analysis made it appear that these fusion proteins were translocated only to the mitochondria. This showed that the native pro-teins were also imported using the co-translational mechanism, just as we found with the EGFP adducts.
The methotrexate experiments with DHFR adducts were designed in order to rule out the possibility that only mitochondrial import was found because of differential rates of import into the organelles. We used methotrexate to bind DHFR in order to inhibit any post-translational mitochondrial import of preALDHsp–DHFR in HeLa cells. The observation that just 70% of the total protein (preALDHsp–DHFR) was processed due to translocation to the mitochondrial matrix was unexpected. The unprocessed protein fraction must have been located in the cytosol, since no unprocessed protein was found in the mitochondria. The drug should not have affected the localization of the protein if only a co-translational import pathway was being followed. If import followed only a post-translational import pathway, then the drug should have completely prevented the protein from being imported to the matrix space, independent of the rate of import into individual organelles. Since the drug did not inhibit the general import machinery, it is suggested that 70% of this preprotein followed the co-translational pathway. The 30% of the DHFR adduct found in the cytosol must have been free to bind methotrexate, and hence could not be imported.
The following argument can be offered to explain why only 70% of the protein was found in mitochondria in the presence of methotrexate, compared with >95% in its absence. First, we can conclude that 70% of the precursor protein was imported by the co-translational mechanism. The other 30% could have gone into the cytosol and initially remained in an unfolded state. A portion of the unfolded protein might use the post-translational pathway to be imported. Any fraction that became stably folded would remain in the cytosol. In the absence of methotrexate, the rate of import of the DHFR adduct seems dominant, as only at most a small percentage of the protein remained in the cytosol. In contrast, when methotrexate was present, the DHFR in the cytosol became folded and was not imported into mitochondria.
When using two localization signals attached to a protein, only mitochondrial localization was found. Based on the methotrexate results, one might have expected to have observed some non-mitochondrial localization, since 30% of the two-leader protein might have followed the post-translational import pathway, as we found with the DHFR adduct. Although we cannot offer a definitive explanation for this difference, we propose that it might be related to DHFR being a cytosolic enzyme. It is known that Hsp70 (heat shock protein 70) is involved in keeping a protein unfolded [31]. We have shown that if the leader sequence from ALDH was fused to the liver cytosolic isoenzyme of ALDH, very little in vitro import occurred [40]. If, however, the first 21 amino acids from the mature portion of the mitochondrial enzyme replaced the first 21 residues of the cytosolic isoenzyme, then import was restored. An explanation for this observation is that a cytosolic factor, perhaps a heat shock protein, may bind to the newly synthesized protein as it is coming off the ribosome [40]. If the protein is one that should remain in the cytosol, the factor that binds to it will differ from that which binds if the protein is destined for the mitochondrial matrix space. In the case of the natural precursor proteins used in the present study, only mitochondrial import was found, consistent with the concept that either a true co-translocation event occurs or the cytosolic factor that might bind to them prevents the proteins from becoming totally free in the cytosol. This binding protein could function in an analogous manner to a docking protein necessary for ER binding [7]. In contrast, when a cytosolic protein such as DHFR is employed, there could be competition between binding factors, such that a portion of protein is found in each subcellular location. Results obtained with EGFP, a non-mammalian cytosolic protein, were identical with those obtained with mitochondrial proteins. It is possible that EGFP is not readily recognized by factors that could facilitate its folding.
The mechanism for co-translocation might have to be altered from a simple model whereby, during synthesis, the leader interacts with the import apparatus, to a model that includes a ‘dockinglike’ factor that actually assists in precursor binding to the mitochondrial membrane. This model supports the recent report that the cytosolic chaperones Hsp90 and Hsp70 deliver the preproteins to the mitochondrial import receptor Tom70 (translocase of the outer membrane of mitochondria 70) [41]. The authors proposed that Hsp90 and Hsp70 dock with Tom70, a mitochondrial outer membrane protein, and that this docking helps preproteins to be translocated subsequently to mitochondria [41].
We previously reported in vivo import via a co-translational pathway when EGFP was the carrier [16]. Here we report that at least 70% of protein is imported by the co-translational mechanism when measured with DHFR. Fujiki and Verner [14,15] showed that the bulk of in vivo mitochondrial import was blocked upon the arrest of cytosolic protein synthesis by cycloheximide. This result can be interpreted to indicate that translation and translocation are tightly coupled. Using a different approach, these authors showed that treatment of yeast cells with a proton ionophore to collapse the mitochondrial membrane potential resulted in the cytosolic accumulation of precursor proteins. After re-establishment of the membrane potential, some, but not all, precursor proteins were imported. This suggested that, for some preproteins, co-translational import might be the only pathway for their import. We cannot state what fractions of the preproteins use the co-translational pathway. The accumulated data, including those from the present study, make it appear that the co-translational pathway is important in the in vivo import of proteins into the mitochondrial matrix of HeLa cells.
Acknowledgments
This work was supported in part by National Institute of Health grants AA10795 and GM 53269. This is journal paper number 17409 from the Purdue University Agriculture Experiment Station.
References
- 1.Holroyd C., Erdmann R. Protein translocation machineries of peroxisomes. FEBS Lett. 2001;501:6–10. doi: 10.1016/s0014-5793(01)02617-5. [DOI] [PubMed] [Google Scholar]
- 2.Subramani S. Convergence of model systems for peroxisome biogenesis. Curr. Opin. Cell Biol. 1996;4:513–518. doi: 10.1016/s0955-0674(96)80029-9. [DOI] [PubMed] [Google Scholar]
- 3.Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol. Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- 4.Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- 5.Schatz G. The protein import system of mitochondria. J. Biol. Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 6.Johnson A. E., van Waes M. A. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- 7.Walter P., Johnson A. E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 8.Pool M. R., Stumm J., Fulga T. A., Sinning I., Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 9.Bacher G., Lutcke H., Jungnickel B., Rapoport T. A., Dobberstein B. Regulation by the ribosome of the GTPase of the signal-recognition particle during protein targeting. Nature (London) 1996;381:248–251. doi: 10.1038/381248a0. [DOI] [PubMed] [Google Scholar]
- 10.Pfanner N., Geissler A. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 11.von Heijne G. Protein targeting signals. Curr. Opin. Cell Biol. 1990;2:604–608. doi: 10.1016/0955-0674(90)90100-s. [DOI] [PubMed] [Google Scholar]
- 12.Schleiff E. Signals and receptors – the translocation machinery on the mitochondrial surface. J. Bioenerg. Biomembr. 2000;32:55–66. doi: 10.1023/a:1005512412404. [DOI] [PubMed] [Google Scholar]
- 13.Harmey M., Hallermayer G., Korb H., Neupert W. Transport of cytoplasmically synthesized proteins into the mitochondria in a cell free system from Neurospora crassa. Eur. J. Biochem. 1977;81:533–544. doi: 10.1111/j.1432-1033.1977.tb11979.x. [DOI] [PubMed] [Google Scholar]
- 14.Fujiki M., Verner K. Coupling of protein synthesis and mitochondrial import in a homologous yeast in vitro system. J. Biol. Chem. 1991;266:6841–6847. [PubMed] [Google Scholar]
- 15.Fujiki M., Verner K. Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast. Evidence for cotranslational import in vivo. J. Biol. Chem. 1993;268:1914–1920. [PubMed] [Google Scholar]
- 16.Ni L., Heard T. S., Weiner H. In vivo mitochondrial import. A comparison of leader sequence charge and structural relationships with the in vitro model resulting in evidence for co-translational import. J. Biol. Chem. 1999;274:12685–12691. doi: 10.1074/jbc.274.18.12685. [DOI] [PubMed] [Google Scholar]
- 17.Eilers M., Schatz G. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature (London) 1986;322:228–232. doi: 10.1038/322228a0. [DOI] [PubMed] [Google Scholar]
- 18.Gruhler A., Arnold I., Seytter T., Guiard B., Schwarz E., Neupert W., Stuart R. A. N-terminal hydrophobic sorting signals of preproteins confer mitochondrial hsp70 independence for import into mitochondria. J. Biol. Chem. 1997;272:17410–17415. doi: 10.1074/jbc.272.28.17410. [DOI] [PubMed] [Google Scholar]
- 19.Rassow J., Guiard B., Wienhues U., Herzog V., Hartl F. U., Neupert W. Translocation arrest by reversible folding of a precursor protein imported into mitochondria. A means to quantitate translocation contact sites. J. Cell Biol. 1989;109:1421–1428. doi: 10.1083/jcb.109.4.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 21.Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 22.Waltner M., Hammen P. K., Weiner H. Influence of the mature portion of a precursor protein on the mitochondrial signal sequence. J. Biol. Chem. 1996;271:21226–21230. [PubMed] [Google Scholar]
- 23.Clark B. J., Waterman M. R. The hydrophobic amino-terminal sequence of bovine 17 alpha-hydroxylase is required for the expression of a functional hemoprotein in COS 1 cells. J. Biol. Chem. 1991;266:5898–5904. [PubMed] [Google Scholar]
- 24.Masaki R., Yamamoto A., Tashiro Y. Microsomal aldehyde dehydrogenase is localized to the endoplasmic reticulum via its carboxyl-terminal 35 amino acids. J. Cell Biol. 1994;126:1407–1420. doi: 10.1083/jcb.126.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moya-Camarena S. Y., Mooré D. J., Mooré D. M. Response of NADH oxidation and growth in K-562 cells to the antitumor sulfonylurea, N-(4-methylphenylsulfonyl)-N(-(4-chlorophenyl)urea ( LY181984) Protoplasma. 1995;188:151–160. [Google Scholar]
- 26.Segade F., Hurlé B., Claudio E., Ramos S., Lazo P. S. Identification of an additional member of the cytochrome c oxidase subunit VIIa family of proteins. J. Biol. Chem. 1996;271:12343–12349. doi: 10.1074/jbc.271.21.12343. [DOI] [PubMed] [Google Scholar]
- 27.Makino Y., Amada K., Taguchi H., Yoshida M. Chaperonin-mediated folding of green fluorescent protein. J. Biol. Chem. 1997;272:12468–12474. doi: 10.1074/jbc.272.19.12468. [DOI] [PubMed] [Google Scholar]
- 28.Maccecchini M. L., Rudin Y., Blobel G., Schatz G. Import of proteins into mitochondria: precursor forms of the extramitochondrially made F1-ATPase subunits in yeast. Proc. Natl. Acad. Sci. U.S.A. 1979;76:343–347. doi: 10.1073/pnas.76.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mihara K., Omura T. Cytosolic factors in mitochondrial protein import. Experientia. 1996;52:1063–1068. doi: 10.1007/BF01952103. [DOI] [PubMed] [Google Scholar]
- 30.Komiya T., Sakaguchi M., Mihara M. Cytoplasmic chaperones determine the targeting pathway of precursor proteins to mitochondria. EMBO J. 1996;15:399–407. [PMC free article] [PubMed] [Google Scholar]
- 31.Terada K., Ohtsuka K., Imamoto N., Yoneda Y., Mori M. Role of heat shock cognate 70 protein in import of ornithine transcarbamylase precursor into mammalian mitochondria. Mol. Cell. Biol. 1995;15:3708–3713. doi: 10.1128/mcb.15.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker K., Guiard B., Rassow J., Sollner T., Pfanner N. Targeting of a chemically pure preprotein to mitochondria does not require the addition of a cytosolic signal recognition factor. J. Biol. Chem. 1992;267:5637–5643. [PubMed] [Google Scholar]
- 33.Heard T. S. Purdue University; 1999. Charge and structural components of mitochondrial leader sequences. Ph.D. Thesis. [Google Scholar]
- 34.Mukhopadhyay A., Heard T. S., Wen X., Hammen P. K., Weiner H. Location of the actual signal in the negatively charged leader sequence involved in the import into the mitochondrial matrix space. J. Biol. Chem. 2003;278:13712–13718. doi: 10.1074/jbc.M212743200. [DOI] [PubMed] [Google Scholar]
- 35.Waltner M., Weiner H. Conversion of a nonprocessed mitochondrial precursor protein into one that is processed by the mitochondrial processing peptidase. J. Biol. Chem. 1995;270:26311–26317. doi: 10.1074/jbc.270.44.26311. [DOI] [PubMed] [Google Scholar]
- 36.Crowley K. S., Payne R. M. Ribosome binding to mitochondria is regulated by GTP and the transit peptide. J. Biol. Chem. 1998;273:17278–17285. doi: 10.1074/jbc.273.27.17278. [DOI] [PubMed] [Google Scholar]
- 37.MacKenzie J. A., Payne R. M. Ribosomes specifically bind to mammalian mitochondrial via protease-sensitive proteins on the outer membrane. J. Biol. Chem. 2004;279:9803–9810. doi: 10.1074/jbc.M307167200. [DOI] [PubMed] [Google Scholar]
- 38.Funfschilling U., Rospert S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell. 1999;10:3289–3299. doi: 10.1091/mbc.10.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marc P., Margeot A., Devaux F., Blugeon C., Corral-Debrinski M., Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J., Weiner H. The N-terminal portion of mature aldehyde dehydrogenase affects protein folding and assembly. Protein Sci. 2001;10:1490–1497. doi: 10.1110/ps.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young J. C., Hoogenraad N. J., Hartl F. U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]