Abstract
The enzymic degradation of insoluble polysaccharides is one of the most important reactions on earth. Despite this, glycoside hydrolases attack such polysaccharides relatively inefficiently as their target glycosidic bonds are often inaccessible to the active site of the appropriate enzymes. In order to overcome these problems, many of the glycoside hydrolases that utilize insoluble substrates are modular, comprising catalytic modules appended to one or more non-catalytic CBMs (carbohydrate-binding modules). CBMs promote the association of the enzyme with the substrate. In view of the central role that CBMs play in the enzymic hydrolysis of plant structural and storage polysaccharides, the ligand specificity displayed by these protein modules and the mechanism by which they recognize their target carbohydrates have received considerable attention since their discovery almost 20 years ago. In the last few years, CBM research has harnessed structural, functional and bioinformatic approaches to elucidate the molecular determinants that drive CBM–carbohydrate recognition. The present review summarizes the impact structural biology has had on our understanding of the mechanisms by which CBMs bind to their target ligands.
Keywords: carbohydrate-binding module (CBM), cellulose-binding domain, lectin, protein–carbohydrate recognition, protein structure
Abbreviations: CBD, cellulose-binding domain; CBM, carbohydrate-binding module; G2M5, 63,64-α-D-galactosylmannopentaose; OB, oligonucleotide/oligosaccharide binding; WGA, wheat germ agglutinin
INTRODUCTION
The non-catalytic polysaccharide-recognizing modules of glycoside hydrolases were originally defined as CBDs (cellulose-binding domains), because the first examples of these protein domains bound crystalline cellulose as their primary ligand [1–3]. Subsequently, the more inclusive term CBM (carbohydrate-binding module) evolved to reflect the diverse ligand specificity of these modules [4]. Many CBMs have now been identified experimentally, and several hundred putative CBMs can be further identified on the basis of amino acid similarity. Similar to the catalytic modules of glycoside hydrolases, CBMs are divided into families based on amino acid sequence similarity. There are currently 39 defined families of CBMs [5] (see http://afmb.cnrs-mrs.fr/CAZY/index.html) and these CBMs display substantial variation in ligand specificity. Thus there are characterized CBMs that recognize crystalline cellulose, non-crystalline cellulose, chitin, β-1,3-glucans and β-1,3-1,4-mixed linkage glucans, xylan, mannan, galactan and starch, while some CBMs display ‘lectin-like’ specificity and bind to a variety of cell-surface glycans. In general, CBMs are appended to glycoside hydrolases that degrade insoluble polysaccharides. Although many of these modules target components of the plant cell wall, several CBM families contain proteins that bind to insoluble storage polysaccharides such as starch and glycogen. Indeed, the structure and biochemistry of several family 20 CBMs, which bind to starch, have been analysed extensively (see [6–13] for examples). Furthermore, numerous crystal structures of starch-modifying enzymes have revealed malto-oligosaccharide-binding sites that are distinct from the substrate-binding cleft, indicating that these enzymes also contain starch-binding CBMs [14–17].
In some CBM families, typically those that recognize crystalline polysaccharides, ligand specificity is invariant, while other families contain proteins that bind to a range of different carbohydrates. Thus CBMs are excellent model systems for studying the mechanism of protein–carbohydrate recognition. Furthermore, this diversity in ligand specificity underpins the exploitation of these protein modules in numerous biotechnological applications [18].
In total, the three-dimensional structures of members from 22 different CBM families are now known, many of which were determined in the last few years (Table 1). The wealth of structural information provided by NMR spectroscopic and X-ray crystallographic studies of CBMs has been invaluable in understanding the biological functions of these proteins, and has provided a foundation for research directed at dissecting the mechanisms by which they bind to oligosaccharides and polysaccharides. The recent elucidation of several CBM structures in complex with oligosaccharide ligands has provided particularly valuable insights into how these proteins recognize their target ligands. By the end of the last millennium, only two structures, representing CBM families 13 and 18, had been solved in complex with ligands by X-ray crystallography. These were the well-known plant-derived lectins ricin toxin B-chain [19] and WGA (wheat germ agglutinin) [20]. The structure of a family 20 starch-binding module in complex with β-cyclodextrin was determined by NMR spectroscopy [9]. Since the beginning of 2001, however, the structures of 15 CBMs, derived from ten different families, have been determined in complex with their oligosaccharide ligands derived both from the three major plant cell-wall polysaccharides, cellulose, xylan and mannan, as well as β-1,3-glucan and starch.
Table 1. CBM structures.
Entries representing structures solved in complex with ligand are shown in bold. PDB, Protein Data Bank.
| Family | Protein | PDB code |
|---|---|---|
| CBM1 | Cellulase 7A (Trichoderma reesei) | 1CBH |
| CBM2 | Xylanase 10A (Cellulomonas fimi) | 1EXG |
| Xylanase 11A (Cellulomonas fimi) | 2XBD | |
| Xylanase 11A (Cellulomonas fimi) | 1HEH | |
| CBM3 | Scaffoldin (Clostridium cellulolyticum) | 1G43 |
| Scaffoldin (Clostridium thermocellum) | 1NBC | |
| Cellulase 9A (Thermobifida fusca) | 1TF4 | |
| CBM4 | Laminarinase 16A (Thermotoga maritima) | 1GUI |
| Cellulase 9B (Cellulomonas fimi) | 1ULO; 1GU3 | |
| Cellulase 9B (Cellulomonas fimi) | 1CX1 | |
| Xylanase 10A (Rhodothermus marinus) | 1K45 | |
| CBM5 | Cellulase 5A (Erwinia chrysanthemi) | 1AIW |
| Chitinase B (Serratia marcescens) | 1E15 | |
| CBM6 | Xylanase 11A (Clostridium thermocellum) | 1UXX |
| Xylanase 11A (Clostridium stercorarium) | 1NAE | |
| Xylanase 11A (Clostridium stercorarium) | 1UY4 | |
| Endoglucanase 5A (Cellvibrio mixtus) | 1UZ0 | |
| CBM9 | Xylanase 10A (Thermotoga maritima) | 1I8A |
| CBM10 | Xylanase 10A (Cellvibrio japonicus) | 1QLD |
| CBM12 | Chitinase Chi1 (Bacillus circulans) | 1ED7 |
| CBM13* | Xylanase 10A (Streptomyces olivaceoviridis) | 1XYF |
| Xylanase 10A (Streptomyces lividans) | 1MC9 | |
| Ricin toxin B-chain (Ricinus communis) | 2AAI | |
| Abrin (Abrus precatorius) | 1ABR | |
| CBM14 | Tachycitin (Tachypleus tridentatus) | 1DQC |
| CBM15 | Xylanase 10C (Cellvibrio japonicus) | 1GNY |
| CBM17 | Cellulase 5A (Clostridium cellulovorans) | 1J83 |
| CBM18* | Agglutinin (Triticum aestivum) | 1WGC |
| Antimicrobial peptide (Amaranthus caudatus) | 1MMC | |
| Chitinase/agglutinin (Urtica dioica) | 1EIS | |
| CBM20* | Glucoamylase (Aspergillus niger) | 1AC0 |
| β-amylase (Bacillus cereus) | 1CQY | |
| CBM22 | Xylanase 10B (Clostridium thermocellum) | 1DYO |
| CBM27 | Mannanase 5A (Thermotoga maritima) | 1OF4 |
| CBM28 | Cellulase 5A (Bacillus sp. 1139) | 1UWW |
| CBM29 | Non-catalytic protein 1 (Pyromyces equi) | 1GWK |
| CBM32 | Sialidase 33A (Micromonospora viridifaciens) | 1EUU |
| Galactose oxidase (Cladobotryum dendroides) | 1GOF | |
| CBM34* | α-Amylase 13A (Thermoactinomyces vulgaris) | 1UH2 |
| Neopullulanase (Geobacillus stearothermophilus) | 1J0H | |
| CBM36 | Xylanase 43A (Paenibacillus polymyxa) | 1UX7 |
* These families contain too many structure entries to list them all so only representatives are given.
This review summarizes our current knowledge of the structure and function of CBMs. The integration of this information attempts to place CBMs in the broader context of carbohydrate-binding proteins and provides new insight into the mechanisms of protein–carbohydrate recognition.
CBM NOMENCLATURE
The modular nature of glycoside hydrolases was first noted in 1986 with a report that showed that small stable polypeptides (approx. 40 amino acids) with cellulose-binding activity could be proteolytically separated from domains displaying cellulase activity [1]. In-depth characterizations of CBMs were first published in 1988 in two separate reports [2,3]. The terminology of CBDs used to refer to these proteins persisted until 1999 at which point it was obvious that a number glycoside-hydrolase-derived families of non-catalytic modules that bound to carbohydrates other than cellulose were being discovered. The term CBM was proposed as a more inclusive term to describe all of the noncatalytic sugar-binding modules derived from glycoside hydrolases [4,5]. The term CBM is now in general use; however, the term CBD appears to have remained in use to describe the subset of CBMs that bind specifically to cellulose.
In keeping with the systematic nomenclature adopted for glycoside hydrolases [21], a similar system for CBMs is being adopted in the literature. At its simplest, a CBM is named by its family, e.g. the family 17 CBM from Clostridium cellulovorans Cel5A would be called CBM17, but one may also include the organism and even the enzyme from which it is derived to improve clarity. Thus this CBM17 may be defined as CcCBM17 or CcCel5ACBM17. If glycoside hydrolases contain tandem CBMs belonging to the same family, a number corresponding to the position of the CBM in the enzyme relative to the N-terminus is included. For example, Clostridium stercorarium contains an enzyme with a triplet of family 6 CBMs. The first CBM is referred to as CsCBM6-1, the second as CsCBM6-2 and the third as CsCBM6-3. This simple and descriptive nomenclature eliminates the need to memorize arbitrary names given to CBMs, as was required previously. Furthermore, its complementarity to the glycoside hydrolase system keeps these two related fields somewhat unified.
CBM BINDING AND POLYSACCHARIDE HYDROLYSIS
CBMs have three general roles with respect to the function of their cognate catalytic modules: (i) a proximity effect, (ii) a targeting function and (iii) a disruptive function.
Through their sugar-binding activity, CBMs concentrate enzymes on to the polysaccharide substrates. It is thought that maintaining the enzyme in proximity with the substrate (i.e. increasing the concentration of the enzyme on the surface of the substrate) leads to more rapid degradation of the polysaccharide [22]. There are numerous examples in the literature where proteolytic excision or genetic truncation of CBMs from the catalytic modules results in significant decreases in the activity of the enzymes on insoluble, but not soluble polysaccharides (see [2,22–29] for examples). It should be pointed out that there are examples of CBMs that have become components of the substrate-binding sites of glycoside hydrolases, and are pivotal to the substrate specificity and mode of action of the cognate enzymes. Thus family 3c CBMs may play a role in the processivity displayed by glycoside hydrolase family 9 ‘endo-processive’ cellulases [30], and a CBM22 was recently shown to change the specificity of a glycoside hydrolase family 10 xylanase such that it displayed primarily β-1,4-β-1,3-glucanase activity [31].
CBMs that bind to the surfaces of crystalline polysaccharides (referred to as Type A modules; see below) can be appended to a variety of glycoside hydrolases. In contrast, CBMs that interact with single polysaccharide chains (Type B, see below) bind to polysaccharides that are the substrates for the cognate catalytic module of the enzyme. For example, cellulases, xylanases and mannanases contain Type B CBMs that bind to cellulose, xylan and mannan respectively. Thus the CBM maintains proximity to the target substrate within complete macromolecular structures, such as the plant cell wall. It is now becoming apparent that this targeting function is even more subtle than the somewhat crude partitioning of enzymes to the different polysaccharides of plant cell walls. The family 9 CBM from xylanase 10A of Thermotoga maritima has a distinct specificity for only the reducing ends of polysaccharides, suggesting the intriguing notion of targeting to damaged regions of plant cell walls [32,33]. An elegant study by Carrard et al. [34] showed that family 1 and family 3 CBMs that were appended to the same catalytic module displayed different capacities to degrade crystalline cellulose, implying that these non-catalytic modules can recognize distinct regions of this otherwise chemically invariant polysaccharide. This work was complemented further by studies demonstrating that examples of CBMs from families 17 and 28 recognize different regions of noncrystalline cellulose, influencing the ability of the enzyme to hydrolyse the polysaccharide [23]. Recent studies using whole plant cell walls are consistent with studies on purified polysaccharides in demonstrating that CBMs that apparently bind to the same polysaccharide display clear differences in specificity ([35]; and H. J. Gilbert, unpublished work). Thus CBMs have significant applications in the production of modified plant fibre by targeting hydrolytic enzymes to specific regions of the cell wall. These modules could also play a prominent role in cell biology where they may be utilized as molecular probes in mapping the glyco-architecture of cells.
Some CBMs appear to have the capability of disrupting polysaccharide structure. This function was first documented for the N-terminal family 2a CBM of Cel6A from Cellulomonas fimi [36]. This CBM appeared to mediate non-catalytic disruption of the crystalline structure of cellulose; furthermore, this disruptive effect enhanced the degradative capacity of the catalytic module. However, this effect has only been observed for one other cellulose-binding CBM [37] so the generality of this phenomenon is uncertain. It has also been noted that the two binding sites of family 20 starch-binding CBMs are required to disrupt the structure of amylose [38]; it remains unclear, however, what influence this has on the catalytic activity of the entire enzyme.
FOLD RELATIONSHIPS AMONG CBMs
The catalytic modules of glycoside hydrolases are classified into 96 different families based on amino acid sequence similarity. These families are grouped into 14 clans or ‘superfamilies’ using the criteria of conservation of the protein fold, catalytic machinery and mechanism of glycosidic bond cleavage. A similar hierarchical classification exists for glycosyltransferases, with both the family and clan groupings predictive of functional and structural features of the members [39,40]. Although fold similarities between CBMs have been demonstrated, and the existence of superfamilies has been suggested [41,42], there are currently no formal ‘super’ groupings of the 39 CBM families. To approach this issue, we manually classified the structures from 22 different CBM families into seven ‘fold families’ (Table 2; Figure 1). To verify these manual structural groupings, DALI searches were used to confirm structural similarities [43].
Table 2. CBM fold families.
| Fold family | Fold | CBM families |
|---|---|---|
| 1 | β-Sandwich | 2, 3, 4, 6, 9, 15, 17, 22, 27, 28, 29, 32, 34, 36 |
| 2 | β-Trefoil | 13 |
| 3 | Cysteine knot | 1 |
| 4 | Unique | 5, 12 |
| 5 | OB fold | 10 |
| 6 | Hevein fold | 18 |
| 7 | Unique; contains hevein-like fold | 14 |
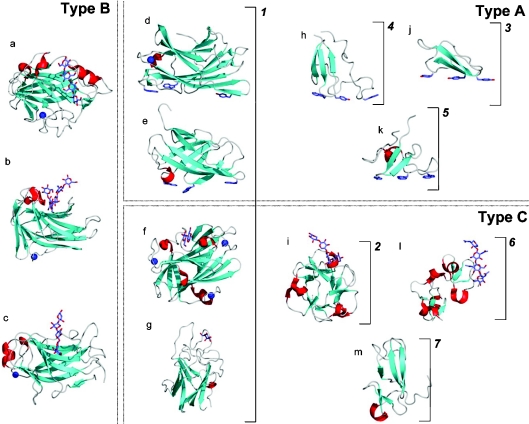
Figure 1. The big picture: CBMs, shown as ribbon structures, grouped as fold families and functional types.
Dotted boxes surround examples of CBMs belonging to the functional Types A, B, and C. Brackets with numbers indicate examples of CBMs belonging to fold families 1–7. CBMs shown are as follows: (a) family 17 CBM, CcCBM17, from Clostridium cellulovorans in complex with cellotetraose (PDB code 1J84 [60]); (b) family 4 CBM, TmCBM4-2, from T. maritima in complex with laminariohexaose (PDB code 1GUI [68]); (c) family 15 CBM, CjCBM15, from Cellvibrio japonicus in complex with xylopentaose (PDB code 1GNY [67]); (d) family 3 CBM, CtCBM3, from Clostridium thermocellum (PDB code 1NBC [52]); (e) family 2 CBM, CfCBM2, from Cellulomonas fimi (PDB code 1EXG [81]); (f) family 9 CBM, TmCBM9-2, from T. maritima in complex with cellobiose (PDB code 1I82 [33]); (g) family 32 CBM, MvCBM32, from Micromonospora viridifaciens in complex with galactose (PDB code 1EUU [65]); (h) family 5 CBM, EcCBM5, from Erwinia chrysanthemi (PDB code 1AIW [82]); (i) family 13 CBM, SlCBM13, from S. lividans in complex with xylopentaose (PDB code 1MC9 [48]); (j) family 1 CBM, TrCBM1, from Trichoderma reesi (PDB code 1CBH [83]); (k) family 10 CBM, CjCBM10, from Cellvibrio japonicus (PDB code 1E8R [84]); (l) family 18 CBM from Urtica dioca in complex with chitotriose (PDB code 1EN2 [85]); (m) family 14 CBM, tachychitin, from Tachypleus tridentatus (PDB code 1DQC [86]). Bound ligands are shown as ‘liquorice’ representations, while bound metal ions are shown as a blue spheres.
The β-sandwich
In terms of both total number of families and entries in databases, the dominant fold among CBMs is the β-sandwich (fold family 1) [44]. This fold comprises two β-sheets, each consisting of three to six antiparallel β-strands. CBMs share this fold with plant legume lectins and animal galectins, pentraxins, spermadhesins, calnexin, and ERGIC-53 (endoplasmic reticulum–Golgi intermediate compartment-53, although no CBM has significant amino acid sequence similarity with these other proteins. With the exception of CBM2a from Cellulomonas fimi xylanase 10A, all of the β-sandwich CBMs have at least one bound metal atom. In most cases, these metal ions appear to be structural; however, the ligand binding of the family 36 CBM from Paenibacillus polymyxa Xyn43A is mediated by a calcium atom [44a]. The ligand-recognition site in the majority of these proteins, including those that bind to crystalline cellulose, is located on the same face of the β-sandwich (Figure 2). In contrast, the CBMs in families 6 and 32, which also adopt the β-sandwich fold, have ligand-binding sites at the edge of the β-sandwich (Figure 2). The large number of CBMs adopting this fold (Table 1) classify it as a CBM superfamily.
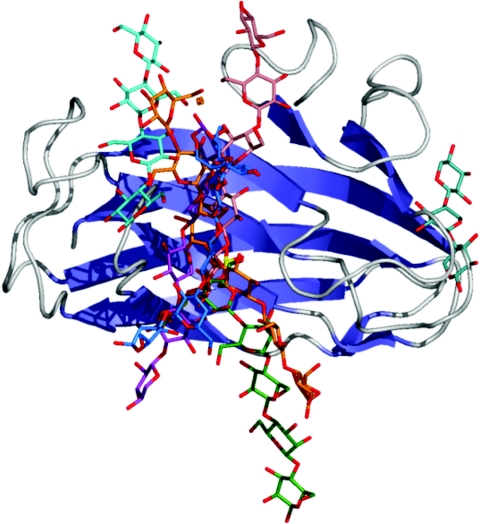
Figure 2. Schematic diagram showing the location of binding sites in the β-sandwich Type B CBMs.
This Figure was produced by overlapping the C-α carbons for all of the β-sandwich Type B CBMs for which ligand bound complexes were available. Only the ribbon structure of the family 4 CBM from the N-terminus of Cel9B from Cellulomonas fimi is shown as a representative β-sandwich Type B CBM. Oligosaccharide ligands are shown in ‘liquorice’ representation and coloured as follows: cyan, cellotetraose from CcCBM17 (PDB code 1J84 [60]); blue, laminariohexaose from TmCBM4-2 (PDB code 1GUI [68]); pink, cellopentaose from CfCBM4-2 (PDB code 1GU3 [68]); marine/aqua, xylotriose from CsCBM6-3 (PDB code 1NAE [61]); orange, cellohexaose from PeCBM29-2 (PDB code 1GWM [69]); green, mannopentaose from TmCBM27 (PDB code 1OF4 [73]); purple, xylopentaose from CjCBM15 (PDB code 1GNY [67]).
The β-trefoil
Second in frequency is the β-trefoil fold (fold family 2) [45], most commonly associated with ricin toxin B-chain [19]. This fold contains 12 strands of β-sheet, forming six hairpin turns. A β-barrel structure is formed by six of the strands, attendant with three hairpin turns. The other three hairpin turns form a triangular cap on one end of the β-barrel called the ‘hairpin triplet’. The subunit of this fold, called here a trefoil domain, is a contiguous amino acid sequence with a four β-strand, two-hairpin structure having a trefoil shape. Each trefoil domain contributes one hairpin (two β-strands) to the β-barrel and one hairpin to the hairpin triplet. The fold of the resulting molecule has a pseudo-3-fold axis [45,46]. The 3-fold symmetry is amenable to the presence of functional carbohydrate-binding sites in each of the three trefoil subdomains, which is exploited by the CBM13 modules of Streptomyces lividans and Streptomyces olivaceoviridis xylanases in order to maintain a reasonably high affinity for β-1,4-linked polymers of xylose [47–50]. The plant lectins with this fold, e.g. ricin toxin B-chain [19], take advantage of β-trefoil multivalency further by having duplicated modules; up to three functional binding sites simultaneously interacting with cell-surface glycans leads to greatly enhanced affinities [51].
‘Cellulose binding’ and OB (oligonucleotide/oligosaccharide binding) folds
The β-sandwich and β-trefoil are relatively small and simple folds that are effective scaffolds into which can be embedded binding specificity for the diverse polysaccharides of plant cell walls, as well as the myriad carbohydrates that make up the ‘glycome’ of plants and animals. In contrast, members of fold families 3–5, which are small 30–60-amino-acid polypeptides containing only β-sheet and coil (Figure 1), show less diversity in their ligand specificities with folds that appear more specialized to the recognition of cellulose and/or chitin. The majority of these CBMs have planar carbohydrate-binding sites comprising aromatic residues, although a notable exception is the family 12 CBMs. The solution NMR structure of this protein reveals no obvious hydrophobic planar surface that could comprise the cellulose-binding site. Interestingly, the ligand-binding site in CBM10 from Cellvibrio japonicus Xyn10A is on a different face to the binding site of other proteins with an OB fold, indicating a degree of convergent evolution between proteins with an OB fold. Fold family 4 is arguably the only other CBM superfamily based on the adoption of this fold by two CBM families.
Hevein fold
Hevein domains are small (approx. 40 amino acids) CBMs originally identified as chitin-binding proteins in plants. The fold comprises predominantly coil, but does have two small β-sheets and a small region of helix (Figure 1). This fold can accommodate a surprisingly extended binding site as exemplified by WGA, which binds optimally to a chitotetrasaccharide. The minimal hevein fold is found in family 18 CBMs and is classified as CBM fold family 6. Family 14 CBMs appear to incorporate aspects of the hevein domain (Figure 1); however, the fusion of this fold with a small β-sheet structure necessitates its classification as a separate fold family.
RELATIONSHIP BETWEEN STRUCTURE AND FUNCTION
Although CBM families can be grouped into fold families based on the conservation of the protein fold, such groupings are not predictive of function. Sufficient diversity exists among fold family members such that functional elements, either specific amino acids or binding-site topographies, are not conserved. Thus predictions of ligand specificity, based solely on possession of a particular fold, must be approached with caution. Another useful classification of CBMs based on structural and functional similarities has been proposed in which these protein modules have been grouped into three types: ‘surface-binding’ CBMs (Type A), ‘glycan-chain-binding’ CBMs (Type B), and ‘small-sugar-binding’ CBMs (Type C). The classification of these CBM types relative to the fold families and sequence families are shown in Table 3.
Table 3. CBM types.
| Type | Fold family | CBM families |
|---|---|---|
| A | 1, 3, 4, 5 | 1, 2a, 3, 5, 10 |
| B | 1 | 2b, 4, 6, 15, 17, 20, 22, 27, 28, 29, 34, 36 |
| C | 1, 2, 6, 7 | 9, 13, 14, 18, 32 |
Type A surface-binding CBMs
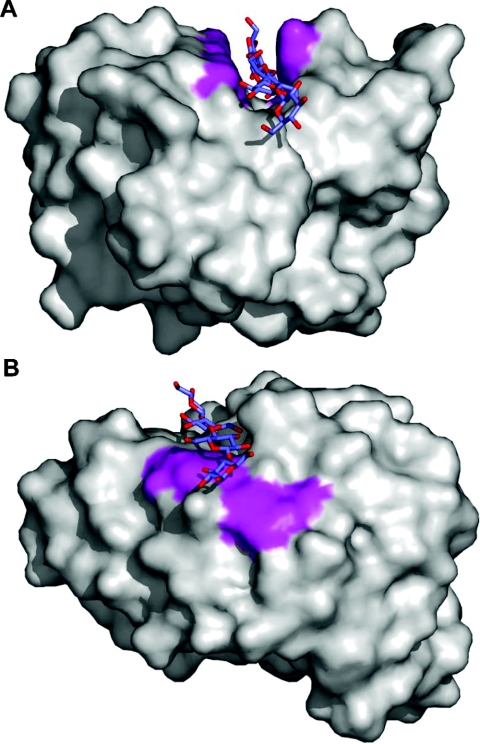
This class of CBM is arguably the most distinct as its properties differ significantly from other types of carbohydrate-binding proteins. It includes members of CBM families 1, 2a, 3, 5 and 10 that bind to insoluble, highly crystalline cellulose and/or chitin. While the prevalence of aromatic amino acid residues in the binding sites of these CBMs is consistent with the majority of carbohydrate-binding proteins, the flat or platform-like binding sites are not (Figure 1; also see Figure 4A). The planar architecture of the binding sites is thought to be complementary to the flat surfaces presented by cellulose or chitin crystals [52,53]. Indeed, there has been considerable controversy regarding the location of the Type A CBM binding site in crystalline cellulose. Tormo et al. [52] proposed that the binding site comprises the hydrophobic 110 face. McLean et al. [53], however, argued that, in perfect cellulose crystals, the surface area presented by the 110 face is too small to account for the binding capacity of CBMs for this ligand, prompting the authors to propose that the binding sites are more likely to comprise the hydrophilic 110 and 010 faces. A recent seminal study by Lehtio et al. [54] has used transmission electron microscopy to probe the location of the CBM-binding site on Valonia crystalline cellulose. They confirmed that both CBM1 and CBM3a bind to the hydrophobic 110 face, and suggest that these regions are often severely disrupted and thus the larger surface area presented by this face accounts for the previous discrepancies concerning the likely binding site and its capacity for ligand.
Figure 4. The three types of binding-site ‘platforms’ formed by aromatic amino acid residues.
(A) The ‘planar’ platform in the family 10 Type A CBM, CjCBM10. (B) The ‘twisted’ platform of the Type B family 29 CBM, PeCBM29-2. (C) The ‘sandwich’ platform of the Type B family 4 CBM, CfCBM4-2. The C-α backbone is shown as grey cylinders, the aromatic amino acid side chains forming the binding sites are shown in grey ‘liquorice’ representations, and the bound oligosaccharides are shown in blue ‘liquorice’ representation.
Type A CBMs show little or no affinity for soluble carbohydrates [55]. The interaction of Type A modules with crystalline cellulose is associated with positive entropy, demonstrating that the thermodynamic forces that drive the binding of CBMs to crystalline ligands are relatively unique among carbohydrate-binding proteins [56]. Creagh et al. [56] argued that the water molecules released from the protein and ligand when CBMs bind to their target carbohydrates increases the entropy of the system, which, in the case of soluble saccharides, is postulated to be more than offset by the conformational restriction of the bound ligand leading to a net reduction in entropy. The molecular basis for the thermodynamic forces that drive protein–carbohydrate interactions remains, however, a highly controversial area, particularly with respect to the role of water molecules and the loss of entropy through conformational restriction.
Type B glycan-chain-binding CBMs
NMR and X-ray crystal structures have revealed that the carbohydrate-binding sites of Type B CBMs are extended (>15 Å; 1 Å=0.1 nm), often described as grooves or clefts, and comprise several subsites able to accommodate the individual sugar units of the polymeric ligand. The binding proficiency of this class of CBM is determined by the degree of polymerization of the carbohydrate ligand; biochemical studies frequently demonstrate increased affinities up to hexasaccharides and negligible interaction with oligosaccharides with a degree of polymerization (DP) of three or less. Thus these CBMs are considered to be ‘chain binders’. The depth of these binding sites varies from very shallow to being able to accommodate the entire width of a pyranose ring (Figure 3). This type of CBM, which currently includes examples from families 2b, 4, 6, 15, 17, 20, 22, 27, 28, 29, 34 and 36 have clearly evolved binding site topographies that are equipped to interact with individual glycan chains rather than crystalline surfaces. As with Type A CBMs, aromatic residues play a pivotal role in ligand binding, and the orientation of these amino acids are key determinants of specificity [57]. In sharp contrast with the Type A CBMs, direct hydrogen bonds also play a key role in the defining the affinity and ligand specificity of Type B glycan chain binders [58–60]. There is currently no evidence, however, indicating that water-mediated hydrogen bonds are critical to the binding of CBMs to their target ligands [58].
Figure 3. Solvent-accessible surface representations of two CBMs showing the depth of binding grooves in Type B CBMs.
(A) Example of a cellopentaose molecule occupying a deep binding groove in CfCBM4-2 (PDB code 1GU3 [68]). (B) Example of a cellotetraose molecule occupying a shallow binding groove in CcCBM17 (PDB code 1J84 [60]). The surfaces created by the aromatic amino acid side chains involved in binding are shown in magenta.
Type C small-sugar-binding CBMs
This unique class of CBM has the lectin-like property of binding optimally to mono-, di- or tri-saccharides, and thus lacks the extended binding-site grooves of Type B CBMs. It should be emphasized, however, that the distinction between Type B and Type C CBMs can be subtle. For example, the Type B CBM6 module of the Clostridium stercorarium xylanase has a very similar fold to the Type C lectin-like CBM32 family [61], but apparently binds longer oligosaccharide ligands. Furthermore, the Cellvibrio mixtus Cel5A CBM6 contains two discrete binding sites that display characteristics of Type B and Type C modules respectively [62,63]. Nevertheless, it is apparent that the hydrogen-bonding network between protein and ligand is more extensive in Type C CBMs than Type B modules, consistent with their lectin-like properties (see below).
The Type C CBMs currently includes examples from families 9, 13, 14, 18 and 32. Members of families 13 (e.g. ricin toxin B-chain), 14 (e.g. tachycitin) and 18 (e.g. WGA) were first discovered as lectins with small-sugar-binding activity and have only subsequently been included as CBMs due to their discovery in a number of glycoside hydrolases. The only characterized member of family 9 is the C-terminal CBM from T. maritima xylanase 10A [32,48,64]. In general, this family of CBMs is found exclusively in xylanases, and this particular CBM from T. maritima has the remarkable property of recognizing the reducing end sugars of xylans and cellulose. Family 32 is a relatively new CBM family whose only currently (partially) characterized member is the C-terminal module from the Micromonospora viridifaciens sialidase [61,65]. This CBM has a very similar fold to the fucose-specific lectin from Anguilla anguilla and appears to bind galactose [61]. Overall, identification and characterization of Type C CBMs is lagging behind Type A and B CBMs, probably due to their limited presence in plant cell wall active glycoside hydrolases. Rather, the Type C CBMs, particularly CBMs from families 13 and 32, appear to be more prevalent in bacterial toxins or enzymes (glycoside hydrolases and glycosyltransferases) that attack eukaryotic cell surfaces or matrix glycans.
STRUCTURAL DETERMINANTS OF CBM BINDING SPECIFICITY
Aromatic amino acid side chains, binding-site topography and ligand conformation
As suggested by the difference between the Type A CBMs and Type B CBMs, binding-site topography is a key determinant of binding specificity. The β-sandwich fold is predominant among CBMs, begging the question of how varied binding-site topography is achieved. The two major factors appear to be the location of aromatic amino acid side chains and loop structures that shape the binding sites to mirror the conformation of the ligand.
The interaction of aromatic amino acid side chains with ligand is ubiquitous to CBM carbohydrate recognition. The side chains of tryptophan, tyrosine and, less commonly, phenylalanine form the hydrophobic platforms in CBM-binding sites, which can be planar, twisted or form a sandwich (Figure 4). As discussed above, planar platforms of aromatic amino acid side chains are a hallmark of the Type A CBMs (Figure 4A). In the binding sites of families 2b, 15, 17, 27, 29, 34 and 36, the apolar platform can be ‘twisted’ due to the rotation of the planes of two to three aromatic amino acid side chains relative to one another (Figure 4B). The aromatic amino acid side chains in the binding site of CBMs often sandwich a sugar unit in the ligand by stacking against the β and α face of the pyranose ring (Figure 4C). This is common to family 4, 6, 9 and 22 CBMs. The sandwich and twisted platforms may be used concurrently.
Either of these two platforms, twisted and sandwich, may be harnessed to accommodate the conformations of soluble oligosaccharide ligands. Fibre diffraction studies of xylan suggested that the polysaccharide chains form a 3-fold helix [66]. This 3-fold helix was confirmed in the X-ray structure of a family 15 CBM, which employed a twisted platform [67]. The same oligosaccharide conformation was observed in two family 6 CBMs, which have a twisted sandwich conformation of aromatic amino acid side chains in their binding sites [61,63]. Somewhat surprisingly, the conformations of cello-oligosaccharides in the X-ray crystal structures of a family 4 (sandwich platform) [68], a family 17 (twisted platform) [60], and a family 29 CBM (twisted platform) [69] revealed a consistent turn in the chain, but neither a 2-nor 3-fold axis, in contrast to the perfect 2-fold helix observed in the chains of crystalline cellulose. The conformations of the bound cello-oligosaccharides did, however, approximate the conformation of a cello-oligosaccharide in solution [68]. Thus CBMs appear, in common with lectins, to have preformed carbohydrate-recognition sites which mirror the solution conformations of their target ligands, thereby minimizing the energetic penalty paid upon binding. The importance of binding-site topography is highlighted by studies of the xylan-specific family 2b CBMs from Cellulomonas fimi. Although similar in sequence to the cellulose-binding family 2a CBMs, the binding sites of these CBMs are formed by two perpendicular tryptophan residues, the angle of which reflects the helical conformation of xylan (Figure 5A). A simple arginine-to-glycine mutation allowed one of the tryptophan residues in the carbohydrate-binding site to lie flat (Figure 5B), mimicking a Type A binding site, and converting the specificity from xylan to crystalline cellulose [57].
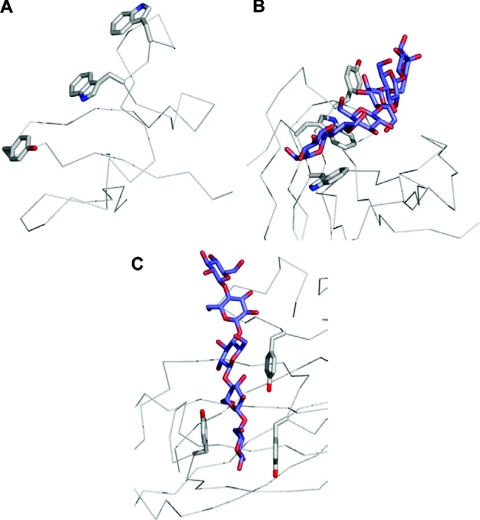
Figure 5. Binding-site topography and oligosaccharide recognition.
Surface representations of the NMR structures of wild-type CfCBM2b-1 (A) and the same protein with an Arg262→Gly mutation (B). The arrow indicates the tryptophan residue that changes conformation due to the mutation. The binding site of CfCBM4-1 extend in a straight path across the face of one of the β-sheets of this CBM creating a linear binding site appropriate for binding cello-oligosaccharides (C). In TmCBM4-2, which has significant sequence identity with CfCBM4-1, two loops are extended (D); one to block an end of the binding site (shown by the arrow labelled 1) and another to accommodate the curvature of the laminarioligosaccharide (shown by the arrow labelled 2). Solvent-accessible surfaces are shown in transparent grey with surfaces created by the aromatic amino acid side chains involved in binding shown in magenta. C-α traces are shown in blue.
Differential loop structure can radically alter the ability of a standard β-sandwich core to present variable carbohydrate-binding sites. This is most clearly evident when comparing the family 4 cellulose-binding CBM, CfCBM4-1, from Cellulomonas fimi and the family 4 β-1,3-glucan-binding CBM, TmCBM4-2, from T. maritima. Both have significant sequence similarity and nearly structurally identical β-sandwich scaffolds. However, insertions in two loops contour the TmCBM4-2 binding site to accommodate the U-shape of laminarioligosaccharides, whereas the binding site of CfCBM4-1 is well suited to a linear cello-oligosaccharide (Figures 5C and 5D) [68].
Hydrogen bonding and calcium
Although the orientation and positioning of the aromatic residues in the binding sites of CBMs is the primary driver of specificity and affinity in these proteins, other interactions, including direct hydrogen bonds and calcium-mediated co-ordination, play a significant role in CBM ligand recognition.
Hydrogen bonds
Carbohydrates are amphipathic molecules that, due to their complement of hydroxy groups, have considerable capacity for hydrogen-bond formation with polar residues in the binding sites of proteins. Indeed, in lectin and other sugar-binding proteins, such as periplasmic monosaccharide transporters, an extensive network of direct and indirect hydrogen bonds is formed between the protein and carbohydrate. A current estimate of direct hydrogen-bonding density in lectins puts this value at approx. 3.4 hydrogen bonds per 100 Å2 of buried polar surface area [70]. In contrast, in CBMs, there is a relative paucity of hydrogen bonds with ligand; approx. 2 hydrogen bonds per 100 Å2 of buried polar surface area (calculated from available X-ray crystal structures of Type B CBMs solved in complex with oligosaccharide ligands). The reason for this difference is not currently known, but may be at least partially driven by the need to accommodate the highly decorated polysaccharides (see below), which are often present in the plant cell wall.
The relative importance of direct hydrogen bonds in the interaction of CBMs with their target sugars varies depending on the ‘Type’. In Type A CBMs, mutation to alanine of polar residues predicted to make direct hydrogen bonds with the crystalline polysaccharide ligands has little effect on affinity, suggesting that, in these proteins, hydrogen bonds play only a minor role in ligand recognition [53]. In Type B and Type C CBMs, replacement of direct hydrogen-bonding resides with alanine can lead to significant losses in affinity from 2-fold [58,60,71] to complete abrogation of binding [59]. However, it must be noted that in some of these cases, it is uncertain if the loss in affinity is solely due to the loss of the hydrogen bond or if subtle structural changes in the binding sites have occurred that are deleterious to ligand binding.
Calcium
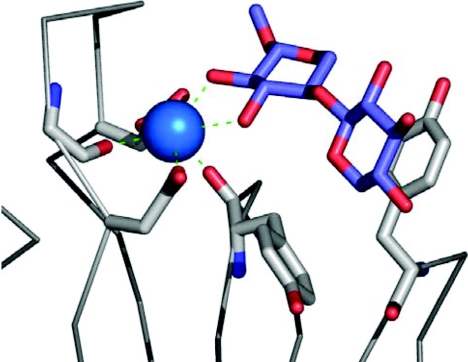
It is well established that calcium plays a significant role in the interaction of many lectins with their target ligands, either by maintaining the binding site in the correct conformation or via direct co-ordination with the carbohydrate itself. Many CBMs are metalloproteins; however, the role of metal ions in CBM-ligand interactions has only recently been discovered. Xylan recognition by the family 35 CBM of the Cellvibrio japonicus enzyme Abf62A is calcium-dependent; however, the structure of this module is unknown [72]. More recently, the recognition of xylo-oligosaccharides by the family 36 CBM from the Pa. polymyxa enzyme Xyn43A was also demonstrated to be calcium-dependent [44a]. The high-resolution X-ray crystal structure of the CBM in complex with calcium and xylotriose showed that a single atom of the bivalent metal made direct interactions with the sugar, thus revealing the basis of its importance in carbohydrate recognition (Figure 6).
Figure 6. The role of calcium in xylan recognition by the family 36 CBM, PpCBM36, from Pa. polymyxa xylanase 43.
The calcium atom bound in the binding site of PpCBM36 is shown as a blue sphere. The amino acid residues involved in co-ordinating the calcium and binding the sugar are shown in grey ‘liquorice’ representation, the bound xylo-oligosaccharide is shown in blue ‘liquorice’ representation and the C-α backbone is represented as grey cylinders.
Binding promiscuity/plasticity
Plant cell-wall polysaccharides, the target ligands of most CBMs, are often extremely heterogeneous. They posses variations in the type and linkage of the backbone saccharides, as well as the presence of an array of different sugar and acetate decorations, depending on the plant species, cell type and differentiation state. Precisely how CBMs contend with this broad diversity while retaining specificity has only recently been elucidated.
Accommodation of polysaccharide side chains
The xylan-binding CBM15 from Cellvibrio japonicus xylanase 10C can interact with both heavily substituted arabinoxylans and relatively unbranched forms of the β-1,4-xylose polymer with equal affinity. Arabinoxylans are thought to retain the 3-fold helical structure determined for linear xylan, with the arabinose groups extending outwards in the same plane as the backbone xylose residues. The complex of CBM15 with xylopentaose provided the first glimpse into how these modules can accommodate the side chains of their target polysaccharides [67]. The protein makes relatively few direct hydrogen bonds with the sugar; six of the ten O-2 and O-3 groups in the xylo-oligosaccharide were solvent-exposed and thus side chains attached at these positions would presumably not interfere with ligand binding. A similar mechanism for the accommodation of the α-1,6-linked galactose side chains of galactomannan was suggested from the complex of CBM29-2 from Piromyces equi with mannohexaose [69]. Here, the O-6 of alternate mannoses in the binding site are solvent exposed, and it was proposed that this would enable the CBM to interact with substituted regions of the polysaccharide, as long as the galactose side chains are not on adjacent backbone sugars. This hypothesis was confirmed by the structural elucidation of the first CBM–decorated-ligand complex. The structure of a family 27 CBM from T. maritima bound to G2M5 (63,64-α-D-galactosylmannopentaose) revealed that the galactose in subsite 4 faces away from the protein and was thus easily accommodated in the binding site, whereas the side chain at subsite 3 was forced into a plane parallel to the mannose backbone by Trp28 (Figure 7) [73]. The energetic penalty caused by this conformational change in the ligand is the likely cause of the large reduction in affinity observed with G2M5 over linear mannopentaose and confirmed the observation from CBM29 that binding to galactomannan would only occur if the substitutions on the polysaccharide were non-adjacent.
Figure 7. The accommodation of galactomannan by a family 27 CBM, TmCBM27.
The overall structure surrounding the binding site of TmCBM27 from T. maritima is shown as a ribbon diagram. The three tryptophan side chains comprising the carbohydrate-binding site are shown in grey ‘liquorice’ representation and are numbered. The ligand, G2M5, a fragment of galactomannan, is shown in ‘liquorice’ representation with mannose residues in blue and galactose residues in yellow. The uppermost galactose residue in the Figure is clearly bent back over the top of the mannose backbone by Trp28, while the other galactose residue extends comfortably away from the mannose backbone.
Backbone heterogeneity
CBMs are also able to maintain a selective flexibility when the target ligand contains a heterogenous backbone. The paradigm for this is CBM29-2 from Pi. equi, which recognizes β-1,4-linked polymers of both mannose and glucose, while retaining the ability to discriminate against a range of other plant structural polysaccharides. The structure of this protein in complex with both manno- and cello-hexaose reveals the basis for this relaxed specificity, which is conferred by the plasticity of the direct interactions the CBM makes with the axial and equatorial 2-hydroxy group of mannose and glucose respectively [69]. This observation is in direct contrast with the mannan-specific family 27 CBM from T. maritima, where the equatorial 2-hydroxy group of glucose would clash with amino acids in two of the protein's five subsites, thus precluding its association with cellulose [73].
CBMs AND MULTIVALENCY
Quiocho [74] classified carbohydrate-binding proteins into two general groups based on their affinity for carbohydrates and their modes of carbohydrate recognition. Group I comprises those proteins that bind carbohydrates tightly (Ka>106 M−1) in binding sites that completely (or nearly so) enclose the carbohydrate ligand. Group II are those proteins that bind carbohydrates more weakly (Ka<106 M−1) in open binding sites that leave significant portions of the carbohydrate ligand exposed to solvent when bound. This latter class of protein–carbohydrate interactions, which includes all CBM–carbohydrate interactions, appears well suited to binding cell-surface glycans, oligosaccharides or polysaccharides that cannot be completely enveloped in the binding site of the protein. These group II carbohydrate-binding proteins may have evolved to have weak binding because this is somehow advantageous to the function of these proteins. Alternatively, the weak binding may be a result of restrictions on the number of direct interactions between the protein and sugar, and incomplete desolvation of the carbohydrate ligand necessitated by the physical aspects of accommodating fragments of much larger, often immobilized, ligands. Nevertheless, these weak interactions are often compensated in nature by the phenomenon of avidity resulting from multivalent interactions. In these cases, multiple clustered carbohydrate-binding sites interact simultaneously with carbohydrate ligands, which present multiple recognition elements, resulting in increased association constants relative to any one of the isolated carbohydrate-binding-site–carbohydrate interactions. Clustered carbohydrate-binding sites can result from a single protein having multiple binding sites, the association of two or more univalent carbohydrate-binding proteins into multivalent quaternary structures, or clustering of receptors, for example, in cell membranes. Multivalent carbohydrate ligands may be in the form of branched saccharides, clustered cell-surface glycans or, as is most relevant to CBMs, polysaccharides. To date, no CBM has been found to form quaternary structures in its natural state. However, multiple CBMs, often tandems, are found frequently in glycoside hydrolases, which effectively become multivalent carbohydrate-binding proteins. The first CBMs in tandem to be investigated were the two family 2b CBMs of Cellulomonas fimi xylanase 11A [75]. While the individual association constants for xylan were low (approx. 104 M−1), the association constant of the CBMs linked in tandem, as they are in their natural state, was approx. 106 M−1. Similar observations have been made with the three family 6 CBMs of the Clostridium stercorarium putative xylanase [76], and the tandem CBM17 and CBM28 modules from Bacillus sp. 1139 Cel5 [23], where the affinities of tandem combinations for polysaccharides relative to the individual modules were increased by anywhere from 10- to 100-fold. Thus this form of multivalency is effectively used by CBMs to overcome relatively weak binding. Interestingly, the appearance of multiple CBMs in glycoside hydrolases appears to occur most frequently in thermo- or hyperthermo-philic enzymes [23]. This may be in response to the need for these proteins to overcome the loss of binding affinity that accompanies most molecular interactions at elevated temperatures [23].
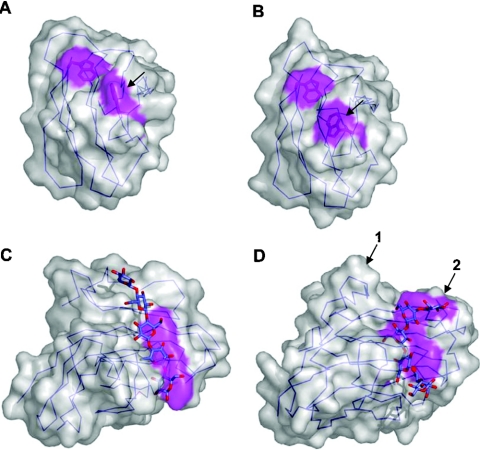
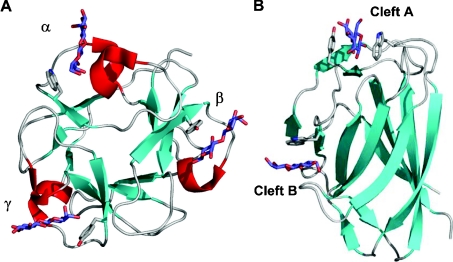
Individual CBM modules occasionally have multiple carbohydrate-binding sites. This was first proposed for the family 13 CBM from S. lividans xylanase 10A on the basis of its similarity to the multivalent ricin toxin B-chain. It was subsequently demonstrated by mutagenesis [47], NMR [49] and X-ray crystallography [48] that this module had three separate binding sites, one in each of its trefoil subdomains (Figure 8A). The presence of multiple binding sites enabled the module to interact simultaneously with multiple binding sites within polymerized xylose to enhance its overall affinity by approx. 10-fold for this polysaccharide [47]. The family 6 CBM from Cellvibrio mixtus endoglucanase 5A also has two binding sites (Figure 8B). The binding site in the so-called ‘cleft A’ can accommodate the chain ends of β-1,4-glucans, β-1,3-glucans and xylans. ‘Cleft B’ binds to internal regions of β-1,4-glucans and mixed β-(1,4)(1,3)-glucans. The ability of both binding sites to recognize cellulose (β-1,4-glucans) results in a multivalent interaction with insoluble cellulose. Studies whereby the affinity of a single binding site for cellulose is removed by site-directed mutagenesis of appropriate residues revealed that the association constants of the individual sites for cellulose were approx. 104 M−1. The affinity of the CBM with both binding sites intact was approx. 105 M−1 demonstrating the co-operativity between the two binding sites [62].
Figure 8. Multivalent CBMs.
The family 13 CBM from S. lividans xylanase 10A, SlCBM13, is shown in complex with three lactose molecules occupying its three binding sites (A), while the family 6 CBM from Cellvibrio mixtus endoglucanase 5A, CmCBM6, is shown in complex with two cellobiose molecules occupying its two binding sites (B). Both are shown as ribbon diagrams with relevant aromatic amino acid side chains in the binding sites shown in grey ‘liquorice’ representation. The three β-trefoil subdomains of SlCBM13 are labelled as α, β and γ, and the two binding sites of CmCBM6 are labelled according to the cleft in which they reside.
Although CBMs harness the advantages of multivalent interactions to enhance their affinity for polymeric substrates, the biological relevance of this to glycoside hydrolase function remains unknown. It is clear that CBMs have three important functions in polysaccharide hydrolysis (see above); the most important of which appears to be to concentrate the enzyme on to the polysaccharide. However, this is based mainly on studies which compare the activity of intact enzymes with enzymes that have had their CBM(s) deleted by genetic engineering. A poorly investigated aspect of CBM biology is how the overall strength of the CBM–polysaccharide interaction may influence the activity of the enzyme: are strong CBM–polysaccharide interactions advantageous?
As mentioned above, CBMs from thermophilic sources often overcome the loss of affinity accompanying binding at elevated temperatures by duplicating CBMs. Individual CBMs can also compensate for this reduction in affinity by having 10–100-fold tighter binding than CBMs from mesophilic organisms when compared at the same temperature (e.g. 25 °C). Precisely how this enhanced affinity is achieved at a structural level is currently unclear; there appears to be no significant differences in terms of binding-site architecture when comparing thermophilic CBMs with their mesophilic counterparts. However, this phenomenon does provide some insight into the importance of affinity to Type B and C CBM function. First, mesophilic CBMs of these types all have approximately the same affinity (Ka of approx. 104 M−1) for model ligands when measured at approx. 25 °C. The affinities of thermophilic and hyperthermophilic Type B and C CBMs for their ligands when calculated by extrapolation or actually measured at the source organisms' growth temperature come out at approx. 104 M−1. Thus there appears to be a ‘baseline’ CBM affinity in the relatively low region of approx. 104 M−1 that suggests ultratight binding is not necessarily an advantage to glycoside hydrolase function. To efficiently hydrolyse the plant cell wall, CBMs are required to mediate close proximity between enzyme and substrate; however, the biocatalysts need to retain the capacity to access the complete plant cell wall, which could be restricted if these modules bound tightly to their ligands. Although Type A CBMs bind tightly to their ligands (interaction of CBM2as with their ligands appear to be irreversible), they are mobile, displaying the capacity to diffuse across the surface of crystalline cellulose, which probably reflects the lack of directionality of hydrophobic stacking interactions [77].
Enhanced binding affinity is the most easily observable consequence resulting from CBMs in tandem. However, there may also be a more subtle reason for the presence of CBM tandems in glycoside hydrolases. As mentioned the CBM17 and CBM28 modules from Bacillus sp. 1139 Cel5 combine in tandem to produce very tight cellulose binding; individually, these CBMs recognize different regions of non-crystalline cellulose [23]. The combined result is tight binding to a very specific region of non-crystalline cellulose, which in turn may influence the activity of the catalytic module. Thus CBMs in tandem may provide another mechanism for fine-tuning the specificity of glycoside hydrolases for their substrate.
CBMs, LECTINS AND CARBOHYDRATE RECOGNITION
CBMs have been considered as contiguous amino acid sequences with discrete folds within the modular structures of carbohydrate-active enzymes and cellulosomal scaffoldins (proteins that mediate the assembly of multiprotein cellulase–hemicellulase complexes). This essentially distinguishes CBMs from lectins, whose carbohydrate-binding activities are generally not associated with catalytic activities of modules within the same polypeptide. Furthermore, lectins are often found as individual entities. This, combined with the specificity of CBMs for plant cell wall glycans rather than the components of complex eukaryotic glycans, is likely to be the reason that the two fields have remained largely insulated from one another. However, in both a structural and functional sense, the distinction between CBMs and lectins is clearly converging.
An entire class of CBMs, the Type C CBMs, appears to share the ability of lectins to recognize small sugars. Furthermore, many of the CBM sequence families that are broadly classified as Type C CBMs are actually families that were initially identified as lectins (i.e. families 13, 14 and 18). This is perhaps most evident with β-trefoil domains (CBM family 13 or fold family 2), of which the classic example is the ricin toxin B-chain lectin. These modules are found as components of plant toxins, insecticidal toxins and bacterial haemolysins, as well as in numerous carbohydrate-active enzymes. Family 18 CBMs contain members which are lectins (e.g. WGA) or whose functions are linked to modules with chitinase activity. Thus, depending on the definition of a lectin, many CBMs are indeed lectins and vice versa.
The explosion in genome sequence data has resulted in the detection of CBM family members that are not directly appended to enzymically active modules. To name a couple, the genome of Mycobacterium tuberculosis appears to harbour a gene encoding an isolated CBM2 module; a number of species also have open reading frames encoding single CBM13 modules. Assuming that the encoded proteins have carbohydrate-binding function, they may, in a sense, be considered lectins.
Lastly, excluding the CBM families that contain bona fide lectins and therefore share both sequence and structure similarity, a number of CBM families share structural similarity with lectins that are unrelated at the amino acid sequence level. As mentioned above, CBM fold family 1 (Table 2) is structurally related to lectins displaying a β-sandwich fold. This is most evident with the family 6 CBMs whose fold is very similar to the A. anguilla fucose-specific lectin despite having no amino acid sequence similarity [61]. Examples from CBM families 32 and 36 are also very similar in structure to the A. anguilla lectin. Furthermore, the locations of metal ions and carbohydrate-binding sites in all of these carbohydrate-binding proteins are well conserved, though amino acid side chains responsible for ligand recognition may not be. Thus there is a significant structural link between the lectins and CBMs. Overall, based on the similarities between the lectin and CBM fields, there is an argument for a unified sequence-based classification of lectin families, as was suggested by Boraston et al. [61], which is so powerful in the bioinformatic analysis of the catalytic modules of carbohydrate-active enzymes.
Despite the growing evidence of overlap between CBMs and lectins, there remains some stark and some subtle differences beyond those already stated. As discussed, the Type A CBMs, although sharing some structural similarity with lectins, have a dramatically different ligand and an apparently different mode of interacting with it. The ligand, insoluble crystalline polysaccharide (i.e. cellulose or chitin), is unique in that it presents a two-dimensional binding ‘surface’ quite different from the three-dimensional arrangement of a soluble glycan. The distinctive thermodynamics of this interaction (discussed above) appear to reflect this difference in modes of recognition.
The Type B CBMs show more subtle differences when compared with lectin–carbohydrate interactions. Studies of lectin–carbohydrate interactions have yielded an average hydrogen-bonding density of 3.45 (±0.52) bonds per 100 Å2 of buried polar surface area. The ΔH for these interactions can be parameterized into ΔHp=46.1 cal/mol/Å2 and ΔHap=−5.8 cal/mol/Å2 (1 cal≡4.184 J), where ΔHp and ΔHap represent the enthalpic contribution of burying polar and apolar surface area respectively [70]. Where data are available, the Type C CBMs agree with this very well, strengthening the lectin-like qualities of Type C CBMs. In contrast, our corresponding preliminary values for Type B CBM–carbohydrate interactions, determined from six CBM–carbohydrate complexes and values measured by isothermal titration calorimetry, are 2.11 (±0.61) hydrogen bonds per 100 Å2 of buried polar surface area, ΔHp=117.2 (±19.4) cal/mol/Å2, ΔHap=−52.3 (±13.9) cal/mol/Å2. Overall, the general thermodynamic signatures of carbohydrate recognition by CBMs and lectins are nearly indistinguishable: dominating favourable changes in enthalpy with partially offsetting unfavourable changes in entropy. However, the parameterized values, although preliminary, suggest that the aetiology of the enthalpic contributions in the two systems are different; mainly, apolar surface area is far more important in Type B CBM–carbohydrate interactions than in lectin–carbohydrate interactions and possibly Type C CBM–carbohydrate interactions. Lectins have been considered the benchmark for studying protein–carbohydrate interactions, but it appears that CBMs provide a new and potentially important set of model systems for studying this phenomenon. Many of the properties of CBMs (e.g. an extended binding site to accommodate long sugar chains) are shared with proteins of clinical relevance (e.g. antibodies [78,79] and viral proteins [80]) making the study of CBMs relevant beyond the recognition of plant cell-wall polysaccharides.
CONCLUSIONS
CBMs play a pivotal role in degradative enzymes that mediate the recycling of photosynthetically fixed carbon in the biosphere. Understanding the structural basis by which CBMs bind to their target ligands provides novel insights into the mechanisms of carbohydrate–protein recognition. The harnessing of this information to inform strategies designed to manipulate carbohydrate-recognition through the use of ligands that act as agonists or antagonists will be of considerable biotechnological importance not only within an industrial context, but also in the generation of novel pharmaceuticals that are designed to modify cell–cell signalling and host–pathogen recognition.
Acknowledgments
We thank Dr Bernard Henrissat, creator and curator of the CAZy website, for many helpful discussions and maintenance of such an outstanding resource. We must also thank Dr Douglas Kilburn and Dr Tony Warren for their important roles in establishing the field of CBM research and for their help and insight.
References
- 1.Van Tilbeurgh H., Tomme P., Claeyssens M., Bhikhabhai R., Pettersson G. Limited proteolysis of the cellobiohydrolase I from Trichoderma reesei. FEBS Lett. 1986;204:223–227. [Google Scholar]
- 2.Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414: analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur. J. Biochem. 1988;170:575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]
- 3.Gilkes N. R., Warren R. A. J., Miller R. C., Jr, Kilburn D. G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J. Biol. Chem. 1988;263:10401–10407. [PubMed] [Google Scholar]
- 4.Boraston A. B., McLean B. W., Kormos J. M., Alam M., Gilkes N. R., Haynes C. A., Tomme P., Kilburn D. G., Warren R. A. J. Carbohydrate-binding modules: diversity of structure and function. In: Gilbert H. J., Davies G. J., Henrissat B., Svensson B., editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: Royal Society of Chemistry; 1999. pp. 202–211. [Google Scholar]
- 5.Coutinho P. M., Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H. J., Davies G. J., Henrissat B., Svensson B., editors. Recent Advances in Carbohydrate Bioengineering. Cambridge: Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 6.Sigurskjold B. W., Svensson B., Williamson G., Driguez H. Thermodynamics of ligand binding to the starch-binding domain of glucoamylase from Aspergillus niger. Eur. J. Biochem. 1994;225:133–141. doi: 10.1111/j.1432-1033.1994.00133.x. [DOI] [PubMed] [Google Scholar]
- 7.Sorimachi K., Jacks A. J., Le Gal-Coeffet M. F., Williamson G., Archer D. B., Williamson M. P. Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy. J. Mol. Biol. 1996;259:970–987. doi: 10.1006/jmbi.1996.0374. [DOI] [PubMed] [Google Scholar]
- 8.Williamson M. P., Le Gal-Coeffet M. F., Sorimachi K., Furniss C. S., Archer D. B., Williamson G. Function of conserved tryptophans in the Aspergillus niger glucoamylase 1 starch binding domain. Biochemistry. 1997;36:7535–7539. doi: 10.1021/bi9702896. [DOI] [PubMed] [Google Scholar]
- 9.Sorimachi K., Le Gal-Coeffet M. F., Williamson G., Archer D. B., Williamson M. P. Solution structure of the granular starch binding domain of Aspergillus niger glucoamylase bound to beta-cyclodextrin. Structure. 1997;5:647–661. doi: 10.1016/s0969-2126(97)00220-7. [DOI] [PubMed] [Google Scholar]
- 10.Southall S. M., Simpson P. J., Gilbert H. J., Williamson G., Williamson M. P. The starch-binding domain from glucoamylase disrupts the structure of starch. FEBS Lett. 1999;447:58–60. doi: 10.1016/s0014-5793(99)00263-x. [DOI] [PubMed] [Google Scholar]
- 11.Paldi T., Levy I., Shoseyov O. Glucoamylase starch-binding domain of Aspergillus niger B1: molecular cloning and functional characterization. Biochem. J. 2003;372:905–910. doi: 10.1042/BJ20021527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikami B., Adachi M., Kage T., Sarikaya E., Nanmori T., Shinke R., Utsumi S. Structure of raw starch-digesting Bacillus cereus β-amylase complexed with maltose. Biochemistry. 1999;38:7050–7061. doi: 10.1021/bi9829377. [DOI] [PubMed] [Google Scholar]
- 13.Lawson C. L., van Montfort R., Strokopytov B., Rozeboom H. J., Kalk K. H., de Vries G. E., Penninga D., Dijkhuizen L., Dijkstra B. W. Nucleotide sequence and X-ray structure of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 in a maltose-dependent crystal form. J. Mol. Biol. 1994;236:590–600. doi: 10.1006/jmbi.1994.1168. [DOI] [PubMed] [Google Scholar]
- 14.Dauter Z., Dauter M., Brzozowski A. M., Christensen S., Borchert T. V., Beier L., Wilson K. S., Davies G. J. X-ray structure of Novamyl, the five-domain “maltogenic” α-amylase from Bacillus stearothermophilus: maltose and acarbose complexes at 1.7 Å resolution. Biochemistry. 1999;38:8385–8392. doi: 10.1021/bi990256l. [DOI] [PubMed] [Google Scholar]
- 15.Abe A., Tonozuka T., Sakano Y., Kamitori S. Complex structures of Thermoactinomyces vulgaris R-47 α-amylase 1 with malto-oligosaccharides demonstrate the role of domain N acting as a starch-binding domain. J. Mol. Biol. 2004;335:811–822. doi: 10.1016/j.jmb.2003.10.078. [DOI] [PubMed] [Google Scholar]
- 16.Geiger J. H. Sugar tongs get a grip on the starch granule in barley α-amylase 1. Structure. 2003;11:903–904. doi: 10.1016/s0969-2126(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 17.Robert X., Haser R., Gottschalk T. E., Ratajczak F., Driguez H., Svensson B., Aghajari N. The structure of barley α-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: a pair of sugar tongs. Structure. 2003;11:973–984. doi: 10.1016/s0969-2126(03)00151-5. [DOI] [PubMed] [Google Scholar]
- 18.Tomme P., Boraston A., McLean B., Kormos J., Creagh A. L., Sturch K., Gilkes N. R., Haynes C. A., Warren R. A. J., Kilburn D. G. Characterization and affinity applications of cellulose-binding domains. J. Chromatogr. B Biomed. Sci. Appl. 1998;715:283–296. doi: 10.1016/s0378-4347(98)00053-x. [DOI] [PubMed] [Google Scholar]
- 19.Rutenber E., Robertus J. D. Structure of ricin B-chain at 2.5 Å resolution. Proteins. 1991;10:260–269. doi: 10.1002/prot.340100310. [DOI] [PubMed] [Google Scholar]
- 20.Bains G., Lee R. T., Lee Y. C., Freire E. Microcalorimetric study of wheat germ agglutinin binding to N-acetylglucosamine and its oligomers. Biochemistry. 1992;31:12624–12628. doi: 10.1021/bi00165a012. [DOI] [PubMed] [Google Scholar]
- 21.Henrissat B., Teeri T. T., Warren R. A. J. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 1998;425:352–354. doi: 10.1016/s0014-5793(98)00265-8. [DOI] [PubMed] [Google Scholar]
- 22.Bolam D. N., Ciruela A., Mcqueen-Mason S., Simpson P., Williamson M. P., Rixon J. E., Boraston A., Hazlewood G. P., Gilbert H. J. Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem. J. 1998;331:775–781. doi: 10.1042/bj3310775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boraston A. B., Kwan E., Chiu P., Warren R. A. J., Kilburn D. G. Recognition and hydrolysis of noncrystalline cellulose. J. Biol. Chem. 2003;278:6120–6127. doi: 10.1074/jbc.M209554200. [DOI] [PubMed] [Google Scholar]
- 24.Charnock S. J., Bolam D. N., Turkenburg J. P., Gilbert H. J., Ferreira L. M., Davies G. J., Fontes C. M. The X6 “thermostabilizing” domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry. 2000;39:5013–5021. doi: 10.1021/bi992821q. [DOI] [PubMed] [Google Scholar]
- 25.Ali M. K., Hayashi H., Karita S., Goto M., Kimura T., Sakka K., Ohmiya K. Importance of the carbohydrate-binding module of Clostridium stercorarium Xyn10B to xylan hydrolysis. Biosci. Biotechnol. Biochem. 2001;65:41–47. doi: 10.1271/bbb.65.41. [DOI] [PubMed] [Google Scholar]
- 26.Zverlov V. V., Volkov I. Y., Velikodvorskaya G. A., Schwarz W. H. The binding pattern of two carbohydrate-binding modules of laminarinase Lam16A from Thermotoga neapolitana: differences in β-glucan binding within family CBM4. Microbiology. 2001;147:621–629. doi: 10.1099/00221287-147-3-621. [DOI] [PubMed] [Google Scholar]
- 27.Gilkes N. R., Kilburn D. G., Warren R. A. J., Miller R. C., Jr Precise excision of the cellulose binding domains form two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J. Biol. Chem. 1988;263:10401–10407. [PubMed] [Google Scholar]
- 28.Maglione G., Matsushita O., Russell J. B., Wilson D. B. Properties of a genetically reconstructed Prevotella ruminicola endoglucanase. Appl. Environ. Microbiol. 1992;58:3593–3597. doi: 10.1128/aem.58.11.3593-3597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall J., Black G. W., Ferreira L. M., Millward-Sadler S. J., Ali B. R., Hazlewood G. P., Gilbert H. J. The non-catalytic cellulose-binding domain of a novel cellulase from Pseudomonas fluorescens subsp. cellulosa is important for the efficient hydrolysis of Avicel. Biochem. J. 1995;309:749–756. doi: 10.1042/bj3090749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakon J., Irwin D., Wilson D. B., Karplus P. A. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat. Struct. Biol. 1997;4:810–818. doi: 10.1038/nsb1097-810. [DOI] [PubMed] [Google Scholar]
- 31.Araki R., Ali M. K., Sakka M., Kimura T., Sakka K., Ohmiya K. Essential role of the family-22 carbohydrate-binding modules for β-1,3→1,4-glucanase activity of Clostridium stercorarium Xyn10B. FEBS Lett. 2004;561:155–158. doi: 10.1016/S0014-5793(04)00160-7. [DOI] [PubMed] [Google Scholar]
- 32.Boraston A. B., Creagh A. L., Alam M. M., Kormos J. M., Tomme P., Haynes C. A., Warren R. A. J., Kilburn D. G. Binding specificity and thermodynamics of a family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A. Biochemistry. 2001;40:6240–6247. doi: 10.1021/bi0101695. [DOI] [PubMed] [Google Scholar]
- 33.Notenboom V., Boraston A. B., Kilburn D. G., Rose D. R. Crystal structures of the family 9 carbohydrate-binding module from Thermotoga maritima xylanase 10A in native and ligand-bound forms. Biochemistry. 2001;40:6248–6256. doi: 10.1021/bi0101704. [DOI] [PubMed] [Google Scholar]
- 34.Carrard G., Koivula A., Soderlund H., Beguin P. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10342–10347. doi: 10.1073/pnas.160216697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCartney L., Gilbert H. J., Bolam D. N., Boraston A. B., Knox J. P. Glycoside hydrolase carbohydrate-binding modules as molecular probes for the analysis of plant cell wall polymers. Anal. Biochem. 2004;326:49–54. doi: 10.1016/j.ab.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 36.Din N., Damude H. G., Gilkes N. R., Miller R. C., Jr, Warren R. A. J., Kilburn D. G. C1-Cx revisited: intramolecular synergism in a cellulase. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11383–11387. doi: 10.1073/pnas.91.24.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao P. J., Chen G. J., Wang T. H., Zhang Y. S., Liu J. Non-hydrolytic disruption of crystalline structure of cellulose by cellulose binding domain and linker sequence of cellobiohydrolase I from Penicillium janthinellum. Shengwu Huaxue Yu Shengwu Wuli Xuebao. 2001;33:13–18. [PubMed] [Google Scholar]
- 38.Giardina T., Gunning A. P., Juge N., Faulds C. B., Furniss C. S., Svensson B., Morris V. J., Williamson G. Both binding sites of the starch-binding domain of Aspergillus niger glucoamylase are essential for inducing a conformational change in amylose. J. Mol. Biol. 2001;313:1149–1159. doi: 10.1006/jmbi.2001.5097. [DOI] [PubMed] [Google Scholar]
- 39.Henrissat B., Davies G. J. Glycoside hydrolases and glycosyltransferases: families, modules, and implications for genomics. Plant Physiol. 2000;124:1515–1519. doi: 10.1104/pp.124.4.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coutinho P. M., Deleury E., Davies G. J., Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 41.Czjzek M., Bolam D. N., Mosbah A., Allouch J., Fontes C. M., Ferreira L. M., Bornet O., Zamboni V., Darbon H., Smith N. L., et al. The location of the ligand-binding site of carbohydrate-binding modules that have evolved from a common sequence is not conserved. J. Biol. Chem. 2001;276:48580–48587. doi: 10.1074/jbc.M109142200. [DOI] [PubMed] [Google Scholar]
- 42.Sunna A., Gibbs M. D., Bergquist P. L. Identification of novel β-mannan- and β-glucan-binding modules: evidence for a superfamily of carbohydrate-binding modules. Biochem. J. 2001;356:791–798. doi: 10.1042/0264-6021:3560791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 44.Richardson J. S. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 44a.Jamal-Talabani S., Boraston A. B., Turkenburg J. P., Tarbouriech N., Ducros V. M., Davies G. J. Ab initio structure determination and functional characterization of CBM36; a new family of calcium-dependent carbohydrate binding modules. Structure. 2004;12:1177–1187. doi: 10.1016/j.str.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 45.Murzin A. G., Lesk A. M., Chothia C. β-Trefoil fold: patterns of structure and sequence in the Kunitz inhibitors interleukins-1β and 1α and fibroblast growth factors. J. Mol. Biol. 1992;223:531–543. doi: 10.1016/0022-2836(92)90668-a. [DOI] [PubMed] [Google Scholar]
- 46.Sweet R. M., Wright H. T., Janin J., Chothia C. H., Blow D. M. Crystal structure of the complex of porcine trypsin with soybean trypsin inhibitor (Kunitz) at 2.6-Å resolution. Biochemistry. 1974;13:4212–4228. doi: 10.1021/bi00717a024. [DOI] [PubMed] [Google Scholar]
- 47.Boraston A. B., Tomme P., Amandoron E. A., Kilburn D. G. A novel mechanism of xylan binding by a lectin-like module from Streptomyces lividans xylanase 10A. Biochem. J. 2000;350:933–941. [PMC free article] [PubMed] [Google Scholar]
- 48.Notenboom V., Boraston A. B., Williams S. J., Kilburn D. G., Rose D. R. High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding. Biochemistry. 2002;41:4246–4254. doi: 10.1021/bi015865j. [DOI] [PubMed] [Google Scholar]
- 49.Scharpf M., Connelly G. P., Lee G. M., Boraston A. B., Warren R. A. J., McIntosh L. P. Site-specific characterization of the association of xylooligosaccharides with the CBM13 lectin-like xylan binding domain from Streptomyces lividans xylanase 10A by NMR spectroscopy. Biochemistry. 2002;41:4255–4263. doi: 10.1021/bi015866b. [DOI] [PubMed] [Google Scholar]
- 50.Fujimoto Z., Kuno A., Kaneko S., Kobayashi H., Kusakabe I., Mizuno H. Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module. J. Mol. Biol. 2002;316:65–78. doi: 10.1006/jmbi.2001.5338. [DOI] [PubMed] [Google Scholar]
- 51.Baenziger J. U., Fiete D. Structural determinants of Ricinus communis agglutinin and toxin specificity for oligosaccharides. J. Biol. Chem. 1979;254:9795–9799. [PubMed] [Google Scholar]
- 52.Tormo J., Lamed R., Chirino A. J., Morag E., Bayer E. A., Shoham Y., Steitz T. A. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 1996;15:5739–5751. [PMC free article] [PubMed] [Google Scholar]
- 53.McLean B. W., Bray M. R., Boraston A. B., Gilkes N. R., Haynes C. A., Kilburn D. G. Analysis of binding of the family 2a carbohydrate-binding module from Cellulomonas fimi xylanase 10A to cellulose: specificity and identification of functionally important amino acid residues. Protein Eng. 2000;13:801–809. doi: 10.1093/protein/13.11.801. [DOI] [PubMed] [Google Scholar]
- 54.Lehtio J., Sugiyama J., Gustavsson M., Fransson L., Linder M., Teeri T. T. The binding specificity and affinity determinants of family 1 and family 3 cellulose binding modules. Proc. Natl. Acad. Sci. U.S.A. 2003;100:484–489. doi: 10.1073/pnas.212651999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagy T., Simpson P., Williamson M. P., Hazlewood G. P., Gilbert H. J., Orosz L. All three surface tryptophans in Type IIa cellulose binding domains play a pivotal role in binding both soluble and insoluble ligands. FEBS Lett. 1998;429:312–316. doi: 10.1016/s0014-5793(98)00625-5. [DOI] [PubMed] [Google Scholar]
- 56.Creagh A. L., Ong E., Jervis E., Kilburn D. G., Haynes C. A. Binding of the cellulose-binding domain of exoglucanase Cex from Cellulomonas fimi to insoluble microcrystalline cellulose is entropically driven. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12229–12234. doi: 10.1073/pnas.93.22.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simpson P. J., Xie H., Bolam D. N., Gilbert H. J., Williamson M. P. The structural basis for the ligand specificity of family 2 carbohydrate-binding modules. J. Biol. Chem. 2000;275:41137–41142. doi: 10.1074/jbc.M006948200. [DOI] [PubMed] [Google Scholar]
- 58.Pell G., Williamson M. P., Walters C., Du H., Gilbert H. J., Bolam D. N. Importance of hydrophobic and polar residues in ligand binding in the family 15 carbohydrate-binding module from Cellvibrio japonicus Xyn10C. Biochemistry. 2003;42:9316–9323. doi: 10.1021/bi0347510. [DOI] [PubMed] [Google Scholar]
- 59.Xie H., Gilbert H. J., Charnock S. J., Davies G. J., Williamson M. P., Simpson P. J., Raghothama S., Fontes C. M., Dias F. M., Ferreira L. M., Bolam D. N. Clostridium thermocellum Xyn10B carbohydrate-binding module 22-2: the role of conserved amino acids in ligand binding. Biochemistry. 2001;40:9167–9176. doi: 10.1021/bi0106742. [DOI] [PubMed] [Google Scholar]
- 60.Notenboom V., Boraston A. B., Chiu P., Freelove A. C., Kilburn D. G., Rose D. R. Recognition of cello-oligosaccharides by a family 17 carbohydrate-binding module: an X-ray crystallographic, thermodynamic and mutagenic study. J. Mol. Biol. 2001;314:797–806. doi: 10.1006/jmbi.2001.5153. [DOI] [PubMed] [Google Scholar]
- 61.Boraston A. B., Notenboom V., Warren R. A. J., Kilburn D. G., Rose D. R., Davies G. Structure and ligand binding of carbohydrate-binding module CsCBM6-3 reveals similarities with fucose-specific lectins and “galactose-binding” domains. J. Mol. Biol. 2003;327:659–669. doi: 10.1016/s0022-2836(03)00152-9. [DOI] [PubMed] [Google Scholar]
- 62.Henshaw J., Bolam D. N., Pires V. M., Czjzek M., Henrissat B., Ferreira L. M., Fontes C. M., Gilbert H. J. The family 6 carbohydrate-binding module CmCBM6-2 contains two ligand-binding sites with distinct specificities. J. Biol. Chem. 2004;279:21552–21559. doi: 10.1074/jbc.M401620200. [DOI] [PubMed] [Google Scholar]
- 63.Pires V. M., Henshaw J., Prates J. A., Bolam D., Ferreira L. M., Fontes C. M., Henrissat B., Planas A., Gilbert H. J., Czjzek M. The crystal structure of the family 6 carbohydrate-binding module from Cellvibrio mixtus lichenase 5A in complex with oligosaccharides reveals two distinct binding sites with different ligand specificities. J. Biol. Chem. 2004;279:21560–21568. doi: 10.1074/jbc.M401599200. [DOI] [PubMed] [Google Scholar]
- 64.Winterhalter C., Heinrich P., Candussio A., Wich G., Liebl W. Identification of a novel cellulose-binding domain within the multidomain 120 kDa xylanase XynA of the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 1995;15:431–444. doi: 10.1111/j.1365-2958.1995.tb02257.x. [DOI] [PubMed] [Google Scholar]
- 65.Gaskell A., Crennell S., Taylor G. The three domains of a bacterial sialidase: a β-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure. 1995;3:1197–1205. doi: 10.1016/s0969-2126(01)00255-6. [DOI] [PubMed] [Google Scholar]
- 66.Atkins E. D. T. Three-dimensional structure, interactions and properties of xylans. In: Visser J., editor. Xylans and Xylanases, vol. 7. Amsterdam: Elsevier; 1992. pp. 39–50. [Google Scholar]
- 67.Szabo L., Jamal S., Xie H., Charnock S. J., Bolam D. N., Gilbert H. J., Davies G. J. Structure of a family 15 carbohydrate-binding module in complex with xylopentaose: evidence that xylan binds in an approximate 3-fold helical conformation. J. Biol. Chem. 2001;276:49061–49065. doi: 10.1074/jbc.M109558200. [DOI] [PubMed] [Google Scholar]
- 68.Boraston A. B., Nurizzo D., Notenboom V., Ducros V., Rose D. R., Kilburn D. G., Davies G. J. Differential oligosaccharide recognition by evolutionarily-related β-1,4 and β-1,3 glucan-binding modules. J. Mol. Biol. 2002;319:1143–1156. doi: 10.1016/S0022-2836(02)00374-1. [DOI] [PubMed] [Google Scholar]
- 69.Charnock S. J., Bolam D. N., Nurizzo D., Szabo L., McKie V. A., Gilbert H. J., Davies G. J. Promiscuity in ligand-binding: the three-dimensional structure of a Piromyces carbohydrate-binding module, CBM29-2, in complex with cello- and mannohexaose. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14077–14082. doi: 10.1073/pnas.212516199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Hernandez E., Hernandez-Arana A. Structural bases of lectin-carbohydrate affinities: comparison with protein-folding energetics. Protein Sci. 1999;8:1075–1086. doi: 10.1110/ps.8.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kormos J., Johnson P. E., Brun E., Tomme P., McIntosh L. P., Haynes C. A., Kilburn D. G. Binding site analysis of cellulose binding domain CBD(N1) from endoglucanse C of Cellulomonas fimi by site-directed mutagenesis. Biochemistry. 2000;39:8844–8852. doi: 10.1021/bi000607s. [DOI] [PubMed] [Google Scholar]
- 72.Bolam D. N., Hefang X., Pell G., Hogg D., Galbraith G., Henrissat B., Gilbert H. J. X4 modules represent a new family of carbohydrate-binding modules that display novel properties. J. Biol. Chem. 2004;279:22953–22963. doi: 10.1074/jbc.M313317200. [DOI] [PubMed] [Google Scholar]
- 73.Boraston A. B., Revett T. J., Boraston C. M., Nurizzo D., Davies G. J. Structural and thermodynamic dissection of specific mannan recognition by a carbohydrate binding module, TmCBM27. Structure. 2003;11:665–675. doi: 10.1016/s0969-2126(03)00100-x. [DOI] [PubMed] [Google Scholar]
- 74.Quiocho F. A. Carbohydrate-binding proteins: tertiary structures and protein-sugar interactions. Annu. Rev. Biochem. 1986;55:287–315. doi: 10.1146/annurev.bi.55.070186.001443. [DOI] [PubMed] [Google Scholar]
- 75.Bolam D. N., Xie H., White P., Simpson P. J., Hancock S. M., Williamson M. P., Gilbert H. J. Evidence for synergy between family 2b carbohydrate binding modules in Cellulomonas fimi xylanase 11A. Biochemistry. 2001;40:2468–2477. doi: 10.1021/bi002564l. [DOI] [PubMed] [Google Scholar]
- 76.Boraston A. B., McLean B. W., Chen G., Li A., Warren R. A. J., Kilburn D. G. Co-operative binding of triplicate carbohydrate-binding modules from a thermophilic xylanase. Mol. Microbiol. 2002;43:187–194. doi: 10.1046/j.1365-2958.2002.02730.x. [DOI] [PubMed] [Google Scholar]
- 77.Jervis E. J., Haynes C. A., Kilburn D. G. Surface diffusion of cellulases and their isolated binding domains on cellulose. J. Biol. Chem. 1997;272:24016–24023. doi: 10.1074/jbc.272.38.24016. [DOI] [PubMed] [Google Scholar]
- 78.Evans S. V., Sigurskjold B. W., Jennings H. J., Brisson J. R., To R., Tse W. C., Altman E., Frosch M., Weisgerber C., Kratzin H. D., et al. Evidence for the extended helical nature of polysaccharide epitopes: the 2.8 Å resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for α-(2→8)-polysialic acid. Biochemistry. 1995;34:6737–6744. doi: 10.1021/bi00020a019. [DOI] [PubMed] [Google Scholar]
- 79.Vyas N. K., Vyas M. N., Chervenak M. C., Johnson M. A., Pinto B. M., Bundle D. R., Quiocho F. A. Molecular recognition of oligosaccharide epitopes by a monoclonal Fab specific for Shigella flexneri Y lipopolysaccharide: X-ray structures and thermodynamics. Biochemistry. 2002;41:13575–13586. doi: 10.1021/bi0261387. [DOI] [PubMed] [Google Scholar]
- 80.Baxa U., Cooper A., Weintraub A., Pfeil W., Seckler R. Enthalpic barriers to the hydrophobic binding of oligosaccharides to phage P22 tailspike protein. Biochemistry. 2001;40:5144–5150. doi: 10.1021/bi0020426. [DOI] [PubMed] [Google Scholar]
- 81.Xu G. Y., Ong E., Gilkes N. R., Kilburn D. G., Muhandiram D. R., Harris-Brandts M., Carver J. P., Kay L. E., Harvey T. S. Solution structure of a cellulose-binding domain from Cellulomonas fimi by nuclear magnetic resonance spectroscopy. Biochemistry. 1995;34:6993–7009. [PubMed] [Google Scholar]
- 82.Brun E., Moriaud F., Gans P., Blackledge M. J., Barras F., Marion D. Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry. 1997;36:16074–16086. doi: 10.1021/bi9718494. [DOI] [PubMed] [Google Scholar]
- 83.Kraulis J., Clore G. M., Nilges M., Jones T. A., Pettersson G., Knowles J., Gronenborn A. M. Determination of the three-dimensional solution structure of the C-terminal domain of cellobiohydrolase I from Trichoderma reesei: a study using nuclear magnetic resonance and hybrid distance geometry-dynamical simulated annealing. Biochemistry. 1989;28:7241–7257. doi: 10.1021/bi00444a016. [DOI] [PubMed] [Google Scholar]
- 84.Raghothama S., Simpson P. J., Szabo L., Nagy T., Gilbert H. J., Williamson M. P. Solution structure of the CBM10 cellulose binding module from Pseudomonas xylanase A. Biochemistry. 2000;39:978–984. doi: 10.1021/bi992163+. [DOI] [PubMed] [Google Scholar]
- 85.Saul F. A., Rovira P., Boulot G., Damme E. J., Peumans W. J., Truffa-Bachi P., Bentley G. A. Crystal structure of Urtica dioica agglutinin, a superantigen presented by MHC molecules of class I and class II. Structure Fold. Des. 2000;8:593–603. doi: 10.1016/s0969-2126(00)00142-8. [DOI] [PubMed] [Google Scholar]
- 86.Suetake T., Tsuda S., Kawabata S., Miura K., Iwanaga S., Hikichi K., Nitta K., Kawano K. Chitin-binding proteins in invertebrates and plants comprise a common chitin-binding structural motif. J. Biol. Chem. 2000;275:17929–17932. doi: 10.1074/jbc.C000184200. [DOI] [PubMed] [Google Scholar]