Abstract
Using EPAC1 (exchange protein directly activated by cAMP 1) as bait in two-hybrid screens of foetal and adult human brain libraries, we identified the LC2 (light chain 2) of MAP1A (microtubule-associated protein 1A) as a protein capable of interaction with EPAC1. We applied an immunoprecipitation assay to demonstrate protein interaction between EPAC1 and LC2 in co-transfected human embryonic kidney 293 cells. EPAC2 also co-immunoprecipitated with LC2 from extracts of rat cerebellum. Immunolocalization in co-transfected human embryonic kidney 293 cells revealed that EPAC1 co-localizes with LC2 throughout the cell body. We found that endogenous EPAC2 is also immunolocalized with LC2 in PC12 cells. Immunolocalization of EPAC1 in transfected COS1 cells showed that EPAC1 is associated with the perinuclear region surrounding the nucleus and filamentous structures throughout the cell. Removal of the cAMP-binding domain of EPAC1 (ΔcAMP-EPAC1) appeared to disrupt targeting of EPAC1 in cells resulting in a more dispersed staining pattern. Using two-hybrid assay, we tested the ability of LC2 to interact with ΔcAMP-EPAC1 and ΔDEP-EPAC1, which lacks a DEP domain (dishevelled, Egl-10 and pleckstrin homology domain). We found that deletion of the cAMP-binding domain inhibited interaction between EPAC1 and LC2 in a two-hybrid assay, but removal of the DEP domain had little effect. LC2 was found to interact with a glutathione-S-transferase-fusion protein of the cAMP-binding domain of EPAC1 in a pull-down assay, but not the DEP, REM (Ras exchange motif) or CAT (catalytic) domains. Together with our two-hybrid results, this suggests that the cAMP-binding domain of EPAC1 mediates interaction with LC2.
Keywords: cAMP-binding domain, cAMP-guanine nucleotide exchange factor (cAMP-GEF), exchange protein directly activated by cAMP (EPAC), microtubule-associated protein 1A (MAP1A), yeast two-hybrid
Abbreviations: CAT domain, catalytic domain; CMV, cytomegalovirus; DEP domain, dishevelled, Egl-10 and pleckstrin homology domain; EPAC, exchange protein directly activated by cAMP; GST, glutathione S-transferase; HA, haemagglutinin; HEK-293 cells, human embryonic kidney 293 cells; LC, light chain; MAP1A, microtubule-associated protein 1A; MCS, multiple cloning site; ONPG, o-nitrophenyl β-D-galactopyranoside; ORF, open reading frame; PKA, cAMP-dependent protein kinase; PKC, protein kinase C; PP1, protein phosphatase 1; REM domain, Ras exchange motif domain; X-Gal, 5-bromo-4-chloroindol-3-yl β-D-galactopyranoside
INTRODUCTION
Many extracellular physiological signalling agents increase the intracellular levels of the ubiquitous second-messenger cAMP [1]. These include immunomodulators, neurotransmitters and hormones, which typically activate the seven-transmembrane domain class of G-protein-coupled receptors [1]. Ligand binding causes activation of heterotrimeric G-proteins that stimulate one or more isoforms of adenylate cyclase to catalyse production of cAMP. Intracellular levels of cAMP are then regulated through the action of cAMP phosphodiesterases, which degrade cAMP to 5′-AMP [2]. Until very recently, the intracellular effects of cAMP were supposed to be transduced solely by PKA (cAMP-dependent protein kinase) in most cell types [3]. Recently, however, there have been consistent reports that certain signalling actions of cAMP are not mediated by PKA [4]. Insight into this very important, new PKA-independent signalling system has arisen from attempts to delineate the pathway by which cAMP activates the small GTPase, Rap1 [5]. These studies implicate direct activation of Rap1 by a recently discovered family of GEFs (guanine nucleotide exchange factors) called cAMP-GEFs or EPAC (exchange protein directly activated by cAMP), with no involvement of PKA [6,7]. Therefore EPACs represent a novel mechanism for governing signalling specificity within the cAMP cascade [6–9].
EPAC1 and EPAC2 are multidomain proteins containing an autoinhibitory cAMP-binding domain that inhibits the catalytic region and a DEP domain (dishevelled, Egl-10 and pleckstrin homology domain), which is supposed to be involved in membrane localization [10]. The presence of cAMP-binding sites in EPAC proteins facilitates their direct activation by cAMP, independent of the activation of PKA [6,7]. EPAC2 has an additional cAMP-binding site in its N-terminus that binds cAMP with low affinity [10]. EPAC1 mRNA is broadly expressed, with particularly high levels occurring in the thyroid, ovary, kidney and certain brain regions, whereas expression of EPAC2 mRNA appears to be restricted to the brain and adrenal glands [6,7].
In the present study, we have identified a novel protein-binding partner for EPAC1 and EPAC2, LC2 (light chain 2) of MAP1A (microtubule-associated protein 1A). The MAP1A/LC2 gene is organized to encode a precursor polypeptide that undergoes proteolytic processing to generate the final 2556-amino-acid MAP1A heavy chain and 249-amino-acid LC2 polypeptide [11]. LC2 appears to serve as a ‘scaffold’ or ‘adaptor’ protein, facilitating the stable interaction of MAP1A with other signal-transduction components or structural proteins [12–16]. In addition, LC2 catalyses the polymerization of microtubules [13]. The ability of EPAC to interact physically with LC2 provides a potential new mechanism for integrating signals from the cAMP cascade to control fundamental cell functions.
EXPERIMENTAL
Materials
Pretransformed MATCHMAKER foetal and human brain cDNA libraries, cloned into pACT2 vector, were purchased from ClonTech (catalogue no. HY4004AH). This vector expresses proteins as fusions with the activation domain of the Saccharomyces cerevisiae GAL4 protein and an HA (haemagglutinin) epitope. PMT2-EPAC1-HA mammalian expression vector encoding the full ORF (open reading frame) of the EPAC1 cDNA (GenBank® accession number XM0389598) was obtained from Professor J. Bos (University of Utrecht, Utrecht, The Netherlands). Anti-FLAG monoclonal and anti-EPAC2 polyclonal antibodies (catalogue no. sc-9383) were purchased from Sigma and Santa Cruz Biotechnology respectively. Anti-LC2 polyclonal antibody and rat LC2 cDNA were gifts from Professor F. Propst (University of Vienna, Vienna, Austria).
Two-hybrid screens
Two-hybrid screen assays were performed using methods described in the ClonTech manual (catalogue no. PT3183-1). EPAC1 (NM_006105) cDNA ORF (amino acids 1–881) was amplified from the PMT2-EPAC1-HA plasmid by PCR using primers (forward: 5′-CTTCCATATGGTGTTGAGAAGGATG-3′; reverse: 5′-GATAGGTCGACTCATGGCTCCAGCTCTCG-3′). The fragment generated was cloned into the NdeI–SalI sites of pGBKT7 (catalogue no. K1612-1, ClonTech) to generate pGBKT7-EPAC1, which encodes a fusion among EPAC1, the Gal4 DNA-binding domain and a c-Myc epitope tag. pGBKT7–EPAC1–ΔDEP and pGBKT7–EPAC1–ΔcAMP, which encode full-length EPAC1-lacking DEP (amino acids 68–144) and cAMP-binding (amino acids 203–323) domains respectively, were generated from pGBKT7-EPAC1 using the Quik Change mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Generation of expression vectors
The full ORF for EPAC1 was cloned by PCR into the EcoRI and XbaI sites of p3xFLAG-myc-CMV-26 (where CMV stands for cytomegalovirus) expression vector (Sigma) to create myc-EPAC1-FLAG. Myc-ΔcAMP-EPAC1-FLAG was generated from myc-EPAC1-FLAG using mutagenesis primers 5′-GGACCTCATCTTTGAGGAGCTGCATGGCAAAGTGGTGCTGGTGCTGG-3′ (forward) and 5′-CCAGCACCAGCACCACTTTGCCATGCAGCTCCTCAAAGATGAGGTCC-3′ (reverse). For simultaneous expression of myc-tagged EPAC1 and LC2 in mammalian cells, we used the pBUDCE4.1 (Invitrogen, Paisley, Renfrewshire, Scotland, U.K.) vector. pBUD-EPAC1/LC2 was generated by subcloning the full ORFs for LC2 and EPAC1 into the EF-1α and CMV MCSs (multiple cloning sites) of pBUDCE4.1 respectively. LC2 was inserted at the XhoI–KpnI site in the EF-1α MCS and EPAC1 was inserted at the HindIII–XbaI site in the CMV MCS.
Generation of pGEX expression vectors
Individual EPAC1 domains were cloned by PCR into the EcoRI–NotI site of pGEX-6P-1. The individual domains generated were the CAT domain (catalytic domain; amino acids 619–881), the cAMP-binding domain (amino acids 199–316), the DEP domain (amino acids 74–140) and the REM domain (Ras exchange motif domain) (amino acids 345–410). When inserted into pGEX-6P-1, each EPAC1 domain is tagged with GST (glutathione S-transferase) to facilitate purification with glutathione–Sepharose as described previously [17].
ONPG (o-nitrophenyl β-D-galactopyranoside) assay
Amplified yeast broths from two-hybrid mating were harvested in 3 ml of Z buffer (113 mM Na2HPO4, 35 mM NaH2PO4, 10 mM KCl and 1 mM MgSO4, pH 7.4) and then permeabilized with 50 μl of 0.1% (w/v) SDS and were then added to the cell suspension and vortex-mixed briefly for 15 s followed by equilibration for 15 min at 30 °C. Then 100 μl of chloroform, followed by 160 μl of ONPG (4 mg/ml solution in Z buffer), was added. After incubation for 30 min, 400 μl from 1 ml of sodium carbonate was added. Cell debris was removed by centrifugation and absorbance A was measured at 420 and 550 nm. The activity of β-galactosidase was calculated using the formula 1000A420–(1.75A550)/time×volume×A600.
Growth of cell lines
COS1 cells were grown in Dulbecco's modified Eagle's medium (Sigma), supplemented with 10% (v/v) heat-inactivated foetal bovine serum (Sigma), 2 mM L-glutamine (Sigma) and 2% (v/v) solution of penicillin–streptomycin (Sigma). PC12 cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) horse serum (Sigma), 5% heat-inactivated foetal bovine serum, 2 mM L-glutamine and 2% solution of penicillin–streptomycin suspended in non-collagen-coated flasks and allowed to attach to collagen-coated coverslips for experiments. These were prepared by adding type I rat tail collagen (Sigma) to 0.1 M acetic acid to obtain 0.1% (w/v) collagen solution. The collagen solution was diluted 1:50 in PBS before use, added to coverslips (1 ml/2.5 cm2) and then incubated for at least 30 min at 37 °C.
Co-immunoprecipitations
Rat cerebellum was collected from two rats and homogenized in 50 mM Tris/HCl (pH 7.4). After homogenization, cell debris was removed by centrifugation at 100000 g for 30 min. For immunoprecipitation of EPAC2 and LC2, 500 μg of cleared extract was mixed with 60 μl of pre-equilibrated Protein G–agarose (Amersham Biosciences, Little Chalfont, Bucks., U.K.) and incubated for 30 min at 4 °C. The beads were removed by centrifugation at 2000 g for 5 min and the cleared lysates were incubated with 8 μl of anti-EPAC2 or 8 μl of LC2 antibody in the presence of Protein G–agarose beads for 3 h at 4 °C. The beads were then collected by centrifugation (2000 g for 5 min) and washed three times with 50 mM Tris/HCl (pH 7.4). Co-immunoprecipitation of EPAC2 with LC2 was analysed by immunoblotting with anti-EPAC2 and anti-LC2 antibodies. For co-immunoprecipitation of EPAC1 and LC2, HEK-293 cells (human embryonic kidney 293 cells) were transfected with pBUD-EPAC1/LC2 and lysed in 55 mM Tris/HCl (pH 7.4), 132 mM NaCl, 22 mM sodium fluoride, 11 mM sodium pyrophosphate, 1.1 mM EDTA and 5.5 mM EGTA containing 1% (v/v) Triton X-100. Immunoprecipitations were then performed as described above using anti-LC2 and anti-myc antibodies.
Immunocytochemistry
Cells were washed with PBS at 4 °C, 36 h after transfection with myc-EPAC1-Flag, and then fixed in a 4% (w/v) paraformaldehyde solution. Cells were then permeabilized in a 0.5% Triton X-100 solution in PBS for 5 min at 4 °C and blocked with goat serum [0.1% (v/v) in PBS] and fish gelatin (0.2% in PBS) for 30 min at room temperature (18 °C). The cells were then co-incubated for 2 h at room temperature with anti-Flag polyclonal antibody, at a dilution of 1:250 in block solution. Cells were then washed three times in PBS and then incubated for 1 h at room temperature with anti-rabbit FITC conjugate (Vector Laboratories, Peterborough, U.K.) diluted 1:200 in block solution. Cells were washed and mounted using Immu-Mount (Shandon Products, Cheshire, U.K.) and visualized by confocal microscopy. PC12 cells were fixed in the same manner and probed with LC2 antiserum followed by anti-mouse IgG Texas Red (Vector Laboratories) and anti-EPAC2 (Santa Cruz Biotechnology) followed by anti-goat IgG FITC conjugate (Vector Laboratories) to detect cellular EPAC2. HEK-293 cells transfected with pBUD-EPAC1/LC2 were probed with anti-LC2 followed by biotin-conjugated, monoclonal horse anti-rabbit IgG (Vector Laboratories) and monoclonal anti-myc antibody, followed by biotin-conjugated monoclonal horse anti-mouse IgG (Vector Laboratories) to detect transfected EPAC1myc. An FITC-conjugated streptavidin tertiary antibody was then added to detect EPAC1myc and a Texas Red-conjugated streptavidin tertiary antibody was used to detect LC2. Stained pBUD-EPAC1/LC2-transfected HEK-293 cells were visualized with a Zeiss Axiovert 135 fluorescent microscope.
In vitro expression of LC2
In vitro transcription translation of pcDNA3-LC2 was performed with the TNT® Coupled Reticulocyte Lysate Systems (Promega, Chilworth, Southampton, U.K.). The transcripts were prepared from the pcDNA3-LC2 plasmid using T7 polymerase (Amersham Biosciences). Incubation was for 90 min at 30 °C. Samples were denatured in SDS sample buffer and analysed by immunoblotting with anti-LC2 antisera.
SDS/PAGE and immunoblotting
SDS/PAGE and immunoblotting were carried out as described previously [17]. In brief, samples were resuspended in Laemmli buffer and boiled for 5 min. Membranes were blocked in 5% (w/v) low-fat milk powder in TBS (10 mM Tris/HCl, pH 7.4 and 150 mM NaCl) for 16 h at 4 °C. They were then incubated with anti-EPAC2 polyclonal antibody or anti-LC2 polyclonal antibody or anti-myc monoclonal antibody (Sigma) diluted to 1:1000 (v/v) in 1% low-fat milk powder in TTBS [TBS plus 0.1% (v/v) Tween 20] for 16 h at 4 °C. Detection of the bound antibody was with anti-goat IgG peroxidase (for EPAC2), anti-rabbit IgG (for LC2) or anti-mouse IgG peroxidase (for myc) secondary antibodies (Sigma) and the enhanced chemiluminescence (ECL®) system (Amersham Biosciences).
Measurement of protein concentrations
Protein concentrations were measured by the method of Bradford, using BSA as a standard [18].
RESULTS
Isolation of EPAC1-interacting proteins with two-hybrid screens
We examined whether specific proteins might bind to EPAC1. For this purpose, a full-length human EPAC1 ORF was used as a ‘bait’ in the two-hybrid screen of a foetal human brain cDNA library and in the second screen of an adult human brain library. We chose to screen these libraries because mRNA for both EPAC1 and EPAC2 are expressed in human brain [6,7]. A large number of cDNA clones were obtained in both screens and are listed in Table 1. These include cDNAs involved in signal transduction, e.g. PKC-potentiated PP1 inhibitory protein and p32-RACK (where PKC stands for protein kinase C, PP1 for protein phosphatase 1 and RACK for receptor for activated C-kinase 1), cytoskeleton (keratin complex 2) and chaperone functions (T-complex polypeptide 1).
Table 1. Results of two-hybrid screen of human adult and foetal brain cDNA libraries using EPAC1 as bait.
| Screen 1 (human foetal brain) | Screen 2 (human adult brain) | |
|---|---|---|
| No. of transformants | 5000000 | 5000000 |
| No. of His+ colonies | 180 | 110 |
| No. of LacZ+ colonies | 120/180 | 90/110 |
| No. of plasmids analysed by sequencing | 30 | 38 |
| No. of different clones present | 3 | 7 |
| Clone identities following BLAST search | Clone E4: NM_002373.3, human MAP1A | Clone A1: NP_503522.1, T-complex polypeptide 1/cpn60, required for assembly of microtubules |
| Clone E8: BT008258.1, human prefoldin 5 | ||
| Clone E10: XM_040708.3, human KIAA1377 protein | Clone C2: NP_03249, keratin complex | |
| Clone D4: AY050668.1, human PKC-potentiated PP1 | ||
| Clone F1: AF300619, p32-RACK | ||
| Clone F2: XM_030426.1, human hypoxia-inducible factor | ||
| Clone F3: NM_002373.3, human MAP1A |
Protein interaction among EPAC1, EPAC2 and LC2
A positive clone was isolated in both of our two-hybrid screens, which encoded a partial cDNA of the ORF for MAP1A [11,19]. MAP1A belongs to a family of proteins that controls the assembly of microtubules during neurogenesis [11]. The MAP1A precursor polypeptide is known to undergo proteolytic processing to generate a final MAP1A heavy chain and a C-terminal LC2 polypeptide [20]. Clone E4, from our foetal brain library screen (Table 1), and clone F3, from our adult brain library screen (Table 1), were comparable cDNAs and encoded an ORF that corresponded to the LC2 of MAP1A. The sequence of the identified clone corresponds to amino acids 1–144 of LC2.
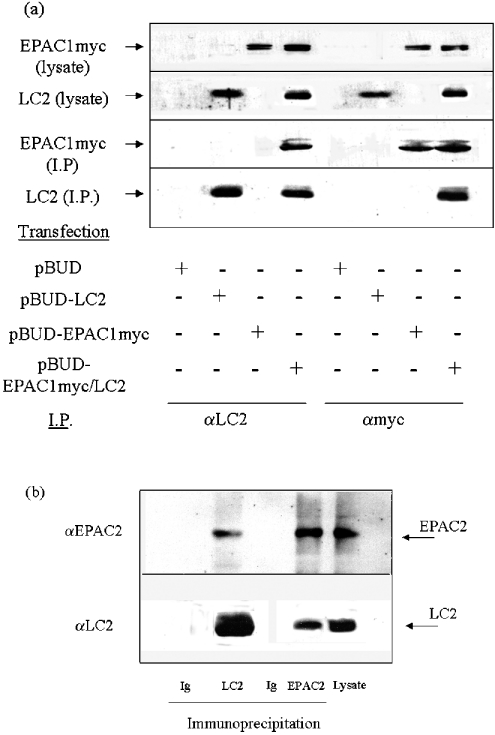
To determine whether endogenous EPAC1 and LC2 interacted in vivo, we immunoprecipitated lysates from pBUD-EPAC1/LC2 HEK-293 cells with anti-myc antibodies or an anti-LC2 antibody, which had been generated previously using a synthetic peptide (CKGPVDRTSRTVPRPR) corresponding to amino acids 2605–2619 of MAP1A [13]. The resulting immunoprecipitates were then subjected to SDS/PAGE and immunoblotting (Figure 1a). As EPAC1 migrates as a 99 kDa protein and LC2 as a 25 kDa protein, we treated the upper half of the blot with the EPAC2 antibody and the lower half with the LC2 antibody. Cell extracts (i.e. not subjected to immunoprecipitation) were immunoblotted as controls (Figure 1a). From this analysis, it was clear that the LC2 antibody not only immunoprecipitated LC2 but also co-immunoprecipitated EPAC1 (Figure 1a). In contrast, we found that the myc antibody not only immunoprecipitated EPAC1 but also co-immunoprecipitated LC2 (Figure 1a).
Figure 1. Co-immunoprecipitation of LC2 with EPAC1 and EPAC2.
(a) HEK-293 cells were transfected with pBUD, pBUD-LC2, pBUD-EPAC1 or pBUD-EPAC1/LC2 vector, which allows co-expression of myc-tagged EPAC1 and LC2. Cell extracts were prepared and immunoprecipitated with IgG, LC2 or myc antibodies, as indicated. Immunoprecipitates and samples of cell extract were then separated by SDS/PAGE and immunoblotted. The upper half of immunoblots was probed with anti-myc antibody and the lower half with anti-LC2. EPAC1 and LC2 were found to precipitate with both myc and LC2 antibodies. (b) Rat cerebellum lysate was subjected to immunoprecipitation with IgG, the EPAC2 antibody or the LC2 antibody. Samples of rat cerebellum lysate and immunoprecipitates were then subjected to SDS/PAGE. The upper half of the gel was immunoblotted with the EPAC2-specific antibody. The lower half of the gel was immunoblotted with the LC2-specific antibody, and the position of the 25 kDa LC2 species is indicated. EPAC2 and LC2 were found to precipitate with both the EPAC2 and LC2 antibodies.
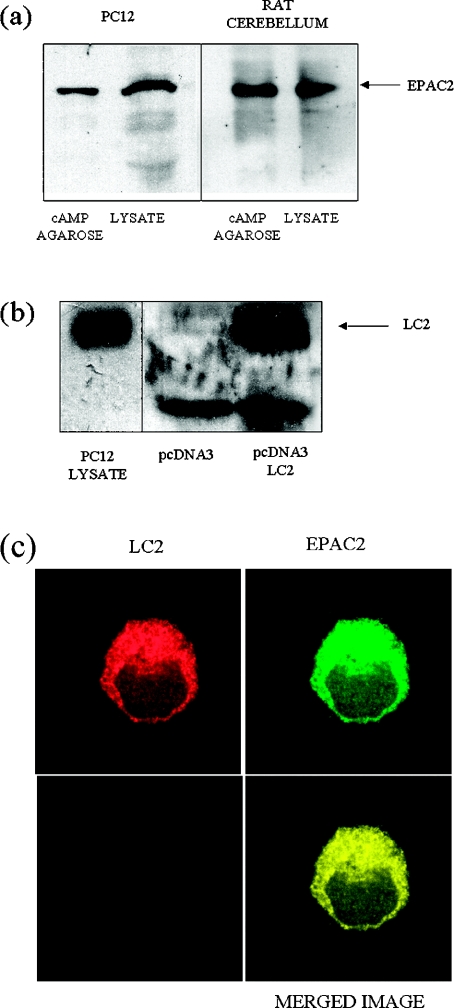
Given the sequence similarity between EPAC1 and EPAC2 [7], we decided to test whether LC2 also interacts with EPAC2. We confirmed rat cerebellum and phaeochromocytoma PC12 cells as a source of endogenously expressed EPAC2 protein by isolating the 120 kDa immunoreactive species with cAMP agarose (Figure 2a) as described previously [7]. We next immunoprecipitated rat cerebellum extracts with the LC2 and EPAC2 antibodies. We found that the EPAC2 antibody not only precipitated EPAC2 but also precipitated LC2 (Figure 1b). In contrast, the LC2 antibody precipitated EPAC2, in addition to LC2 (Figure 1b).
Figure 2. Expression and immunolocalization of EPAC2 and LC2 in PC12 cells.
(a) Lysates were prepared from rat cerebellum (right panel) and PC12 cells (left panel) and then precipitated with cAMP-agarose beads. Samples of cell lysate and cAMP-agarose bead precipitates were separated by SDS/PAGE and probed with an anti-EPAC2 polyclonal antibody. Immunoblotting revealed the presence a major immunoreactive species of approx. 120 kDa (EPAC2) within PC12 and rat cerebellum cell lysates and associated with the cAMP-agarose beads. (b) Recombinant LC2 was synthesized from pcDNA3-LC2 by coupled, in vitro transcription/translation (see the Experimental section) and separated by SDS/PAGE together with PC12 cell lysate. A control transcription/translation reaction was also subjected to SDS/PAGE, using pcDNA3 as a template. Immunoblots were probed with the LC2 antibody, demonstrating the presence of LC2 in PC12 lysates, which co-migrated with recombinant LC2 at 25 kDa. (c) Laser scans of confocal optical sections were taken through PC12 cells that had been probed with an LC2-specific polyclonal antibody (detected with an anti-rabbit IgG Texas Red conjugate) and an EPAC2-specific polyclonal antibody (detected with an anti-goat FITC conjugate). The intracellular distributions of EPAC2 and LC2 were very similar, as indicated by the yellow colour of overlaid images.
Given the ability of EPAC1 and EPAC2 to interact with LC2, we investigated whether EPAC2 localizes with LC2 in neuronal PC12 cells using a confocal scanning microscopy approach. We identified LC2 in PC12 extracts with the anti-peptide antibody and this co-migrated with recombinant LC2 synthesized in vitro (Figure 2b). LC2 immunofluorescence (red) was evident in a filamentous network surrounding the nucleus of the cell (Figure 2c). The distribution of EPAC2 (green) was remarkably similar to that of LC2 and, indeed, overlay of the images gave an essentially uniform yellow coloration, suggesting that both LC2 and EPAC2 are highly co-localized in PC12 cells.
Localization of EPAC1 and ΔcAMP-EPAC1 in transfected cells
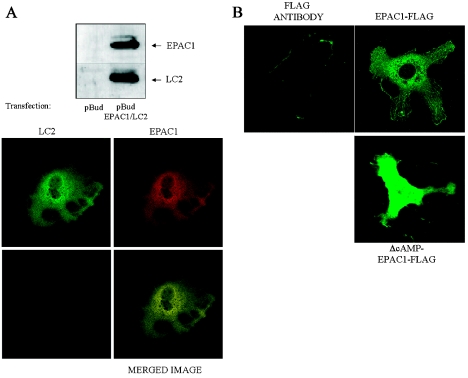
Results so far indicate that LC2 can interact with EPAC1 and EPAC2, perhaps through regions common to both proteins. We next investigated whether EPAC1 and LC2 interact in intact cells. We transfected HEK-293 cells with pBUD-EPAC1/LC2, which facilitated co-expression of myc-tagged EPAC1 and LC2 as evidenced by immunoblotting cell extracts (Figure 3A, upper panel). An immunofluorescent microscope was used to analyse cells probed with anti-myc (FITC; green) and anti-LC2 (Texas Red; red) antibodies to detect transfected EPAC1 and LC2 respectively. Immunofluorescence was present throughout the cell body for both LC2 and EPAC1 (Figure 3A, lower panel). Overlay of the two images gave a strong yellow coloration throughout the cell (Figure 3A, lower panel) indicating co-localization of EPAC1 and LC2.
Figure 3. Localization of EPAC1 in COS1 cells and immunolocalization of EPAC1 with LC2 in HEK-293 cells.
(A) HEK-293 cells were transfected with pBUD-EPAC1/LC2 vector, which allows co-expression of myc-tagged EPAC1, which can be detected by an anti-myc antibody, and LC2, detected with the LC2 antibody, as shown in the upper panel. Individual cells transfected with pBUD-EPAC1/LC2 were probed with a myc-specific monoclonal antibody to detect transfected EPAC1 (detected with an anti-mouse IgG Texas Red conjugate) and the LC2-specific polyclonal antibody to detect transfected LC2 (detected with an anti-goat FITC conjugate), and then analysed with an immunofluorescent microscope (lower panel). The LC2 and EPAC1 frames are given together as a superimposed image shown in the panel on the bottom right. In all the treatments applied, the immunofluorescent staining patterns of both LC2 and EPAC1 were very similar in all the images taken. In each case, this yielded a uniform yellow image indicative of a high degree of co-localization seen in the three different transfection studies. Analyses were repeated using different combinations of fluorescent-labelled antisera with similar results. (B) COS1 cells were transfected with myc-EPAC1-FLAG or constitutively active myc-ΔcAMP-EPAC1-FLAG (which lacks the cAMP-binding domain, amino acids 203–323) and then probed with an anti-Flag polyclonal antibody, secondary labelled with an anti-rabbit FITC conjugate, to detect myc-EPAC1-FLAG. A series of 0.25 μm optical sections were then captured using a laser scanning confocal microscope. The green-coloured immunofluorescent-staining pattern of myc-EPAC1-FLAG or constitutively active myc-ΔcAMP-EPAC1-FLAG was different in that myc-EPAC1-FLAG displayed a more diffuse staining pattern within the cell, demonstrating interaction with cytoskeletal components. The localization of myc-ΔcAMP-EPAC1-FLAG was more widespread within the cell and no longer seemed to be associated with cytoskeletal components.
We decided to examine the intracellular localization of EPAC1 and compare it with ΔcAMP-EPAC1, which lacks the cAMP-binding domain and has been reported to be constitutively active [6]. COS7 cells were transiently transfected with a cDNA-expressing myc and FLAG-tagged EPAC1 (FLAG-EPAC1-myc) or FLAG-ΔcAMP-EPAC1-myc. Cells were then fixed and incubated with an anti-Flag polyclonal antibody and visualized with FITC-conjugated anti-rabbit IgG, which gives a green coloration.
A punctate-staining pattern surrounding the nucleus was observed in EPAC1-transfected COS7 cells. Immunoreactivity was also observed to be associated with fibrous structures throughout the cell body (Figure 3B). A perinuclear-staining pattern was also seen in cells transfected with ΔcAMP-EPAC1; however, less immunoreactivity appeared to be associated with fibrous structures in cells (Figure 4c). These results suggest that EPAC1 can associate with the cytoskeleton of cells and that the cAMP-binding domain of EPAC1 might play a role in governing this association.
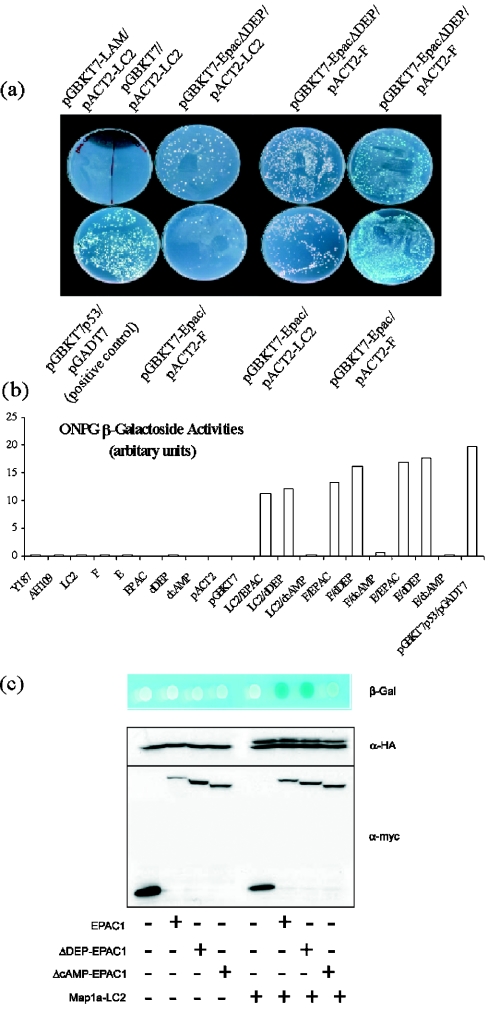
Figure 4. LC2 interacts with EPAC1 and EPAC1-ΔDEP, but not with EPAC1-ΔcAMP.
(a) AH109 yeast were transformed with the bait vector pGBKT7 or pGBKT7–EPAC1, pGBKT7–EPAC1–ΔDEP or pGBKT7-LAM (negative control), which express the full-length EPAC1 cDNA, EPAC1 cDNA lacking the DEP domain (amino acids 68–144; pGBKT7–EPAC1–ΔDEP) or laminin cDNA (pGBKT7-LAM) respectively, as an in-frame fusion with the Gal4 DNA-binding domain (BD). These were then mated with Y187 yeast that had been transformed with the library vector pACT2 or pACT2 containing either rat LC2 cDNA or clone F3 (F in the Figure) as in-frame fusions with the Gal4 activation domain (AD) in the combinations indicated in the Figure. As a positive control, AH109 yeast containing pGBKT7-p53 was mated with Y187 yeast containing the pGADT7 vector (positive control). After 2–3 days of incubation at 30 °C, the mated yeast was then plated on -Ade/-His/-Leu/-Trp plates to test for positive interaction between EPAC1-fusion protein (EPAC1-BD or EPAC1ΔDEP-BD) and the DNA activation domain (LC2-AD or F-AD). Growth of mated yeast on -Ade/-His/-Leu/-Trp plates indicates positive BD–AD protein interaction. In some cases, X-Gal was added to plates (+X-Gal) to check for expression and activation of α-galactosidase, a further determinant of BD–AD protein interaction, indicated by blue colour formation by mated colonies. (b) Yeast cells derived from the indicated mating pairs (pGBKT7 or pGBKT7–EPAC1, pGBKT7–EPAC1–ΔDEP or pGBKT7–EPAC1–ΔcAMP mated with pACT2 or pACT2-LC2 or pACT2-E4 or pACT2-F3) were grown overnight. Next day, cells were collected by centrifugation, resuspended in assay buffer and A600 measured. Following this, β-galactosidase substrate was added and the A values were measured at 420 and 550 nm. (c) AH109 cells were co-transformed with the empty bait vector, pGBKT7 or pGBKT7 containing EPAC1, EPAC1-ΔDEP or EPAC1-ΔcAMP and the empty ‘library vector’, pACT2 or pACT2-LC2. After 2–3 days, cells were spotted on to -Ade/-His/-Leu/-Trp agar plates containing X-Gal to check for activation of β-galactosidase expression. To check that the ratio of expression levels of EPAC1-BD, EPAC1-ΔDEP-BD and EPAC1-ΔcAMP-BD to E4-AD was equal, protein extracts were extracted from co-transformed yeast. Protein extracts were separated by SDS/PAGE and immunoblotted with an anti-HA-epitope monoclonal antibody (to detect E4-AD) and an anti-myc-epitope monoclonal antibody (to detect EPAC1-BD, EPAC1-ΔDEP-BD and EPAC1-ΔcAMP-BD). In all experiments, EPAC1-ΔcAMP-BD showed reduced interaction with E4.
cAMP-binding domain of EPAC1 is required for protein interaction with LC2
Given the suggestion that the cAMP-binding domain of EPAC1 might determine the intracellular localization of EPAC1, we decided to investigate the role of the EPAC1 cAMP-binding domain in protein interactions with LC2. We also investigated the role of the EPAC1 DEP domain in mediating the interaction with LC2, since deletion of this domain has been reported to disrupt interaction of EPAC1 with cell membranes [21].
We performed two-hybrid interaction assays on selective media using the LC2, clone F3 isolated from our two-hybrid screen and a full-length rat LC2 cDNA, which corresponds to amino acids 2554–2774 of rat MAP1A [13]. We found that diploid yeast transformed with plasmids encoding rat LC2 and lamin were unable to propagate on selective media, indicating that these two protein species did not interact (negative control; Figure 4a). As a positive control, we used yeast transformed with p53 and SV40 (simian-virus-40) large T-antigen, which grew vigorously on selective media, indicating positive protein interaction (Figure 4a). Yeast transformed with EPAC1 or EPAC1 lacking its DEP domain (EPAC1-ΔDEP), together with LC2 were capable of growing on selective media, indicating positive protein interaction (Figure 5A). These results indicate that the DEP domain of EPAC1 does not contribute to protein interactions between LC2 and EPAC1. We tested a second indicator of positive protein interaction between EPAC1, EPAC1-ΔDEP and LC2 clone F3, namely β-galactosidase-driven blue-colour colony formation. Mating pairs of EPAC1 or EPAC1-ΔDEP with clone F3 gave blue-colour colonies when treated with the β-galactosidase substrate, X-Gal (5-bromo-4-chloroindol-3-yl β-D-galactopyranoside), indicating strong, positive protein interaction (Figure 4a).
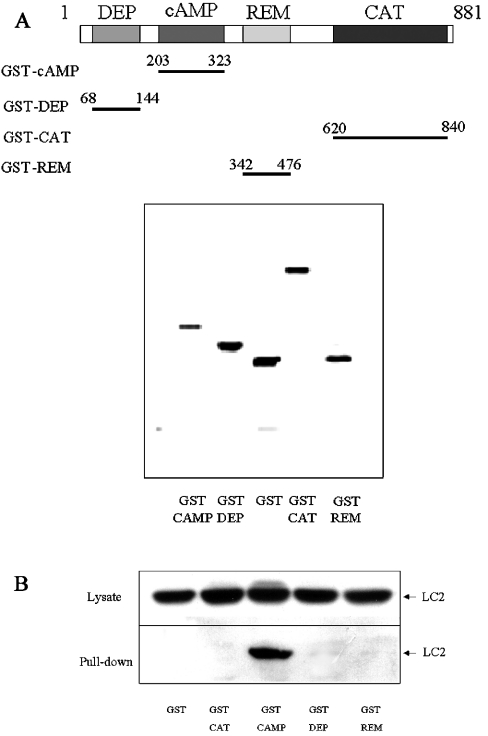
Figure 5. LC2 interacts with the cAMP-binding domain of EPAC1.
(A) Individual EPAC1 DEP (amino acids 68–144), REM (amino acids 342–476), cAMP binding (amino acids 203–323) and CAT (amino acids 620–840) domains were subcloned into pGEX6X, which expresses each domain as an in-frame fusion with GST, thereby facilitating purification of recombinant protein chimaeras from bacteria using glutathione (GSH)–Sepharose. To check the purity of isolated recombinant chimaeras, GST-DEP, GST-REM, GST-cAMP and GST-CAT were separated by SDS/PAGE and stained with Coomassie Blue (lower panel). The position of the domains in the primary structure of EPAC1 is shown diagrammatically in the upper panel. (B) To check for the ability of GST-fusion proteins to interact with LC2, PC12 Triton X-100 lysates were used as a source of soluble LC2 protein. PC12 lysates were precipitated with GST-DEP, GST-REM, GST-cAMP or GST-CAT. Cell lysates and precipitates were then separated by SDS/PAGE, followed by immunoblotting with the LC2 polyclonal antibody. In all experiments, LC2 was found to interact with GST-cAMP but not with GST-DEP, GST-REM or GST-CAT.
We were able to quantify the extent of protein interaction in mated yeast using an ONPG assay, which measures β-galactosidase activity (Figure 4b and the Experimental section). Strikingly, we found that EPAC1-ΔcAMP consistently failed to interact with LC2 in the ONPG assay. To investigate this result further, we tested the ability of EPAC1, EPAC1-ΔDEP and EPAC1-ΔcAMP to interact with the LC2 clone E4 in co-transformed yeast, using α-galactosidase-mediated blue-colour formation as an indicator of positive protein interaction (Figure 4c). Protein extracts were prepared from yeast and immunoblotted with anti-myc and anti-HA epitope antibodies to detect EPAC1, EPAC1-ΔDEP and EPAC1-ΔcAMP and LC2 clone E4 and ensure equal protein expression (Figure 4c). In agreement with our results from the ONPG assay, we found that EPAC1 and EPAC1-ΔDEP interacted strongly with MAP1A-LC2, as indicated by blue-colour formation, whereas EPAC1-ΔcAMP interacted only weakly (Figure 4c).
These results suggested that either LC2 interacts with EPAC1 principally through the cAMP-binding domain or LC2 does not interact with ‘active’ EPAC1, as EPAC1-ΔcAMP is constitutively active [6]. To investigate this, we prepared GST-fusion proteins of the EPAC1 domains, REM, DEP, cAMP-binding domain and CAT, and purified them from bacteria (Figure 5A). We then tested the ability of the GST-fusion proteins to interact with LC2 in cell lysates from PC12 cells (Figures 5B). Our results demonstrated that only GST-cAMP was capable of interacting with LC2 in pull-down assay, whereas GST-REM, GST-DEP and GST-CAT failed to interact. This, together with our two-hybrid analysis, suggests that the principal determinant in EPAC1 for interaction with LC2 is the cAMP-binding domain.
DISCUSSION
Our results implicate an association between EPAC proteins and MAP1A-LC2, which may be central to cell-growth mechanisms in general. Our identification of MAP1A-LC2 as a protein-binding partner for EPAC suggests that the EPAC protein may be functionally involved in microtubule assembly. It has been known for some time that cAMP promotes the assembly of microtubules and microfilaments [22]. Indeed, antibodies directed against cAMP and type II regulatory subunits of PKA localize these molecules to the mitotic spindle in mitotic cells [23]. The mitotic spindle is a macromolecular structure, which is required to segregate duplicated chromosomes into the two daughter cells during cell division. Throughout mitosis, the distribution of cAMP and PKA closely resembles that of tubulin, a conserved essential protein required for assembly and function of the mitotic spindle in humans and yeast [23]. The association between specific components of the cAMP-signalling system and the mitotic spindle suggests that cAMP-dependent regulation of spindle proteins, such as those of microtubules, may play a fundamental role in the regulation of spindle assembly and chromosome motion.
This is particularly exciting in the light of recent results from Qiao et al. [21], which demonstrate EPAC1 in association with the nuclear membrane during interphase, and the mitotic spindle during the M-phase, of the COS7 cell cycle. Using EPAC1/tubulin immunolocalization studies, we also demonstrate an intimate association between EPAC1 and microtubular structures in HEK-293 cells (results not shown). Indeed, MAP1A is known to promote nucleation and elongation of tubulin [24]. MAP1A promotes incorporation of tubulin dimers at low GTP concentrations and promotes the formation of oligomers at high GTP concentrations [24]. Catalysis of microtubule polymerization is also a function that has been independently attributed to LC2 [13].
The physiological implications of EPAC–MAP1A-LC2 interaction may also be related to the ability of MAP1A to serve as a ‘scaffold’ or ‘adaptor’ protein that mediates the recruitment of EPAC1 into a protein complex. A single ‘scaffold’ or adaptor protein may interact with many different proteins, and all of them could be recruited into a protein complex. For example, A-kinase anchoring proteins can interact with PKA, PKC and PP1 [25]. Since the interaction between LC2 and EPAC1 requires the cAMP-binding domain of EPAC, it can be argued that the protein complex formed is analogous to the interaction between PKA and A-kinase anchoring proteins, which is mediated by the cAMP-binding regulatory subunit of PKA [26]. LC2-mediated recruitment of EPAC1 to MAP1A may lead to the regulation of other signalling proteins within, or within the immediate vicinity, of this protein complex. Indeed, it has been demonstrated that MAP1A is a target for phosphorylation by casein kinase 2 [27] and insulin-stimulated kinases [28].
Acknowledgments
This work was funded by a Biotechnology and Biological Sciences Research Council (BBSRC; grant number C158009) grant awarded to S.J.Y. C.J.R. is a BBSRC research student.
References
- 1.Kopperud R., Krakstad C., Selheim F., Doskeland S. O. cAMP effector mechanisms. Novel twists for an ‘old’ signaling system. FEBS Lett. 2003;546:121–126. doi: 10.1016/s0014-5793(03)00563-5. [DOI] [PubMed] [Google Scholar]
- 2.Houslay M. D. Adaptation in cyclic AMP signalling processes: a central role for cyclic AMP phosphodiesterases. Semin. Cell Dev. Biol. 1998;9:161–167. doi: 10.1006/scdb.1997.0221. [DOI] [PubMed] [Google Scholar]
- 3.Engh R. A., Bossemeyer D. The protein kinase activity modulation sites: mechanisms for cellular regulation – targets for therapeutic intervention. Adv. Enzyme Regul. 2001;41:121–149. doi: 10.1016/s0065-2571(00)00010-8. [DOI] [PubMed] [Google Scholar]
- 4.Medina D. L., Santisteban P. Thyrotropin-dependent proliferation of in vitro rat thyroid cell systems. Eur. J. Endocrinol. 2000;143:161–178. doi: 10.1530/eje.0.1430161. [DOI] [PubMed] [Google Scholar]
- 5.Stork P. J., Schmitt J. M. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12:258–266. doi: 10.1016/s0962-8924(02)02294-8. [DOI] [PubMed] [Google Scholar]
- 6.de Rooij J., Zwartkruis F. J., Verheijen M. H., Cool R. H., Nijman S. M., Wittinghofer A., Bos J. L. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature (London) 1998;396:474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki H., Springett G. M., Mochizuki N., Toki S., Nakaya M., Matsuda M., Housman D. E., Graybiel A. M. A family of cAMP-binding proteins that directly activate Rap1. Science. 1998;282:2275–2279. doi: 10.1126/science.282.5397.2275. [DOI] [PubMed] [Google Scholar]
- 8.Rehmann H., Rueppel A., Bos J. L., Wittinghofer A. Communication between the regulatory and the catalytic region of the cAMP-responsive guanine nucleotide exchange factor Epac. J. Biol. Chem. 2003;278:23508–23514. doi: 10.1074/jbc.M301680200. [DOI] [PubMed] [Google Scholar]
- 9.Rehmann H., Prakash B., Wolf E., Rueppel A., De Rooij J., Bos J. L., Wittinghofer A. Structure and regulation of the cAMP-binding domains of Epac2. Nat. Struct. Biol. 2003;10:26–32. doi: 10.1038/nsb878. [DOI] [PubMed] [Google Scholar]
- 10.Bos J. L., de Rooij J., Reedquist K. A. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell. Biol. 2001;2:369–377. doi: 10.1038/35073073. [DOI] [PubMed] [Google Scholar]
- 11.Langkopf A., Hammarback J. A., Muller R., Vallee R. B., Garner C. C. Microtubule-associated proteins 1A and LC2. Two proteins encoded in one messenger RNA. J. Biol. Chem. 1992;267:16561–16566. [PubMed] [Google Scholar]
- 12.Azuma Y., Dasso M. The role of Ran in nuclear function. Curr. Opin. Cell Biol. 2000;12:302–307. doi: 10.1016/s0955-0674(00)00093-4. [DOI] [PubMed] [Google Scholar]
- 13.Noiges R., Eichinger R., Kutschera W., Fischer I., Nemeth Z., Wiche G., Propst F. Microtubule-associated protein 1A (MAP1A) and MAP1B: light chains determine distinct functional properties. J. Neurosci. 2002;22:2106–2114. doi: 10.1523/JNEUROSCI.22-06-02106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahana J. A., Cleveland D. W. Beyond nuclear transport. Ran-GTP as a determinant of spindle assembly. J. Cell Biol. 1999;146:1205–1210. doi: 10.1083/jcb.146.6.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalab P., Pu R. T., Dasso M. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- 16.Nishimoto T. Upstream and downstream of ran GTPase. Biol. Chem. 2000;381:397–405. doi: 10.1515/BC.2000.052. [DOI] [PubMed] [Google Scholar]
- 17.Yarwood S. J., Steele M. R., Scotland G., Houslay M. D., Bolger G. B. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J. Biol. Chem. 1999;274:14909–14917. doi: 10.1074/jbc.274.21.14909. [DOI] [PubMed] [Google Scholar]
- 18.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 19.Garner C. C., Garner A., Huber G., Kozak C., Matus A. Molecular cloning of microtubule-associated protein 1 (MAP1A) and microtubule-associated protein 5 (MAP1B): identification of distinct genes and their differential expression in developing brain. J. Neurochem. 1990;55:146–154. doi: 10.1111/j.1471-4159.1990.tb08832.x. [DOI] [PubMed] [Google Scholar]
- 20.Fink J. K., Jones S. M., Esposito C., Wilkowski J. Human microtubule-associated protein 1a (MAP1A) gene: genomic organization, cDNA sequence, and developmental- and tissue-specific expression. Genomics. 1996;35:577–585. doi: 10.1006/geno.1996.0400. [DOI] [PubMed] [Google Scholar]
- 21.Qiao J., Mei F. C., Popov V. L., Vergara L. A., Cheng X. Cell cycle-dependent subcellular localization of exchange factor directly activated by cAMP. J. Biol. Chem. 2002;277:26581–26586. doi: 10.1074/jbc.M203571200. [DOI] [PubMed] [Google Scholar]
- 22.Seite R., Leonetti J., Luciani-Vullet J., Vio M. Cyclic AMP and ultrastructural organization of the nerve cell nucleus: stimulation of nuclear microtubules and microfilaments assembly in sympathetic neurons. Brain Res. 1977;124:41–51. doi: 10.1016/0006-8993(77)90862-9. [DOI] [PubMed] [Google Scholar]
- 23.Browne C. L., Lockwood A. H., Su J. L., Beavo J. A., Steiner A. L. Immunofluorescent localization of cyclic nucleotide-dependent protein kinases on the mitotic apparatus of cultured cells. J. Cell Biol. 1980;87:336–345. doi: 10.1083/jcb.87.2.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedrotti B., Islam K. Purified native microtubule associated protein MAP1A: kinetics of microtubule assembly and MAP1A/tubulin stoichiometry. Biochemistry. 1994;33:12463–12470. doi: 10.1021/bi00207a013. [DOI] [PubMed] [Google Scholar]
- 25.Pawson T., Scott J. D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 26.Newlon M. G., Roy M., Morikis D., Carr D. W., Westphal R., Scott J. D., Jennings P. A. A novel mechanism of PKA anchoring revealed by solution structures of anchoring complexes. EMBO J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Avila J., Ulloa L., Gonzalez J., Moreno F., Diaz-Nido J. Phosphorylation of microtubule-associated proteins by protein kinase CK2 in neuritogenesis. Cell. Mol. Biol. Res. 1994;40:573–579. [PubMed] [Google Scholar]
- 28.Erickson A. K., Ray L. B., Sturgill T. W. Microtubule-associated protein 1A is the fibroblast HMW MAP undergoing mitogen-stimulated serine phosphorylation. Biochem. Biophys. Res. Commun. 1990;166:827–832. doi: 10.1016/0006-291x(90)90884-p. [DOI] [PubMed] [Google Scholar]