Abstract
ECP (eosinophil cationic protein) is a major component of eosinophil granule proteins, and is used as a clinical biomarker for asthma and allergic inflammatory disease. ECP has been implicated in damage to the cell membrane of many tissue types, but the mechanism is not well known. In the present study, mECP–eGFP–6H, a recombinant fusion protein containing mature ECP (mECP), enhanced green fluorescence protein (eGFP) and a His6 tag (6H), has been expressed, purified and added to GH3 neuroendocrine cells to study the internalization ability of ECP. We found that mECP–eGFP–6H entered into GH3 neuroendocrine cells and inhibited the growth of the cells with an IC50 of 0.8 μM. By yeast two-hybrid screening and immunoprecipitation, we have identified a specific protein–protein interaction between mECP and CPE (carboxypeptidase E), a well characterized metalloprotease. Further in vivo yeast two-hybrid screening has also revealed that residues 318–387 located in a region of unknown function in mature CPE are indispensable for association with mECP. In addition, the uptake of mECP–eGFP–6H is suppressed by dominant-negative expression of the recycling defect mutant pre-pro-HA–CPES471A,E472A in GH3 cells, suggesting that the entry of mECP–eGFP–6H is associated with the recycling of CPE in GH3 cells. Taken together, we have demonstrated that CPE possesses a novel function to facilitate the entry of ECP to neuroendocrine cells, and such an endocytotic process allows the cytotoxic ECP to inhibit growth of the target cells.
Keywords: carboxypeptidase E, cell entry, endocytosis, eosinophil cationic protein (ECP), RNase
Abbreviations: AD, activation domain of Gal4; BD, DNA-binding domain of Gal4; BS-RNase, bovine seminal RNase; CPB (etc.), carboxypeptidase B (etc.); CPEc, clone of CPE containing the last 363 amino acids; mCPE, mature CPE; ECP, eosinophil cationic protein; hECP, human ECP; mECP, mature ECP; EDN, eosinophil-derived neurotoxin; FBS, foetal bovine serum; eGFP, enhanced green fluorescent protein; 6H, His6 tag; HA, haemagglutinin; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; RITC, rhodamine B isothiocyanate; SD, synthetic dropout
INTRODUCTION
ECP (eosinophil cationic protein) is one of the major components of the eosinophilic granules and has a molecular mass in the range 16–21.4 kDa due to varying degrees of glycosylation. It is classified as a member of the RNase A superfamily and exhibits differential biological effects both in vivo and in vitro [1]. Among eight human RNases, ECP is most similar to EDN (eosinophil-derived neurotoxin). ECP is bactericidal [2] and helminthotoxic [3,4], and is cytotoxic to tracheal epithelium [5,6]. When intrathecally injected into rabbits, ECP or EDN elicit the Gordon phenomenon, a syndrome manifested by ataxia, muscular rigidity, paralysis and tremor that may lead to death, and, most of the time, the characteristic Purkinje cell toxicity occurs [7,8]. Although the mechanism of its cytotoxicity is not completely understood, it is suggested that the pore-forming activity of ECP, but not the RNase activity, plays a key role in destabilizing lipid membranes of the target cells [9,10]. This is consistent with data showing that the cytotoxicity of ECP is greater than that of EDN [11].
It has been reported that ECP exhibits growth-inhibitory effects on several cell types [12,13]. Despite the high degree of sequence similarity between ECP and other members of the RNase A superfamily, ECP exhibits weak, but definite, RNase activity. ECP is able to internalize the cytosol effectively and escape from proteolytic attack. It is accumulated in the cytosol to large excess over the concentration of RNase inhibitors, such that degradation of cytosolic RNA molecules occurs [13]. The cellular internalization of proteins often occurs via a specific energy-dependent endocytosis pathway, through either clathrin-coated or non-clathrin-coated endosomes. Maeda et al. [12] suggested that ECP may interact preferentially with a receptor or binding protein on the cell surface [12]. Hence ECP appears to enter cells via energy-dependent endocytosis. However, no ECP-interacting protein involved in this endocytotic process has ever been identified.
In the present study, we have assessed the effect of the mECP–eGFP–6H fusion protein (where mECP is mature ECP, eGFP is enhanced green fluorescent protein and 6H is a His6 tag) on the growth of the mammalian neuroendocrine cell line, GH3. The results show that mECP–eGFP–6H enters the cell, whereas RNase A does not. In addition, mECP–eGFP–6H inhibits growth of GH3 cells with an IC50 of 0.8 μM. Using mECP as a bait to investigate its interacting proteins via a yeast two-hybrid system, we found that ECP interacted with the transmembrane, lipid-raft-associated pro-hormone-sorting receptor, CPE (carboxypeptidase E). Blocking the maturation of CPE-associated secretory granules and the dominant-negative expression of CPES473A,E474A, a recycling-defect mutant, in the GH3 cell line both interfered with the internalization of mECP–eGFP–6H, indicating that interaction between mECP and CPE plays a major role in internalization of mECP to the neuroendocrine cells. Our results provide the first demonstration of a non-surface-receptor-mediated mechanism responsible for the entry of cytotoxic RNase into the target cells.
EXPERIMENTAL
Cell culture and transfection
GH3 cells were obtained from the American Type Culture Collection (Manassas, VA, U.S.A.), and were cultured in Ham's F12K medium (Sigma) containing 15% (v/v) horse serum and 2.5% (v/v) FBS (foetal bovine serum), penicillin/streptomycin and incubated at 37 °C under 5% CO2. Transient transfections were performed using TransFast™ (Promega), according to the manufacturer's protocol. In brief, GH3 cells were subcultured 1 day before transfection. Subcultured GH3 cells were harvested by trypsin/EDTA treatment, collected by centrifugation at 500 g for 10 min at 25 °C, and then washed twice with PBS. Finally, the cells were resuspended in serum-free Ham's F12K medium, and the cell number was counted using the dye-exclusion haemocytometer procedure with 0.4% (w/v) Trypan Blue in HBSS (Hanks balanced salt solution). GH3 cells (2×106) were transfected with 2 μg of DNA, and were incubated at 37 °C under 5% CO2 for 48 h to express the target protein.
Preparation of recombinant mECP–eGFP–6H and eGFP–6H fusion proteins
hECP (human ECP) cDNA was isolated and expressed in an Escherichia coli T7 expression system by a modified method as described previously [14]. The eGFP gene was cloned into pET23b(+) vector between the HindIII and XhoI sites to generate eGFP–pET23b(+). The mECP gene was subsequently introduced between the NdeI and HindIII sites to generate mECP–eGFP–pET23b(+). Both recombinant DNAs were transformed into E. coli BL21(DE3) cells for protein expression. Volumes of 1 ml of overnight culture of each transformant were inoculated in 1 litre of LB (Luria–Bertani) containing 200 μg/ml carbenicillin (Sigma), and grown overnight at 20 °C with shaking. IPTG (isopropyl β-D-thiogalactoside) (Sigma) was added to a final concentration of 2 mM, and the bacteria were harvested 48 h after induction. Recombinant mECP–eGFP–6H and eGFP–6H proteins in the soluble portion of bacterial cell lysates were subjected to purification by His-Bind® affinity column chromatography (Novagen).
MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide] assay for GH3 cell growth
The effect of mECP–eGFP–6H on the growth of GH3 cell line was assessed by a colorimetric assay using MTT by a modified method as described previously [12]. Cells were plated into 96-well plates in Ham's F12K medium containing 15% (v/v) horse serum and 5% (v/v) FBS at 500 cells/well for 24 h before addition of mECP–eGFP–6H at the indicated concentration (0–2.5 μM). At 3 days after plating, the medium was replaced with fresh medium containing mECP–eGFP–6H at the same concentration. After a further 3 days of cultivation, MTT (5 mg/ml in PBS) was added, and cell growth was monitored by measuring the D570.
Uptake of mECP–eGFP–6H into GH3 cells
GH3 cells were cultured as described above. mECP–eGFP–6H (0.5 μM) was added directly to the culture medium in the presence and absence of 15 mM NH4Cl or 5 μM forskolin, and incubated for different durations. Cells were collected by centrifugation at 1500 g for 5 min and washed twice with PBS, then treated with cell-lysis buffer for further analysis. For fluorescence-microscopy experiments, bovine RNase A (Sigma) was labelled with RITC (rhodamine B isothiocyanate; Sigma) as described previously [15]. GH3 cells were seeded on to a six-well plate and grown on coverslips at 40000 cells/well for 24 h. mECP–eGFP–6H, eGFP–6H or RITC-labelled RNase A was added separately to the culture medium to a final concentration of 0.5 μM and incubated for 15 min–3 h. After incubation, cells were washed twice with ice-cold PBS, fixed with 4% (w/v) formaldehyde solution at room temperature (25 °C) for 15 min, and the nuclei were stained with 300 nM DAPI (4′,6-diamidino-2-phenylindole; Molecular Probes), before imaging by fluorescence microscopy.
Yeast two-hybrid assay
Yeast two-hybrid screening was performed by using the MATCHMAKER Two-Hybrid System 3 (Clontech) according to the manufacturer's instructions. For bait vector construction, the human mECP gene (mECP) was amplified by PCR with 5′-CATATGGTTCCAAAACTG-3′ as the forward primer and 5′-GAATTCTTAGATGGTGGTATC-3′ as the reverse primer. The PCR fragments were cloned into the NdeI and EcoRI sites of pGBKT7 (Clontech). The human brain cDNA library (Clontech) was screened for mECP-associated proteins. mECP-bait construct alone did not show autonomous transcriptional activation in the AH109 yeast strain. Thus yeast cells were co-transformed with 0.1 mg of cDNA library plasmids and 0.2 mg of mECP-bait plasmids, according to a basic lithium acetate protocol. The transformants were grown on SD-Ade-His-Leu-Trp (where SD is synthetic dropout) plates to select for protein–protein interactions. The β-galactosidase assay was performed further to test LacZ reporter gene expression of the clones selected. The library plasmids were then isolated from the positive selections and retransformed into yeast AH109 together with the mECP-bait plasmid for β-galactosidase activity assay. The ade2+/his3+/lacZ+ library plasmids containing the candidate genes were transformed into E. coli Top10F' cells, and the plasmid DNAs were prepared for sequencing. For quantitative and qualitative assays for specific interaction between mECP and the candidate prey on the AD (activation domain of Gal4), the mECP-bait and prey constructs were co-transformed into yeast host AH109. Clones containing different candidates were scraped and cultured on SD-Ade-His-Leu-Trp plates for qualitative assay of protein–protein interactions. For liquid-culture β-galactosidase assay, a single colony was cultured in SD-Leu-Trp medium until the D600 reached 0.6. The assay was performed in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, pH 7.0, 10 mM KCl and 1 μM MgSO4); 0.64 mg/ml ONPG (o-nitrophenyl β-D-galactopyranoside, Sigma) was used as substrate and the D420 values of samples after 400 min of incubation at 30 °C were measured.
Constructions of CPE deletion mutants
To identify the region of CPE for interaction with mECP, various deletions were constructed. The candidate gene CPE154–476–pACT2 was subcloned into AD vector (pGADT7), and seven serial C-terminal deletions of CPE were generated by a standard nested deletion method. Briefly, two restriction enzyme sites, BamHI and SacI, were introduced at the 3′-end of CPE154–476 DNA fragment on the CPE154–476–pGADT7 clone to generate CPE154–476,BamHI,SacI–pGADT7. CPE154–476,BamHI,SacI–pGADT7 was double digested by BamHI and SacI restriction enzymes, then treated with ExoIII nuclease at 25 °C, and samples were taken at 30 s intervals. After ExoIII digestion, DNAs were treated with S1 nuclease and Klenow enzyme for 15 min at 37 °C. Finally, the treated DNA products were ligated by T4 DNA ligase at room temperature for 1 h, before the self-ligated DNAs were transformed into Top10F' for screening.
Immunoprecipitation and Western blot analysis
Equal amounts (1 μg) of either purified eGFP–6H or mECP–eGFP–6H fusion proteins were added to the binding buffer (PBS containing 5 mM EDTA and 0.5% Nonidet P40) containing 100 μg of GH3 cell lysates and incubated at 4 °C overnight. Subsequently, eGFP–6H and mECP–eGFP–6H were subjected to immunoprecipitation from the mixture by adding 1 μg of polyclonal anti-eGFP antibody (Clontech). Protein A–agarose beads (Amersham Biosciences) were added to the cell lysate and then incubated at 4 °C overnight on a rotating apparatus. The beads were collected by centrifugation at 8000 g and washed twice with PBS. Finally, the pellet was resuspended in 1×SDS/PAGE sample buffer for further analysis. Monoclonal anti-CPE antibody (CashmereBiotech, Taiwan) and monoclonal anti-ECP109 antibody (BCRC 69019) were used in Western blotting.
Construction of pre-pro-HA–CPES471A,E472A mutation
Human pre-pro-HA–CPES471A,E472A point mutation was made by a two-stage PCR method. First, 5′-GAATTCATGGCCGGGCGCGGACGG-3′ and 5′-AGCGTAATCTGGTACGTCGTACGCCGGCGCCGCCGCATGC-3′ were used to amplify the pre-pro-HA fragment. Subsequently, 5′-TACGACGTACCAGATTACGCTCTGCAGCAAGAGGACGGC-3′ and 5′-CTCGAGTTAAAAATTTAAAGTTGCTGCCATCATTTTCC-3′ were used to amplify the HA–CPES471A,E472A fragment. The HA (haemagglutinin) tag epitope region is underlined and S471A,E472A (Ser471→Ala and Glu472→Ala) point mutations are displayed in italics. Finally, the pre-pro-HA and HA–CPES471A,E472A fragments were used as PCR template to generate the pre-pro-HA–CPES471A,E472A. The final PCR fragment was cloned between the EcoRI and XhoI sites of pcDNA3 (Invitrogen). The sequence of the plasmid cDNA was verified by sequencing analysis.
RESULTS
Preparation of recombinant mECP–eGFP–6H and eGFP–6H
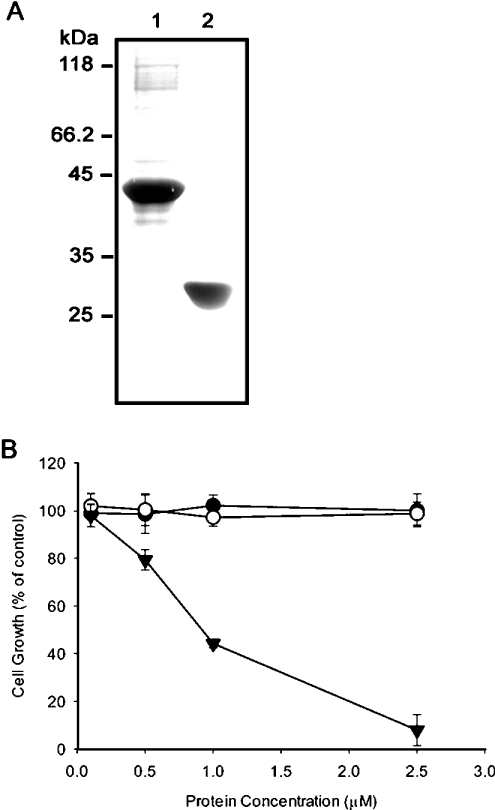
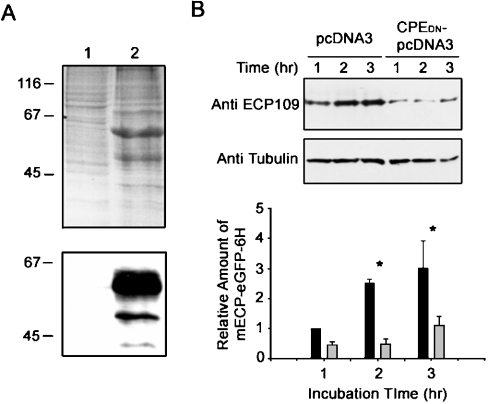
mECP was fused with eGFP, expressed as mECP–eGFP–6H fusion protein in E. coli cells, and the soluble portion was purified by His-Bind® Resin (Novagen). The molecular mass of mECP–eGFP–6H was determined to be 42 kDa. The 27 kDa recombinant eGFP–6H was obtained in a similar manner (Figure 1A). The RNase activity of purified recombinant mECP–eGFP–6H was assessed with yeast RNA by the perchloric and precipitation method [16]. It was determined to be about 100 times lower than RNase A.
Figure 1. Suppression of GH3 cell growth in the presence of mECP–eGFP–6H.
(A) The molecular mass of mECP–eGFP–6H fusion protein (lane 1) and eGFP-6H (lane 2) are about 42 kDa and 27 kDa respectively. (B) Growth of GH3 cells was monitored by the MTT assay; the percentages of viable cells at various concentrations of RNase A (•), eGFP-6H (○) and mECP–eGFP–6H (▾) were plotted. Each experiment was carried out in triplicate and results are means±S.D.
mECP–eGFP–6H inhibits the growth of neuroendocrine cells
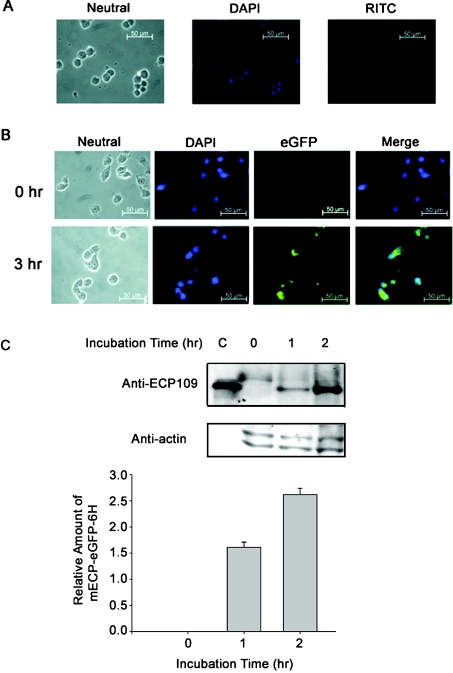
In the presence of 2.5 μM mECP–eGFP–6H, the growth of GH3 cells was suppressed completely, whereas no inhibitory effect was observed upon treatment with either RNase A or eGFP–6H (Figure 1B). This result indicated clearly that only the recombinant mECP–eGFP–6H could suppress the growth of GH3 cells. Neither eGFP–6H nor RNase A was able to interfere with the growth of GH3 cells. To monitor the internalization of the RNases, the GH3 cells were incubated with RITC-labelled RNase A or mECP–eGFP–6H at 37 °C for 3 h. No RITC-labelled RNase A was detected in the GH3 cells after 3 h of incubation, suggesting that RNase A could not be internalized (Figure 2A). However, GH3 cells exhibited increased levels of fluorescence labelling from mECP–eGFP–6H, and most likely located in the cytoplasm rather than the nuclei (Figure 2B). This result indicated that only the recombinant mECP–eGFP–6H could enter the GH3 cells. The cell lysates collected from reactions treated with mECP–eGFP–6H for 0, 1 or 2 h were analysed further by SDS/PAGE and Western blotting. Figure 2(C) revealed that the relative amount of mECP–eGFP–6H taken up by GH3 cells after 2 h of treatment was approx. 1.7-fold higher than that of 1 h treatment. The increasing amount of mECP–eGFP–6H accumulated in GH3 cells is consistent with inhibition of cell growth.
Figure 2. GH3 cells treated with RITC-labelled mECP–eGFP–6H and RITC-labelled RNase A.
GH3 cells were seeded on to six-well plates and grown on coverslips at 40000 cells/well. After 24 h, 0.5 μM mECP–eGFP–6H and RITC-labelled RNase A were added to the culture medium respectively, and the cells were fixed and detected by fluorescence microscopy at the time indicated and nuclei of the cells were stained with DAPI (4′,6-diamidino-2-phenylindole; blue). (A) GH3 cells treated with RITC-labelled RNase A for 3 h. No RITC-labelled RNase A was detected. (B) GH3 cells treated with mECP–eGFP–6H for 0 and 3 h. mECP–eGFP–6H was visualized (green). (C) Relative amounts of mECP–eGFP–6H in GH3 cells at different incubation time were detected by Western blot employing monoclonal anti-ECP109 antibody.
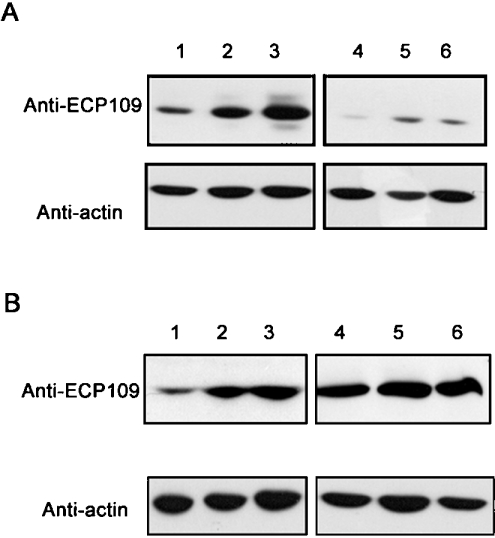
Secretagogues and lysosomotrophic agents affect mECP–eGFP–6H uptake into cells
To examine further whether the entry of mECP–eGFP–6H to GH3 cells is associated with the endocytosis and regulated secretion pathway, the cells were pre-treated with secretagogue (5 μM forskolin) or lysosomotrophic (15 mM NH4Cl) agents in the culture medium for 1 h before the addition of 0.5 μM mECP–eGFP–6H fusion protein. Figure 3(A) shows that the relative amount of mECP–eGFP–6H in GH3 cells was decreased upon treatment with NH4Cl, presumably due to blockage of the endosomal acidification and the secretion of vesicles. One the other hand, pre-treatment with forskolin resulted in a 2- to 3-fold increase of mECP–eGFP–6H accumulation, as shown in Figure 3(B). These results indicate that the acidic vesicles are involved in the internalization of mECP–eGFP–6H.
Figure 3. Effects on mECP–eGFP–6H uptake into GH3 cells by lysosomotrophic and secretagogue agents.
Lanes 1, 2, 3 in (A) and (B) were GH3 cells treated with medium containing 0.5 μM mECP–eGFP–6H for 1, 2 and 3 h respectively. (A) GH3 cells were pre-treated with 15 mM NH4Cl in culture medium for 1 h at 37 °C under 5% CO2, and then treated with 0.5 μM mECP–eGFP–6H fusion protein for 1 h (lane 4), 2 h (lane 5) and 3 h (lane 6). (B) GH3 cells were pre-treated with 5 μM forskolin in culture medium for 1 h at 37 °C under 5% CO2, and then treated with 0.5 μM mECP–eGFP–6H fusion protein for 1 h (lane 4), 2 h (lane 5) and 3 h (lane 6). The uptake of mECP–eGFP–6H was detected by Western blotting using anti-ECP109 monoclonal antibody.
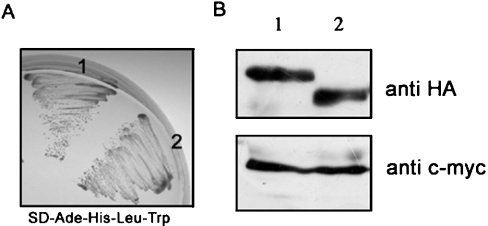
CPE interacts with mECP in yeast cells
To investigate which cellular proteins interact with mECP, a yeast two-hybrid screening system was constructed consisting of a human brain cDNA library and hECP as bait. This screen identified one clone containing the last 363 amino acids of human CPE (CPEc). The interaction was tested against ECP and human RNase A (used as a negative control) in a growth assay. In the absence of adenine and histidine, only the yeast expressing ECP could grow under such stringent conditions. The full-length human mCPE (mature CPE) was cloned further to examine its ability to interact with ECP. Figure 4(A) shows that mCPE binds to mECP with a similar affinity as that of CPEc, indicating that the N-terminal 114 amino acids of CPE do not affect its interaction with mECP. The expression of mECP–BD (where BD is DNA-binding domain of Gal4) and CPE–AD fusion proteins in yeast cells were detected further by Western blotting with anti-c-myc and anti-HA antibodies respectively. The molecular masses of mECP–BD, mCPE–AD and CPEc–AD fusion proteins expressed in yeast cells were estimated to be 34 kDa, 65 kDa and 53 kDa respectively (Figure 4B).
Figure 4. Yeast two-hybrid screen using mECP as bait and a human brain cDNA library identified CPEc.
mECP was cloned into bait vector pGBKT7. Yeast strain AH109 was co-transformed with mCPE–pGADT7 or CPEc–pGADT7. (A) Transformants were spread on to SD-Ade-His-Leu-Trp medium plates. Vigorous growth was observed for cells co-expressing the mECP bait plasmid and two CPE clones at the high-strength selection plate. 1, mCPE; 2, CPEc. (B) The expression of mECP–BD and CPE–AD fusion proteins in yeast cells was detected by Western blotting with anti-c-myc and anti-HA antibody respectively. mCPE–AD (upper panel, lane 1), CPEc–AD (upper panel, lane 2) and mECP–BD (lower panel, lanes 1 and 2) fusion proteins were expressed in yeast cells.
mECP is associated with CPE in vitro
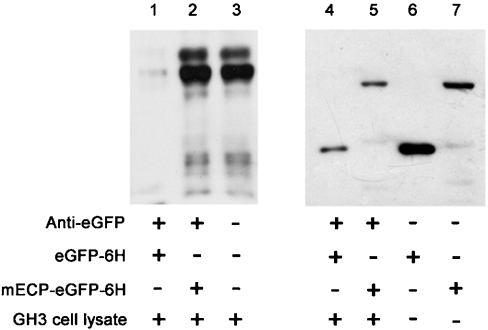
Direct interaction between CPE and ECP was confirmed further by immunoprecipitation using purified mECP–eGFP–6H fusion protein and GH3 cell lysates containing endogenous CPE. Equal amounts (1 μg) of either purified eGFP–6H or mECP–eGFP–6H fusion protein were added to the binding buffer containing 1 mg of GH3 cell lysates and incubated at 4 °C overnight. Subsequently, eGFP–6H and mECP–eGFP–6H were subjected to immunoprecipitation from the mixture by a polyclonal anti-eGFP antibody. The immunocomplexes were subjected to Western blot analysis with anti-CPE antibody, which specifically recognized the N-terminal region of CPE. No CPE protein was detected in the former experiment (Figure 5, lane 1), whereas a 50 kDa CPE protein was specifically detected in the latter experiment (Figure 5, lane 2). These results provide additional evidence of physical interaction between mECP and CPE in vitro.
Figure 5. ECP is associated with CPE in vitro.
The direct interaction between CPE and ECP was confirmed further by immunoprecipitation using purified mECP–eGFP–6H fusion protein and GH3 cell lysates with monoclonal anti-CPE antibody (lanes 1–3) or anti-His6 antibody (lanes 4–7). The cell lysates were collected, and 100 μg of lysate proteins were pre-mixed with 1 μg of eGFP–6H (lane 1) or 1 μg of mECP–eGFP–6H fusion protein (lane 2) in immunoprecipitation buffer. Lane 3 is GH3 cell lysate loading control. Immunoprecipitation was performed using a rabbit polyclonal anti-eGFP antibody and Protein A–Sepharose, subjecting samples to SDS/PAGE and transferring on to PVDF membrane. Lanes 4 and 5, immunoprecipitation efficiency control of mECP–eGFP–6H or eGFP–6H with anti-eGFP antibody respectively. Lanes 6 and 7, the input protein of mECP–eGFP–6H or eGFP–6H.
The region from 318 to 387 of mCPE is indispensable for association with mECP in yeast
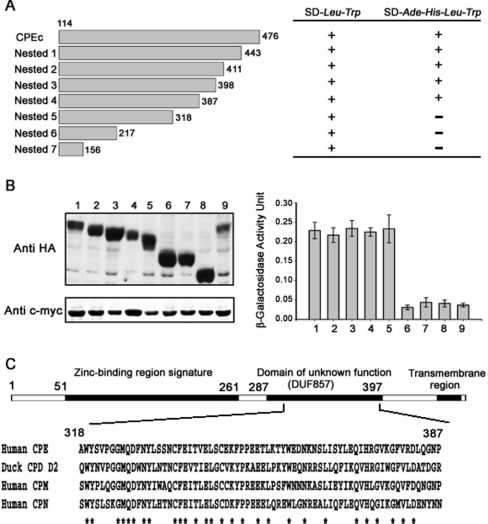
To determine which region in mCPE is responsible for the interaction with mECP, seven deletion mutants of CPEc were constructed by the nested deletion method (Figure 6A), and the binding of mECP to these truncated CPE mutants was examined in yeast cells. The reporter gene expression assays and cell growth results indicated that the C-terminally truncated forms of CPEc-nested-5 (amino acids 114–318), CPEc-nested-6 (amino acids 114–217) and CPEc-nested-7 (amino acids 114–156) could not interact with mECP in yeast cells. However, the reporter gene expression was activated by the CPEc-nested-4 (amino acids 114–387) containing a domain (amino acids 318–387) of unknown function. β-galactosidase activity assay was used to monitor Gal4 reporter gene expression, and it was found that no activity difference existed among the nested deletion mutants 1–4, whereas mutants 5–7 all lost enzymic activity (Figure 6B, right-hand panel). Primary sequence alignment of human CPE and its duck orthologue protein, CPD domain II, as well as two human orthologue proteins, CPM (carboxypeptidase M) and CPN (carboxypeptidase N), reveals that the mECP-associating region containing residues 318–387 of mCPE is highly conserved (Figure 6C).
Figure 6. Investigation of the interacting region on CPE.
(A) CPEc–pGADT7 was used as template to generate seven nested deletion mutants (left-hand panel). The numbers at both ends of each fragment represent the positions of the terminal amino acid residues in the CPE deletion mutants. Interactions were indicated by growth (+) or no growth (−) on SD-Ade-His-Leu-Trp medium plates (right-hand panel). (B) The expression of AD–CPEc and AD-nested CPE deletion mutant fusion proteins was detected by Western blotting using anti-HA antibody (upper left panel). Lanes 1 and 9, AD–CPEc; lanes 2–8: nested 1–7 respectively. The expression of BD–mECP (lanes 1–8) or BD–hRNase A (lane 9) fusion proteins was detected by Western blotting with anti-c-myc antibody (lower left panel). The interacting strength between mECP and CPEc nested deletion mutants was monitored with the Gal4 reporter gene expression by β-galactosidase activity assay (right-hand panel). (C) Amino acid sequence alignment of human CPE 318–387 with duck CPD domain II (CPD D2; Protein Data Bank code 1QMU), human CPM (EC 3.4.17.12) and human CPN (EC 3.4.17.3).
The uptake of mECP–eGFP–6H fusion protein is blocked by dominant-negative expression of the pre-pro-HA–CPES471A,E472A in GH3 cells
To examine further the effect of the uptake of mECP–eGFP–6H, GH3 cells were transiently transfected with pre-pro-HA–CPES471A,E472A–pcDNA3 or pcDNA3 vector, and incubated to overexpress the recombinant protein. After 48 h of incubation, both clones were incubated with 0.5 μM mECP–eGFP–6H for an additional 1–3 h. The expression of the recycling-defect protein pre-pro-HA–CPES471A,E472A was examined further by SDS/PAGE and Western blotting using anti-HA antibody (Figure 7A). The results demonstrated that the dominant-negative pre-pro-HA–CPES471A,E472A was successfully expressed in GH3 cells and significantly decreased the uptake of mECP–eGFP–6H fusion protein into the GH3 cells. On the other hand, not much difference in the internalization of mECP–eGFP–6H could be observed in the cells transfected with pcDNA3 only (Figure 7B).
Figure 7. Internalization of mECP–eGFP–6H fusion protein is prevented by expression of the pre-pro-HA–CPES471A,E472A cytoplasmic tail mutation protein in GH3 cells.
(A) pcDNA3 vector (lane 1) and pre-pro-HA–CPES471A,E472A–pcDNA3 (lane 2) were transfected into GH3 cells. The expression of recombinant protein was analysed by SDS/PAGE (upper panel) or Western blotting using monoclonal anti-CPE antibody (lower panel). (B) GH3 cells transfected with pcDNA3 or pre-pro-HA–CPES471A,E472A–pcDNA3 (CPEDN–pcDNA3) were incubated with 0.5 μM mECP–eGFP–6H for the indicated times. The mECP–eGFP–6H fusion proteins in GH3 cells were detected by Western blotting employing anti-ECP109 monoclonal antibody (top panel). The relative amount of mECP–eGFP–6H was quantitatively analysed with α-tubulin as an internal control (bottom panel). Black bars show results of GH3 cells transfected with pcDNA3 vector only. Grey bars show results of GH3 cells transfected with pre-pro-HA–CPES471A,E472A–pcDNA3.
DISCUSSION
The results of the present study show that mECP not only gets internalized into GH3 neuroendocrine cells, but also inhibits the growth of the cells. This phenomenon is consistent with previously reported growth-inhibitory effects of ECP on mammalian leukaemia, epidermoid carcinoma and breast carcinoma cell lines [12,13]. We found that the growth of GH3 cells was inhibited completely by mECP–eGFP–6H at a concentration of 2.5 μM, with an IC50 of 0.8 μM. Both values are lower than those for other cell lines used in previous studies under similar conditions, indicating that neuroendocrine cells are particularly sensitive to mECP.
The entry of mECP–eGFP–6H into the GH3 cells has been shown to be associated with the secretion cycle of the regulated secretory granules, indicating that mECP may interact with the component proteins within the secretory granules. Yeast two-hybrid library screening and immunoprecipitation experiments provide two lines of evidence that mECP interacts specifically with CPE. CPE was first purified from bovine brain and is also called enkephalin convertase or CPH. It is a CPB-like metalloenzyme associated with the biosynthesis of many peptide neurotransmitters and hormones. The role of CPE in the removal of C-terminal basic residues exposed by the endoproteases is known to be necessary for the processing of a large number of the protein precursors [17]. Previous studies suggested that CPE expressed in breast cancer and small-cell carcinoma of the lung cells was necessary to process neuropeptides for autocrine loops [18,19]. To investigate further the critical role that CPE plays in different cells, in terms of facilitating internalization of recombinant ECP, we have examined the expression of CPE in mammalian cell lines such as K562 and A431 by Western blot analysis. The results clearly reveal that CPE is expressed at different levels in these cell lines (results not shown), meaning that the expression of CPE in these cells may be associated with the ECP-uptake effect. Several metallocarboxypeptidases have now been demonstrated to be multifunctional. For example, duck gp180 (glycoprotein 180), which has CPD activity, binds to preS envelope protein of duck hepatitis B virus particles [20], and human ACE2 (angiotensin-converting enzyme 2) is a receptor for SARS (severe acute respiratory syndrome) coronavirus [21]. The present study indicates that membrane-bound CPE facilitates the entry of recombinant ECP to cells. To our knowledge, this is the first report showing that a non-surface-receptor protein is directly responsible for the internalization of recombinant mECP.
Further analysis of CPE deletion clones employing the yeast two-hybrid system has revealed that residues 318–387 in mCPE are indispensable for association with mECP. This region is highly conserved in all regulatory carboxypeptidase members and is located in a domain of previously unknown function. Our results provide the first evidence for the biological function of this particular domain. Both CPE and CPD are major peptide-processing enzymes that are present in the secretory pathway of neuroendocrine cells and can recycle from the cell surface. However, they are enriched in different parts of the pathway [22]. The subcellular localization of the former is in the regulated secretory vesicles, but the latter is in the trans-Golgi network [22,23]. The mECP-interacting region on CPE is highly conserved with the second domain of CPD, indicating that specific protein–protein interaction between mECP and CPE may also occur between mECP and CPD. Since CPD is a carboxypeptidase that is widely distributed in different tissues, and not only limited to neuroendocrine tissues, such an interaction may contribute to the broad role of CPD in processing proteins that transit the secretory pathway.
RNases constitute a large superfamily spanning many species. In addition to simple RNase activity, some members of the RNase A superfamily have cytotoxic activity towards tumour cell lines. For example, BS (bovine seminal)-RNase, onconase, human EDN with four extra residues SLHV (Ser-Leu-His-Val) at the N-terminus and hECP all showed cytotoxicity to different mammalian cell lines [24–26]. hECP is one member of the human RNase A family, and the growth-suppression effect of several types of mammalian cell lines has been well studied. The growth-inhibitory effect of ECP is cytostatic, but not cytotoxic, mainly due to its extraordinary stability among human RNases. Previous studies suggest that onconase, angiogenin and BS-RNase can bind to the specific receptor-like site in the plasma membrane, but no specific receptor protein has yet been identified [27]. In addition, the internalization mechanism of α-sarcin, onconase and toxic G88R RNase A has been demonstrated to be clathrin-independent and dynamin-independent [28,29]. Hence researchers have suggested that ECP may be located in the vesicles or endosomes, and that there are cell-surface receptors for ECP [12,13].
We have demonstrated for the first time that hECP interacts with CPE, a metallocarboxypeptidase widely distributed in the central nervous system. Previous studies have demonstrated that the C-terminal 25 amino acids of CPE are critical for the membrane binding and recycling of CPE [30,31]. The S471A and E472A point mutations on CPE block the interaction between CPE and ARF6 (ADP-ribosylation factor 6) and abolish the recycling of CPE from plasma membrane to the trans-Golgi network in a clathrin-independent manner [31]. Although the primary structure of CPE shows high identity with the second domain of CPD, our results show that the uptake of mECP–eGFP–6H is suppressed by dominant-negative expression of the pre-pro-HA–CPES471A,E472A recycling-defect mutant in GH3 cells, suggesting that uptake of mECP–eGFP–6H is mainly associated with the recycling of CPE in GH3 cells. Therefore it is expected that cells containing a sufficient amount of CPE can be potential target cells of mECP.
In the present study, we have demonstrated that specific interaction between mECP and CPE, the recycling lipid raft-associated pro-hormone-processing enzyme, plays a critical role in the entry of mECP to the neuroendocrine cells and causes the cell-growth-inhibitory effect of the cells. The entry of mECP is associated with the recycling of CPE, and is consistent with the hypothesis that extracellular RNase uptake is via a clathrin-independent endocytosis pathway. Furthermore, this special cell-entry mechanism through CPE may provide clues for the loss of Purkinje cells in the cerebellum found with the Gordon phenomenon. Although CPE is identified to be a key player in the molecular mechanism of internalization and cytostatic effect of mECP, it still requires further investigation to search for other component proteins responsible for different cytotoxic RNases.
Acknowledgments
We thank Dr E.N.G. Marsh, Dr Y.-D. Liao, Dr Y.-K. Lai, Dr W.-G. Chou and Dr L.-Y. Lin for critical comments. This work was supported by National Science Council, R. O. C. grants, NSC 91-2321-B-007-002 to Dr C.Y. Tang (who kindly provided equipment for the present study) and NSC 92-3112-B-007-001 to M. D.-T. C. Ministry of Education (MOE) program for promoting academic excellence of university under the grant number 89-B-FA04-1-4.
References
- 1.Rosenberg H. F. The eosinophil ribonucleases. Cell. Mol. Life Sci. 1998;54:795–803. doi: 10.1007/s000180050208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehrer R. I., Szklarek D., Barton A., Ganz T., Hamann K. J., Gleich G. J. Antibacterial properties of eosinophil major basic protein and eosinophil cationic protein. J. Immunol. 1989;142:4428–4434. [PubMed] [Google Scholar]
- 3.Hamann K. J., Gleich G. J., Checkel J. L., Loegering D. A., McCall J. W., Barker R. L. In vitro killing of microfilariae of Brugia pahangi and Brugia malayi by eosinophil granule proteins. J. Immunol. 1990;144:3166–3173. [PubMed] [Google Scholar]
- 4.Hamann K. J., Barker R. L., Loegering D. A., Gleich G. J. Comparative toxicity of purified human eosinophil granule proteins for newborn larvae of Trichinella spiralis. J. Parasitol. 1987;73:523–529. [PubMed] [Google Scholar]
- 5.Motojima S., Frigas E., Loegering D. A., Gleich G. J. Toxicity of eosinophil cationic proteins for guinea pig tracheal epithelium in vitro. Am. Rev. Respir. Dis. 1989;139:801–805. doi: 10.1164/ajrccm/139.3.801. [DOI] [PubMed] [Google Scholar]
- 6.Fredens K., Dybdahl H., Dahl R., Baandrup U. Extracellular deposit of the cationic proteins ECP and EPX in tissue infiltrations of eosinophils related to tissue damage. APMIS. 1988;96:711–719. doi: 10.1111/j.1699-0463.1988.tb00934.x. [DOI] [PubMed] [Google Scholar]
- 7.Fredens K., Dahl R., Venge P. The Gordon phenomenon induced by the eosinophil cationic protein and eosinophil protein X. J. Allergy Clin. Immunol. 1982;70:361–366. doi: 10.1016/0091-6749(82)90025-2. [DOI] [PubMed] [Google Scholar]
- 8.Durack D. T., Sumi S. M., Klebanoff S. J. Neurotoxicity of human eosinophils. Proc. Natl. Acad. Sci. U.S.A. 1979;76:1443–1447. doi: 10.1073/pnas.76.3.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young J. D., Peterson C. G., Venge P., Cohn Z. A. Mechanism of membrane damage mediated by human eosinophil cationic protein. Nature (London) 1986;321:613–616. doi: 10.1038/321613a0. [DOI] [PubMed] [Google Scholar]
- 10.Rosenberg H. F. Recombinant human eosinophil cationic protein: ribonuclease activity is not essential for cytotoxicity. J. Biol. Chem. 1995;270:7876–7881. doi: 10.1074/jbc.270.14.7876. [DOI] [PubMed] [Google Scholar]
- 11.Barker R. L., Loegering D. A., Ten R. M., Hamann K. J., Pease L. R., Gleich G. J. Eosinophil cationic protein cDNA: comparison with other toxic cationic proteins and ribonucleases. J. Immunol. 1989;143:952–955. [PubMed] [Google Scholar]
- 12.Maeda T., Kitazoe M., Tada H., de Llorens R., Salomon D. S., Ueda M., Yamada H., Seno M. Growth inhibition of mammalian cells by eosinophil cationic protein. Eur. J. Biochem. 2002;269:307–316. doi: 10.1046/j.0014-2956.2001.02653.x. [DOI] [PubMed] [Google Scholar]
- 13.Maeda T., Mahara K., Kitazoe M., Futami J., Takidani A., Kosaka M., Tada H., Seno M., Yamada H. RNase 3 (ECP) is an extraordinarily stable protein among human pancreatic-type RNases. J. Biochem. (Tokyo) 2002;132:737–742. doi: 10.1093/oxfordjournals.jbchem.a003281. [DOI] [PubMed] [Google Scholar]
- 14.Mallorqui-Fernandez G., Pous J., Peracaula R., Aymami J., Maeda T., Tada H., Yamada H., Seno M., de Llorens R., Gomis-Ruth F. X., Coll M. Three-dimensional crystal structure of human eosinophil cationic protein (RNase 3) at 1.75 Å resolution. J. Mol. Biol. 2000;300:1297–1307. doi: 10.1006/jmbi.2000.3939. [DOI] [PubMed] [Google Scholar]
- 15.Hiratsuka T. Selective fluorescent labeling of the 50-, 26-, and 20-kilodalton heavy chain segments of myosin ATPase. J. Biochem. (Tokyo) 1987;101:1457–1462. doi: 10.1093/oxfordjournals.jbchem.a122015. [DOI] [PubMed] [Google Scholar]
- 16.Futami J., Seno M., Kosaka M., Tada H., Seno S., Yamada H. Recombinant human pancreatic ribonuclease produced in E coli: importance of the amino-terminal sequence. Biochem. Biophys. Res. Commun. 1995;216:406–413. doi: 10.1006/bbrc.1995.2638. [DOI] [PubMed] [Google Scholar]
- 17.Manser E., Fernandez D., Loo L., Goh P. Y., Monfries C., Hall C., Lim L. Human carboxypeptidase E: isolation and characterization of the cDNA, sequence conservation, expression and processing in vitro. Biochem. J. 1990;267:517–525. doi: 10.1042/bj2670517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du J., Keegan B. P., North W. G. Key peptide processing enzymes are expressed by breast cancer cells. Cancer Lett. 2001;165:211–218. doi: 10.1016/s0304-3835(01)00409-8. [DOI] [PubMed] [Google Scholar]
- 19.North W. G., Du J. Key peptide processing enzymes are expressed by a variant form of small-cell carcinoma of the lung. Peptides. 1998;19:1743–1747. doi: 10.1016/s0196-9781(98)00130-2. [DOI] [PubMed] [Google Scholar]
- 20.Eng F. J., Varlamov O., Fricker L. D. Sequences within the cytoplasmic domain of gp180/carboxypeptidase D mediate localization to the trans-Golgi network. Mol. Biol. Cell. 1999;10:35–46. doi: 10.1091/mbc.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W., Moore M. J., Vasilieva N., Sui J., Wong S. K., Berne M. A., Somasundaran M., Sullivan J. L., Luzuriaga K., Greenough T. C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature (London) 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varlamov O., Wu F., Shields D., Fricker L. D. Biosynthesis and packaging of carboxypeptidase D into nascent secretory vesicles in pituitary cell lines. J. Biol. Chem. 1999;274:14040–14045. doi: 10.1074/jbc.274.20.14040. [DOI] [PubMed] [Google Scholar]
- 23.Varlamov O., Eng F. J., Novikova E. G., Fricker L. D. Localization of metallocarboxypeptidase D in AtT-20 cells. Potential role in prohormone processing. J. Biol. Chem. 1999;274:14759–14767. doi: 10.1074/jbc.274.21.14759. [DOI] [PubMed] [Google Scholar]
- 24.Rybak S. M., Newton D. L. Natural and engineered cytotoxic ribonucleases: therapeutic potential. Exp. Cell Res. 1999;253:325–335. doi: 10.1006/excr.1999.4718. [DOI] [PubMed] [Google Scholar]
- 25.Kim J. S., Soucek J., Matousek J., Raines R. T. Mechanism of ribonuclease cytotoxicity. J. Biol. Chem. 1995;270:31097–31102. doi: 10.1074/jbc.270.52.31097. [DOI] [PubMed] [Google Scholar]
- 26.Bracale A., Spalletti-Cernia D., Mastronicola M., Castaldi F., Mannucci R., Nitsch L., D'Alessio G. Essential stations in the intracellular pathway of cytotoxic bovine seminal ribonuclease. Biochem. J. 2002;362:553–560. doi: 10.1042/0264-6021:3620553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leland P. A., Raines R. T. Cancer chemotherapy – ribonucleases to the rescue. Chem. Biol. 2001;8:405–413. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olmo N., Turnay J., Gonzalez de Buitrago G., Lopez de Silanes I., Gavilanes J. G., Lizarbe M. A. Cytotoxic mechanism of the ribotoxin α-sarcin: induction of cell death via apoptosis. Eur. J. Biochem. 2001;268:2113–2123. doi: 10.1046/j.1432-1327.2001.02086.x. [DOI] [PubMed] [Google Scholar]
- 29.Haigis M. C., Kurten E. L., Abel R. L., Raines R. T. KFERQ sequence in ribonuclease A-mediated cytotoxicity. J. Biol. Chem. 2002;277:11576–11581. doi: 10.1074/jbc.M112227200. [DOI] [PubMed] [Google Scholar]
- 30.Varlamov O., Fricker L. D. The C-terminal region of carboxypeptidase E involved in membrane binding is distinct from the region involved with intracellular routing. J. Biol. Chem. 1996;271:6077–6083. doi: 10.1074/jbc.271.11.6077. [DOI] [PubMed] [Google Scholar]
- 31.Arnaoutova I., Jackson C. L., Al-Awar O. S., Donaldson J. G., Loh Y. P. Recycling of Raft-associated prohormone sorting receptor carboxypeptidase E requires interaction with ARF6. Mol. Biol. Cell. 2003;14:4448–4457. doi: 10.1091/mbc.E02-11-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]