Summary
Recent studies have highlighted the essential role of RNA splicing, a key mechanism of alternative RNA processing, in establishing connections between genetic variations and disease. Genetic loci influencing RNA splicing variations show considerable influence on complex traits, possibly surpassing those affecting total gene expression. Dysregulated RNA splicing has emerged as a major potential contributor to neurological and psychiatric disorders, likely due to the exceptionally high prevalence of alternatively spliced genes in the human brain. Nevertheless, establishing direct associations between genetically altered splicing and complex traits has remained an enduring challenge. We introduce Spliced-Transcriptome-Wide Associations (SpliTWAS) to integrate alternative splicing information with genome-wide association studies to pinpoint genes linked to traits through exon splicing events. We applied SpliTWAS to two schizophrenia (SCZ) RNA-sequencing datasets, BrainGVEX and CommonMind, revealing 137 and 88 trait-associated exons (in 84 and 67 genes), respectively. Enriched biological functions in the associated gene sets converged on neuronal function and development, immune cell activation, and cellular transport, which are highly relevant to SCZ. SpliTWAS variants impacted RNA-binding protein binding sites, revealing potential disruption of RNA-protein interactions affecting splicing. We extended the probabilistic fine-mapping method FOCUS to the exon level, identifying 36 genes and 48 exons as putatively causal for SCZ. We highlight VPS45 and APOPT1, where splicing of specific exons was associated with disease risk, eluding detection by conventional gene expression analysis. Collectively, this study supports the substantial role of alternative splicing in shaping the genetic basis of SCZ, providing a valuable approach for future investigations in this area.
Keywords: alternative RNA splicing, TWAS, neurological disorders, genetic variations, RNA binding proteins, fine mapping
RNA splicing plays a critical role in linking genetic variations to diseases such as schizophrenia. We introduce SpliTWAS, integrating alternative splicing information with GWAS. Applied to two schizophrenia cohorts, SpliTWAS identifies disease relevant genes and demonstrates the substantial role of alternative splicing in shaping the genetic basis of schizophrenia.
Introduction
Understanding the molecular mechanisms that underlie genetic associations identified through genome-wide association studies (GWASs) remains a major challenge despite substantial efforts invested in this area. Genetic variants linked to complex traits often reside within large blocks of linkage disequilibrium (LD) in non-coding regions, making it difficult to determine causal variants and their functional implications. Approaches that integrate GWASs with transcriptomic studies, such as transcriptome-wide association studies (TWASs), enabled significant advances in prioritizing candidate causal genes and tissues underlying GWAS loci.1,2,3,4,5,6,7 However, these efforts focused on total gene expression not investigating other molecular mechanisms, such as alternative splicing, that can regulate RNA processing and alter total gene expression levels, all of which may be influenced by genetic variants and contribute to disease risks.8,9,10 Genetic regulation of alternative splicing and other RNA processing steps can broaden our understanding of GWAS risk loci.
Alternative splicing refers to the alternative inclusion of exons, sometimes introns, in the mature mRNA.11 Over 90% of human genes undergo alternative splicing, resulting in multiple transcript isoforms from the same gene locus.12 As a result, multiple protein-coding and non-coding transcripts can be generated from the same gene, expanding the complexity and diversity of both the transcriptome and proteome. Alternative splicing is highly tissue specific; in particular, the brain expresses the most alternatively spliced genes of all human tissues, likely contributing to the complexity of this organ.13 RNA splicing is tightly regulated by a combination of trans-acting factors and cis-regulatory elements. Approximately half of the required information14 for exon recognition during splicing is determined by the consensus 5′ and 3′ splice sites. Exons and introns contain auxiliary splicing elements of 6–8 nucleotides that either enhance or repress the use of associated splice sites,14 with existing studies showing an enrichment of these auxiliary elements near exon-intron boundaries. However, our current understanding of cis-acting splicing elements remains incomplete. Given its regulation by cis-elements, splicing can be significantly altered by genetic variations. Indeed, it has been estimated that around 35% of disease-causing point mutations disrupt splicing, over a third of which reside in consensus splice sites and known exonic or intronic auxiliary splicing elements.15,16,17 Splicing quantitative trait loci studies have discovered that genetic variants that alter splicing ratios are associated with phenotypic traits to an equal or even greater degree than those that affect gene expression.10 Variants affecting splicing are notably different from those affecting gene expression, representing a unique set of genetic variants that have not been thoroughly studied yet in relationship with disease risk.
Although the most-often used approach to infer relationships between genetic variants and splicing is quantitative trait loci (QTL)-based methods,10,18 allele-specific RNA processing can also be used to capture genetically modulated splicing events.9,19,20 In addition, machine-learning-based methods have been developed to predict the impact of genetic variants on splicing.8,21 These methods greatly facilitated a better understanding of RNA regulation, specifically the genetic regulation of RNA splicing. However, the above approaches focus on linking genotypes to the molecular phenotypes of splicing and cannot directly infer the disease relevance of such relationships. To fill in this gap, we propose SpliTWAS, a framework that associates the genetically driven complexity of alternative RNA processing to disease. We applied SpliTWAS to two different RNA-sequencing (RNA-seq) datasets together with large-scale GWASs to characterize the role of genetically driven alternative splicing in schizophrenia (SCZ). We discovered splicing-specific SCZ associations missed by traditional TWAS approaches at both gene and exon levels. SpliTWAS identified more SCZ-associated genes in comparison with conventional TWAS, with a small overlap, emphasizing the distinctive role of splicing in genetic associations. The associations provided further insight into how the dysregulation of RNA splicing caused by genetic variants may contribute to the pathogenesis of SCZ.
Material and methods
Datasets
In this study, we utilized two datasets from the PsychENCODE consortium, BrainGVEX and CommonMind (CMC) (https://www.synapse.org, BrainGVEX [syn3270015], CMC [syn22344687]). Postmortem human brain samples (dorsolateral prefrontal cortex [DLPFC]) were collected. RNA-seq and genotype data were generated independently by each participating site and subsequently underwent unified processing by a central analysis core (https://www.synapse.org/). For both datasets, individuals were selected based on their ancestry and availability of both RNA-seq and genotype data. Only individuals from European ancestry were included to avoid the confounding effects of genetic architectures. A total of 769 samples were finally retained (344 from BrainGVEX, 425 from CMC). CMC had an almost even split between cases and controls with 214 controls and 211 SCZ individuals, with 173 females and 252 males and an overall mean age of 68 years. BrainGVEX had a higher proportion of cases than controls with 91 cases and 253 controls, a distribution in gender of 111 females and 213 males with an overall mean age of 65 years. Genotype data from whole genome sequencing was further processed with PLINK v1.9, removing variants with Hardy-Weinberg equilibrium (HWE) p < 10−6, minor allele frequency (MAF)< 0.01, or missingness rate >0.05, and removing samples with missingness rate >0.1 across typed variants or missingness rate >0.5 on any individual chromosome. For GWAS summary statistics, we utilized the Psychiatric Genomics Consortium (PGC) SCZ GWAS, number of cases = 34,241 and number of controls = 45,604. LD information was obtained from the 1000 Genomes European reference.
PSI calculation and processing
Alternative splicing was quantified via the percent-spliced-in (PSI) value of each exon. PSI indicates the degree of exon inclusion in the transcript population of a gene, summarizing alternative splicing events across individual exons without prior knowledge of the underlying composition of the transcripts. Our approach uses the PSI calculation as defined in Schafer et al.22 Generally, PSI is calculated based on two read categories, inclusion reads (IRs) and exclusion reads (ERs). IR is a read that overlaps the exon of interest completely or by a minimum number of nucleotides (i.e., overhang, 8 by default). An ER is a read that fully skips the exon. Minimum cutoffs for the reads are IR + ER ≥ 10 or IR ≥ 2.
The IR and ER counts were then normalized based on the length of the exon, overhang and read, as follows:
PSI was then calculated as
Following the calculation of PSI, we further eliminated any exons that did not have a PSI value at least 40% of samples due to poor read coverage and that had little to no variation across individuals (SD ≤ 0.005 of PSI value). Next, we imputed missing PSI values for each exon as the mean of the computed PSIs across all samples.
S-value transformation
PSI values typically follow a bimodal distribution centered around 0 and 1, which violates the assumption of a Gaussian distribution made by the regression schemes (below). To address this problem, we conducted a logit transformation on the PSI values, naming the transformed metric S values. The transformation is as follows:
where α = 0.001, in order to accommodate cases where PSI was 1. Additionally, all PSI values of 0 were replaced with alpha to avoid improper mathematical operations.
Predictive models of alternative splicing
We first surveyed the cis-heritability of alternative splicing for individual exons. SNP heritability estimation assesses the maximum theoretically possible accuracy of linear prediction based on a corresponding set of SNPs. In order to perform heritability estimation, we first harmonized the SNPs between the GWAS summary statistics, LD reference panel, and genotype data. Second, we regressed the following covariates (age, pH, RNA integrity number [RIN], sex, post-mortem interval [PMI], and 15 genotype principal components [PCs]) against the S values. Heritability estimation was then performed using the restricted maximum likelihood (REML) algorithm implemented in GCTA. In line with previous studies,1,2,23 heritability estimates were allowed to converge outside the expected 0–1 variance bounds to achieve unbiased mean estimates. The candidate genomic window for heritability estimation was defined as the exon plus a cis-window surrounding it (1,10, or 100 kb). Only exons with positive heritability estimates and a nominal p value < 0.05 were deemed heritable. Following heritability estimation, SNPs within the heritability window (cis-SNPs) were utilized to build predictive models of S values by using the following prediction schemes: top1 or top cis-expression QTL (eQTL), best linear unbiased prediction (BLUP), bayesian sparse linear mixed model (BSLMM), least absolute shrinkage and selection operator (LASSO), and elastic net models. Top1 or top cis-eQTL utilizes only the single most significantly associated SNP in the training set as the predictor. BLUP estimates the causal effect sizes of all SNPs in the locus jointly using a single variance component. BSLMM estimates the underlying effect size distribution and then fits all SNPs in the window jointly. Each model’s prediction accuracy was evaluated by a 5-fold cross validation in a random sampling of the top 1,000 most heritable exons. R2 between the predicted and true S values was used to determine accuracy, models with an R2 ≤ 0.01 were filtered out and the remaining were used to perform summary-based imputation.
Alternative splicing imputation and association testing with summary statistics
Summary-based imputation allows us to perform splicing-trait associations with a significant gain in statistical power, compared to associations at individual level, due to the large sample size used to derive summary statistics. Imputation on GWAS summary statistics was performed using the ImpG-Summary algorithm, as used in FUSION.1,24 The imputation was carried out as follows, let Z be a vector of standardized effects sizes (Z scores) of SNP-trait associations obtained from the GWAS summary statistics at a given locus. Then the Z score of the imputed exon usage, for each exon, is a linear combination of the elements of Z and W, where W is the vector of weights for each SNP obtained in the heritability estimation. It follows that the imputed Z score between exon usage and trait (WZ) has variance ; therefore, we used as the imputation Z score of the cis-genetic effect on the trait. Σs,s was obtained from the European 1000 Genomes LD reference panel. Exons were considered significantly associated with the trait at a Bonferroni-corrected p value < 0.05. Additionally, we deemed genes with at least one significantly associated exon as having a significant splicing-trait association.
Following the association tests, all significant results were subject to a joint analysis to determine which exons provide independent signals and which ones were being driven by shared genetic predictors. Additionally, we performed conditional analysis where GWAS associations were conditioned on significant SpliTWAS exon associations to determine how much of the GWAS association signal remained after the splicing association was removed.
Fine mapping features
To fine map the genetic features, we implemented a modified approach of the probabilistic fine-mapping procedure, FOCUS. Traditionally, gene-level association statistics from TWAS are used to create TWAS-significant regions, where multiple associations are present, which span large genomic regions due to the large heritability windows (e.g., ∼1 Mb). For SpliTWAS associations, one of the main assumptions is that the relevant variants and features are in much closer proximity than in the case of gene expression. Therefore, we built our risk regions for fine mapping using a ±10 kb heritability window to the associated genes. If the window overlapped with another significantly associated gene, the regions were combined and extended, an approach akin to building LD blocks. Once the risk regions were built, FOCUS was applied using the exon-splicing association statistics for each gene within the defined risk region. The association statistics were used regardless of significance level. Using posterior inclusion probabilities (PIPs) from the FOCUS results, credible gene sets were computed by generalizing the concept of credible SNP sets from SNP fine mapping.25,26 We computed the 90% credible set for each of the risk regions of association, with the fine-mapped feature being exons. Similarly to our approach with gene level associations, a gene was deemed as putative causal if it possessed at least one fine-mapped exon.
Enrichment of SNPs within RBP binding sites and binding levels
To determine the enrichment of splicing-trait-associated SNPs in RNA-binding protein (RBP) binding sites, we overlapped the SNPs with RBP binding peaks of 150 RBPs generated by the ENCODE consortium using enhanced crosslinking and immunoprecipitation (eCLIP) data from HepG2 and K562 cell lines.27 To ameliorate ascertainment bias, we sampled at random the same number of control SNPs as cis-SNPs with their MAF and distance to transcription start site (TSS) matched with the SNPs in the query. The sampling strategy was repeated 1,000 times to reach an empirical distribution of the number of control SNPs overlapping with each RBP. Fold enrichment for each RBP was computed as the ratio between the proportion of overlapping associated SNPs over the proportion of overlapping control SNPs. Wilcoxon’s signed-rank test was then used to assess whether the splicing-associated SNPs proportion was significantly greater than the control SNPs proportion.
Furthermore, we investigated the effect of each cis-SNP on RBP-RNA interaction. To this avail, we used the DeepRiPe method, a multitask and multimodal deep neural network approach trained via cross-linking immunoprecipitation (CLIP) data. SNPs for this analysis were selected if they fell within ±500 bases from either of the splice sites of the associated exons. We then calculated the difference between the DeepRipe-predicted binding scores of the reference and variant allele. This difference is denoted by:
As controls, similar to the analysis for RBP binding enrichment, we matched the investigated SNPs with control SNPs and selected the same number of control SNPs as cis-SNPs. This procedure was repeated 1,000 times. Wilcoxon’s signed-rank test was then used to determine whether the binding disruption by splicing-associated SNPs was significantly higher than that of control SNPs.
GO analysis
To investigate the relevance of the associated genes in biological processes, gene ontology (GO) analysis was performed using associated gene sets for BrainGVEX and CMC independently. To account for tissue-specific expression for each query gene, a random control gene was chosen to match gene expression level and gene length (±10% relative to that of query gene). For each query and control gene, their respective gene ontology term was downloaded from Ensembl via biomaRt. A Gaussian distribution was fit to each ontology term by sampling 10,000 sets of control genes. The enrichment p value of the gene ontology term from the query genes was calculated using this distribution. Terms were deemed significant based on a false discovery rate (FDR) <0.05 cutoff. We then applied rrvgo to the significant GO terms to group them by semantic similarity (using default parameters). rrvgo assigns parent terms to each group based on the GO term that has the most significant enrichment p value. Groups with five or more GO terms were then selected and visualized accordingly.
Results
Overview of SpliTWAS
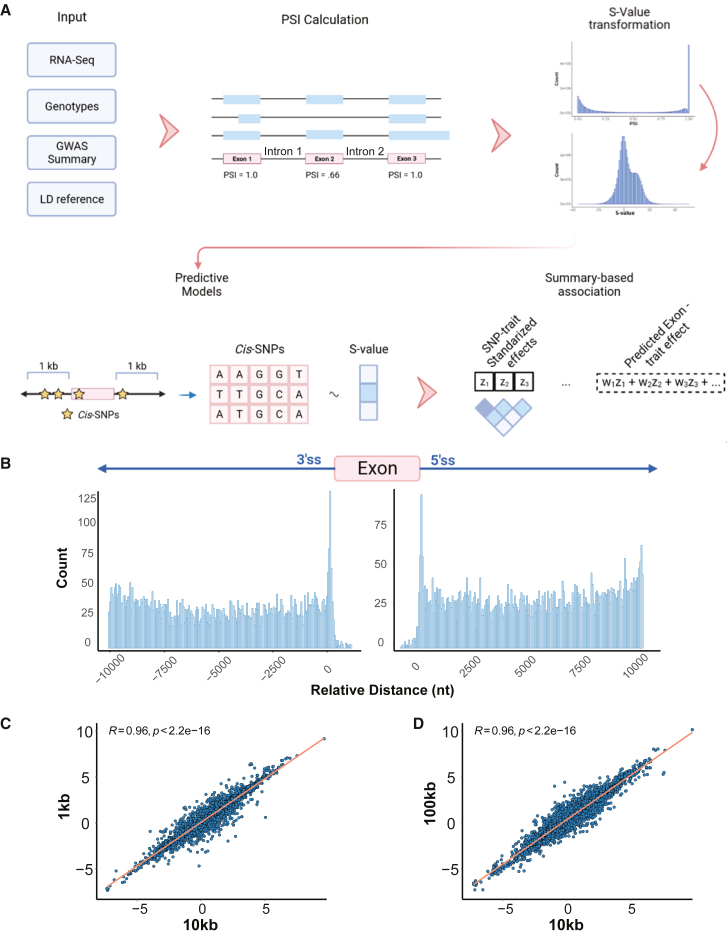
SpliTWAS examines alternative splicing patterns to identify splicing-trait associations, which are then fine mapped at both the gene and exon levels. The degree of alternative splicing of an exon is quantified through PSI,22 which indicates the level of exon inclusion in the transcript population of a gene, allowing an exon-centric view of the transcriptome. The SpliTWAS workflow begins with the calculation of PSI via RNA-seq reads including or excluding an exon (Figure 1A). PSI values are then logit-transformed to yield an S value, similar to the transformation performed for methylation values,28 which better aligns with the Gaussian distribution assumptions made by downstream steps. The S value is then corrected for covariates, such as age, sex, PMI, and RIN. In order to infer the genetic components of splicing, heritability estimates are calculated using the REML algorithm implemented in GCTA for each exon29 (see material and methods). Next, SpliTWAS builds predictive models of alternative splicing (i.e., S value) for each exon using a local window centered around each exon. Multiple types of models are constructed (see material and methods) and model performance was assessed via 5-fold cross-validation, requiring an adjusted R2 > 0.1 between the observed and predicted exon usage, the best model is then chosen for imputation.
Figure 1.
SpliTWAS overview and heritability window evaluation
(A) Schematic of the spliTWAS framework, exon-level alternative splicing quantification, training of predictive models based on cis-genetic locus, and the indirect estimation of association between predicted alternative splicing and trait at the exon level.
(B) Enrichment of trait-associated SNPs relative to the 3′ and 5′ splice sites (10 kb heritability window).
(C) Correlation of Z scores of nominal associations (p < 0.05) using the 10 or 1 kb window.
(D) Similar to (C) but for the 10 vs. 100 kb window.
Exon imputation models are then used to impute alternative splicing into GWAS data to estimate the association between splicing and the trait of interest (Figure 1A). If individual-level genotypes are available, splicing can be directly modeled based on the genotypes, and imputed into the GWAS associations. If only GWAS summary statistics are available, imputation is performed using a reference panel of alternative splicing predictive models. The estimated association between alternative splicing and trait is represented as a linear combination of SNP-trait standardized effects sizes and splicing-related weights per SNP, while accounting for LD among SNPs1 (see material and methods). The linear combination of weights and Z scores of individual SNPs results in a singular Z score for the splicing-trait association. Multiple hypothesis testing is accounted for and corrected through family-wise error control. Significant associations are then subject to a probabilistic fine-mapping strategy where the correlation among associations is used to assign a probability for each exon in the risk region to explain the observed association signal.30 Thus, by leveraging the unique biological signal of alternative splicing, SpliTWAS is able to identify disease-relevant trait-associations at the exon level that would have been obfuscated by traditional TWAS approaches.
Application of SpliTWAS to RNA-seq of brain samples in SCZ cohorts
In this study, the SpliTWAS framework was applied to two different RNA-seq datasets, BrainGVEX and CMC. For each dataset, RNA-seq from frozen DLPFC samples was used for PSI quantification, and genotype information was used to prune out individuals of non-European ancestry. A total of 344 and 425 samples were retained for the BrainGVEX and CMC cohorts, respectively. Each dataset was used to build predictive models of alternative splicing. The models were then integrated with summary statistics from the PGC SCZ GWAS,31 derived from 79,845 individuals (34,241 cases and 45,604 controls).
Predictive models were built through cis-SNPs, which were defined as SNPs that reside close to the predicted targets, in our case, exons. Traditionally, a 500 kb or 1 Mb window was used to define cis-SNPs for TWAS. However, known cis-regulatory elements of splicing are typically located relatively close to the 3′ and 5′ splice sites (ss) of their target exons.32 To determine a more suitable window size, we first applied SpliTWAS to the BrainGVEX dataset using three different window sizes flanking exons, 1, 10, and 100 kb, respectively (Table S1). We then investigated the distance of lead SNPs with respect to the 3′ and 5′ ss of exons for each window size. As shown in Figures 1B and S1, lead SNPs were enriched in regions close to the 3′ and 5′ ss. This splice site enrichment relative to the proximal intronic regions was most prominent for the 10 kb window size (Figure 1B), although results from the three alternative window sizes followed the same trend (Figure S1). Importantly, since splice sites are the most crucial cis-elements for splicing, the SNP enrichment near the splice sites supports the biological relevance of the genetic signal captured in our models.
Next, we asked if the alternative window sizes were equivalent in capturing predictive and association signals. To this end, we compared the number of predictive models as well as the correlation of Z scores of associations across the different windows. This analysis showed that the overall Z scores were correlated significantly across all three windows for commonly identified splicing events (Figure 1C), suggesting that most of the signal is captured even with a small window size (1 kb) for these events. Nonetheless, the 1-kb window size yielded a smaller number of overall predictive models (Figure S2). Interestingly, using 10-kb windows did not significantly reduce the number of heritable exons, mean heritability, or predictive models compared to 100-kb windows (Figures S2 and S3). Thus, we chose to use a window size of 10 kb hereafter, as it captures the most amount of signal without introducing considerable noise from distal SNPs. Furthermore, when comparing heritability estimates from GCTA and LDAK, we found a significant degree of correlation. Similarly, we observed a significant correlation between the heritability estimates and the R2 of the predictive models across both methods (Figure S4).
SpliTWAS identified exons and genes with relevance to SCZ
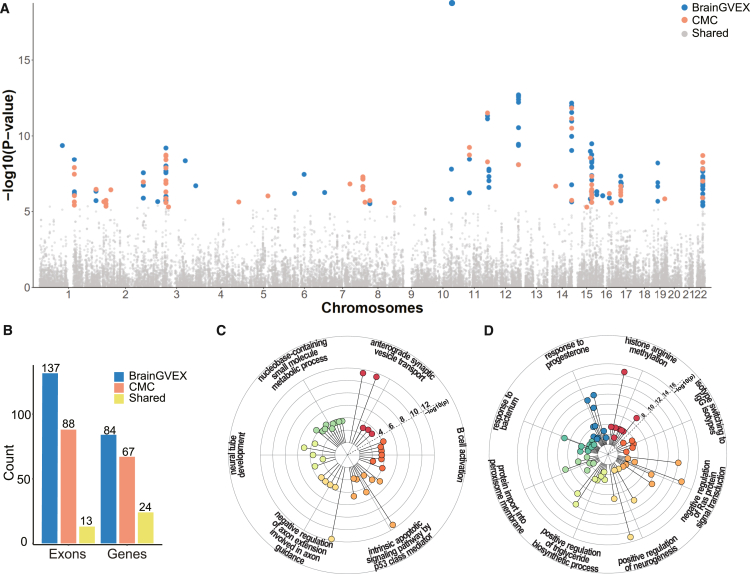
Overall, we tested ∼270,000 exons, of which 19,361 and 17,149 were heritable, and 12,860 and 6,682 passed the 5-fold cross-validation cutoff for the BrainGVEX and CMC datasets, respectively (Table S1). All exons that passed the cross-validation cutoff underwent association testing with SCZ (Figure 2A). 137 and 88 trait-associated exons (in 84 and 67 genes) reached genome-wide significance level (Bonferroni-corrected p value <0.05) in the BrainGVEX and the CMC datasets (Figure 2B; Table S2). Among the trait-associated exons or genes, 13 exons and 24 genes were shared between the two cohorts (p = 0.036 and p = 0.006, hypergeometric test) (Figure 2B). Thirty-seven SpliTWAS-associated genes overlap a catalog of 321 high-confidence SCZ risk genes, which were discovered through gene regulatory networks and a deep-learning approach.33 Interestingly, perhaps unsurprisingly, while a smaller proportion of trait-associated exons were shared between the two datasets, a higher proportion of shared genes (containing different trait-associated exons) was observed, indicating that trait-associated splicing may affect multiple exons of a gene. While the CMC cohort had a larger sample size, a lower number of genome-wide associations was identified both at the exon and gene levels. Comparing both the R2 performance and the overall Z score distribution between both cohorts, we observed that the BrainGVEX models outperformed the CMC models in both metrics (Figure S5). This difference is likely due to a higher degree of heterogeneity among samples in the CMC cohort.
Figure 2.
SpliTWAS identifies many exons and genes enriched in relevant biological pathways to SCZ
(A) Genome-wide splicing association results using both the BrainGVEX and CMC datasets imputed into the PGC SCZ GWAS summary statistics. Significant results are colored according to datasets and each dot represents an exon.
(B) Number of significant associations per dataset and shared associations by the two datasets at the exon and gene level. Genes with at least one significantly associated exon are included.
(C) Gene ontology (GO) analysis of the significantly associated genes obtained via the BrainGVEX data. GO terms were pruned and clustered using rrvgo; statistical significance for enrichment of each term is displayed as the radius of the lollipop.
(D) Similar to (C) but for the CMC data.
We next examined the biological functions enriched in the SpliTWAS genes. Both sets showed enrichment in GO terms related to neuronal function and development, immune cell activation and regulation, cellular transport, and metabolic processes (Figures 2C and 2D). Specifically, MDA-5 signaling, IL-10 regulation, T cell, and B cell activation are known to contribute to inflammation and immune dysregulation, which are important aspects of SCZ.34,35,36,37,38 Ras proteins have been shown to be crucial for regulating neuronal morphology, axon guidance, and dendritic spine formation.39,40,41,42 Additionally, proper regulation of apoptosis is crucial during neurodevelopment for sculpting neuronal circuits and eliminating excess or aberrant neurons.43,44 The combined disruption of apoptotic regulation and Ras proteins impact neuronal connectivity, synaptic plasticity, and overall brain development. In addition, cellular transport is also critical to neurodevelopment, abnormalities of which during critical periods of neurodevelopment may lead to altered synaptic connections, cellular metabolism, organelle function, and neuronal connectivity.45,46,47,48,49,50 Together, these results provide evidence that genetically modulated RNA splicing events identified by SpliTWAS may have close relevance to SCZ.
SpliTWAS uncovered more biologically plausible associations than TWAS
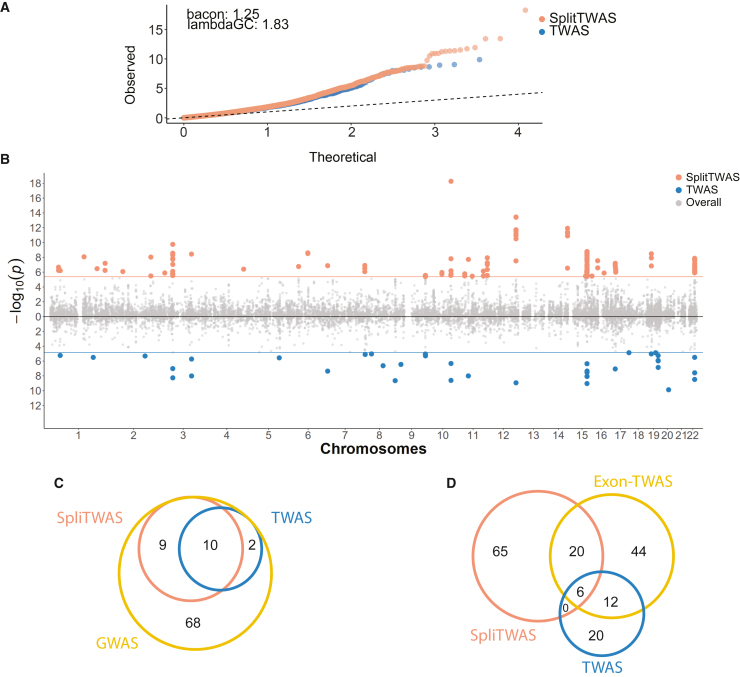
Next, we examined whether splicing-driven trait associations provide unique signals relative to the traditional TWAS analysis. For comparison between the two approaches, we focused on the BrainGVEX cohort. To enable a fair comparison, we used a window size of 100 kb for both approaches. We first examined the possible existence of test statistic inflation.23,51 The quantile-quantile plots of trait-association p values did not show significant differences between SpliTWAS and TWAS (Figure 3A). We also observed similar inflation values, lambdaGC and bacon,51 of the p values for the two methods (Figure 3A), indicating that the test statistic of SpliTWAS is not inflated compared to TWAS.
Figure 3.
SpliTWAS uncovers more trait associations than TWAS without higher inflation
(A) Q-Q plot of test statistics for all tested exons in SpliTWAS and genes in TWAS.
(B) Miami plot of associations for SpliTWAS and TWAS, respectively; cutoff represents Bonferroni-corrected p < 0.05.
(C) Overlap between SpliTWAS significant associations and GWAS risk loci.
(D) Overlap between SpliTWAS, exon-TWAS, and TWAS.
Although their p value distributions were not significantly different, SpliTWAS captured a higher number of genes with significant associations than TWAS (91 vs. 38) and a slightly higher number of associations due to the larger heritability window (100 kb). (Figure 3B). Note that the significance cutoff was the same for the two methods (Bonferroni-corrected p < 0.05). Additionally, we compared the standardized effect sizes (Z scores) for shared genes in SpliTWAS and TWAS associations at a nominal p value <0.05. A significant positive correlation was observed for the set of genes with concordant direction (Figure S6A) indicating that splicing changes may drive gene expression associations detected by TWAS, at least for some genes. Similarly, we see that some genes have opposite directions in their effect sizes, suggesting a different effect depending on the molecular phenotype surveyed. Thus, investigating genetic associations via SpliTWAS provides a refined view of the genetic basis of SCZ.
We next evaluated the overlap of trait associations discovered by SpliTWAS and TWAS. Among the 89 GWAS risk loci for SCZ, SpliTWAS identified 41 gene associations for 19 (∼21%) risk loci, while TWAS identified associations for 19 genes and 12 (∼13.4%) loci (Figure 3C). Thus, SpliTWAS explained more GWAS risk loci than TWAS. Interestingly, 10 risk loci were shared between SpliTWAS and TWAS-based associations, which is a significant overlap (p = 0.0076, hypergeometric test). For these loci, it is possible that both gene expression and splicing contribute independently to disease risk. Alternatively, due to the relatedness of the two molecular traits, some of these loci may be primarily driven by one molecular process, for example, splicing, which affects the observed values of gene expression. Indeed, the latter may be true for four risk loci where the same gene was detected with significant association by both TWAS and SpliTWAS. Additionally, although only a small number of GO terms were enriched among TWAS-uncovered genes (Figure S7), similar processes (related to metabolic and cellular transport) were found for SpliTWAS genes. Together, these results support the significance of examining contributions to GWAS risk loci from multiple molecular perspectives.
For completeness we also compared SpliTWAS to an exon-expression TWAS (exonTWAS); exonTWAS is similar to a conventional TWAS but utilizes exon expression as the molecular phenotype (see material and methods). In addition, we compared SpliTWAS to LeafCutter paired with FUSION as an alternative method based on splicing. As expected, the overlap between associated genes identified by SpliTWAS and TWAS was small, whereas a larger overlap was observed between genes from SpliTWAS and exonTWAS and between exonTWAS and conventional TWAS (Figure 3D). These observations indicate that exon expression is substantially confounded by gene expression levels. In comparison, PSI values represent a normalized metric that is less correlated with gene expression and therefore more appropriate to quantify and study splicing (Figure S8). Similarly, there was a large overlap between SpliTWAS results and LeafCutter compared to TWAS, with both splicing-based approaches having a significantly higher number of associated genes (Figure S9). Interestingly, the overlap of associated genes identified by both splicing methods is significant but lower than expected, with more genes being uniquely associated to each method than shared. Overall, the above results showed that SpliTWAS captured independent signals relative to general gene/exon expression, revealing unique and significant contributions of alternative splicing to SCZ risk.
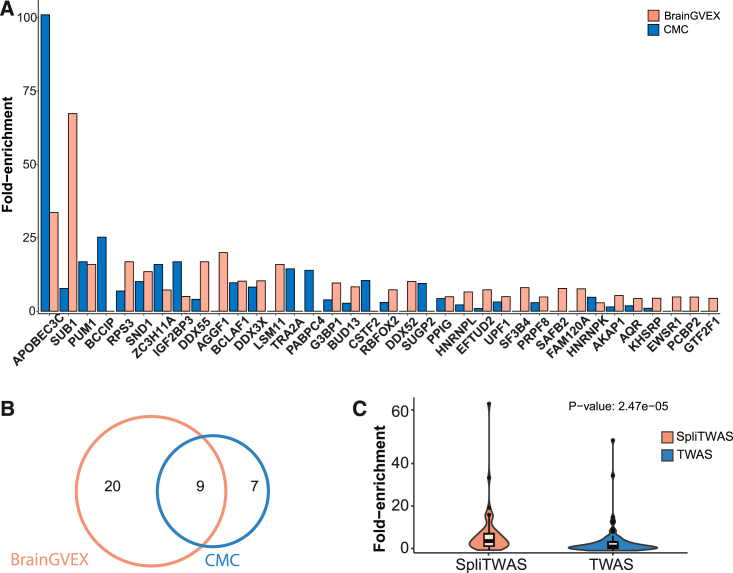
SpliTWAS variants are likely to disrupt protein-RNA interactions
It is well established that pre-mRNA splicing is regulated by a myriad of cis-elements interacting with trans-factors, primarily RBPs. Thus, to investigate the potential role of RBPs in disrupting trait-associated exons, we extracted variants from significantly associated exons. The variants were then matched to random controls (material and methods). Using eCLIP-seq data of 150 RBPs generated by the ENCODE consortium,49 we overlapped both the query variants and the matched controls to the binding sites of the 150 RBPs. We observed significant fold enrichment of trait-associated variants overlapping the binding sites of 36 RBPs when compared to the matched controls (material and methods; Figures 4A and 4B). The top RBPs, such as SUB1 and PUM1, are known to affect splicing.52,53,54,55,56 Curiously, the most highly enriched protein, APOBEC3C, is best known for DNA editing with potential roles in RNA editing.57,58 It is likely that the C to U RNA editing by APOBEC3C may lead to changes in RNA sequence and subsequent alternative splicing patterns. Alternatively, APOBEC3C may be a splicing factor resulting from its RNA binding capacity.
Figure 4.
SpliTWAS-associated variants potentially disrupt protein-RNA interactions
(A) Enrichment of SpliTWAS variants in RBP binding sites relative to random control SNPs.
(B) Overlap of significantly enriched RBPs between the two cohorts.
(C) Fold enrichment of SpliTWAS and TWAS SNPs across RBP binding peaks. Wilcoxon’s signed-rank test was used to determine statistical significance.
As a control, we compared the fold enrichment of all RBPs overlapping SpliTWAS-identified SNPs and those resulting from conventional TWAS analysis. SNPs obtained from SpliTWAS showed a significantly higher mean fold enrichment than those from TWAS (Figure 4C). This observation confirms the expectation that SpliTWAS-identified variants are enriched with those involved in splicing regulation, often executed by RBPs, whereas TWAS-identified variants likely reflect regulation at the level of transcription (and/or RNA stability).
As an alternative approach, we examined the potential impact of SpliTWAS variants on RBP binding using the DeepRiPe method, a multitask and multimodal deep neural network approach trained via CLIP data.59 To quantify whether a SNP disrupts the binding of a specific RBP, we calculated the difference between the DeepRiPe-predicted binding scores of the reference and variant allele, which is denoted as delta binding (material and methods). Compared to random controls, SpliTWAS variants from the BrainGVEX and CMC datasets significantly disrupted 16 and 13 RBP binding, respectively, with 12 RBPs common to the two cohorts (Figures S10 and S11; Table S3). Among the significant RBPs, NKRF is a transcriptional repressor that binds to specific DNA sequences and regulates gene expression.60 While it is primarily associated with transcriptional regulation, it has been reported to interact with splicing factors and influence alternative splicing events.61,62 DDX3X is an RNA helicase involved in RNA splicing, translation, and other RNA-related processes.63,64,65,66,67 Dysregulation of DDX3X has been associated with altered gene expression of synaptic proteins and synaptic dysfunction.68 Together, the above results show that trait-associated variants may be enriched with functional ones that alter RBP-RNA interactions.
SpliTWAS highlights the role of splicing in SCZ
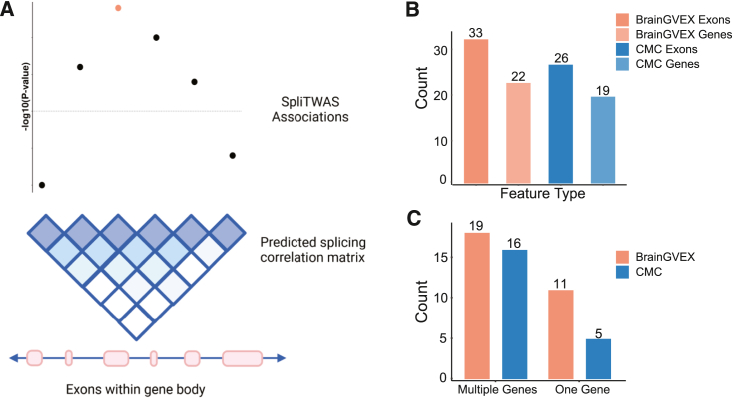
We next sought to infer putative causal genes and exons for SCZ based on the genome-wide association signals from SpliTWAS. For this purpose, we used FOCUS (fine mapping of causal gene sets) that leverages the correlation structure between associations due to LD and the prediction weights between the genetics and molecular traits.30 FOCUS estimates sets of genes that contain the causal genes. We adapted FOCUS to perform fine mapping at an exon level and utilized the alternative splicing prediction weights to explain SpliTWAS-associated signals (Figure 5A). We applied FOCUS to the 84 and 67 genes with at least one significant trait-associated exon for BrainGVEX and CMC, respectively. Using the estimated PIPs, credible gene sets at a 90% confidence level were computed, yielding a total of 19 and 15 credible sets, including 33 and 26 putative casual exons in 22 and 19 genes, respectively (Figure 5B; 48 exons and 36 genes total combining the two datasets). Furthermore, 11 of the 19 and 5 of the 15 credible sets contained a single putative causal gene, providing a much more refined look into alternative splicing-trait association (Figure 5C).
Figure 5.
Fine mapping on the exon level
(A) Schematic of the fine-mapping approach to examine the correlation between exon-level SpliTWAS associations within a gene body.
(B) Number of fine-mapped exons and genes for each cohort.
(C) Number of credible sets (90% confidence level) with multiple genes and a single putative causal gene.
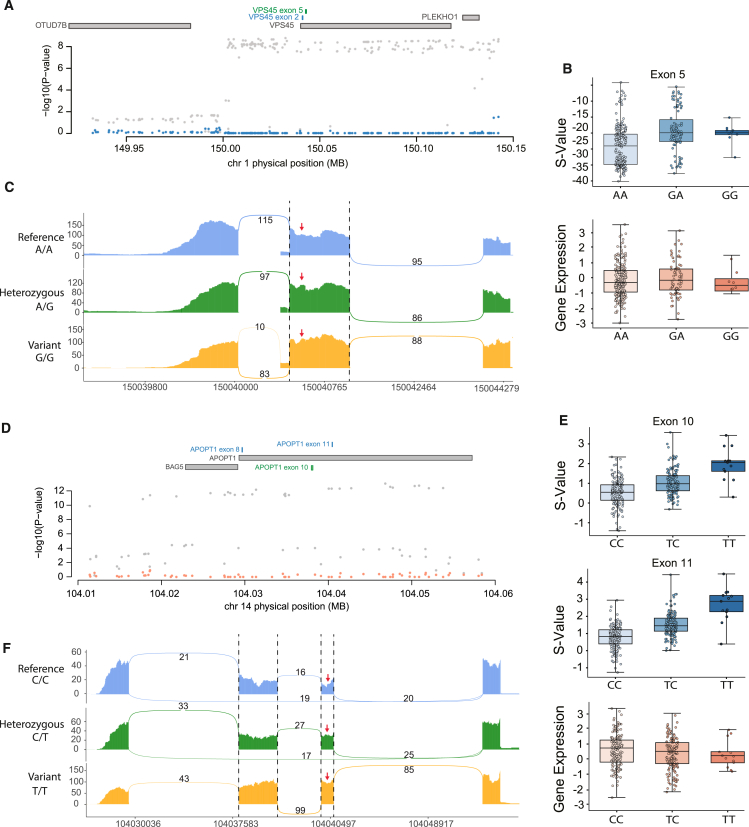
We highlight two examples with significant exon-trait associations that are not captured by gene expression. First, we detected a significant association with the exons of VPS45 (Figure 6A), which encodes a protein involved in intracellular trafficking and membrane fusion processes. It functions in the endosomal sorting pathway and is essential for proper vesicle transport.69,70,71 The endosomal network regulates synaptic vesicle pools, receptor endocytosis, recycling, and degradation. Disruptions in endocytic trafficking can greatly impact postsynaptic function and plasticity.48,72,73,74,75 Interestingly, when we conditioned the GWAS association on risk variants driving splicing of the exons in VPS45, we observed that the signal was largely abolished, suggesting that the original association was exclusively explained by splicing of the highlighted exon 5 (green) (Figure 6A). Consistent with the above data, splicing levels of the exon showed differences in samples with differing genotypes, while there is no observable stratification for gene expression (Figures 6B and 6C).
Figure 6.
SpliTWAS implicates specific exons of VPS45 and APOPT1 with SCZ
(A) Conditional analysis of the PGC SCZ GWAS on the splicing effect of the highlighted (green) exon. Gray dots represent the original GWAS signal. Blue dots represent the signal conditioned on the splicing association.
(B) Top: S values of the exon highlighted in (A) (green) in samples with different genotypes at the SpliTWAS-associated variant. Bottom: expression (RPKM) of VPS45 in samples with different genotypes at the SpliTWAS-associated variant.
(C) Example read distribution plots for the highlighted exon in A (green) in 3 samples with different genotypes at the SpliTWAS-associated variant.
(D–F) Similar to (A–C) but for exons 10 and 11 of APOPT1.
Similarly, we identified a strong disease association of multiple exons in APOPT1, a gene encoding a mitochondrial protein that induces apoptotic cell death.76,77 Recent evidence has linked mitochondrial dysfunction and apoptosis with SCZ and other neuropsychiatric disorders.49,78,79,80 In the case of APOPT1, we also observed an almost complete elimination of GWAS signal when conditioning on splicing of the associated exons (Figure 6D). Furthermore, both exons showed splicing differences according to genotype, suggesting that they may be regulated concordantly (Figures 6E and 6F). In contrast, APOPT1 expression did not change depending on genotype. The above refined look at associations captured by SpliTWAS further demonstrates that splicing is a large contributor to the GWAS signal of the risk region, which would be otherwise missed by traditional TWAS approaches.
Discussion
We present SpliTWAS, an approach that integrates alternative splicing information with genetics-trait associations to identify genes associated with a trait through specific exon splicing events. Furthermore, we extend the application of FOCUS30 toward an exon-centric approach to probabilistically fine map exons and genes. This framework is applicable to any complex trait but particularly is useful for those associated with abundant occurrences of alternative splicing.
Previous studies have shed light on the critical role of alternative splicing as a fundamental molecular mechanism contributing to the complexity of traits, offering insights beyond gene expression levels.4,9,10,18 Unfortunately, quantifying isoforms using short-read RNA-seq poses computational and technical challenges, leading to limitations in accuracy and consistency across different analytical approaches. Importantly, isoform-based analyses often fail to establish direct connections with splicing regulatory mechanisms since investigations into splicing regulation necessitate a granular examination of individual exons and splicing events.
Additionally, studies utilizing LeafCutter intron excision events, paired with methods such as FUSION and S-PrediXcan, represent an alternative approach for splicing-based TWAS. However, intron excision rate may not be easily interpretable toward alternative splicing mechanisms. In addition, these approaches typically include 500 kb or 1 Mb heritability windows and standardized and quantile-normalized data to satisfy the assumption of a Gaussian distribution made by the linear regression schemes. In contrast, SpliTWAS utilizes PSI, a widely used splicing metric that is directly related to levels of alternative splicing. In addition, SpliTWAS affords the advantage of using heritability windows that are much closer to the cis-regulatory elements of splicing, which are typically located close to the 3′ and 5′ splice sites of target exons. Furthermore, we apply a logit transformation to the PSI values, inspired by the methylation field, which helps ameliorate the heteroscedasticity of the PSI values, while preserving the relationship between nearby values and providing a suitable distribution for the regression analysis within the framework.
Another advantage of SpliTWAS also stems from its exon-centric nature. By centering on exonic regions, our approach circumvents the requirement for prior knowledge regarding the specific composition of transcripts.22 This feature enables the detection of genetically driven splicing events that would otherwise be obscured if relying solely on gene or isoform expression analyses or intron-centric approaches. Furthermore, by integrating alternative splicing models with GWAS, we directly infer a biologically interpretable relationship between genetically influenced exon splicing patterns and complex traits. Thus, SpliTWAS can offer valuable unique insights into the molecular mechanisms underlying the interplay between genetic variations, splicing regulation, and phenotypic outcomes.
The application of SpliTWAS to SCZ data has provided compelling evidence supporting the involvement of splicing regulation as a candidate mechanism underlying certain disease-associated loci. Our findings not only validated known SCZ disease genes31,33,36,75 but also uncovered potential targets that contribute to our understanding of SCZ pathogenesis. Notably, genes such as VPS45 and APOPT1, with established functions highly relevant to SCZ, emerged as previously undiscovered candidates associated with the disease. These findings emphasize the application of SpliTWAS in identifying genetic variants and regulatory mechanisms that may have remained elusive using conventional approaches.
Furthermore, our analysis reveals that splicing events associated with SCZ exhibited enrichment in pathways related to cellular transport, neuronal development, and immune responses.34,35,43,44,45,46 These pathways have been extensively implicated in the pathophysiology of SCZ, reinforcing the notion that these processes play critical roles in disease development and progression. Connecting to the disruption of multiple RNA regulators, we provided prioritization of genetic variants and regulatory mechanisms driving the observed genetic associations.
While SpliTWAS enables the exploration of connections between splicing and complex traits, it is not without limitations. Like many other genome-wide association strategies, SpliTWAS focuses on common variants without capturing the potential impact of rare variants on disease susceptibility. Given that splicing patterns can exhibit tissue-specific variations, establishing biologically meaningful associations via SpliTWAS requires RNA-seq data from tissues relevant to the trait of interest. Furthermore, it’s worth noting that, in certain instances, the presence of high correlation among exon associations within the same gene can pose challenges in pinpointing the causal exon responsible for the associated traits. Efforts in accounting for these correlations at a gene level have recently been published and could be applied at the exon level.81,82 Lastly, using an out-of-sample LD panel could potentially lead to inflated type 1 error; recent works have also started to tackle this issue.25,26
In summary, our method, SpliTWAS, establishes connections between genetics, splicing dysregulation, and complex traits, providing valuable insights into the genetic basis and underlying molecular mechanisms of SCZ. Furthermore, SpliTWAS is generally applicable for investigating diverse traits and enables opportunities to unveil molecular mechanisms underlying other complex traits.
Data and code availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental information. SpliTWAS software is freely available at https://github.com/gxiaolab/SpliTWAS.
Acknowledgments
We thank members of the Xiao and Pasaniuc laboratories for helpful discussions and comments on this work. We thank the PsychENOCDE and CommonMind consortia for making available the RNA-seq data used in this study. This work was supported in part by grants from the National Institutes of Health (R01MH123177 and R01AG05476 to X.X.). J.L.H. was supported by the NIH T32HL139450. K.A. was supported by the University of California-Historically Black Colleges and Universities (HBCUs) Fellowship. M.C. was supported by the NIH T32LM012424.
Author contributions
J.L.H., B.P., and X.X. designed the study. J.L.H, K.A., J.D., M.C., A.B., and G.Q.-V. developed methods and performed analyses. J.L.H. and X.X. drafted the manuscript. All authors provided critical revisions.
Declaration of interests
The authors declare no competing interests.
Published: June 25, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2024.06.001.
Contributor Information
Bogdan Pasaniuc, Email: pasaniuc@ucla.edu.
Xinshu Xiao, Email: gxxiao@ucla.edu.
Supplemental information
References
- 1.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., Jansen R., de Geus E.J.C., Boomsma D.I., Wright F.A., et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J., et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu L., Shi W., Long J., Guo X., Michailidou K., Beesley J., Bolla M.K., Shu X.-O., Lu Y., Cai Q., et al. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat. Genet. 2018;50:968–978. doi: 10.1038/s41588-018-0132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gusev A., Mancuso N., Won H., Kousi M., Finucane H.K., Reshef Y., Song L., Safi A., Schizophrenia Working Group of the Psychiatric Genomics Consortium. McCarroll S., et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018;50:538–548. doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbeira A.N., Pividori M., Zheng J., Wheeler H.E., Nicolae D.L., Im H.K. Integrating predicted transcriptome from multiple tissues improves association detection. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1007889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y., Li M., Lu Q., Weng H., Wang J., Zekavat S.M., Yu Z., Li B., Gu J., Muchnik S., et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat. Genet. 2019;51:568–576. doi: 10.1038/s41588-019-0345-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L., et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1–20. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K.C., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R., et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347 doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amoah K., Hsiao Y.-H.E., Bahn J.H., Sun Y., Burghard C., Tan B.X., Yang E.-W., Xiao X. Allele-specific alternative splicing and its functional genetic variants in human tissues. Genome Res. 2021;31:359–371. doi: 10.1101/gr.265637.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 13.Johnson M.B., Kawasawa Y.I., Mason C.E., Krsnik Z., Coppola G., Bogdanović D., Geschwind D.H., Mane S.M., State M.W., Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manning K.S., Cooper T.A. The roles of RNA processing in translating genotype to phenotype. Nat. Rev. Mol. Cell Biol. 2017;18:102–114. doi: 10.1038/nrm.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 16.Lim K.H., Ferraris L., Filloux M.E., Raphael B.J., Fairbrother W.G. Vol. 108. 2011. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes; pp. 11093–11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X., Hurst L.D. Determinants of the Usage of Splice-Associated cis-Motifs Predict the Distribution of Human Pathogenic SNPs. Mol. Biol. Evol. 2016;33:518–529. doi: 10.1093/molbev/msv251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takata A., Matsumoto N., Kato T. Genome-wide identification of splicing QTLs in the human brain and their enrichment among schizophrenia-associated loci. Nat. Commun. 2017;8:1–11. doi: 10.1038/ncomms14519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G., Bahn J.H., Lee J.-H., Peng G., Chen Z., Nelson S.F., Xiao X. Identification of allele-specific alternative mRNA processing via transcriptome sequencing. Nucleic Acids Res. 2012;40:e104. doi: 10.1093/nar/gks280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsiao Y.-H.E., Bahn J.H., Lin X., Chan T.-M., Wang R., Xiao X. Alternative splicing modulated by genetic variants demonstrates accelerated evolution regulated by highly conserved proteins. Genome Res. 2016;26:440–450. doi: 10.1101/gr.193359.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 22.Schafer S., Miao K., Benson C.C., Heinig M., Cook S.A., Hubner N. Alternative Splicing Signatures in RNA-seq Data: Percent Spliced in (PSI) Curr. Protoc. Hum. Genet. 2015;87:11.16.1–11.16.14. doi: 10.1002/0471142905.hg1116s87. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharya A., Hirbo J.B., Zhou D., Zhou W., Zheng J., Kanai M., Global Biobank Meta-analysis Initiative. Pasaniuc B., Gamazon E.R., Cox N.J. Best practices for multi-ancestry, meta-analytic transcriptome-wide association studies: Lessons from the Global Biobank Meta-analysis Initiative. Cell Genom. 2022;2 doi: 10.1016/j.xgen.2022.100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasaniuc B., Zaitlen N., Shi H., Bhatia G., Gusev A., Pickrell J., Hirschhorn J., Strachan D.P., Patterson N., Price A.L. Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics. 2014;30:2906–2914. doi: 10.1093/bioinformatics/btu416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C., Zhang Z., Yang X., Zhao B. 2023. Large-scale Imputation Models for Multi-Ancestry Proteome-wide Association Analysis. [Google Scholar]
- 26.Xue H., Shen X., Pan W. Causal Inference in Transcriptome-Wide Association Studies with Invalid Instruments and GWAS Summary Data. J. Am. Stat. Assoc. 2023;118:1525–1537. doi: 10.1080/01621459.2023.2183127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Nostrand E.L., Freese P., Pratt G.A., Wang X., Wei X., Xiao R., Blue S.M., Chen J.-Y., Cody N.A.L., Dominguez D., et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020;583:711–719. doi: 10.1038/s41586-020-2077-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du P., Zhang X., Huang C.-C., Jafari N., Kibbe W.A., Hou L., Lin S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinf. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mancuso N., Freund M.K., Johnson R., Shi H., Kichaev G., Gusev A., Pasaniuc B. Probabilistic fine-mapping of transcriptome-wide association studies. Nat. Genet. 2019;51:675–682. doi: 10.1038/s41588-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z., Burge C.B. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D., Liu S., Warrell J., Won H., Shi X., Navarro F.C.P., Clarke D., Gu M., Emani P., Yang Y.T., et al. Comprehensive functional genomic resource and integrative model for the human brain. Science. 2018;362 doi: 10.1126/science.aat8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sadler A.J. The role of MDA5 in the development of autoimmune disease. J. Leukoc. Biol. 2018;103:185–192. doi: 10.1189/jlb.4MR0617-223R. [DOI] [PubMed] [Google Scholar]
- 35.Na K.-S., Jung H.-Y., Kim Y.-K. The role of pro-inflammatory cytokines in the neuroinflammation and neurogenesis of schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2014;48:277–286. doi: 10.1016/j.pnpbp.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 36.Jauhar S., Johnstone M., McKenna P.J. Schizophrenia. Lancet. 2022;399:473–486. doi: 10.1016/S0140-6736(21)01730-X. [DOI] [PubMed] [Google Scholar]
- 37.van Mierlo H.C., Broen J.C.A., Kahn R.S., de Witte L.D. B-cells and schizophrenia: A promising link or a finding lost in translation? Brain Behav. Immun. 2019;81:52–62. doi: 10.1016/j.bbi.2019.06.043. [DOI] [PubMed] [Google Scholar]
- 38.Steiner J., Jacobs R., Panteli B., Brauner M., Schiltz K., Bahn S., Herberth M., Westphal S., Gos T., Walter M., et al. Acute schizophrenia is accompanied by reduced T cell and increased B cell immunity. Eur. Arch. Psychiatr. Clin. Neurosci. 2010;260:509–518. doi: 10.1007/s00406-010-0098-x. [DOI] [PubMed] [Google Scholar]
- 39.Hall A., Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harbor Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y.E., Baek S.T. Neurodevelopmental Aspects of RASopathies. Mol. Cell. 2019;42:441–447. doi: 10.14348/molcells.2019.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryu H.-H., Lee Y.-S. Cell type-specific roles of RAS-MAPK signaling in learning and memory: Implications in neurodevelopmental disorders. Neurobiol. Learn. Mem. 2016;135:13–21. doi: 10.1016/j.nlm.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Nussinov R., Tsai C.-J., Jang H. Neurodevelopmental disorders, immunity, and cancer are connected. iScience. 2022;25 doi: 10.1016/j.isci.2022.104492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth K.A., D’Sa C. Apoptosis and brain development. Ment. Retard. Dev. Disabil. Res. Rev. 2001;7:261–266. doi: 10.1002/mrdd.1036. [DOI] [PubMed] [Google Scholar]
- 44.Kuan C.Y., Roth K.A., Flavell R.A., Rakic P. Mechanisms of programmed cell death in the developing brain. Trends Neurosci. 2000;23:291–297. doi: 10.1016/s0166-2236(00)01581-2. [DOI] [PubMed] [Google Scholar]
- 45.Maday S., Twelvetrees A.E., Moughamian A.J., Holzbaur E.L.F. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sleigh J.N., Rossor A.M., Fellows A.D., Tosolini A.P., Schiavo G. Axonal transport and neurological disease. Nat. Rev. Neurol. 2019;15:691–703. doi: 10.1038/s41582-019-0257-2. [DOI] [PubMed] [Google Scholar]
- 47.Chen X.-J., Xu H., Cooper H.M., Liu Y. Cytoplasmic dynein: a key player in neurodegenerative and neurodevelopmental diseases. Sci. China Life Sci. 2014;57:372–377. doi: 10.1007/s11427-014-4639-9. [DOI] [PubMed] [Google Scholar]
- 48.Ferguson S.M. Axonal transport and maturation of lysosomes. Curr. Opin. Neurobiol. 2018;51:45–51. doi: 10.1016/j.conb.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bülow P., Patgiri A., Faundez V. Mitochondrial protein synthesis and the bioenergetic cost of neurodevelopment. iScience. 2022;25 doi: 10.1016/j.isci.2022.104920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandopulle M.S., Lippincott-Schwartz J., Ward M.E. RNA transport and local translation in neurodevelopmental and neurodegenerative disease. Nat. Neurosci. 2021;24:622–632. doi: 10.1038/s41593-020-00785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Iterson M., van Zwet E.W., BIOS Consortium. Heijmans B.T. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol. 2017;18:19. doi: 10.1186/s13059-016-1131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moehle E.A., Braberg H., Krogan N.J., Guthrie C. Adventures in time and space: splicing efficiency and RNA polymerase II elongation rate. RNA Biol. 2014;11:313–319. doi: 10.4161/rna.28646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García A., Collin A., Calvo O. Sub1 associates with Spt5 and influences RNA polymerase II transcription elongation rate. Mol. Biol. Cell. 2012;23:4297–4312. doi: 10.1091/mbc.E12-04-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de la Mata M., Alonso C.R., Kadener S., Fededa J.P., Blaustein M., Pelisch F., Cramer P., Bentley D., Kornblihtt A.R. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Bedi K., Magnuson B.R., Narayanan I., Paulsen M., Wilson T.E., Ljungman M. Co-transcriptional splicing efficiencies differ within genes and between cell types. RNA. 2021;27:829–840. doi: 10.1261/rna.078662.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Markus M.A., Yang Y.H.J., Morris B.J. Transcriptome-wide targets of alternative splicing by RBM4 and possible role in cancer. Genomics. 2016;107:138–144. doi: 10.1016/j.ygeno.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Smith H.C., Bennett R.P., Kizilyer A., McDougall W.M., Prohaska K.M. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 2012;23:258–268. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma S., Patnaik S.K., Taggart R.T., Kannisto E.D., Enriquez S.M., Gollnick P., Baysal B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat. Commun. 2015;6:6881. doi: 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghanbari M., Ohler U. Deep neural networks for interpreting RNA-binding protein target preferences. Genome Res. 2020;30:214–226. doi: 10.1101/gr.247494.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dreikhausen U., Hiebenthal-Millow K., Bartels M., Resch K., Nourbakhsh M. NF-kappaB-repressing factor inhibits elongation of human immunodeficiency virus type 1 transcription by DRB sensitivity-inducing factor. Mol. Cell Biol. 2005;25:7473–7483. doi: 10.1128/MCB.25.17.7473-7483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coccia M., Rossi A., Riccio A., Trotta E., Santoro M.G. Vol. 114. 2017. Human NF-κB repressing factor acts as a stress-regulated switch for ribosomal RNA processing and nucleolar homeostasis surveillance; pp. 1045–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexandrova J., Piñeiro D., Jukes-Jones R., Mordue R., Stoneley M., Willis A.E. Full-length NF-κB repressing factor contains an XRN2 binding domain. Biochem. J. 2020;477:773–786. doi: 10.1042/BCJ20190733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Soto-Rifo R., Rubilar P.S., Limousin T., de Breyne S., Décimo D., Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shih J.-W., Tsai T.-Y., Chao C.-H., Wu Lee Y.-H. Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene. 2008;27:700–714. doi: 10.1038/sj.onc.1210687. [DOI] [PubMed] [Google Scholar]
- 65.Lee C.-S., Dias A.P., Jedrychowski M., Patel A.H., Hsu J.L., Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lai M.-C., Lee Y.-H.W., Tarn W.-Y. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Geissler R., Golbik R.P., Behrens S.-E. The DEAD-box helicase DDX3 supports the assembly of functional 80S ribosomes. Nucleic Acids Res. 2012;40:4998–5011. doi: 10.1093/nar/gks070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Distler U., Schumann S., Kesseler H.-G., Pielot R., Smalla K.-H., Sielaff M., Schmeisser M.J., Tenzer S. Proteomic Analysis of Brain Region and Sex-Specific Synaptic Protein Expression in the Adult Mouse Brain. Cells. 2020;9 doi: 10.3390/cells9020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cowles C.R., Emr S.D., Horazdovsky B.F. Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J. Cell Sci. 1994;107:3449–3459. doi: 10.1242/jcs.107.12.3449. [DOI] [PubMed] [Google Scholar]
- 70.Bryant N.J., Piper R.C., Gerrard S.R., Stevens T.H. Traffic into the prevacuolar/endosomal compartment of Saccharomyces cerevisiae: a VPS45-dependent intracellular route and a VPS45-independent, endocytic route. Eur. J. Cell Biol. 1998;76:43–52. doi: 10.1016/S0171-9335(98)80016-2. [DOI] [PubMed] [Google Scholar]
- 71.Vilboux T., Lev A., Malicdan M.C.V., Simon A.J., Järvinen P., Racek T., Puchalka J., Sood R., Carrington B., Bishop K., et al. A congenital neutrophil defect syndrome associated with mutations in VPS45. N. Engl. J. Med. 2013;369:54–65. doi: 10.1056/NEJMoa1301296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang C., Wang X. Lysosome biogenesis: Regulation and functions. J. Cell Biol. 2021;220 doi: 10.1083/jcb.202102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rizzoli S.O., Betz W.J. Synaptic vesicle pools. Nat. Rev. Neurosci. 2005;6:57–69. doi: 10.1038/nrn1583. [DOI] [PubMed] [Google Scholar]
- 74.Toribio V., Yáñez-Mó M. Tetraspanins interweave EV secretion, endosomal network dynamics and cellular metabolism. Eur. J. Cell Biol. 2022;101 doi: 10.1016/j.ejcb.2022.151229. [DOI] [PubMed] [Google Scholar]
- 75.Schmidt-Kastner R., van Os J., Esquivel G., Steinbusch H.W.M., Rutten B.P.F. An environmental analysis of genes associated with schizophrenia: hypoxia and vascular factors as interacting elements in the neurodevelopmental model. Mol. Psychiatr. 2012;17:1194–1205. doi: 10.1038/mp.2011.183. [DOI] [PubMed] [Google Scholar]
- 76.Yasuda O., Fukuo K., Sun X., Nishitani M., Yotsui T., Higuchi M., Suzuki T., Rakugi H., Smithies O., Maeda N., Ogihara T. Apop-1, a novel protein inducing cyclophilin D-dependent but Bax/Bak-related channel-independent apoptosis. J. Biol. Chem. 2006;281:23899–23907. doi: 10.1074/jbc.M512610200. [DOI] [PubMed] [Google Scholar]
- 77.Brischigliaro M., Corrà S., Tregnago C., Fernandez-Vizarra E., Zeviani M., Costa R., De Pittà C. Knockdown of APOPT1/COA8 Causes Cytochrome c Oxidase Deficiency, Neuromuscular Impairment, and Reduced Resistance to Oxidative Stress in Drosophila melanogaster. Front. Physiol. 2019;10:1143. doi: 10.3389/fphys.2019.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rajasekaran A., Venkatasubramanian G., Berk M., Debnath M. Mitochondrial dysfunction in schizophrenia: pathways, mechanisms and implications. Neurosci. Biobehav. Rev. 2015;48:10–21. doi: 10.1016/j.neubiorev.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 79.Prabakaran S., Swatton J.E., Ryan M.M., Huffaker S.J., Huang J.T.-J., Griffin J.L., Wayland M., Freeman T., Dudbridge F., Lilley K.S., et al. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol. Psychiatr. 2004;9:684. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- 80.Glantz L.A., Gilmore J.H., Lieberman J.A., Jarskog L.F. Apoptotic mechanisms and the synaptic pathology of schizophrenia. Schizophr. Res. 2006;81:47–63. doi: 10.1016/j.schres.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 81.Liu L., Yan R., Guo P., Ji J., Gong W., Xue F., Yuan Z., Zhou X. Conditional transcriptome-wide association study for fine-mapping candidate causal genes. Nat. Genet. 2024;56:348–356. doi: 10.1038/s41588-023-01645-y. [DOI] [PubMed] [Google Scholar]
- 82.Zhao S., Crouse W., Qian S., Luo K., Stephens M., He X. Adjusting for genetic confounders in transcriptome-wide association studies improves discovery of risk genes of complex traits. Nat. Genet. 2024;56:336–347. doi: 10.1038/s41588-023-01648-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental information. SpliTWAS software is freely available at https://github.com/gxiaolab/SpliTWAS.