Abstract
DEC1 (differentially expressed in chondrocytes 1) and DEC2 are E-box-binding transcription factors and exhibit a circadian expression pattern. Recently, both proteins were found to repress the Clock/Bmal1-activated E-box promoters (e.g. mPer1). Yeast two-hybrid assay detected interactions between Bmal1 and DECs. It was hypothesized that DEC-mediated repression on the mPer1 promoter is achieved by binding to E-box elements and interacting with Bmal1. In the present study, we report that E-box binding rather than Bmal1 interaction is responsible for the observed repression. In the absence of Clock/Bmal1, both DEC1 and DEC2 markedly repressed the mPer1 promoter reporter; however, DNA-binding mutants showed no repressive activity. Similarly, DEC1, but not its DNA-binding mutants, repressed the Clock/Bmal1-induced activation. In addition, DEC1R58P, a DNA-binding mutant with Bmal1 interactivity, repressed neither the mPer1 reporter directly nor the Clock/Bmal1-induced activation, providing direct evidence that DNA binding, rather than Bmal1 interactions, is responsible for the repression on the mPer1 promoter. Furthermore, disruption of the Sp1 site in the proximal promoter of mPer1 increased the repression of DEC1 proteins. Previous studies with mouse DEC2 showed that this factor interacts with Sp1. These findings suggest that DEC proteins regulate the expression of mPer1 through E-box binding and Sp1 interaction. Alterations on circadian systems are increasingly recognized as important risk factors for disease initiation and progression, and the expression of Dec genes is rapidly induced by environmental stimuli and is highly increased in tumour tissues. Therefore de-regulated expression of DEC genes probably alters normal circadian rhythms and contributes significantly to the pathogenesis of many diseases including cancer.
Keywords: Bmal1, circadian rhythm, Clock, DEC1, Per1
Abbreviations: bHLH, basic helix–loop–helix; DEC, differentially expressed in chondrocytes; DEC1-M, DEC1 without a DNA-binding domain; EMSA, electrophoretic mobility-shift assay; HCE-1, human carboxylesterase-1; HEK-293T cells, human embryonic kidney 293T cells; luc, luciferase; SCN, suprachiasmatic nucleus
INTRODUCTION
Circadian rhythms are considered to play critical roles in adapting to daily environmental changes (e.g. light/dark) and co-ordinating such physiological processes as the sleep–wake cycle, endocrine secretion, liver metabolism and renal activity [1–7]. Recent studies have linked disruption of normal circadian rhythms to therapeutic failures and acceleration of disease process (e.g. malignancy) [1,6,8–10]. In mammals, there are central and peripheral circadian clocks [6,11–13]. The central clock is located in the SCN (suprachiasmatic nucleus) of the hypothalamus, which operates autonomously and can be entrained by environmental cues, in particular in light–dark cycles [2,3,9,11,14–16]. The SCN receives photic input signals through the retinohypothalamic tract and generates rhythms, which subsequently synchronize multiple peripheral clocks through neural and humoral signalling. The amplitude of the peripheral oscillators is generally less persistent and robust [1]. Interestingly, the peripheral clocks do not always depend on the SCN. For example, restricted feeding induces a shift on the liver rhythm by 10 h, whereas the rhythmicity in the SCN remains phase-locked to the light–dark cycle [17].
It is generally accepted that circadian rhythms are generated and co-ordinated by several transcription–translation feedback loops composed of positive and negative elements [5,18–22]. Bmal1 and Clock are well-characterized positive elements, whereas Per and Cry proteins act as negative regulators. Clock and Bmal1 form heterodimers, which transcriptionally activate the expression of target genes (e.g. Per1) through E-box elements in the promoters. Per proteins form homodimers or heterodimerize with Cry proteins. The resultant protein complexes interact with Clock–Bmal1 dimers, resulting in the removal of positive regulatory activity mediated by Clock–Bmal1. In the last few years, several new members of the clock gene family [e.g. DEC (differentially expressed in chondrocytes) proteins] have been identified, resulting in the increased complexity of the circadian regulatory loops [23–26]. Clock genes generally exhibit a circadian expression pattern or circadian-dependent regulatory activity. In addition, the cross species of these genes share an unusually high degree of sequence identity in both coding and regulatory regions, providing a molecular explanation for the conserved evolution associated with circadian rhythms among living organisms [27].
The Per1 gene has been studied extensively and recognized to play a central role in conveying the light-entraining information to the central clock [27–31]. In the Per1 promoter (approx. 5.0 kb upstream), several potential regulatory elements are identified including five E-boxes, four cAMP-response elements and a single Sp1-binding site [27,29,32]. Both E-boxes and cAMP-response elements mediate stimulation on the Per1 promoter, although it is not clear whether the cAMP-response elements are involved in circadian regulation [18,27]. The expression of Per1 is regulated by the feedback loop of Clock/Bmal1 (stimulation) and Per/Cry (attenuation) through the E-boxes. Recently, repressive transcription factors, DECs, have been shown to repress Clock/Bmal1-induced activation of the mPer1 (mouse Per1) promoter [24]. DEC proteins belong to a new and structurally distinct class of bHLH (basic helix–loop–helix) transcription factors and have been shown to interact with class B E-box [33,34]. Yeast two-hybrid experiments have demonstrated that DEC proteins interact directly with Bmal1 [24]. Therefore it has been hypothesized that DEC proteins repress Clock/Bmal1-induced activation through two distinct mechanisms: competing for E-box binding and interacting with Bmal1.
The present study was performed to determine whether these two mechanisms operate independently of each other. DEC1 mutants were prepared selectively to disrupt DNA binding without altering interactivity with Bmal1. In the absence of Clock/Bmal1, both DEC1 and DEC2 markedly repressed the mPer1 promoter; however, DNA-binding mutants showed no repressive activity. Similarly, DEC1, but not its DNA-binding mutants, repressed the Clock/Bmal1-induced activation. In addition, DEC1R58P, a DNA-binding mutant with Bmal1 interactivity, repressed neither the mPer1 reporter directly nor the Clock/Bmal1-induced activation, providing direct evidence that DNA binding, rather than Bmal1 interactions, is responsible for the repression on the mPer1 promoter. Furthermore, disruption of the Sp1 site in the proximal promoter of mPer1 increased the repression of DEC1 proteins. These findings provide molecular details on how DECs are involved in the circadian regulation.
MATERIALS AND METHODS
Plasmid
Expression constructs of Bmal1 and Clock, tagged by a Flag or HA, were gifts from Dr M. P. Antoch of the Cleveland Clinic [35]. DEC1 deletion or substitution mutants were described elsewhere [34]. The DEC1 promoter reporter [DEC1-luc (luciferase)] was prepared by inserting an approx. 1.0 kb upstream sequence (based on putative translation starting codon) into the pGL3 basic vector through blunt ligation. This fragment, containing a single E-box, was generated by PCR with primers 5′-TTGGCCGTCGGCCCGCTTCCCATG-3′ and 5′-CAAGTTGAGAGTGGCGCATAAC-3′. To prepare the Flag-tagged DEC2 construct, the DEC2 image clone from Openbiosystems was digested by EcoRI and XbaI, blunted and reinserted into the Flag-CMV2 vector (Sigma) pretreated with EcoRV and subsequently with alkaline phosphatase (Promega). The DEC2 promoter reporter (DEC2-luc) was described elsewhere [34]. The mPer1 promoter reporter (mPer1-luc), which contained approx. 6.7 kb of mPer1 promoter and 5′-untranslated region sequence, was a gift from Dr J. S. Takahashi of Northwestern University [32]. The 5′-deletion mutant of mPer1 reporter (mPer1E4-luc) was prepared by inserting NheI–HindIII fragment into the pGL3 basic vector. This fragment was generated by PCR with primers 5′-CAGAGCTAGCACTAGTCACCAAGTAG-3′ and 5′-AGCTGAGGGTCAAAGCTTGC-3′. All constructs were subjected to sequence analysis.
Co-transfection experiment
Co-transfection experiments were performed with HEK-293T cells (human embryonic kidney 293T cells), which have been shown to be valid models for the regulation studies on the transcription and translation of clock genes [35]. Cells were plated in 24-well plates in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum at a density of 1.6×105 cells/well. Transfection experiments were performed using lipofection with lipofectAMINE™ as described previously [34]. Transfection mixtures generally contained DEC1, DEC2, Clock/Bmal1 or a corresponding mutant construct (100 ng), reporter plasmid (50 ng) and the pRL-null Renilla plasmid (5 ng) unless otherwise specified. Vector plasmid was used to equalize the amount of plasmid DNA for each transfection. The transfected cells were cultured for an additional 24 h, washed once with PBS and resuspended in passive lysis buffer (Promega). The lysed cells were subjected to two cycles of freezing and thawing. The reporter enzyme activities were assayed with a dual-luciferase reporter assay system (Promega). This system contained two substrates, which were used to determine the activity of two luciferases sequentially. The firefly luciferase activity, which represented the reporter gene activity, was initiated by mixing an aliquot of lysates (20 μl) with Luciferase Assay Reagent II. Then the firefly luminescence was quenched and the Renilla luminescence was simultaneously activated by adding Stop & Glo Reagent to the sample wells. The firefly luminescence signal was normalized based on the Renilla luminescence signal.
In vivo protein–protein cross-linking by formaldehyde
HEK-293T cells were cultured in 6-well plates and transfected with DEC1 (0.2 μg), Flag-Bmal1 (0.8 μg) or both, and vector plasmid was used to equalize the amount of plasmid DNA for each transfection. In some cases, DEC1 was replaced by a mutant construct. After 24 h incubation, cells were washed initially with serum-free medium and subsequently with PBS. To each well, 3.6 ml of PBS was added and then 0.4 ml of formaldehyde (100 mM) for 10 min [36]. The cross-linking was terminated by the addition of glycine to a final concentration of 100 mM. The cells were then washed twice with PBS and lysed with SDS/PAGE sample buffer. The lysates were analysed by Western blotting for the presence of cross-linked DEC1–Bmal1.
EMSA (electrophoretic mobility-shift assay)
HEK-293T cells were plated in six-well plates and transfected with Flag-DEC1 (1 μg/well), DEC1R58P (1 μg/well) or Flag-DEC2 expression construct (1 μg/well) for overnight. Nuclear extracts were prepared with a nuclear extraction kit (Active Motif). Nuclear proteins (10 μg) were incubated with radiolabelled double-stranded oligonucleotides (5′-AAGCTTTAGCCACGTGACAGTGAGGG-3′) in a final volume of 10 μl containing 1×DNA-binding buffer (Promega). For competition experiments, nuclear extracts were first incubated with excess unlabelled probe (50×) and then mixed with the radiolabelled probe. For supershift assays, an anti-Flag antibody was added after the nuclear extracts were incubated with the radiolabelled probe. The protein–DNA complexes were resolved on 6% polyacrylamide and visualized by Typhoon imager. The procedure used for EMSA is the same as described in detail previously [34].
Co-immunoprecipitation
HEK-293T cells were plated in six-well plates and transfected with DEC1 or DEC1R58P (0.75 μg) along with Flag-Bmal1 or Flag-DEC2 (0.25 μg). As controls for co-immunoprecipitation, a Flag construct was replaced with the empty vector. The vector was used to equalize the amount of constructs in each well. After 24 h incubation, total cell lysates were prepared with an extraction kit (Active Motif). The lysates containing equal amounts of total protein (200 μg) were incubated with anti-Flag M2 affinity beads (Sigma) overnight. The beads were washed eight times with washing buffer (50 mM Tris/HCl, pH 7.4 and 150 mM NaCl). The precipitates were analysed for the presence of DEC1 or DEC1R58P by Western blots with an anti-DEC1 antibody. The samples (5 μg) before immunoprecipitation were also analysed by Western blotting with either anti-DEC1 or anti-Flag antibody.
Site-directed mutagenesis
The mPer1E4Sp1(–)-luc, lacking the Sp1 site in the proximal promoter of the mPer1, was prepared with a Quik Change site-directed mutagenesis kit (Stratagene) [34]. Complementary oligonucleotides (5′-GATCCTTAGCCAACCGAGATCGATGCCTGCGGCTCTTCG-3′) were synthesized to target this region. To perform the substitutions, the primers were annealed to the mPer1E4-luc and subjected to a thermocycler for a total of 15 cycles. The resultant PCR-amplified constructs were then digested with DpnI to remove the non-mutated parent constructs. The mutated PCR-amplified constructs were used to transform Epicurian Coli XL1-Blue cells. The resultant mutated construct was subjected to sequence analysis to confirm that the desired mutation was correct without secondary mutations.
Other analyses
The anti-DEC1 antibody against a peptide derived from the C-terminus was described elsewhere [37]. Protein concentration was determined with the BCA assay (Pierce), with BSA as the standard. Results are expressed as means±S.D. for at least four separate experiments, except where results of blots are shown, in which case a representative experiment is depicted in the Figures.
RESULTS
DEC1 and DEC2 repress the mPer1 promoter
DEC1 and DEC2 are E-box-binding transcription factors and exhibit a circadian expression pattern [24,25,34,38]. Recently, both proteins were found to repress Clock/Bmal1-induced activation of an mPer1 element reporter (E-box) and a human DEC1 promoter reporter. Yeast two-hybrid experiments detected interactions between Bmal1 and DECs [24]. Therefore it has been hypothesized that DEC proteins repress the mPer1 promoter by two distinct mechanisms: (i) binding to E-box elements in the mPer1 promoter and leading to transcriptional repression, and (ii) interacting with Bmal1 and interfering with Clock/Bmal1-induced transactivation. To examine whether these two mechanisms operate independently, we first examined the ability of DEC proteins to repress directly the promoter of mPer1 in the absence of Clock/Bmal1. For comparison, DEC1 and DEC2 promoter reporters were also included. The mPer1 reporter (6.75 kb) contained five E-box elements [32], the DEC1-luc (approx. 1.1 kb) contained a single E-box [25] and the DEC2-luc contained two E-boxes [34]. Co-transfection experiments were performed with a reporter, along with a DEC construct or Clock/Bmal1. To reveal the quantitative relationship, various amounts of the expression constructs were used in the transfection assays.
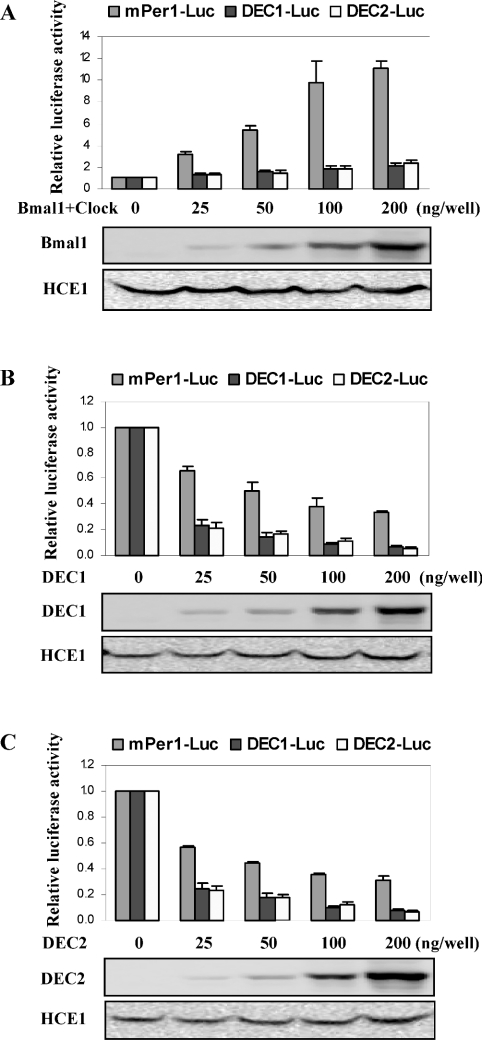
Results on the activation of these reporters by Clock/Bmal1 are summarized in Figure 1(A). Clock/Bmal1 markedly transactivated the mPer1 reporter by as many as 10-fold (Figure 1A), although the activation magnitude was lower than that on an mPer1 E-box element reporter [24]. The overall activation was proportionately increased as higher levels of Clock/Bmal1 were present. In contrast, the activation on the DEC1-luc and DEC2-luc reporters was very minimal, and the maximum activation for both reporters was only approx. 2-fold, and occurred when the highest amount of Clock/Bmal1 was used (Figure 1A). We next examined the repressive activity of DEC1 and DEC2 towards these reporters. As shown in Figures 1(B) and 1(C), DEC1 and DEC2 exhibited a comparable potency on repressing all the three reporters, and the repressive effect increased when larger amounts of DEC1 or DEC2 were used. Overall, the mPer1 reporter was markedly less sensitive than the DEC reporters. The mPer1-luc reporter was repressed only moderately and the maximum repression was approx. 60% (200 ng). In contrast, DEC1-luc and DEC2-luc reporters were repressed to a much higher extent. As much as 70% repression was detected on the DEC reporters when only as little as 25 ng of DEC1 or DEC2 construct (the lowest amount) was used in the transfection experiments (Figures 1B and 1C). It should be emphasized that some co-transfection experiments were performed with a cytomegalovirus-driven construct encoding HCE-1 (human carboxylesterase-1) [39]. Apparently, neither Clock/Bmal1 nor DECs affected the expression of HCE-1, suggesting that the observed transactivation (Clock/Bmal1) or repression (DECs) is specific to E-box containing promoters (e.g. mPer1).
Figure 1. Regulation of mPer1-luc, DEC1-luc and DEC2-luc by Clock/Bmal1, DEC1 and DEC2.
(A) Activation of mPer1-luc, DEC1-luc and DEC2-luc by Clock/Bmal1. HEK-293T cells were cultured in 24-well plates at approx. 80% confluence and transfected with a reporter construct (50 ng), Clock/Bmal1 (0–200 ng each) and the pRL-null Renilla (5 ng). Vector plasmid was used to equalize the amount of plasmid DNA for each transfection. The transfected cells were cultured for 24 h, collected with PBS and resuspended in passive lysis buffer. The reporter enzyme activities were assayed with a dual-luciferase reporter assay system. The firefly luminescence signal was normalized based on the Renilla luminescence signal, and ratios are calculated based on the normalized firefly luminescence signal in the presence versus the absence of Clock/Bmal1. (B) Repression of mPer1-luc, DEC1-luc and DEC2-luc by DEC1. Cells were transfected with a reporter construct (50 ng), DEC1 (0–200 ng each) and the pRL-null Renilla (5 ng). The reporter enzyme activities were assayed with a dual-luciferase reporter assay system. The firefly luminescence signal was normalized based on the Renilla luminescence signal. The signal in the absence of DEC1 was recoded as 100%. (C) Repression of mPer1-luc, DEC1-luc and DEC2-luc by DEC2. Cells were transfected with a reporter construct (50 ng), DEC2 (0–200 ng each) and the pRL-null Renilla (5 ng). The reporter enzyme activities were assayed with a dual-luciferase reporter assay system and expressed as described in (B). Cell lysates (5 μg) were analysed for the abundance of transfected genes with anti-DEC1 (DEC1) or anti-Flag antibody (Bmal1 and DEC2). All experiments were performed in triplicate and selected co-transfection experiments were performed with a construct (100 ng) encoding HCE-1.
The DNA-binding domain is required for DEC1 to regulate the mPer1 promoter
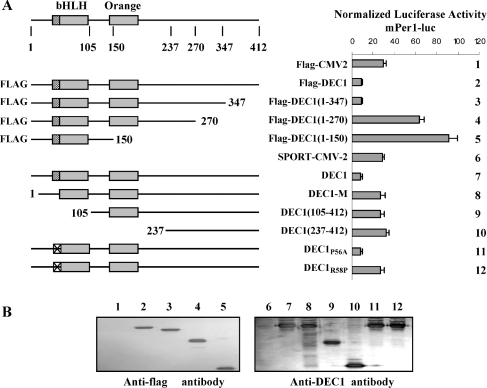
The repression of DECs on the mPer1 reporter provides direct evidence that these transcription factors are regulators of the Per1 promoter, presumably by interacting with the E-box elements. To test this possibility, we examined DEC1 DNA-binding mutants for their repressive ability towards this reporter. Several other mutants with certain functional domains deleted were also included. As shown in Figure 2(A), substitution mutant DEC1R58P showed no effects towards the mPer1 reporter, whereas DEC1P56A repressed this reporter comparably as the wild-type DEC1. We previously reported that DEC1P56A, but not DEC1R58P, binds to the E-box in the DEC2 proximal promoter [34]. The inability of DEC1R58P to repress the mPer1 promoter provided further evidence that DNA binding is required for DEC1 to repress the mPer1 promoter. Consistent with this finding, the N-terminal deletion mutants [DEC1-M (containing no DNA-binding domain), DEC1105–412 and DEC1237–412] contained no DNA-binding domain and failed to repress this reporter (Figure 2A). The C-terminal deletion mutants, on the other hand, varied markedly from mutant to mutant. DEC11–347 repressed the reporter similarly as DEC1, whereas mutants DEC11–270 and DEC11–150 markedly transactivated the mPer1 reporter (2–3-fold; Figure 2A). All C-terminal mutants were previously shown to bind to E-box element [34]. The expression levels of these mutants were comparable based on Western-blot analysis (Figure 2B).
Figure 2. Essentiality of DNA binding for DEC1 to repress the mPer1 promoter.
(A) Co-transfection experiment. HEK-293T cells were cultured in 24-well plates and transfected with DEC1 or a DEC1 mutant (100 ng), the mPer1-luc (50 ng) and the pRL-null Renilla (5 ng). Vector plasmid was used to equalize the amount of plasmid DNA for each transfection. After a 24 h incubation, cells were collected and analysed for luciferase activities. Similarly, firefly luminescence signal was normalized based on the Renilla luminescence signal. Orange, orange domain. (B) Immunoblot analysis. The cell lysates (5 μg) from the cells used for reporter activity were analysed for the expression of DEC1 or its mutants by anti-Flag (C-terminal deletion mutants) or anti-DEC1 antibody (all others) as described in the legend for Figure 1. All experiments were performed in triplicate.
The DNA-binding domain is required to repress Clock/Bmal1-induced transactivation
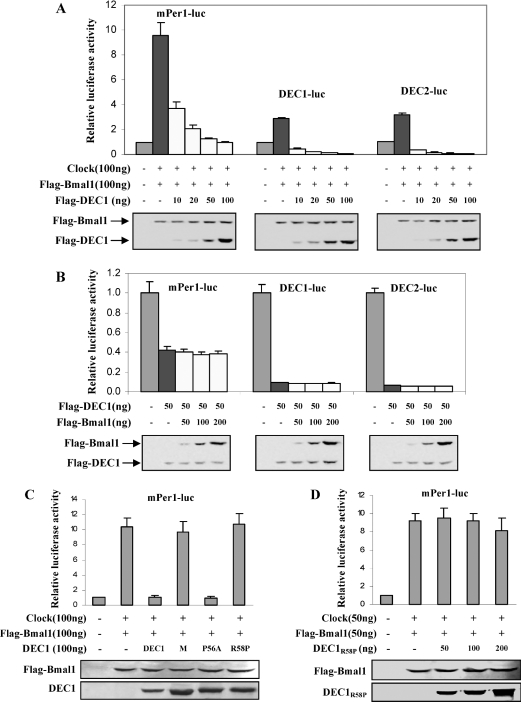
The study with DEC1 mutants clearly demonstrated that the integrity of the DNA-binding domain is essential for DEC1 to regulate the mPer1 reporter. However, it remains to be determined whether the repression of Clock/Bmal1-induced activation requires the intact DNA-binding domain as well. We first examined whether DEC1 actually represses Clock/Bmal1-induced activation on the mPer1 promoter reporter, in particular, whether the repression is similar towards all the three promoter reporters (mPer1-luc, DEC1-luc and DEC2-luc). Co-transfection experiments were performed with the same amount of Clock/Bmal1 in the presence or absence of various amounts of DEC1. As shown in Figure 3(A), the Clock/Bmal1-induced activation of all three reporters was repressed by DEC1 in a dose-dependent manner. However, the decreased magnitude varied markedly between the mPer1 reporter and the DEC reporters. When DEC1 was assayed at 10 ng (the lowest amount), the Clock/Bmal1-induced activation on the mPer1 reporter was reduced by 60%. However, the same amount of DEC1 construct completely abolished the Clock/Bmal1-induced activation on the DEC reporters. In fact, the overall activities of the DEC reporters in the DEC1-transfected cells were even lower than the activity in the vector-transfected cells, suggesting that DNA-binding mechanism rather than interaction with Bmal1 plays a determinant role in regulating these reporters, particularly the DEC1 and DEC2 reporters.
Figure 3. Repression of Clock/Bmal1-induced transactivation.
(A) Repression of Clock/Bmal1-induced transactivation by DEC1. HEK-293T cells were cultured in 24-well plates and transfected with a reporter construct (50 ng), Clock/Bmal1 (100 ng each, Flag-tagged Bmal1), Flag-DEC1 construct (0–100 ng) and pRL-null (5 ng). Vector plasmid was used to equalize the amount of plasmid DNA for each transfection. After 24 h incubation, cells were collected and reporter enzyme activities were assayed with a dual-luciferase reporter assay system as described above. Cell lysates (5 μg) were analysed by Western blotting for the abundance of the transfected genes. (B) Effects of Bmal1 on the DEC1-mediated repression. Cells were transfected with a promoter reporter (50 ng) or along with DEC1 (50 ng) and increasing amounts of Bmal1 (0–200 ng). The DEC1 and Bmal1 constructs were Flag-tagged so that the intracellular levels of both DEC1 and Bmal1 could be monitored simultaneously. After 24 h incubation, cells were collected and reporter enzyme activities were assayed with a dual-luciferase reporter assay system. Cell lysates (5 μg) were analysed by Western blotting for the abundance of the transfected genes. (C) Repression of Clock/Bmal1-induced transactivation of Per1 by DEC1 mutants. Cells were transfected with mPer1-luc (50 ng), Clock/Bmal1 (100 ng each), pRL-null (5 ng) and DEC1 or a DEC1 mutant (100 ng). After 24 h incubation, cells were collected and reporter enzyme activities were assayed with a dual-luciferase reporter assay system and normalized as described above. Cell lysates (5 μg) were analysed by Western blotting for the abundance of the transfected genes. All experiments were performed in triplicate.
We next tested whether Bmal1 alters DEC1-mediated transcriptional activity. Cells were transfected with a promoter reporter or along with DEC1 (50 ng) and increasing amounts of Bmal1 (0–200 ng). The DEC1 and Bmal1 constructs were Flag-tagged so that the intracellular levels of both DEC1 and Bmal1 could be monitored simultaneously. As expected, DEC1 alone repressed all reporters (mPer1-luc, DEC1-luc and DEC2-luc) with the mPer1 being repressed to a lesser extent (Figure 3B). Co-transfection of Bmal1, however, caused no changes on the magnitude of the repression even in the cells that expressed markedly higher levels of Bmal1 than DEC1 (>3-fold; Figure 3B). The inability of the excessive Bmal1 to modulate DEC1-mediated repression provides further evidence that interactions with Bmal1 play an insignificant role in both DEC1-mediated direct repression and DEC1-mediated attenuation on Clock/Bmal1-induced transactivation.
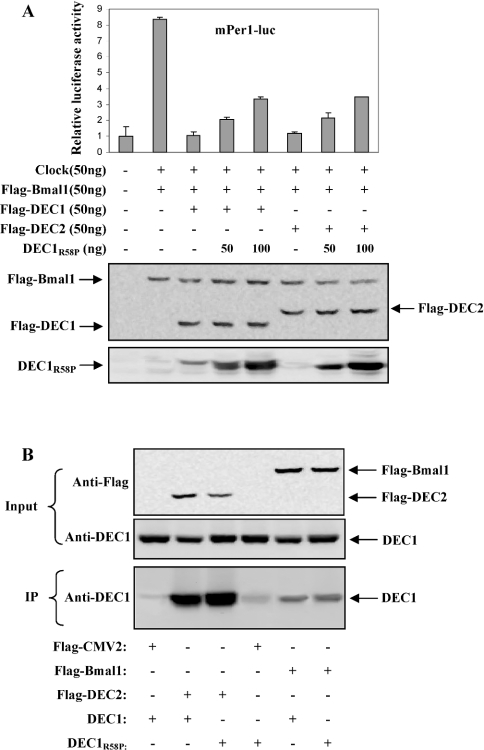
To establish further the necessity of DNA binding in repressing Clock/Bmal1-induced transactivation, DEC1 mutants (defective of DNA binding) were tested for the repressive activity. Similarly, co-transfection experiments were performed with the mPer1 reporter and Clock/Bmal1 in the presence or absence of DEC1 or a DNA-binding mutant. As shown in Figure 3(C), Clock/Bmal1 markedly transactivated the reporter, and the activation was completely attenuated by DEC1 and DEC1P56A. However, DNA-binding mutants, DEC1-M and DEC1R58P, caused no changes on the Clock/Bmal1-induced activation. To determine whether greater amounts of the DNA-binding mutants are required to repress effectively Clock/Bmal1 activity, co-transfection experiments were performed with increasing amounts of DEC1R58P. As shown in Figure 3(C), higher amounts of the DEC1R58P construct resulted in higher expression levels of the corresponding protein; however, the repression on Clock/Bmal1 was very minimal even with the highest amount of DEC1R58P (approx. 10%).
DNA binding and Bmal1 interaction can be separated
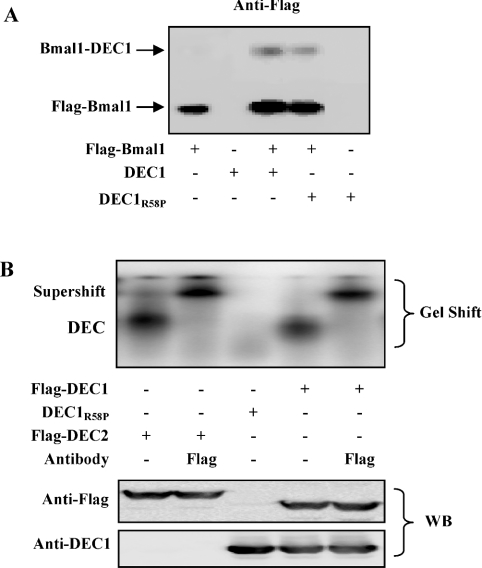
The inability of DEC1R58P to repress Clock/Bmal1-induced activation points to two important possibilities: (i) repression of Clock/Bmal1-induced activation by DEC proteins is exclusively achieved by an E-box-binding mechanism, and/or (ii) this substitution mutant no longer interacts with Bmal1. To test the second possibility, intracellular cross-linking experiments were performed. Cells were transfected with Bmal1 (Flag-tagged) and DEC1 or DEC1R58P and then subjected to cross-linking by formaldehyde [37]. The concentration of formaldehyde was chosen so that only limited cross-linking took place. Cell extracts were prepared and analysed by Western blots. As expected, the anti-Flag antibody detected abundant expression of Bmal1 (Figure 4A). In addition, an extra protein band with a combined molecular mass of Bmal1 and DEC1 was also detected. Based on the immunostaining intensities, the abundance of this high molecular mass was approx. 5% of the levels of the Bmal1 monomer. More importantly, this higher-molecular-mass band appeared only when cells were co-transfected with Flag-tagged Bmal1 and DEC1 or DEC1R58P. These findings suggest that both DEC1 and DEC1R58P bind to Bmal1 but the interactions are very minimal.
Figure 4. In vivo protein–protein cross-linking by formaldehyde and EMSA analysis.
(A) In vivo protein–protein cross-linking by formaldehyde. HEK-293T cells were cultured in six-well plates and transfected with DEC1 or DEC1R58P (0.2 μg/per well), Flag-Bmal1 (0.8 μg/well) or both. After 24 h incubation, cells were washed with serum-free medium and subsequently with PBS. To each well, 3.6 ml of PBS was added and then 0.4 ml of formaldehyde (100 mM) for 10 min. The cross-linking was terminated by the addition of glycine to a final concentration of 100 mM. The cell lysates (20 μg) were analysed by Western blotting for the presence of crosslinked DEC1-Bmal1. (B) EMSA. HEK-293T cells were plated in six-well plates and transfected with Flag-DEC1 (1 μg/well), DEC1R58P (1 μg/well) or Flag-DEC2 expression construct (1 μg/well) overnight. Nuclear extracts were prepared with a nuclear extraction kit (Active Motif). Nuclear proteins (10 μg) were incubated with 32P-labelled oligonucleotides (5′-AAGCTTTAGCCACGTGACAGTGAGGG-3′) in a final volume of 10 μl containing 1× DNA-binding buffer. For competition experiments, nuclear extracts were first incubated with excess unlabelled probe (50×) and then mixed with the radiolabelled probe (results not shown). For supershift assays, an anti-Flag antibody was added after the nuclear extracts were incubated with the radiolabelled probe. The protein–DNA complexes were resolved on 6% polyacrylamide and visualized by Typhoon imager. The cell lysates (20 μg) were analysed by Western blotting for the expression of transfected genes.
We next tested whether DEC1 and DEC1R58P bind directly to the E-box elements present in the mPer1 promoter. We and other investigators have previously demonstrated that DEC proteins bind to class B E-box element (CACGTG) with higher affinity [33,34]. However, the E-box elements in the mPer1 promoter are flanked by different nucleotides, which may alter the binding reactivity towards DEC proteins [27]. To establish definitively that DEC1 and DEC2 actually bind to the E-box elements in the mPer1 promoter, we performed EMSA with a clustered E-box element (three adjacent E-boxes) [24] or a single E-box element (E-box 3) [27]. As shown in Figure 4(B), both proteins bound to the E-box 3 element, and the shifted band was completely supershifted by an anti-Flag antibody. In contrast, the mutant DEC1R58P did not show any binding activity (Figure 4B), although it was expressed comparably as DEC1 or DEC2 (Figure 4B, bottom panel). Similar binding was detected with the clustered E-box element, and non-labelled oligonucleotides effectively competed for the binding (results not shown).
DEC1R58P negatively interferes with DEC-mediated repression on the mPer1 promoter
The DEC1R58P had the intact bHLH domain, which is known to mediate dimerization (either homodimers or heterodimers) [40]. Therefore it was assumed that this mutant negatively interferes with DEC-mediated transcriptional regulatory activity. To test this hypothesis, co-transfection experiments were conducted with a DEC construct and Clock/Bmal1 in the presence or absence of DEC1R58P. Consistent with the data described in Figure 3(A), both DEC1 and DEC2 effectively repressed the Clock/Bmal1-induced activation on the mPer1 reporter (Figure 5A). The repression, however, was partially reversed by the presence of DEC1R58P. The reversed magnitude was moderate (approx. 30%) and similar towards both DEC1 and DEC2 (Figure 5A). Based on Western-blot analysis (DEC1R58P migrated slightly faster than Flag-tagged DEC1), the level of DEC1R58P was markedly higher than that of DEC1, particularly when 100 ng of DEC1R58P was used in the transfection experiments (approx. 2-fold; Figure 5A, bottom panel). Based on sequence alignment analysis, DEC1 and DEC2 are identical in the bHLH domain, suggesting that DEC1R58P forms dimers with DEC1 and DEC2 to a comparable extent.
Figure 5. Dominant interfering regulation on DEC-mediated repression and co-immunoprecipitation.
(A) Effect of DNA-binding mutant on the DEC1-mediated repression. HEK-293T cells were cultured in 24-well plates and transfected with the mPer1-luc reporter (50 ng) and the pRL-null Renilla (5 ng) along with Flag-DEC1 or Flag-DEC2 in the presence or absence of DEC1R58P. Vector plasmid was used to equalize the amount of plasmid DNA for each transfection. After 24 h incubation, cells were collected and analysed for luciferase activities. Similarly, firefly luminescence signal was normalized based on the Renilla luminescence signal. Cell lysates (10 μg) were analysed by Western blotting for the abundance of transfected genes. (B) DEC1 differentially interacts with DEC2 and Bmal1. HEK-293T cells were cultured in six-well plates and transfected with Flag-Bmal1 or Flag-DEC2 (250 ng/well) along with DEC1 or DEC1R58P (750 ng/well). After 24 h incubation, whole cell lysates were prepared and subjected to co-immunoprecipitation (200 μg of total protein) by Anti-Flag affinity gel. The immunoprecipitates and input (10 μg) were analysed for the presence of DEC1 by Western blotting with anti-DEC1 antibody.
We next performed co-immunoprecipitation experiments to determine whether DEC1 and DEC2 actually form heterodimers. Cells were transfected with DEC1 or DEC1R58P along with the Flag-tagged DEC2 construct. For comparison, the Flag-tagged DEC2 construct was replaced with the Flag-tagged Bmal1 construct. Lysates were prepared and subjected to co-immunoprecipitation with an anti-Flag antibody, and the precipitates were analysed for the presence of DEC1. As shown in Figure 5(B, bottom panel), immunoprecipitation of DEC2 resulted in the abundant co-precipitation of DEC1 and DEC1R58P. The relative levels between DEC1 and DEC1R58P in the precipitates were comparable based on the immunostaining intensities (Figure 5B), suggesting that the substitution of Arg-58 causes no changes in dimerization. DEC1 and DEC1R58P were also precipitated by Flag-Bmal1; however, the amount of DEC1 or DEC1R58P precipitated by Bmal1 was rather minimal. Based on the immunostaining intensities, the precipitated DEC1 or DEC1R58P by Bmal1 was only approx. 10% of that by DEC2 (Figure 5B). Western-blot analyses detected comparable levels of DEC1, Flag-tagged Bmal1 or DEC2, excluding the possibility that differences in the expression levels were contributing factors to the markedly less precipitated DEC1 or DEC1R58P by Bmal1 (Figure 5B, top and middle panels).
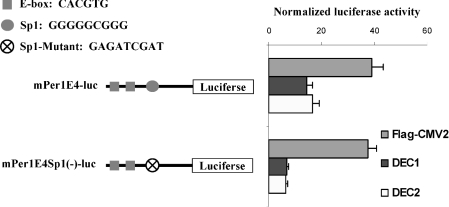
Disruption of the Sp1 site increases DEC-mediated repression
In addition to the E-boxes, several other response elements are also located in the mPer1 promoter and some of them have been found to mediate regulation in response to various stimuli [27,29,32]. One of the sites is the Sp1 site, which is located closely to the last E-box [27]. This site was investigated further because mSharp1 (mouse DEC2) has been shown to interact with Sp1 and represses the promoter of Stra13 (mouse DEC1 [26]). Site-directed mutagenesis was performed to disrupt the Sp1 site selectively. The resultant mutant was tested for the responsiveness to DEC1 and DEC2. Surprisingly, instead of relieving repression, disruption of the Sp1 site caused further repression, suggesting that this Sp1 site actually supports activation of the mPer1 promoter. In the cells co-transfected with the Flag vector, similar luciferase activities were detected with both wild-type (mPer1 E4-luc) and Sp1-mutated reporters [mPer1 E4Sp1(–)-luc (Figure 6)], suggesting that the Sp1 site is not involved in the basal transcription.
Figure 6. Disruption of Sp1 increases DEC-mediated repression.
HEK-293T cells were cultured in 24-well plates and transiently transfected with mPer1E4-luc or Sp1-disrupted mutant mPer1E4Sp1(1)-luc (50 ng) and the pRL-null Renilla (5 ng) along with DEC1, DEC2 or vector (100 ng). After 24 h incubation, cells were collected and analysed for luciferase activities. Similarly, firefly luminescence signal was normalized based on the Renilla luminescence signal. All experiments were performed in triplicate.
DISCUSSION
DEC proteins belong to a new and structurally distinct class of the bHLH transcription factors [34,38]. Both DEC1 and DEC2 are expressed in a circadian fashion and have been found to repress Clock/Bmal1-induced activation of clock gene mPer1 [24]. Based on reporter and yeast two-hybrid experiments, it has been hypothesized that DEC-mediated repression on the mPer1 promoter is achieved by binding to E-box elements and interacting with Bmal1 [24]. In the present study, we report that E-box binding rather than interaction with Bmal1 is responsible for the observed repression. Several lines of evidence support this conclusion. First, in the absence of Clock/Bmal1, both DEC1 and DEC2 markedly repress the mPer1 promoter reporter; however, DNA-binding mutants show no repressive activity (Figures 1 and 2). Secondly, the mPer1 promoter reporter is significantly activated by Clock/Bmal1, and the Clock/Bmal1-induced activation is effectively attenuated by DECs but not the DNA-binding mutants (Figure 3B). Finally, DEC1R58P, a DNA-binding mutant that retains the ability to interact with Bmal1, has no repressive activity towards either the mPer1 reporter directly or the Clock/Bmal1-induced transactivation (Figures 2A and 3B), providing direct evidence that DNA binding rather than interactions with Bmal1 is responsible for the repression on the Clock/Bmal1-induced transactivation. These results also suggest that DECs effectively compete with Clock/Bmal1 for binding to E-box elements in the mPer1 promoter.
The insignificant effect of Bmal1–DEC interactions on the Clock/Bmal1-induced activation is probably due to insufficient amount of Bmal1–DEC complexes formed under normal physiological conditions or even in the transfected cells. In the present study, we have demonstrated that DEC proteins prefer to form DEC1–DEC2 heterodimers (presumably homodimers as well) instead of forming Bmal1–DEC1 complexes (approx. 20:1; Figure 5B). It is probable that the DEC dimers no longer interact with Bmal1 or to a markedly lesser extent. DEC1 and DEC2 are highly identical in the bHLH domain (differing by only two residues) but rather diverse in the rest of the proteins (<40%) [38]; therefore, interactions of DECs with Bmal1 are presumably mediated through the bHLH motif. Dimerization through the bHLH motif masks the bHLH interface and prevents the dimers from interacting with Bmal1. In support of this possibility, yeast two-hybrid assays establish that the interactions with Bmal1 occur in the N-terminal half of DECs (where the bHLH motif is located) [24], and intracellular cross-linking experiments detect a protein complex with a molecular mass of Bmal1 plus DEC monomer but not dimer (Figure 4A). Finally, Bmal1 normally heterodimerizes with Clock, a protein that is twice as large as Bmal1 or DEC [19–22]. Thus the Bmal1–Clock complex has a markedly larger size than Bmal1, and is probably less interactive with DECs, although it remains to be determined whether Bmal1 forms a complex preferably with Clock over DECs.
It is interesting to note that the substitution mutant DEC1R58P reverses the DEC1-mediated repression to a rather moderate extent (approx. 30%; Figure 5A). Based on Western-blot analyses, the level of DEC1R58P in the transfected cells was much higher than that of DEC1 (approx. 2-fold), excluding the possibility that the limited interfering activity of DEC1R58P is due to poor expression (Figure 5A, bottom panel). We have previously made similar observations with this mutant on the DEC2-luc reporter [34]. In contrast, DEC1-M, a mutant that lacks the entire DNA domain, is much more effective in terms of interfering with the activity of DEC1 [34]. Both DEC1-M and DEC1R58P have no DNA-binding ability and form dimers with DEC proteins (Figure 5B) [34]. It is probable that DEC1–DEC1R58P dimers have some DNA-binding activity, whereas DEC1–DEC1-M dimers no longer bind to DNA elements. As a result, DEC1–DEC1R58P dimers have partial activity of DEC1–DEC1 dimers. Alternatively, DEC1–DEC1R58P dimers bind to E-box as tightly as DEC1–DEC1 dimers; however, DEC1–DEC1R58P complexes position differently on the DNA molecule from DEC1–DEC1 dimers, and hence are less repressive.
DNA binding is essential for DECs to regulate the mPer1 promoter but not sufficient to determine the transcriptional magnitude and the nature. The C-terminal deletion mutants (DEC11–347, DEC11–270 and DEC11–150), for example, exhibit opposing activities towards the mPer1 reporter (Figure 2A), although they all bind to the E-box element to a similar extent [34]. DEC11–347 represses the reporter, whereas DEC11–270 and DEC11–150 activate it. As a matter of fact, DEC11–347 exhibits a similar repressive potency as DEC1, suggesting that the C-terminal 65 residues are dispensable for the repressive activity. The region encoding residues 150–347 contains multiple helical structures and is required to exert maximum repressive activity in both native and chimaeric status, and deletion of this region decreases repressive activity towards the DEC2 promoter reporter [34,41]. Based on the fact that DEC11–150 contains minimal sequence that is known to be repressive [34,41], it is probable that the mPer1 promoter is under a highly repressive status, presumably through the E-box elements. Binding of these mutants (DNA binding with no transcription regulatory activity) actually de-represses these elements and, thus, has transcription-activation activity. In support of the negative interfering mechanism, we have shown previously that DEC11–150 effectively attenuates the repression of DEC1 on the DEC2 promoter [34].
The mPer1 and DEC reporters all contain functional E-box elements and respond to the transcription regulation mediated by Clock/Bmal1 and DEC proteins; however, they differ significantly in responding to these transcription factors. The mPer1 reporter is markedly more sensitive than DEC reporters in responding to Clock/Bmal1-mediated activation (10-fold versus 2-fold; Figure 1A), whereas the opposite is true in responding to DEC-mediated repression (60% versus 90%; Figures 1B and 1C). Even with DEC proteins, the mPer1 and DEC reporters respond differentially to DEC1 mutants. For example, DEC11–150 activates the mPer1 reporter (Figure 2A), but has no activity towards the DEC2 reporter [34]. In contrast, DEC1R58P shows no activity towards the mPer1 reporter (Figure 2A), whereas it moderately represses the DEC2 reporter [34]. Recently, it has been shown that Clock differentially regulates the expression of mPer1 and DEC1 [4]. In mice that express a truncated Clock (a dominant negative), the peak expression levels of mPer1 are decreased; in contrast, the peak levels of DEC1 are slightly increased, although phase shifts occur in these mice with the expression of both genes [4].
The transcription regulation of Per1 and DEC genes appears to be determined collectively by a group of transcription factors including Clock/Bmal1 and DECs. In the present study, we have demonstrated that the Sp1 site in the mPer1 proximal promoter is functionally active, and disruption of this element results in increased repression in response to DEC1 (Figure 6). Recently, transfection of Sp1 has been shown to increase the activity of a Stra13 promoter reporter (mouse DEC1), and the increased activity is reduced by mSharp1, the mouse counterpart of DEC2 [26]. The repression of mSharp1 on Sp1-mediated activation involves physical interactions, although it remains to be determined whether the Sp1–mSharp1 complex retains the ability to bind to the Sp1 DNA element. Nevertheless, these findings suggest that DEC proteins regulate the expression of mPer1 probably through two distinct mechanisms: binding directly to E-box elements and interacting with Sp1.
In summary, DEC proteins are newly identified members of the molecular clock family and have been hypothesized to regulate the expression of clock gene mPer1 through direct E-box binding and/or interacting with Bmal1, a central positive element in the core circadian feedback loop [24]. In the present study, we report that E-box binding, but not the interaction with Bmal1, is responsible for the repression on the Clock/Bmal1-induced activation of the mPer1 promoter. We have also demonstrated that the Sp1 element in the mPer1 proximal promoter is functionally important, and DEC proteins probably form complexes with Sp1, and thus alter the Sp1-mediated regulation on the expression of Per1 as well. These findings provide molecular details on how DECs are involved in the circadian regulation. Emerging evidence suggests that alterations on circadian systems are important risk factors for disease initiation and progression [1], and the expression of DEC genes is rapidly induced by many environmental stimuli and highly elevated in tumour tissues (e.g. DEC1) [34,37,41,42]. Therefore de-regulated expression of DEC genes probably alters normal circadian rhythms and contributes significantly to the pathogenesis of many diseases including cancer.
Acknowledgments
We thank Dr J.S. Takahashi and Dr M.P. Antoch for providing constructs. This work was partially supported by NIH grants R01GM61988 and RR16457.
References
- 1.Canaple L., Kakizawa T., Laudet V. The days and nights of cancer cells. Cancer Res. 2003;63:7545–7552. [PubMed] [Google Scholar]
- 2.Griffin E. A., Jr, Staknis D., Weitz C. J. Light-independent role of Cry1 and Cry2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa T., Hirayama J., Kobayashi Y., Todo T. Zebrafish CRY represses transcription mediated by CLOCK-BMAL heterodimer without inhibiting its binding to DNA. Genes Cells. 2002;7:1073–1086. doi: 10.1046/j.1365-2443.2002.00579.x. [DOI] [PubMed] [Google Scholar]
- 4.Oishi K., Miyazaki K., Kadota K., Kikuno R., Nagase T., Atsumi G., Ohkura N., Azama T., Mesaki M., Yukimasa S., et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J. Biol. Chem. 2003;278:41519–41527. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 5.Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 6.Cermakian N., Boivin D. B. A molecular perspective of human circadian rhythm disorders. Brain Res. Brain Res. Rev. 2003;42:204–220. doi: 10.1016/s0165-0173(03)00171-1. [DOI] [PubMed] [Google Scholar]
- 7.Bittman E. L., Doherty L., Huang L., Paroskie A. Period gene expression in mouse endocrine tissues. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003;285:R561–R569. doi: 10.1152/ajpregu.00783.2002. [DOI] [PubMed] [Google Scholar]
- 8.Li J. J. Circadian variation in myocardial ischemia: the possible mechanisms involving in this phenomenon. Med. Hypotheses. 2003;61:240–243. doi: 10.1016/s0306-9877(03)00154-3. [DOI] [PubMed] [Google Scholar]
- 9.Chilov D., Hofer T., Bauer C., Wenger R. H., Gassmann M. Hypoxia affects expression of circadian genes PER1 and CLOCK in mouse brain. FASEB J. 2001;15:2613–2622. doi: 10.1096/fj.01-0092com. [DOI] [PubMed] [Google Scholar]
- 10.Lévi F. Circadian chronotherapy for human cancer. Lancet Oncol. 2001;2:307–315. doi: 10.1016/S1470-2045(00)00326-0. [DOI] [PubMed] [Google Scholar]
- 11.Boivin D. B., James F. O., Wu A., Cho-Park P. F., Xiong H., Sun Z. S. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–4145. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 12.LeSauter J., Yan L., Vishnubhotla B., Quintero J. E., Kuhlman S. J., McMahon D. G., Silver R. A short half-life GFP mouse model for analysis of suprachiasmatic nucleus organization. Brain Res. 2003;964:279–287. doi: 10.1016/s0006-8993(02)04084-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shieh K. R. Distribution of the rhythm-related genes rPERIOD1, rPERIOD2, and rCLOCK, in the rat brain. Neuroscience. 2003;118:831–843. doi: 10.1016/s0306-4522(03)00004-6. [DOI] [PubMed] [Google Scholar]
- 14.Matsuo T., Yamaguchi S., Mitsui S., Emi A., Shimoda F., Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 15.Paul K. N., Fukuhara C., Tosini G., Albers H. E. Transduction of light in the suprachiasmatic nucleus: evidence for two different neurochemical cascades regulating the levels of Per1 mRNA and pineal melatonin. Neuroscience. 2003;119:137–144. doi: 10.1016/s0306-4522(03)00098-8. [DOI] [PubMed] [Google Scholar]
- 16.Asai M., Yamaguchi S., Isejima H., Jonouchi M., Moriya T., Shibata S., Kobayashi M., Okamura H. Visualization of mPer1 transcription in vitro: NMDA induces a rapid phase shift of mPer1 gene in cultured SCN. Curr. Biol. 2001;11:1524–1527. doi: 10.1016/s0960-9822(01)00445-6. [DOI] [PubMed] [Google Scholar]
- 17.Challet E., Caldelas I., Graff C., Pevet P. Synchronization of molecular clock by light- and food-related cues in mammals. Biol. Chem. 2003;384:711–719. doi: 10.1515/BC.2003.079. [DOI] [PubMed] [Google Scholar]
- 18.Travnickova-Bendova Z., Cermakian N., Reppert S. M., Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okano T., Sasaki M., Fukada Y. Cloning of mouse BMAL2 and its daily expression profile in the suprachiasmatic nucleus: a remarkable acceleration of Bmal2 sequence divergence after Bmal gene duplication. Neurosci. Lett. 2001;300:111–114. doi: 10.1016/s0304-3940(01)01581-6. [DOI] [PubMed] [Google Scholar]
- 20.Etchegaray J. P., Lee C., Wade P. A., Reppert S. M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature (London) 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 21.Jung H., Choe Y., Kim H., Park N., Son G. H., Khang I., Kim K. Involvement of CLOCK:BMAL1 heterodimer in serum-responsive mPer1 induction. Neuroreport. 2003;14:15–19. doi: 10.1097/00001756-200301200-00003. [DOI] [PubMed] [Google Scholar]
- 22.Hirayama J., Nakamura H., Ishikawa T., Kobayashi Y., Todo T. Functional and structural analyses of cryptochrome. Vertebrate CRY regions responsible for interaction with the CLOCK:BMAL1 heterodimer and its nuclear localization. J. Biol. Chem. 2003;278:35620–35628. doi: 10.1074/jbc.M305028200. [DOI] [PubMed] [Google Scholar]
- 23.Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell (Cambridge, Mass.) 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 24.Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature (London) 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 25.Kawamoto T., Noshiro M., Sato F., Maemura K., Takeda N., Nagai R., Iwata T., Fujimoto K., Furukawa M., Miyazaki K., et al. A novel autofeedback loop of Dec1 transcription involved in circadian rhythm regulation. Biochem. Biophys. Res. Commun. 2004;313:117–124. doi: 10.1016/j.bbrc.2003.11.099. [DOI] [PubMed] [Google Scholar]
- 26.Azmi S., Sun H., Ozog A., Taneja R. mSharpq/DEC2, a basic helix-loop-helix protein functions as a transcriptional repressor of E-box activity and Stra13 expression. J. Biol. Chem. 2003;278:20098–20109. doi: 10.1074/jbc.M210427200. [DOI] [PubMed] [Google Scholar]
- 27.Hida A., Koike N., Hirose M., Hattori M., Sakaki Y., Tei H. The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics. 2000;65:224–233. doi: 10.1006/geno.2000.6166. [DOI] [PubMed] [Google Scholar]
- 28.Kriegsfeld L. J., Korets R., Silver R. Expression of the circadian clock gene period 1 in neuroendocrine cells: an investigation using mice with a Per1::GFP transgene. Eur. J. Neurosci. 2003;17:212–220. doi: 10.1046/j.1460-9568.2003.02431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi S., Mitsui S., Miyake S., Yan L., Onishi H., Yagita K., Suzuki M., Shibata S., Kobayashi M., Okamura H. The 5′ upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Curr. Biol. 2000;10:873–876. doi: 10.1016/s0960-9822(00)00602-3. [DOI] [PubMed] [Google Scholar]
- 30.Motzkus D., Maronde E., Grunenberg U., Lee C. C., Forssmann W., Albrecht U. The human PER1 gene is transcriptionally regulated by multiple signaling pathways. FEBS Lett. 2000;486:315–319. doi: 10.1016/s0014-5793(00)02315-2. [DOI] [PubMed] [Google Scholar]
- 31.Kojima S., Hirose M., Tokunaga K., Sakaki Y., Tei H. Structural and functional analysis of 3′ untranslated region of mouse Period1 mRNA. Biochem. Biophys. Res. Commun. 2003;301:1–7. doi: 10.1016/s0006-291x(02)02938-8. [DOI] [PubMed] [Google Scholar]
- 32.Wilsbacher L. D., Yamazaki S., Herzog E. D., Song E. J., Radcliff L. A., Abe M., Block G., Spitznagel E., Menaker M., Takahashi J. S. Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice in vivo. Proc. Natl. Acad. Sci. U.S.A. 2002;99:489–499. doi: 10.1073/pnas.012248599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.St-Pierre B., Flock G., Zacksenhaus E., Egan S. E. Stra13 homodimers repress transcription through class B E-box elements. J. Biol. Chem. 2002;277:46544–46551. doi: 10.1074/jbc.M111652200. [DOI] [PubMed] [Google Scholar]
- 34.Li Y., Xie M., Song X., Gragen S., Sachdeva K., Wan Y., Yan B. DEC1 negatively regulates the expression of DEC2 through binding to the E-box in the proximal promoter. J. Biol. Chem. 2003;278:16899–16907. doi: 10.1074/jbc.M300596200. [DOI] [PubMed] [Google Scholar]
- 35.Kondratov R. V., Chernov M. V., Kondratova A. A., Gorbacheva V. Y., Gudkov A. V., Antoch M. P. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakkenist C. J., Kastan M. B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature (London) 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Zhang H., Xie M., Hu M., Ge S., Yang D., Wan Y., Yan B. DEC1/STRA13/SHARP2 is abundantly expressed in colon carcinoma, antagonizes serum deprivation induced apoptosis and selectively inhibits the activation of procaspases. Biochem. J. 2002;367:413–422. doi: 10.1042/BJ20020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujimoto K., Shen M., Noshiro M., Matsubara K., Shingu S., Honda K., Yoshida E., Suardita K., Matsuda Y., Kato Y. Molecular cloning and characterization of DEC2, a new member of basic helix-loop-helix proteins. Biochem. Biophys. Res. Commun. 2001;280:164–171. doi: 10.1006/bbrc.2000.4133. [DOI] [PubMed] [Google Scholar]
- 39.Xie M., Yang D., Liu L., Xue B., Yan B. Rodent and human carboxylesterases: immuno-relatedness, overlapping substrate specificity, differential sensitivity to serine inhibitors, and tumor-related expression. Drug Metab. Dispos. 2002;30:541–547. doi: 10.1124/dmd.30.5.541. [DOI] [PubMed] [Google Scholar]
- 40.Massari M. E., Murre C. Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boudjelal M., Taneja R., Matsubara S., Bouillet P., Dollè P., Chambon P. Overexpression of Strat13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997;11:2052–2065. doi: 10.1101/gad.11.16.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossner M. J., Dörr J., Gass P., Schwab M. H., Nave K. A. SHARPs: mammalian enhancer-of-split and hairy-related proteins coupled to neuronal stimulation. Mol. Cell. Neurosci. 1997;9:460–475. doi: 10.1006/mcne.1997.0640. [DOI] [PubMed] [Google Scholar]