Abstract
The nuclear bile acid receptor FXR (farnesoid X receptor) is one of the key factors that suppress bile acid biosynthesis in the liver. PGC-1α [PPARγ (peroxisome-proliferator-activated receptor γ) co-activator-1α] is known to control energy homoeostasis in adipose tissue, skeletal muscle and liver. We performed cell-based reporter assays using the expression system of a GAL4–FXR chimaera, the ligand-binding domain of FXR fused to the DNA-binding domain of yeast GAL4, to find the co-activators for FXR. We found that the transcriptional activation of a reporter plasmid by a GAL4–FXR chimaera was strongly enhanced by PGC-1α, in a ligand-dependent manner. Transcriptional activation of the SHP (small heterodimer partner) gene by the FXR–RXRα (retinoid X receptor α) heterodimer was also enhanced by PGC-1α in the presence of CDCA (chenodeoxycholic acid). Co-immunoprecipitation and pull-down studies using glutathione S-transferase–PGC-1α fusion proteins revealed that the ligand-binding domain of FXR binds PGC-1α in a ligand-influenced manner both in vivo and in vitro. Furthermore, our studies revealed that SHP represses its own transcription, and the addition of excess amounts of PGC-1α can overcome the inhibitory effect of SHP. These observations indicate that PGC-1α mediates the ligand-dependent activation of FXR and transcription of SHP gene.
Keywords: bile acid, farnesoid X receptor (FXR), fasting, nuclear receptor, peroxisome-proliferator-activated receptor-γ co-activator-1α (PGC-1α), transcriptional co-activator
Abbreviations: CDCA, chenodeoxycholic acid; CYP7A1, cholesterol 7α-hydroxylase; DBD, DNA-binding domain; DCA, deoxycholic acid; DMEM, Dulbecco's modified Eagle's medium; EYFP, enhanced yellow fluorescent protein; FCS, foetal calf serum; FXR, farnesoid X receptor; GST, glutathione S-transferase; HNF-4α, hepatocyte nuclear factor 4α; HRP, horseradish peroxidase; LBD, ligand-binding domain; LCA, lithocholic acid; LRH-1, liver receptor homologue-1; PEPCK, phosphoenolpyruvate carboxykinase; PGC-1α, peroxisome-proliferator-activated receptor γ co-activator-1α; PPARγ, peroxisome-proliferator-activated receptor γ; RXRα, retinoid X receptor α; SHP, small heterodimer partner; SRC1, steroid receptor co-activator 1
INTRODUCTION
FXR (farnesoid X receptor) is a nuclear receptor for bile acids [1–3]. In the liver, cholesterol is converted into bile acids by several routes. CYP7A1 (cholesterol 7α-hydroxylase) catalyses the rate-limiting step in the classical pathway [4,5]. It is generally thought that activation and repression of CYP7A1 gene expression are modulated by multiple and redundant pathways [6–14]. Among them, two FXR-dependent pathways that suppress CYP7A1 gene expression have been reported [9,10,14]. One pathway involves FXR, SHP (small heterodimer partner), LRH-1 (liver receptor homologue-1), and CYP7A1. In this pathway, FXR activates the expression of SHP, which is a nuclear receptor without a DBD (DNA-binding domain), and represses the transcription of nuclear receptors. LRH-1 is a positive regulator of CYP7A1, and its transcriptional activity is repressed by SHP. It is thought that the cascade involving FXR, SHP, LRH-1 and CYP7A1, in the presence of bile acids, especially CDCA (chenodeoxycholic acid), suppresses the conversion of cholesterol into bile acids [9,10]. The other pathway, which was recently discovered, involves FXR, fibroblast growth factor-19, c-Jun N-terminal kinase and CYP7A1 [14]. The expression levels of SHP and CYP7A1 reportedly exhibit a significant inverse relationship [15]. This study supports the model that activated FXR represses CYP7A1 via SHP [15], although alternative SHP-independent pathways may regulate CYP7A1, as suggested by experiments using mice deficient in SHP [13]. Similar inverse relationships were reportedly observed between SHP and CYP7A1, using mice deficient in FXR [16] and using synthetic agonists [17]. Notably, there have been several reports of decrease of enzyme activity of CYP7A1 and transcriptional repression of CYP7A1 [18–21] during starvation. It is known that the levels of CYP7A1 enzyme activity, as well as the amounts of its mRNA and protein are reduced in the livers of starved animals. The lowest level of CYP7A1 mRNA in starved animals was 20–30 times lower than the highest level in normal animals [20]. On the other hand, several studies have recently demonstrated that the level of CYP7A1 expression increased in the starved conditions [22–24].
PGC-1α [PPARγ (peroxisome-proliferator-activated receptor γ) co-activator-1α] was cloned in 1998 [25]. Interactions of PGC-1α with several receptors have subsequently been reported, such as with thyroid-hormone receptor β [26], retinoic-acid receptor α [27], oestrogen receptor α [28], glucocorticoid receptor [29], PPARα [30], constitutive androstane receptor [31] and HNF-4α (hepatocyte nuclear factor 4α) [32,33]. PGC-1α is expressed in response to environmental stimuli, and induces gene expression that stimulates mitochondrial oxidative metabolism in brown fat, fibre-type switching in skeletal muscle and gluconeogenesis in liver [34]. Notably, fasting induces PGC-1α in the liver to modulate gluconeogenesis [32]. PEPCK (phosphoenolpyruvate carboxykinase) is the rate-limiting enzyme in gluconeogenesis, to produce glucose from non-carbohydrate precursors. The association of PGC-1α with HNF-4α is an important event in the regulation of PEPCK transcription in gluconeogenesis upon fasting [32]. It was reported that cholesterol catabolism to bile acids and gluconeogenesis are controlled in a co-ordinated manner [23]. Bile acids themselves have capacities to suppress transcription of CYP7A1 and PEPCK by modulating the assembly of transcription factors including nuclear receptors and their co-activators [23]. Recently, Zhang et al. [35] reported that PGC-1α interacts with the DBD of FXR in a ligand-independent manner and enhances the transcription of FXR target genes.
In contrast, in the present paper, we report that the LBD (ligand-binding domain) of FXR fused to the DBD of yeast GAL4 (GAL4–FXR chimaera)-mediated transcription is effectively enhanced by PGC-1α in a ligand-dependent manner. We show that FXR–LBD and PGC-1α interact physically in a ligand-dependent manner both in vivo and in vitro. Furthermore, we demonstrate that full-length FXR–RXRα (retinoid X receptor α) heterodimer-mediated transcription of the SHP gene is enhanced by PGC-1α in a ligand-dependent manner. This transcription is repressed by the product protein, SHP, and excess amounts of PGC-1α overcome the repression by SHP.
EXPERIMENTAL
Plasmids
The plasmids pCMX-flag-mPGC-1α, pCMX-flag-mPGC-1β, and UASx4-tk Luc were kindly provided by Dr A. Kakizuka (Kyoto University, Kyoto, Japan) [36]. The plasmids pOZ-Tip60 and pOZ-P/CAF (p300/cAMP-response-element-binding-protein-binding protein) were from Dr T. Ikura (Hiroshima University, Hiroshima, Japan). The plasmids pSG5-SRC1a and pSG5-SRC1e were kindly provided by Dr S. Salam (Queen Charlottes & Chelsea Hospital, London, U.K.). The plasmids pCMX-hFXR, pCMX-hRXRα and pGL3-hSHP(569)-Luc were kindly provided by Dr D. J. Mangelsdorf (University of Texas Southwestern Medical Center, Dallas, TX, U.S.A.). The cDNA encoding the human FXR–LBD (amino acids 192–472) was obtained by PCR from a human liver cDNA library (OriGene Technologies, Rockville, MD, U.S.A.). The PCR fragment was cloned into pET28a, and the sequence was confirmed. The NdeI/XhoI fragment was excised, blunted, and ligated to EcoRI/SalI-digested and blunted pCMX-GAL4-DBD. The construction of the bacterial expression plasmids pGEX-PGC-1-full, pGEX-PGC-1-Nhe/Xba, and pGEX-PGC-1-Xba was described elsewhere [31]. For the construction of the expression plasmid pCMX-SHP, human SHP cDNA was prepared by a similar PCR method and was cloned into the pCMX vector.
Cell culture and transient transfections
COS-7 cells were maintained in DMEM (Dulbecco's modified Eagle's Medium) (Invitrogen) supplemented with 10% FCS (foetal calf serum) (Thermo Trace, Melbourne, Australia). At 1 day before transfection, the cells were plated in Phenol-Red-free DMEM (Invitrogen) containing 10% charcoal/dextran-treated FCS (Sigma). Transfection of cells was performed in 24-well plates, according to the procedure described previously [31]. Transient co-transfections using the UASx4-tk Luc reporter were performed with pEYFP-C1 as an internal control, the GAL4–DBD or GAL4–FXR chimaera expression construct, and the co-activator expression construct. The plasmid pcDNA3 was added to compensate for the total amount of DNA transfected in each assay. Transient co-transfections using the pGL3-hSHP(569)-Luc reporter were performed with pEYFP-C1, pCMX-hFXR, pCMX-hRXRα, pCMX-flag-mPGC-1α and pcDNA3. A plasmid pCMX was used for control assays. After 6 h of transfection, ligands were added for 20 h. Then, fluorescence of EYFP (enhanced yellow fluorescent protein) expressed in COS-7 cells in a 24-well plate was measured with a microplate fluorescence reader, model FL500 (BioTek Instruments, Winooski, VT, U.S.A.) to examine the transfection efficiency. Then the cells were washed with PBS and lysed with PicaGene lysis buffer (Toyo Ink, Tokyo, Japan). After the cell debris was removed by centrifugation for 14000 g for 2 min, the luciferase activity was measured using a luminometer, model TD-20/20 (Promega). Luciferase activities were normalized to the EYFP fluorescence value and are referred to as relative light units (RLU) as previously described [31]. All transfection experiments were repeated at least three times, and the data represent the means±S.E.M.
Immunoprecipitation and Western blot analysis
COS-7 cells (2×106) were transiently transfected with pCMX-His-FXR192-472 (6 μg) and pCMX (6 μg), or with pCMX-flag-mPGC-1α (6 μg) and pCMX (6 μg), or with pCMX-His-FXR192-472 (6 μg) and pCMX-flag-mPGC-1α (6 μg) mediated by calcium phosphate using ProFection Mammalian Transfection Systems (Promega). After 48 h, cells were treated with CDCA (100 μM) or DMSO (0.1%) for an additional 16 h. Cells were then washed and detached. The cell pellet was resuspended in the modified RIPA buffer reported by De Fabiani et al. [23] with protease inhibitors in the presence or absence of CDCA (100 μM) for 10 min. Cell debris was removed by centrifugation at 14000 g for 10 min, and the supernatants were subjected to anti-FLAG M2–agarose (Sigma) for 6 h at 4 °C. Agarose beads were washed three times with the modified RIPA buffer [23] in the presence or absence of CDCA, divided in two, and subjected to Western blotting. His-tagged FXR was detected by a nickel-activated derivative of HRP (horseradish peroxidase) (India His-Probe–HRP; Pierce). FLAG-tagged PGC-1α was detected by anti-FLAG M2–peroxidase conjugate (Sigma).
Pull-down assay
The GST (glutathione S-transferase)–PGC-1α fusion proteins were expressed in BL21(STAR) bacteria, as previously described [31]. The GST–PGC-1α fusion proteins bound to glutathione–Sepharose CL4B beads were used for protein–protein interaction assays between FXR and PGC-1α. The N-terminal His-tagged, human FXR–LBD (amino acids 217–472) was expressed in BL21(STAR) bacteria, and was purified by Ni-affinity and ion-exchange chromatography. A 30 μl aliquot of a 50% slurry of GST–PGC-1α fusion protein bound to glutathione beads was suspended in 100 μl of binding buffer (20 mM phosphate buffer, pH 7.5, 100 mM NaCl, 0.1 mM EDTA and 1 mM 2-mercaptoethanol) with protease inhibitors, in the presence or absence of ligand. The His-tagged FXR–LBD protein was added to the suspended GST–PGC-1α fusion proteins, and the mixture was incubated at 4 °C for 30 min. The beads were sedimented and washed five times in wash buffer [20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 0.1 mM EDTA, 0.1% (v/v) Nonidet P40 and 10% (v/v) glycerol] with or without ligand. An equal volume of reducing sample buffer was added, and the mixture was boiled for 3 min. The samples were divided in two, and analysed by SDS/PAGE (15% gel) and Western blotting. The presence of the retained FXR–LBD protein with a His tag was detected using India His-Probe–HRP.
RESULTS
PGC-1α activates FXR-mediated transcription in a ligand-dependent manner
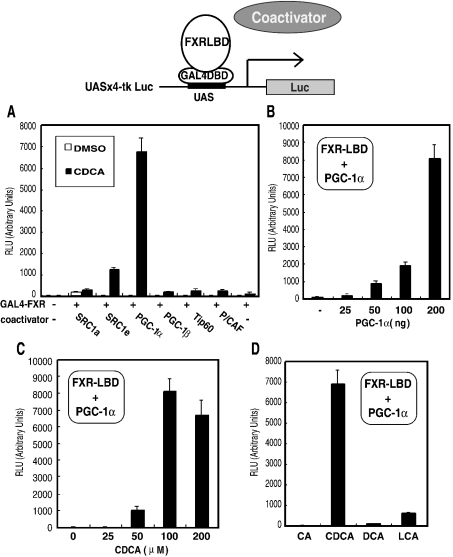
To search for co-activators of FXR, we used a chimaeric protein composed of the FXR–LBD (amino acids 192–472) fused to the yeast GAL4–DBD (GAL4–FXR chimaera). Transient transfection assays were performed in COS-7 cells, using a luciferase reporter gene with the GAL4–FXR chimaera and several nuclear receptor co-activators. Among the six co-activators that we examined, we found that PGC-1α markedly enhanced the transcriptional activity of the GAL4–FXR chimaera in the presence of 100 μM CDCA, producing an increase of approx. 50-fold as compared with that in the absence of co-activators (Figure 1A). SRC1e (steroid receptor co-activator 1e) also enhanced the FXR-mediated transcription to a lesser extent in the presence of 100 μM CDCA, with an approx. 10-fold enhancement as compared with that in the absence of co-activators (Figure 1A). The other co-activators, SRC1a, PGC-1β, Tip60 and P/CAF, showed an approx. 2-fold enhancement (Figure 1A). Conversely, in the absence of CDCA, transcriptional activation by the co-activators was hardly observed, except for SRC1a (open bar in Figure 1A).
Figure 1. PGC-1α enhances the transcriptional activation of GAL4–FXR most effectively, among SRC1a, SRC1e, PGC-1α, PGC-1β, Tip60 and P/CAF, in a ligand-dependent manner.
(A) Transfections of COS-7 cells and measurements of luciferase activity were carried out as described in the Experimental section using pCMX-GAL4-FXR (200 ng), UASx4-tk Luc reporter construct (400 ng), co-activator expression construct (200 ng) and pEYFP-C1 (200 ng), in the absence (open bars) or presence (closed bars) of 100 μM CDCA. (B) The pCMX-flag-PGC-1α expression plasmid used for transfection was at amounts of between 0 and 200 ng, and similar experiments were performed. (C) The transfection was carried out using pCMX-GAL4-FXR (200 ng), UASx4-tk Luc reporter construct (400 ng) or pCMX-flag-PGC-1α (200 ng) in the presence of increasing amounts of CDCA from 0 to 200 μM. (D) Similar experiments were carried out in the presence of 100 μM CA (cholic acid), CDCA, DCA or LCA.
The co-activation of GAL4–FXR chimaera-mediated transcription by PGC-1α was analysed by co-transfection of increasing amounts of the PGC-1α expression plasmids (0, 25, 50, 100 and 200 ng). With 200 ng of the PGC-1α expression plasmid, the GAL4–FXR chimaera-mediated transcription was enhanced approx. 60-fold, as compared with that in the absence of PGC-1α (Figure 1B). We also performed transient transfection assays with the GAL4–FXR chimaera and PGC-1α in the presence of increasing amounts of CDCA (0, 25, 50, 100 and 200 μM). The GAL4–FXR chimaera and PGC-1α-mediated transcription of the reporter gene was activated most in the presence of 100 μM CDCA (Figure 1C). We next examined the ability of different physiologically relevant bile acids, such as CA (cholic acid), DCA (deoxycholic acid) and LCA (lithocholic acid), to activate transcription of the reporter gene with the GAL4–FXR chimaera and PGC-1α, and compared these results with that obtained with CDCA. The secondary bile acids, DCA and LCA, were both moderate activators of the reporter gene in the presence of the GAL4–FXR chimaera and PGC-1α (Figure 1D). These observations (Figure 1) demonstrate that GAL4–FXR chimaera-mediated transcription is strongly co-activated by PGC-1α in a ligand-dependent manner.
PGC-1α activates FXR–RXRα heterodimer-mediated transcription of SHP in a ligand-dependent manner
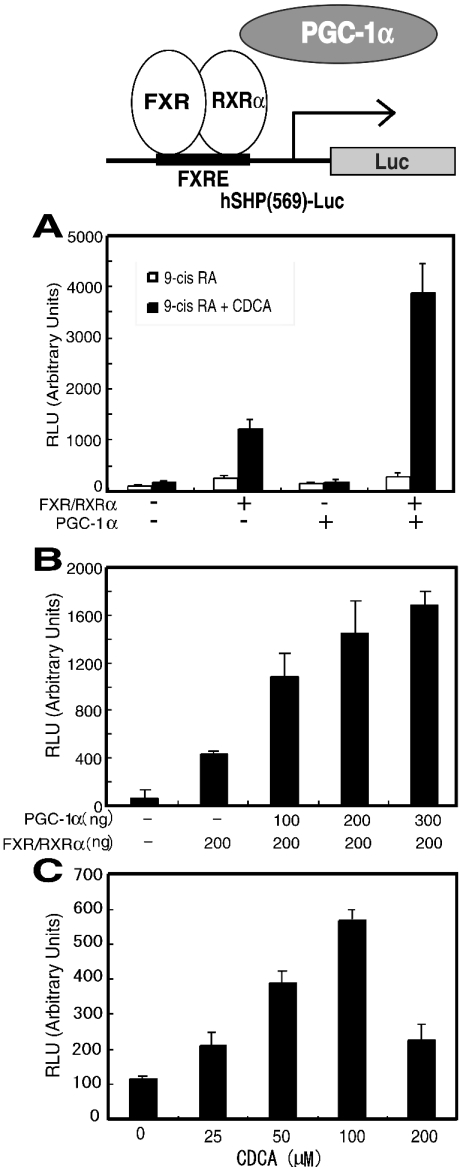
FXR regulates gene transcription as a heterodimer with RXRα. We next sought to determine whether the FXR–RXRα heterodimer-mediated transcriptional activation is enhanced by PGC-1α. FXR-mediated transcriptional activation has been reported for several genes [9,10,14,35,37–41]. Among them, we chose the SHP gene promoter for the cell-based assays. The reporter construct that we used contains the SHP promoter region (bases −569 to +10) and the coding region of the luciferase reporter gene [10]. This reporter gene was transiently transfected into COS-7 cells together with the FXR and RXRα expression vectors, and/or the PGC-1α expression vector, and the cells were treated with 9-cis-retinoic acid alone or with both 9-cis-retinoic acid and CDCA. As shown in Figure 2(A), reporter gene transcriptional activity by FXR and RXRα in the presence of 100 μM CDCA was enhanced approx. 6-fold, as compared with that in the absence of FXR and RXRα, or that with PGC-1α alone. In the presence of 100 μM CDCA, the addition of the PGC-1α expression vector resulted in an approx. 3-fold enhancement of FXR–RXRα-mediated transcription of the reporter gene, as compared with that without PGC-1α (Figure 2A, closed bars). In the absence of CDCA, the enhancement of FXR–RXRα-mediated transcription by PGC-1α was low (Figure 2A, open bars). The co-activation of FXR–RXRα-mediated transcription by PGC-1α was analysed by co-transfection of increasing amounts of PGC-1α (0, 100, 200 and 300 ng). The FXR–RXRα-mediated transcription was enhanced with increasing amounts of the transfected PGC-1α expression vector (Figure 2B). We also performed transient transfection assays with FXR–RXRα and PGC-1α in the presence of increasing amounts of CDCA (0, 25, 50, 100 and 200 μM). The FXR–RXRα heterodimer- and PGC-1α-mediated transcription of the SHP gene was activated most in the presence of 100 μM CDCA (Figure 2C). These observations suggest that the SHP gene is regulated by the FXR–RXRα heterodimer, and that PGC-1α effectively enhances FXR–RXRα-mediated transcription of the SHP gene in a ligand-dependent manner.
Figure 2. PGC-1α enhances FXR–RXRα heterodimer-mediated transcription in a ligand-dependent manner.
(A) Transfections and measurements of luciferase activities were carried out using pCMX-FXR (200 ng), pCMX-RXRα (200 ng), pGL3-hSHP(569)-Luc (400 ng), pCMX-flag-PGC-1α (200 ng) and pEYFP-C1 (200 ng), in the presence of either 1 μM 9-cis-retinoic acid (open bars) or 100 μM CDCA and 1 μM 9-cis-retinoic acid (closed bars). (B) The experiments were carried out using pCMX-FXR (100 ng), pCMX-RXRα (100 ng), pGL3-hSHP(569)-Luc (200 ng), pEYFP-C1 (100 ng) and increasing amounts of pCMX-flag-PGC-1α (100, 200 and 300 ng), in the presence of 100 μM CDCA and 1 μM 9-cis-retinoic acid. (C) The experiments were carried out using pCMX-FXR (100 ng), pCMX-RXRα (100 ng), pGL3-hSHP(569)-Luc (200 ng), pEYFP-C1 (100 ng) and pCMX-flag-PGC-1α (100 ng) in the presence of increasing amounts of CDCA (25, 50, 100 and 200 μM) and 1 μM 9-cis-retinoic acid.
SHP inhibits PGC-1α co-activation of the FXR–RXRα heterodimer
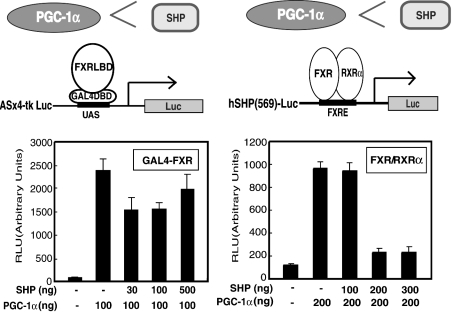
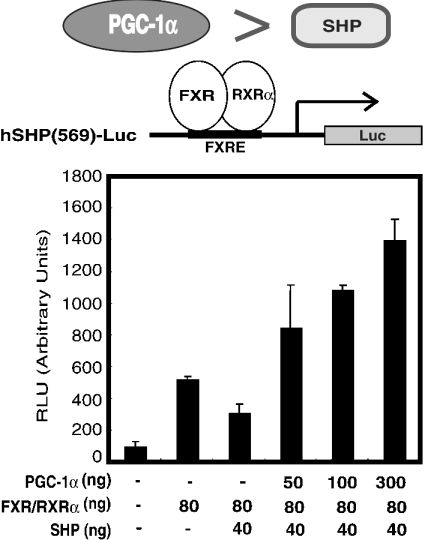
SHP is thought to be a common repressor of nuclear receptors involved in bile acid biosynthesis. Thus we next examined whether or not SHP would suppress the ability of PGC-1α to potentiate the ligand-induced activity of FXR. GAL4–FXR chimaera-mediated transcriptional activity was effectively enhanced when PGC-1α was co-expressed in COS-7 cells (Figure 3A). However, clear inhibition was not observed by adding increasing amounts of the SHP expression plasmid (30, 100 and 500 ng) to the cells transfected with the expression plasmids for the GAL4–FXR chimaera and PGC-1α (Figure 3A). In contrast, the FXR–RXRα heterodimer-mediated transcription activated by PGC-1α was clearly inhibited by adding increasing amounts of SHP (200 and 300 ng) (Figure 3B). The inhibitory effect of SHP (40 ng) on FXR–RXRα-mediated transcription of SHP was overcome by the addition of increasing amounts of PGC-1α (50, 100 and 300 ng) (Figure 4).
Figure 3. SHP clearly inhibits co-activation of FXR–RXRα heterodimer-mediated transcription by PGC-1α, in contrast with its effect on GAL4–FXR-mediated transcriptional co-activation by SHP.
(A) Transfections and measurements of luciferase activities were carried out using pCMX-GAL4-FXR (100 ng), UASx4-tk Luc reporter construct (200 ng), pCMX-flag-PGC-1α (100 ng), pEYFP-C1 (100 ng) and increasing amounts of pCMX-SHP (30, 100 and 500 ng) in the presence of 100 μM CDCA. (B) Similar experiments were carried out using pCMX-FXR (100 ng), pCMX-RXRα (100 ng), pGL3-hSHP(569)-Luc (200 ng), pCMX-flag-PGC-1α (200 ng), pEYFP-C1 (100 ng) and increasing amounts of pCMX-SHP (100, 200 and 300 ng) in the presence of 1 μM 9-cis-retinoic acid and 100 μM CDCA.
Figure 4. The inhibitory effect of SHP on FXR–RXRα-mediated transcription is overcome by the addition of increasing amounts of PGC-1α.
The experiments were carried out using pCMX-FXR (40 ng), pCMX-RXRα (40 ng), pEYFP-C1 (100 ng), pCMX-SHP (0 or 40 ng) and increasing amounts of pCMX-flag-PGC-1α (50, 100 and 300 ng) in the presence of 1 μM 9-cis-retinoic acid and 100 μM CDCA.
Structural changes of FXR–RXRα heterodimer proteins by the addition of CDCA
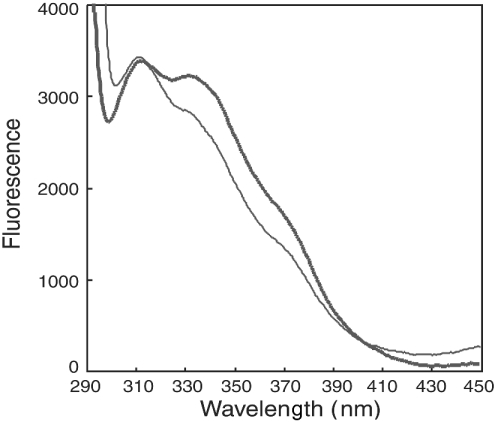
Since FXR has two tryptophan residues at positions 454 and 469 around the region for activation function 2, we expected to observe a spectral change in the intrinsic fluorescence, due to the structural change caused by the addition of CDCA. The fluorescence spectra of the proteins composed of FXR–RXRα-LBD heterodimer were measured in the presence and absence of CDCA. As shown in Figure 5, the fluorescence spectrum of the FXR–RXRα heterodimer in the absence of CDCA differed from that in the presence of 100 μM CDCA. These observations suggest that the addition of CDCA induces a structural change in the FXR–RXRα heterodimer proteins. It probably results from helix 12 adopting the active form, as reported previously [42].
Figure 5. The intrinsic fluorescence spectrum of the FXR–RXRα heterodimer is changed by the addition of CDCA.
The intrinsic fluorescence spectra of FXR–RXRα–LBD heterodimer proteins were measured, in the absence (thick line) or presence of 100 μM CDCA (thin line), on a Hitachi F-4500 fluorescence spectrophotometer. The excitation wavelength was 280 nm, and the emission was monitored from 280 to 350 nm.
The FXR–LBD and PGC-1α proteins interact directly in vivo and in vitro in a ligand-influenced manner
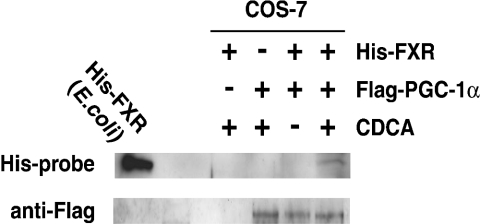
We next examined whether or not the FXR and PGC-1α proteins interact directly in vivo. A co-immunoprecipitation experiment was performed using His-tagged FXR (amino acids 192–472) and FLAG-tagged PGC-1α-transfected COS-7 cell extract in the presence or absence of CDCA with an anti-FLAG antibody-conjugated affinity gel. A band corresponding to FXR protein was observed only in the presence of FXR, PGC-1α and CDCA, but was not observed in the absence of CDCA using His-probe (Figure 6). Migration of this band was almost the same as that of a band from the His-tagged FXR protein (amino acids 192–472) expressed in bacteria (Figure 6). An anti-FLAG antibody was used to reveal the presence of PGC-1α in the immunoprecipitate. The bands corresponding to PGC-1α protein were observed in the presence and absence of CDCA (Figure 6). These results showed that FXR and PGC-1α interacted in vivo in a ligand-dependent manner.
Figure 6. FXR interacts with PGC-1α in vivo in a ligand-dependent manner.
COS-7 cells were transfected with two expression plasmids of pCMX, pCMX containing the coding sequence for the expression of His-tagged FXR (192–472), and that for the expression of FLAG-tagged PGC-1α, and subsequently treated with CDCA (100 μM) or DMSO (0.1%). Total cell extracts were prepared as described in the Experimental section, and subjected to immunoprecipitation using an anti-FLAG antibody-conjugated affinity gel in the presence or absence of CDCA. FXR was detected by Western blotting using a His-Probe. PGC-1α was detected using an anti-FLAG antibody.
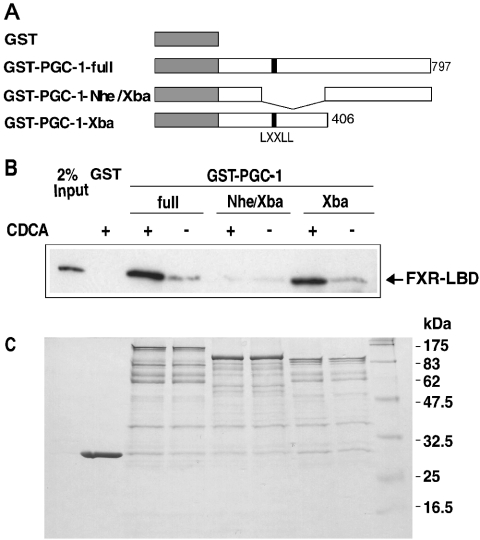
We next examined whether the FXR–LBD protein and PGC-1α protein interact directly in vitro. GST and three fusion proteins, GST–PGC-1-full, GST–PGC-1-Nhe/Xba and GST–PGC-1-Xba, were each expressed in bacteria, and then were immobilized on glutathione–Sepharose beads. GST–PGC-1-full is a fusion protein composed of GST and the full-length PGC-1α (amino acids 1–797). The GST–PGC-1-Nhe/Xba protein lacks the region corresponding to amino acid residues 87–406, which contains the LXXLL motif sequence for interactions with nuclear receptors. GST–PGC-1-Xba is a fusion protein composed of GST and the C-terminal-half deletion mutant of PGC-1α (amino acids 1–406) as shown in Figure 7(A). Western blotting analyses revealed that the immobilized GST–PGC-1-full and GST–PGC-1-Xba proteins strongly interact with the His-tagged FXR–LBD protein in the presence of CDCA (Figure 7B), whereas weak interactions between the FXR–LBD protein and each GST–PGC-α 1α fusion protein were observed in the absence of CDCA. Interaction between the LXXLL-motif-deletion mutant, GST–PGC-1-Nhe/Xba and FXR–LBD was weak in both the presence and absence of CDCA. No interaction was observed between the FXR–LBD protein and GST (Figure 7B). These results demonstrate that the FXR–LBD protein interacts directly with PGC-1α, and CDCA increases the association between the FXR and PGC-1α proteins.
Figure 7. FXR interacts with PGC-1α in a ligand-influenced manner.
(A) Schematic representation of the GST-fused PGC-1α full-length protein (1–797) and two deletion mutants (Δ87–406 and Δ406–797), which were used for pull-down experiments. (B) Western blot. GST pull-downs were performed as described in the Experimental section, using GST alone, GST-PGC-1-full (1–797), GST-PGC-1-Nhe/Xba (Δ87–406) or GST-PGC-1-Xba (1–406) with an N-terminal His-tagged FXR–LBD protein (217–472). FXR was detected using a His-Probe. The CDCA-dependent interaction between FXR (217–472) and PGC-1α was confirmed by repeated assays. (C) SDS/PAGE. A half portion of each sample was analysed for loading controls.
DISCUSSION
Among the nuclear receptor co-activators that we studied, PGC-1α strongly increased GAL4–FXR chimaera-mediated transcription, in a strict, ligand-dependent, manner (Figure 1). Accelerated activation of GAL4–FXR by PGC-1α was observed by the increase of PGC-1α (Figure 1B) or CDCA (Figure 1C), whereas activation of FXR–RXRα was not accelerated (Figures 2B and 2C). These results possibly suggest that interaction between GAL4–FXR chimaera and PGC-1α might be different from that between FXR–RXRα and PGC-1α. Enhancement of the protein–protein interaction between FXR and PGC-1α by CDCA (Figure 7) suggests that the CDCA-induced structural change of FXR (Figure 5) is involved in the co-activation of FXR-mediated transcription by PGC-1α.
Zhang et al. [35] reported that FXR–DBD and PGC-1α interact in a ligand-independent manner. The region corresponding to the DBD of FXR is sufficient for the interaction with PGC-1α (1–400) [35]. We showed that PGC-1α (1–406) interacts with FXR–LBD (217–472) in a ligand-dependent manner (Figure 7). These results probably suggest that the DBD of FXR interacts basically with PGC-1α in the absence or ineffective concentrations of ligands, and the interaction between the LBD of FXR and PGC-1α takes part in the presence of effective concentrations of ligands. Multiple interaction sites seem to be present between FXR and PGC-1α.
It is interesting to note that PGC-1α is induced in liver by fasting [32], and that CYP7A1 expression is reduced during starvation [18–21]. Since the reduction of CYP7A1 is probably regulated mainly by FXR and SHP [9,10,15], it is possible that CYP7A1 repression during starvation [18–21] could be induced by PGC-1α coupling to FXR. De Fabiani et al. [23] recently reported that bile acids suppress CYP7A1 and PEPCK transcription by impairing co-activator recruitment and by inducing dissociation of nuclear receptors and co-activators. Based on this observation, the regulation of bile acids in hepatocytes seems to be required for the activation of gluconeogenesis. CYP7A1 repression during starvation [18–21] is probably programmed for the activation of gluconeogenesis.
Among the target genes of FXR [9,10,14,37–41], we chose the SHP gene promoter for the reporter construct. SHP is now considered to be a common repressor for nuclear receptors, including LRH-1 and HNF-4α, in regulating bile acid synthesis genes, including CYP7A1 and CYP8B1 [43]. Our assays demonstrated that PGC-1α enhances the FXR–RXRα-mediated transcription of the SHP gene approx. 3-fold, in a ligand-dependent manner (Figure 2A). We have no evidence that FXR-mediated transcription of the SHP gene is enhanced by fasting; however, it is quite likely that SHP activation by FXR is involved in the CYP7A1 repression caused by starvation [18–21] because of the significant inverse relationship between the expression level of SHP and that of CYP7A1 reported previously [15–17]. Meanwhile, Shin et al. [22] reported different, yet convincing, results that CYP7A1 transcription was induced by fasting and stimulated by PGC-1α. Similar results were reported by De Fabiani et al. [23] and Hunt et al. [24]. In their studies, SHP was not induced and the FXR level was not reported, suggesting that the inhibitory effect on the CYP7A1 promoter through the FXR–SHP pathway had not occurred, or was circumvented by other positive elements. CYP7A1 transcription might be differently regulated in different species, because the repression of CYP7A1 expression by fasting was observed in rat and piglet [18–21], while the studies reporting activation of CYP7A1 were performed in mice [22–24]. CYP7A1 gene expression seems to be intricately tuned with nuclear receptors [44] and undefined mediators.
SHP represses the transcription of nuclear receptors by forming a heterodimer, or by competing with co-activators [45,46]. The results shown in Figure 3 suggest that SHP probably prevents FXR–RXRα heterodimer formation in the transcription. Since FXR-mediated transcriptional co-activation of the SHP gene by PGC-1α is ligand-dependent (Figures 2A and 2C), and is inhibited by the protein product SHP (Figure 3B), the physiological level of SHP is probably regulated positively by the amounts of bile acids, and negatively by that of the protein product SHP. PGC-1α can overcome the inhibitory effect of SHP as shown in Figure 4. PGC-1α, induced in response to environmental stimuli, could force FXR–RXRα-mediated SHP transcriptional machinery to produce SHP and repress the CYP7A1 gene. This might correspond to the marked decrease of CYP7A1 expression during starvation [18–21]. On the other hand, the co-activation of the orphan receptor HNF-4α by PGC-1α is powerful and ligand-independent [47], and probably depends on the amount of PGC-1α protein induced in response to environmental stimuli [34].
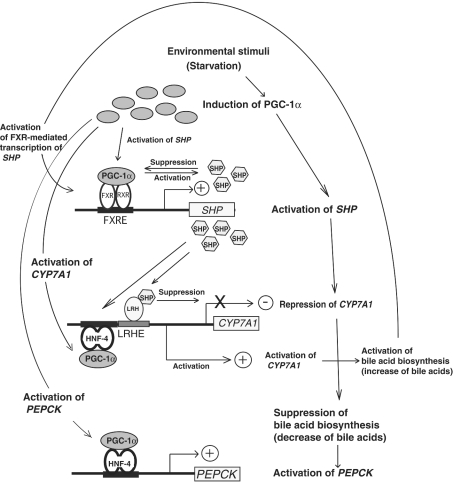
Since bile acid synthesis is the major cholesterol excretion route from hepatocytes, expression of the rate-limiting enzyme CYP7A1 is tightly regulated through multiple pathways to keep adequate levels of cellular cholesterol. When hepatocytes are exposed to the stressful conditions such as starvation or elevated cAMP, PGC-1α is induced in the liver [32]. The induced PGC-1α could co-activate FXR–RXRα-mediated SHP transcription, HNF-4α-mediated CYP7A1 transcription [22] and HNF-4α-mediated PEPCK transcription [32] as shown in Figure 8. Co-activation of HNF-4α-mediated CYP7A1 transcription by PGC-1α is followed by the further activation of bile acid biosynthesis and the increase of bile acid content. In the presence of bile acids with adequate concentrations, co-activation of FXR–RXRα-mediated SHP transcription by PGC-1α produces the SHP protein, inhibits the activity of LRH-1, and represses CYP7A1 transcription [9,10] (Figure 8). The presence of this re-suppression by PGC-1α through the FXR–SHP pathway underlies that the bile acid pool must be kept small to promote and provide an adequate condition for the activation of PEPCK transcription mediated by HNF-4α and PGC-1α as reported by De Fabiani et al. [23] (Figure 8).
Figure 8. A model for the relation between FXR-mediated transcription to regulate the bile acid biosynthesis and PGC-1α-mediated transcription.
A model is presented for the possible relation among the induction of PGC-1α by the environmental stimuli such as starvation [32], co-activation of FXR-mediated SHP transcription by PGC-1α, transcriptional repression of CYP7A1 by the suppression of LRH-1 by SHP [9,10] and suppression of bile acid biosynthesis, co-activation of HNF-4α-mediated CYP7A1 transcription [43] by PGC-1α and activation of bile acid biosynthesis, and activation of PEPCK by the coupling of HNF-4α and PGC-1α [32].
In conclusion, our study showed that the nuclear receptor FXR is efficiently co-activated by PGC-1α in a ligand-dependent manner. Our observations suggest that PGC-1α functions as a ligand-dependent co-activator of FXR.
Acknowledgments
We gratefully acknowledge Dr D.J. Mangelsdorf for the gifts of plasmids pCMX-hFXR, pCMX-hRXRα and pGL3-hSHP (569)-Luc. We also gratefully acknowledge Dr S. Salam for the gifts of plasmids pSG5-SRC1a and pSG5-SRC1e. We thank Dr Y. Suzuki and Dr E. Moriyoshi for helpful comments. This study was supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO).
References
- 1.Makishima M., Okamoto A. Y., Repa J. J., Tu H., Learned R. M., Luk A., Hull M. V., Lustig K. D., Mangelsdorf D. J., Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 2.Parks D. J., Blanchard S. G., Bledsoe R. K., Chandra G., Consler T. G., Kliewer S. A., Stimmel J. B., Willson T. M., Zavacki A. M., Moore D. D., Lehmann J. M. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 3.Wang H., Chen J., Hollister K., Sowers L. C., Forman B. M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 4.Russell D. W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003;72:137–174. doi: 10.1146/annurev.biochem.72.121801.161712. [DOI] [PubMed] [Google Scholar]
- 5.Russell D. W., Setchell K. D. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- 6.Wang L., Lee Y. K., Bundman D., Han Y., Thevananther S., Kim C. S., Chua S. S., Wei P., Heyman R. A., Karin M., Moore D. D. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 7.Stravitz R. T., Vlahcevic Z. R., Gurley E. C., Hylemon P. B. Repression of cholesterol 7α-hydroxylase transcription by bile acids is mediated through protein kinase C in primary cultures of rat hepatocytes. J. Lipid Res. 1995;36:1359–1369. [PubMed] [Google Scholar]
- 8.Antes T. J., Chen J., Cooper A. D., Levy-Wilson B. The nuclear matrix protein CDP represses hepatic transcription of the human cholesterol-7α hydroxylase gene. J. Biol. Chem. 2000;275:26649–26660. doi: 10.1074/jbc.M002852200. [DOI] [PubMed] [Google Scholar]
- 9.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 10.Lu T. T., Makishima M., Repa J. J., Schoonjans K., Kerr T. A., Auwerx J., Mangelsdorf D. J. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 11.Miyake J. H., Wang S. L., Davis R. A. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7α-hydroxylase. J. Biol. Chem. 2000;275:21805–21808. doi: 10.1074/jbc.C000275200. [DOI] [PubMed] [Google Scholar]
- 12.De Fabiani E., Mitro N., Anzulovich A. C., Pinelli A., Galli G., Crestani M. The negative effects of bile acids and tumor necrosis factor-α on the transcription of cholesterol 7α-hydroxylase gene (CYP7A1) converge to hepatic nuclear factor-4: a novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J. Biol. Chem. 2001;276:30708–30716. doi: 10.1074/jbc.M103270200. [DOI] [PubMed] [Google Scholar]
- 13.Kerr T. A., Saeki S., Schneider M., Schaefer K., Berdy S., Redder T., Shan B., Russell D. W., Schwarz M. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holt J. A., Luo G., Billin A. N., Bisi J., McNeill Y. Y., Kozarsky K. F., Donahee M., Wang D. Y., Mansfield T. A., Kliewer S. A., et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu G., Pan L. X., Li H., Forman B. M., Erickson S. K., Shefer S., Bollineni J., Batta A. K., Christie J., Wang T. H., et al. Regulation of the farnesoid X receptor bile acid repression. J. Biol. Chem. 2002;276:41690–41699. doi: 10.1074/jbc.M209176200. [DOI] [PubMed] [Google Scholar]
- 16.Kok T., Hulzebos C. V., Wolters H., Havinga R., Agellon L. B., Stellaard F., Shan B., Schwarz M., Kuipers F. Enterohepatic circulation of bile salts in farnesoid X receptor-deficient mice: efficient intestinal bile salt absorption in the absence of ileal bile acid-binding protein. J. Biol. Chem. 2003;278:41930–41937. doi: 10.1074/jbc.M306309200. [DOI] [PubMed] [Google Scholar]
- 17.Dussault I., Beard R., Lin M., Hollister K., Chen J., Xiao J. H., Chandraratna R., Forman B. M. Identification of gene-selective modulators of the bile acid receptor FXR. J. Biol. Chem. 2003;278:7027–7033. doi: 10.1074/jbc.M209863200. [DOI] [PubMed] [Google Scholar]
- 18.Lewis D. S., Oren S., Wang X., Moyer M. L., Beitz D. C., Knight T. J., Mott G. E. Developmental changes in cholesterol 7α- and 27-hydroxylases in the piglet. J. Anim. Sci. 2000;78:943–951. doi: 10.2527/2000.784943x. [DOI] [PubMed] [Google Scholar]
- 19.Bjorkhem I., Akerlund J. E. Studies on the link between HMG-CoA reductase and cholesterol 7α-hydroxylase in rat liver. J. Lipid Res. 1988;29:136–143. [PubMed] [Google Scholar]
- 20.Noshiro M., Nishimoto M., Okuda K. Rat liver cholesterol 7α-hydroxylase. Pretranslational regulation for circadian rhythm. J. Biol. Chem. 1990;265:10036–10041. [PubMed] [Google Scholar]
- 21.Nguyen L. B., Shefer S., Salen G., Ness G., Tanaka R. D., Packin V., Thomas P., Shore V., Batta A. Purification of cholesterol 7α-hydroxylase from human and rat liver and production of inhibiting polyclonal antibodies. J. Biol. Chem. 1990;265:4541–4546. [PubMed] [Google Scholar]
- 22.Shin D. J., Campos J. A., Gil G., Osborne T. F. PGC-1α activates CYP7A1 and bile acid biosynthesis. J. Biol. Chem. 2003;278:50047–50052. doi: 10.1074/jbc.M309736200. [DOI] [PubMed] [Google Scholar]
- 23.De Fabiani E., Mitro N., Gilardi F., Caruso D., Galli G., Crestani M. Coordinated control of cholesterol catabolism to bile acids and of gluconeogenesis via a novel mechanism of transcription regulation linked to the fasted-to-fed cycle. J. Biol. Chem. 2003;278:39124–39132. doi: 10.1074/jbc.M305079200. [DOI] [PubMed] [Google Scholar]
- 24.Hunt M. C., Yang Y. Z., Eggertsen G., Carneheim C. M., Gafvels M., Einarsson C., Alexson S. E. The peroxisome proliferator-activated receptor α (PPARα) regulates bile acid biosynthesis. J. Biol. Chem. 2000;275:28947–28953. doi: 10.1074/jbc.M002782200. [DOI] [PubMed] [Google Scholar]
- 25.Puigserver P., Spiegelman B. M. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y., Delerive P., Chin W. W., Burris T. P. Requirement of helix 1 and the AF-2 domain of the thyroid hormone receptor for coactivation by PGC-1. J. Biol. Chem. 2002;277:8898–8905. doi: 10.1074/jbc.M110761200. [DOI] [PubMed] [Google Scholar]
- 27.Delerive P., Wu Y., Burris T. P., Chin W. W., Suen C. S. PGC-1 functions as a transcriptional coactivator for the retinoid X receptors. J. Biol. Chem. 2002;277:3913–3917. doi: 10.1074/jbc.M109409200. [DOI] [PubMed] [Google Scholar]
- 28.Tcherepanova I., Puigserver P., Norris J. D., Spiegelman B. M., McDonnell D. P. Modulation of estrogen receptor-α transcriptional activity by the coactivator PGC-1. J. Biol. Chem. 2000;275:16302–16308. doi: 10.1074/jbc.M001364200. [DOI] [PubMed] [Google Scholar]
- 29.Knutti D., Kaul A., Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol. Cell. Biol. 2000;20:2411–2422. doi: 10.1128/mcb.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vega R. B., Huss J. M., Kelly D. P. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiraki T., Sakai N., Kanaya E., Jingami H. Activation of orphan nuclear constitutive androstane receptor requires subnuclear targeting by peroxisome proliferator-activated receptor-γ coactivator-1α: a possible link between xenobiotic response and nutritional state. J. Biol. Chem. 2003;278:11344–11350. doi: 10.1074/jbc.M212859200. [DOI] [PubMed] [Google Scholar]
- 32.Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature (London) 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 33.Rhee J., Inoue Y., Yoon J. C., Puigserver P., Fan M., Gonzalez F. J., Spiegelman B. M. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Castellani L. W., Sinal C. J., Gonzalez F. J., Edwards P. A. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18:157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamei Y., Ohizumi H., Fujitani Y., Nemoto T., Tanaka T., Takahashi N., Kawada T., Miyoshi M., Ezaki O., Kakizuka A. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ananthanarayanan M., Balasubramanian N., Makishima M., Mangelsdorf D. J., Suchy F. J. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J. Biol. Chem. 2001;276:28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- 38.Grober J., Zaghini I., Fujii H., Jones S. A., Kliewer S. A., Willson T. M., Ono T., Besnard P. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene: involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J. Biol. Chem. 1999;274:29749–29754. doi: 10.1074/jbc.274.42.29749. [DOI] [PubMed] [Google Scholar]
- 39.Song C. S., Echchgadda I., Baek B. S., Ahn S. C., Oh T., Roy A. K., Chatterjee B. Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid X receptor. J. Biol. Chem. 2001;276:42549–42556. doi: 10.1074/jbc.M107557200. [DOI] [PubMed] [Google Scholar]
- 40.Zhao A., Lew J. L., Huang L., Yu J., Zhang T., Hrywna Y., Thompson J. R., Pedro N. D., Blevins R. A., Peláez F., Wright S. D., Cui J. Human kininogen gene is transactivated by the farnesoid X receptor. J. Biol. Chem. 2003;278:28765–28770. doi: 10.1074/jbc.M304568200. [DOI] [PubMed] [Google Scholar]
- 41.Urizar N. L., Dowhan D. H., Moore D. D. The farnesoid X-activated receptor mediates bile acid activation of phospholipid transfer protein gene expression. J. Biol. Chem. 2000;275:39313–39317. doi: 10.1074/jbc.M007998200. [DOI] [PubMed] [Google Scholar]
- 42.Bourguet W., Germain P., Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol. Sci. 2000;21:381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- 43.Zhang M., Chiang J. Y. Transcriptional regulation of the human sterol 12α-hydroxylase gene (CYP8B1): roles of hepatocyte nuclear factor 4α in mediating bile acid repression. J. Biol. Chem. 2001;276:41690–41699. doi: 10.1074/jbc.M105117200. [DOI] [PubMed] [Google Scholar]
- 44.Chiang J. Y., Kimmel R., Stroup D. Regulation of cholesterol 7α-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRα) Gene. 2001;262:257–265. doi: 10.1016/s0378-1119(00)00518-7. [DOI] [PubMed] [Google Scholar]
- 45.Johansson L., Bavner A., Thomsen J. S., Farnegardh M., Gustafsson J. A., Treuter E. The orphan nuclear receptor SHP utilizes conserved LXXLL-related motifs for interactions with ligand-activated estrogen receptors. Mol. Cell. Biol. 2000;20:1124–1133. doi: 10.1128/mcb.20.4.1124-1133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee Y. K., Dell H., Dowhan D. H., Hadzopoulou-Cladaras M., Moore D. D. The orphan nuclear receptor SHP inhibits hepatocyte nuclear factor 4 and retinoid X receptor transactivation: two mechanisms for repression. Mol. Cell. Biol. 2000;20:187–195. doi: 10.1128/mcb.20.1.187-195.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borgius L. J., Steffensen K. R., Gustafsson J. A., Treuter E. Glucocorticoid signaling is perturbed by the atypical orphan receptor and corepressor SHP. J. Biol. Chem. 2002;277:49761–49766. doi: 10.1074/jbc.M205641200. [DOI] [PubMed] [Google Scholar]