Abstract
20α-Hydroxysteroid dehydrogenase (20α-HSD), which metabolizes progesterone to an inactive steroid in the corpus luteum of mice and rats but not of humans, is thought to play a crucial role in shortening the oestrous cycles in these rodent species. We determined the nucleotide sequence of the 5′-flanking region of the mouse 20α-HSD gene, and examined its promoter activity using a rat luteinized granulosa cell culture. A reporter assay, using reporter constructs of various lengths of the 5′-flanking region, revealed that the region between −83 and 60 bp upstream of the transcription start site was essential for transcriptional activity. Furthermore, mutational analysis demonstrated that a putative Sp1 site in this region was critical to the expression of the reporter gene. Electrophoretic mobility-shift assays showed that the interaction of proteins in a nuclear extract from rat luteinized granulosa cells with this region was inhibited by a competitor having the wild-type Sp1 sequence in its promoter, but not a mutated Sp1 sequence. Supershift analysis confirmed that Sp1 and Sp3 were present in the nuclear extract of these cells, and that these factors bound to the element. Finally, promoter activity was elevated by the co-transfection of an Sp1 expression vector, and, to a lesser extent, by an Sp3 expression vector, supporting further the involvement of these factors in the expression of the 20α-HSD gene.
Keywords: 20α-hydroxysteroid dehydrogenase, ovary, promoter, Sp1, Sp3
Abbreviations: DMEM, Dulbecco's modified Eagle's medium; EMSA, electrophoretic mobility-shift assay; FBS, fetal bovine serum; hCG, human chorionic gonadotropin; 20α-HSD, 20α-hydroxysteroid dehydrogenase; PGF2α, prostaglandin F2α; PMSG, pregnant mare's serum gonadotropin; PRL, prolactin; STAT, signal transducers and activators of transcription; TESS, Transcription Element Search Software
INTRODUCTION
20α-Hydroxysteroid dehydrogenase (20α-HSD) is an enzyme that converts progesterone into a biologically inactive steroid, 20α-dihydroprogesterone. In rats, 20α-HSD is expressed in the corpus luteum, and increases in level during functional luteolysis, such as in oestrous cycles and at the end of pregnancy [1,2]. Because there is a good correlation between the rise in 20α-HSD expression and the decrease in serum progesterone [2–4], it is considered that 20α-HSD plays an crucial role in the regulation of progesterone secretion in these rodent species.
Progesterone production in the rodent corpus luteum is regulated by hormones, including PRL (prolactin) and PGF2α (prostaglandin F2α), which have luteotropic and luteolytic functions respectively [5]. Studies using genetically engineered mice lacking receptors for these hormones have shown that they play essential roles in the completion of pregnancy [6,7], one of which is the regulation of 20α-HSD expression. PRL suppresses 20α-HSD expression, and progesterone secretion is maintained in the first half of pregnancy, whereas PGF2α stimulates 20α-HSD expression at the end of pregnancy [8,9] and thereby decreases progesterone secretion.
20α-HSD belongs to the aldo–keto reductase superfamily, which consists of monomeric NAD(P)(H)-dependent oxido-reductases with a variety of substrate specificities, including steroids, prostaglandins, carbohydrates and xenobiotics in the environment [10], and it is supposed that these members are derived from a common ancestral gene. Therefore the regulation of 20α-HSD is also an interesting issue from the point of view that 20α-HSD is the enzyme which has acquired a specific role in the reproductive strategy of rats and mice through the process of molecular evolution.
From this background, we investigated the mechanisms of 20α-HSD expression in luteal cells using the mouse 20α-HSD gene 5′-flanking region, which was cloned by screening of the 129 SvJ mouse genomic library [11]. We show here that a GC box in the 5′-flanking region of the mouse 20α-HSD gene is essential for its promoter activity, and that Sp1 and Sp3 are involved in the expression of the 20α-HSD gene in the corpus luteum.
MATERIALS AND METHODS
Reagents
Restriction enzymes, T4 DNA ligase and Klenow enzyme were purchased from Takara (Shiga, Japan). Oligonucleotides were obtained from Sigma Genosys Japan (Hokkaido, Japan). DpnI was obtained from New England Biolabs (Beverly, MA, U.S.A.). PMSG (pregnant mare's serum gonadotropin) and hCG (human chorionic gonadotropin) were purchased from Teikoku Zoki (Tokyo, Japan). DMEM (Dulbecco's modified Eagle's medium)/F-12 (1:1, v/v), FBS (fetal bovine serum), penicillin–streptomycin and Fungizone were obtained from Invitrogen (Carlsbad, CA, U.S.A.). Hepes and Mops were purchased from Dojin Chemicals (Kumamoto, Japan). Protease inhibitor cocktail was purchased from Sigma (St Louis, MO, U.S.A.). Poly(dI-dC) was obtained from Amersham Biosciences (Little Chalfont, Bucks., U.K.). Antibodies against Sp1 (PEP2) and Sp3 (D-20) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). pCMV-Sp1 and pCMV-Sp3 expression vectors were kindly provided by Dr G. Suske (University of Marburg, Germany). The other reagents were purchased from Wako (Osaka, Japan).
Determination of the sequence of the mouse 20α-HSD gene 5′-flanking region
The λFIX II mouse genomic library was screened, and 20α-HSD genomic clones containing the 5′-flanking region of the gene were isolated as described previously [11]. The 5′-flanking region of the mouse 20α-HSD gene was excised by restriction enzymes and subcloned into pBluescript SK plasmid (Stratagene, La Jolla, CA, U.S.A.). The nucleotide sequences were determined by digesting further with the appropriate restriction enzymes, subcloning into plasmids, and then sequencing.
DNA sequence analysis
The 5′-flanking sequences of the mouse 20α-HSD gene were compared with those of the rat 20α-HSD gene (accession no. AF084365) with DNASIS software (Hitachi, Tokyo, Japan). Transcription Element Search Software (TESS; http://www.cbil.upenn.edu/tess/) was used to analyse the 5′-flanking region of the mouse 20α-HSD gene.
Construction of luciferase reporter vectors
DNA fragments of sizes 2.6, 2.2, 1.7, 1.1 and 0.6 kb were excised from the pBluescript SK plasmid with EcoRV for the 3′ end, and with appropriate enzymes (KpnI, SalI, BamHI, BglII or SalI respectively) for the 5′ end. They were ligated to a pGL3-Basic luciferase reporter vector (Promega, Madison, WI, U.S.A.) and digested with NcoI, followed by a fill-in reaction and appropriate enzymes (KpnI, XhoI or BglII). DNA fragments of nt −255/+58, −132/+58, −103/+58, −92/+58, −83/+58, −60/+58 and −35/+58 were amplified by PCR using a 0.6 kb construct as a template, with primers as shown in Table 1. They were subcloned into pGEM-T vector (Promega), sequenced, digested with SalI and EcoRI and ligated to the vectors prepared from the 0.6 kb construct.
Table 1. Oligonucleotides used for construction of reporter vectors and EMSA.
| Use | Sense/antisense | Nucleotide sequence |
|---|---|---|
| pGL3-255 | Sense | 5′-CAATCCCTCGGTCTCTCATA-3′ |
| pGL3-132 | Sense | 5′-TGAGAATAAACGGTCTGGTG-3′ |
| pGL3-103 | Sense | 5′-TCCTCACAAGTAGAATGCTG-3′ |
| pGL3-92 | Sense | 5′-AGAATGCTGTAAGAGCTCAC-3′ |
| pGL3-83 | Sense | 5′-TAAGAGCTCACTCCGCCCCC-3′ |
| pGL3-60 | Sense | 5′-CCATTGACTGCTGATTACTC-3′ |
| pGL3-35 | Sense | 5′-CTTGGGGCTATAATTTGCAC-3′ |
| pGL3-255∼pGL3-35 | Antisense | 5′-CTTTATGTTTTTGGCGTCTTCC-3′ |
| pGL3-255(delSp1) | Sense | 5′-CCATTGACTGCTGATTACTC-3′ |
| pGL3-255(delSp1) | Antisense | 5′-GAGCTCTTACAGCATTCTAC-3′ |
| pGL3-255(mutSp1) | Sense | 5′-AGAGCTCACTCATAACCCGTGCCAT-3′ |
| pGL3-255(mutSp1) | Antisense | 5′-ATGGCACGGGTTATGAGTGAGCTCT-3′ |
| EMSA probe | Sense | 5′-GCTGTAAGAGCTCACTCCGCCCCCGTGCCATTGACTG-3′ |
| EMSA probe | Antisense | 5′-GCAGTCAATGGCACGGGGGCGGAGTGAGCTCTTACAG-3′ |
| Competitor | ||
| Wild-type | Sense | 5′-AGAGCTCACTCCGCCCCCGTGCCAT-3′ |
| Wild-type | Antisense | 5′-ATGGCACGGGGGCGGAGTGAGCTCT-3′ |
| Mutant | Sense | 5′-AGAGCTCACTCATAACCCGTGCCAT-3′ |
| Mutant | Antisense | 5′-ATGGCACGGGTTATGAGTGAGCTCT-3′ |
Introduction of mutations in a putative Sp1-binding site
Mutations of a putative Sp1-binding site were introduced by PCR using the primers shown in Table 1. PCR was performed with Pfu Turbo DNA polymerase (Stratagene) and pGEM-T, into which the DNA fragment of nt −255/+58 was subcloned as a template. For construction of the deletion mutant, primers were phosphorylated at the 5′ ends with T4 polynucleotide kinase before the reaction, and the PCR product was self-ligated with T4 DNA ligase. After reaction with DpnI for digestion of the template plasmid, the solution was used for transformation and the mutant plasmids were obtained. They were sequenced and subcloned into the luciferase vector, as described above.
Rat luteinized granulosa cell culture
Twenty 6-day-old immature female Wistar–Imamichi rats housed in our laboratory were injected subcutaneously with 50 units of PMSG, and 42 h later intraperitoneally with 10 units of hCG. After 6 h, the rats were killed by decapitation, and the ovaries were isolated and collected in DMEM/F12 (1:1, v/v) without FBS. In a clean bench, the follicles were punctured with a 27-gauge needle, and the luteinized granulosa cells were recovered. The cells were mechanically dispersed by pipetting, filtered through a nylon mesh, centrifuged and suspended in DMEM/F12 (1:1, v/v) with 5% (v/v) FBS. Luteinized granulosa cells were cultured on a collagen type I-coated dish (Iwaki, Chiba, Japan) at a density of 3×105 cells/ml. After a 24 h incubation, the medium was changed and the cells were cultured for an appropriate time [24, 48 and 84 h for Northern hybridization, 48 h for transfection, and 24–48 h for EMSA (electrophoretic mobility-shift assay) experiments respectively]. Cells (6×105 cells/well in 6-well plates, 1.5×105 cells/well in 24-well plates and 106 cells/well in 60 mm plates) were used for Northern hybridization, transfection and EMSA respectively.
Northern hybridization
Cells were washed with PBS, and total RNA was extracted with TRIzol® Reagent (Invitrogen), according to the manufacturer's instructions. RNA was precipitated with 2-propanol and dissolved in diethylpyrocarbonate-treated water. RNA (5–10 μg) was fractionated by electrophoresis in a 1% (w/v) agarose gel with Mops/formalin, and stained with ethidium bromide to confirm that equal amounts of RNA were loaded in each lane. After the gels were washed with distilled water, RNA was transferred to a Biodyne B nylon membrane (Pall, East Hills, NY, U.S.A.). Full-length rat 20α-HSD cDNA [12] was labelled with [α-32P]dCTP using a Random Primed Labeling Kit (Roche, Mannheim, Germany) and purified with a ProbeQuant G-50, allowed to proceed in 5×Denhardt's solution (where 1×Denhardt's solution is 0.02% Ficoll 400/0.02% polyvinylpyrrolidone/0.02% BSA), 6×SSC (where 1×SSC is 0.15 M NaCl/0.015 M sodium citrate), 0.5% SDS, 50% formamide and 100 μg/ml denatured fragmented salmon sperm DNA containing [α-32P]dCTP-labelled probe (1×106 c.p.m./ml) at 42 °C overnight. Membranes were washed with 2×SSC/0.1% SDS at 55 °C, and exposed to an X-ray film (Kodak, Tokyo, Japan).
Transfection experiments
Samples (0.75 μg) of each experimental vector plus 0.15 μg of pRL-TK control vector (Promega) were mixed with 3 μl of FuGENE6 Transfection Reagent (Roche) in 100 μl of Opti-MEM (Invitrogen) and incubated at room temperature (≈25 °C) for 15 min, after which the FuGENE6–DNA complex was added to each well containing cells that were pre-incubated at 37 °C for 24 h followed by exchange of medium with 500 μl of Opti-MEM. Following incubation at 37 °C for 3 h, the medium was changed to DMEM/F12 (1:1) containing 5% (v/v) FBS. After incubation for 48 h, luciferase activity was measured using a Dual Luciferase Assay Kit (Promega) and a Mini Lumat LB9506 luminometer (Berthold, Bad Wildbad, Germany), according to the manufacturer's instructions.
In co-transfection experiments, a pCMV-Sp1 or pCMV-Sp3 expression vector in addition to the promoter construct containing 250 bp sequences was transfected together with the pCMV empty vector to adjust the total amount to 0.25 μg/well. Therefore the total amount of transfected DNA, including pRL-TK, was 1.15 μg/well.
Preparation of nuclear extracts
After washing with PBS, cells were collected with a cell scraper, recovered in a microtube and pelleted by centrifugation. Cells were suspended in 400 μl of buffer A [10 mM Hepes/KOH (pH 7.8)/10 mM KCl/0.1 mM EDTA/0.1% Nonidet P40/1 mM dithiothreitol/protease inhibitor cocktail (1:1000, v/v)], vortex-mixed and centrifuged at 1900 g (5000 rev./min) for 1 min at 4 °C. The supernatant was removed, and the pellet was resuspended in 50 μl of buffer B [50 mM Hepes/KOH (pH 7.8)/420 mM KCl/0.1 mM EDTA/5 mM MgCl2/2% (v/v) glycerol/1 mM dithiothreitol/protease inhibitor cocktail (1:1000, v/v)], followed by incubation for 30 min at 4 °C. with vortex-mixing every 10 min. After centrifugation at 17400 g (15000 rev./min) for 15 min at 4 °C, the supernatant was recovered and then used for EMSA.
EMSA
Sets of complementary oligonucleotides for probe, wild-type and mutant competitors (Table 1) were annealed in Tris/EDTA buffer by boiling for 5 min, followed by gradual cooling. The oligonucleotides for the probe were designed to have 5′-cohesive ends when annealed. The oligonucleotides for probe were radiolabelled with [α-32P]dCTP and Klenow enzyme, followed by purification with a ProbeQuant G-50 Micro Column (Amersham Biosciences). A DNA–protein-binding reaction was allowed to proceed in a volume of 25 μl, consisting of 5 μl of 5× binding buffer [50 mM Hepes/KOH (pH 7.8)/250 mM KCl/5 mM EDTA/25 mM MgCl2/50% (v/v) glycerol], 1.5 μl of 2 μg/μl poly(dI-dC), 5 μl of nuclear extract and 1 μl of the labelled probe (104 c.p.m./μl; specific radioactivity, 104 c.p.m./nmol) with or without 2 μl of 4 μM wild-type or mutant competitor for 15 min at room temperature. For supershift analysis, 2 μg of rabbit polyclonal antibodies against Sp1 and/or Sp3 was added and incubated for 1 h at 4 °C, before the addition of the probe. The reaction mixtures were then run on a 4% Tris/borate/EDTA polyacrylamide gel, which was then exposed to X-ray film at −80 °C.
Data analysis
All experiments were repeated at least three times and representative results are shown in the Figures. For the luciferase assay, results are expressed as means±S.E.M. Differences in luciferase activity were tested by a one-way analysis of variance followed by Sheffe's multiple comparison test, with STATVIEW 4.0 (Abacus Concepts, Berkeley, CA, U.S.A.), and differences were considered to be significant at P<0.05.
RESULTS
Determination of the sequences and analysis of putative transcription-factor-binding sites of the mouse 20α-HSD gene 5′-flanking region
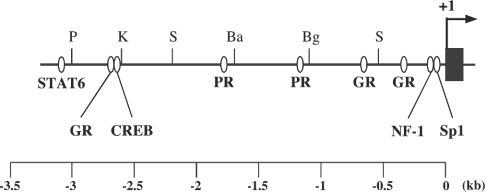
Although the draft sequences of the mouse genome have been already reported, to exclude ambiguity of the sequence, the sequences of the 5′-flanking region of the mouse 20α-HSD gene were determined for up to approx. 4.2 kb using the λFIX II genomic clones containing the mouse 20α-HSD gene. The sequence identity between that of the present study and the mouse genome databases was 99.8%. Computer analysis using TESS revealed putative transcription-factor-binding sites, including a single STAT6 (signal transducers and activators of transcription protein 6) binding site at −3101 to −3093 bp, three GREs (glucocorticoid-response elements) at −2710 to −2692, −673 to −664 and −333 to −322 bp, a single CRE (cAMP-response element) at −2612 to −2605 bp, two PREs (progesterone-response elements) at −1806 to −1792 and −1175 to −1163 bp, and single NF-1 (nuclear factor-1) and Sp1-binding sites at −123 to −109 and −75 to −63 bp respectively (Figure 1).
Figure 1. Putative transcription-factor-binding sites in the 5′-flanking region of the mouse 20α-HSD gene.
Sequences approx. 3.2 kb upstream from the transcription start site of the mouse 20α-HSD gene were analysed with TESS (see the Materials and methods section). Putative binding sites for STAT6, glucocorticoid receptor (GR), progesterone receptor (PR), NF-1 and Sp1, as well as restriction sites for PstI (P), KpnI (K), Sau3AI (S), BamHI (Ba) and BglII (Bg), are shown. The arrow indicates the transcription start site, which is designated as +1. Exon 1 is represented by a dark box.
Promoter activities of various lengths of the mouse 20α-HSD gene 5′-flanking region
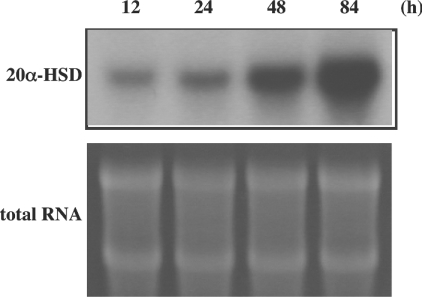
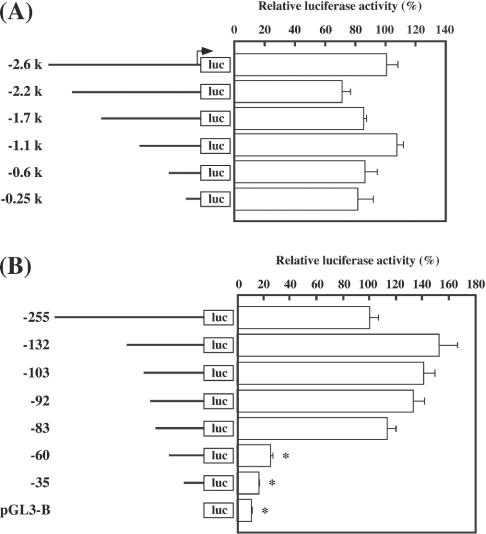
We used primary cultured rat luteinized granulosa cells to study the mechanisms of 20α-HSD expression, because this system expresses endogenous 20α-HSD mRNA and successfully reproduces the regulation of the gene by PRL and PGF2α [8,9]. As shown in Figure 2, the amount of 20α-HSD mRNA increased spontaneously in a time-dependent manner in these cells, which seems to reflect the spontaneous rise in 20α-HSD expression in the luteal tissue of rats during the oestrous cycle. Both an inhibition and a stimulation of 20α-HSD mRNA expression by PRL and PGF2α respectively were confirmed to verify the culture system (results not shown). To identify the regulatory region crucial to transcriptional activity of the gene, promoter activities of various lengths of the 5′-flanking region were measured. A series of deleted constructs from 2.6 to 0.5 kb in length had similar promoter activities (Figure 3A); however, when the region from −83 to −61 bp was deleted, a remarkable fall in promoter activity was observed (Figure 3B).
Figure 2. Expression of the endogenous 20α-HSD mRNA in rat luteinized granulosa cells.
After 24 h of culture, the medium was changed and incubated further for 12, 24, 48 and 84 h. Total RNA was extracted and used for Northern hybridization with rat 20α-HSD cDNA as a probe.
Figure 3. Promoter activities of the serially deleted 5′-flanking region of the mouse 20α-HSD gene.
DNA segments [−2.6, −2.2, −1.7, −1.1, −0.6 and −0.25 kb (A) and −255, −132, −103, −92, −83, −60 and −35 kb (B)] of the 5′-flanking region of the mouse 20α-HSD gene were fused to a luciferase reporter gene (luc), and rat luteinized granulosa cells were transfected with the constructs. Results (means±S.E.M. for triplicate wells) are presented as a percentage of relative luciferase activity of −2.6 kb (A) or −255 kb (B) construct. Promoter activity of pGL3-Basic is also shown. *P<0.05 compared with the −255-luc construct.
Analyses of the putative Sp1-binding site
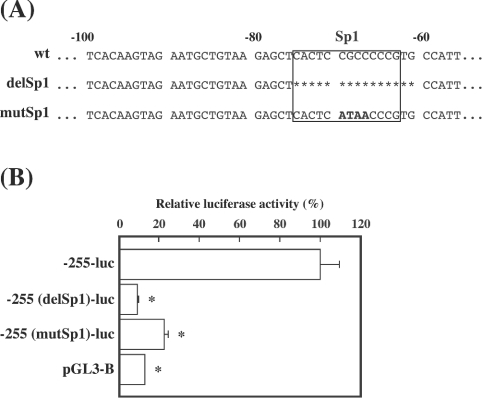
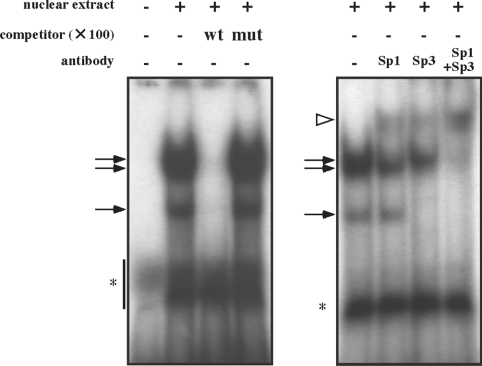
As the putative Sp1 element was found between −83 and −61 bp, we examined the effect of deleted ([−255(delSp1)-luc] or substituted [−255(mutSp1)-luc] mutations of the element on promoter activity in the next experiment (Figure 4A). Compared with the wild-type −255 bp construct, mutant constructs showed markedly reduced promoter activities, similar to those of pGL3-Basic used as a negative control (Figure 4B). To confirm binding of certain nuclear factors to wild-type, but not mutated, Sp1 elements, EMSA was performed with nuclear extracts prepared from rat luteinized granulosa cells. There were three bands in the presence of the nuclear extract, the patterns of which were similar to those in other reports [13]. Addition of an unlabelled wild-type competitor resulted in the disappearance of the bands, whereas addition of an unlabelled mutant competitor had no effect (Figure 5, left panel). To confirm further that the shifted bands contained Sp1 family molecules, a supershift experiment with antibodies against Sp1 and Sp3 was carried out. As shown in Figure 5 (right panel), pre-incubation of the antibody against both Sp1 and Sp3 produced a supershift band, in accordance with disappearance of the uppermost band and the two lower bands respectively. Almost all of the three bands disappeared upon addition of antibodies against Sp1 and Sp3 in combination, and a dense supershifted band appeared.
Figure 4. Effect of mutations in the putative Sp1 site of the 5′-flanking region of the mouse 20α-HSD gene on promoter activity.
(A) Sequences of the construct in which 15 nt containing the putative Sp1 site [−255(delSp1)-luc] were deleted (shown by the asterisks) and of the mutated construct in which CGCC (−70 to −67 bp) was changed to ATAA [−255(mutSp1)-luc] (shown in boldface lettering), as well as sequences of the wild-type construct (wt), are shown. (B) Promoter activities of the mutant constructs, as well as the wild-type construct, are shown. Relative luciferase activity (means±S.E.M. for triplicate wells) is presented as a percentage of a wild-type −255-luc construct. Promoter activity of pGL3-Basic is also shown. *P<0.05 compared with the −255-luc construct.
Figure 5. EMSA with nuclear extract from rat luteinized granulosa cells.
The radiolabelled oligonucleotides and nuclear extracts were incubated with or without a 100-fold molar excess amount of wild-type (wt) or mutant (mut) competitor. For the supershift experiment, antibodies against Sp1 and/or Sp3 were mixed before addition of the probe. Arrows and a white arrowhead indicate shifted bands and a supershifted band respectively. Asterisks indicate non-specific signals.
Effects of co-transfection with Sp1 and/or Sp3 expression vectors on promoter activity
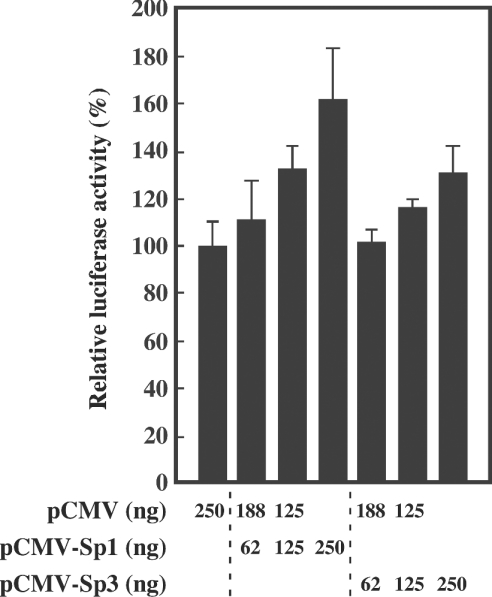
Finally, the involvement of Sp1 and Sp3 in the expression of the 20α-HSD gene was examined by co-transfection experiments. Rat luteinized granulosa cells were co-transfected with −255-luc constructs together with expression vectors pCMV-Sp1 and/or pCMV-Sp3, and the effect on promoter activity was assessed. Co-transfection of both pCMV-Sp1 and pCMV-Sp3 enhanced promoter activity in a dose-dependent manner (Figure 6).
Figure 6. Effect of co-transfection with Sp1 or Sp3 expression vector on the promoter activity.
Luteinized granulosa cells were co-transfected with the luciferase constructs, together with pCMV and pCMV-Sp1 or pCMV-Sp3 in different ratios in a total amount of 250 ng/well. Relative luciferase activity (means±S.E.M. for triplicate wells) is presented as a percentage of that co-transfected with pCMV alone.
DISCUSSION
The present study demonstrated that the expression of the 20α-HSD gene in the luteal cells required Sp1 and/or Sp3, which are known as ubiquitous transcription factors that bind to the GC/GT-box element found in 5′-flanking regions of many constitutively expressing and inducible genes [14]. Considering that 20α-HSD is not expressed in undifferentiated rat granulosa cells, in which Sp1 and Sp3 are expressed [13], it seems that the tissue specificity of 20α-HSD expression is not determined by these factors, but by other transcription factors and/or specific chromatin structures suitable for transcriptional activation in luteal cells, which may be achieved during the process of luteinization. Whatever the mechanism of the tissue specificity of 20α-HSD, our present results clearly indicate that either Sp1 or Sp3 is indispensable for the regulation of 20α-HSD gene expression.
The Sp1 family consists of four members: Sp1, Sp2, Sp3, and Sp4. Sp1 and Sp3 have been extensively studied, and are known to be co-expressed in several tissues and to interact with an identical consensus sequence. There are reports that Sp1 is phosphorylated [15] and glycosylated [16], whereas Sp3 is acetylated at a site known as an inhibitory domain [17]. Although the general role of these modifications in Sp1- or Sp3-mediated transcriptional regulation is not clear, it seems reasonable to consider the involvement of such modifications in the hormonal regulation of the 20α-HSD gene.
Another possibility is that hormones regulate the expression of the 20α-HSD gene by modifying the expression level of Sp1 and Sp3. For example, it has been reported that the level of the Sp3 protein is reduced in the interferon-γ-mediated suppression of macrophage lipoprotein lipase gene transcription. In contrast, the DNA-binding activity of Sp1 is decreased without affecting the protein levels by the cytokine [18]. It would be necessary to examine whether the level of Sp1 or Sp3 is regulated by hormones such as PGF2α, PRL or other factors which regulate the expression of the 20α-HSD gene.
In the present study, both Sp1 and Sp3 elevated the promoter activity of the 5′-flanking region of the mouse 20α-HSD gene. The role of Sp3 is complicated by the fact that Sp3 functions as a transcriptional repressor, as well being as an activator of Sp1-mediated transcription [19,20]. The role of Sp3 seems to depend on the promoter sequence or on cellular context. The present results indicate that Sp3 is a transcriptional activator for the 20α-HSD gene in luteal cells, but a role as a transcriptional repressor due to its relatively weak effect on transcriptional activity is also possible by increasing the expression ratio of Sp3 to Sp1. Further investigations are necessary to clarify the precise role of Sp1 and Sp3 in the hormonal regulation of the 20α-HSD gene.
In conclusion, the present study has demonstrated that Sp1 family molecules play a significant role in the expression of the 20α-HSD gene in luteal cells. Further characterization of the 5′-flanking region of the 20α-HSD gene, including comparative analyses between species, will be useful in understanding the general mechanisms of tissue-specific expression, and for providing an insight into the molecular evolution, as well as the regulatory mechanisms, of 20α-HSD gene expression. The present study is a prerequisite for such comparative analyses.
References
- 1.Wiest W. G., Wilcox R. B., Kirschbaum T. H. Rat ovarian 20α-hydroxysteroid dehydrogenase: effects of estrogens and pituitary gonadotrophins. Endocrinology. 1963;73:588–595. doi: 10.1210/endo-73-5-588. [DOI] [PubMed] [Google Scholar]
- 2.Wiest W. G., Kidwell W. R., Balogh K., Jr Progesterone catabolism in the rat ovary: a regulatory mechanism for progestational potency during pregnancy. Endocrinology. 1968;82:844–859. doi: 10.1210/endo-82-4-844. [DOI] [PubMed] [Google Scholar]
- 3.Hashimoto I., Wiest W. G. Luteotropic and luteolytic mechanisms in rat corpora lutea. Endocrinology. 1969;84:886–892. doi: 10.1210/endo-84-4-886. [DOI] [PubMed] [Google Scholar]
- 4.Hashimoto I., Henricks D. M., Anderson L. L., Melampy R. M. Progesterone and pregn-4-en-20α-ol-3-one in ovarian venous blood during various reproductive states in the rat. Endocrinology. 1968;82:333–341. doi: 10.1210/endo-82-2-333. [DOI] [PubMed] [Google Scholar]
- 5.Stocco C., Callegari E., Gibori G. Opposite effect of prolactin and prostaglandin F2α on the expression of luteal genes as revealed by rat cDNA expression array. Endocrinology. 2001;142:4158–4161. doi: 10.1210/endo.142.9.8493. [DOI] [PubMed] [Google Scholar]
- 6.Ormandy C. J., Camus A., Barra J., Damotte D., Lucas B., Buteau H., Edery M., Brousse N., Babinet C., Binart N., Kelly P. A. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev. 1996;11:167–178. doi: 10.1101/gad.11.2.167. [DOI] [PubMed] [Google Scholar]
- 7.Sugimoto Y., Yamasaki A., Segi E., Tsuboi K., Aze Y., Nishimura T., Oida H., Yoshida N., Tanaka T., Katsuyama M., et al. Failure of parturition in mice lacking the prostaglandin F receptor. Science. 1997;277:681–683. doi: 10.1126/science.277.5326.681. [DOI] [PubMed] [Google Scholar]
- 8.Albarracin C. T., Parmer T. G., Duan W. R., Nelson S. E., Gibori G. Identification of a major prolactin-regulated protein as 20α-hydroxysteroid dehydrogenase: coordinate regulation of its activity, protein content, and messenger ribonucleic acid expression. Endocrinology. 1994;134:2453–2460. doi: 10.1210/endo.134.6.8194472. [DOI] [PubMed] [Google Scholar]
- 9.Stocco C. O., Zhong L., Sugimoto Y., Ichikawa A., Lau L. F., Gibori G. Prostaglandin F2α-induced expression of 20α-hydroxysteroid dehydrogenase involves the transcription factor NUR77. J. Biol. Chem. 2000;275:37202–37211. doi: 10.1074/jbc.M006016200. [DOI] [PubMed] [Google Scholar]
- 10.Jez J. M., Bennett M. J., Schlegel B. P., Lewis M., Penning T. M. Comparative anatomy of the aldo–keto reductase superfamily. Biochem. J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishida M., Hirabayashi K., Suzuki M., Yamanouchi K., Nishihara M. Cloning and chromosomal localization of mouse 20α-hydroxysteroid dehydrogenase gene. J. Reprod. Dev. 2003;49:79–85. doi: 10.1262/jrd.49.79. [DOI] [PubMed] [Google Scholar]
- 12.Miura R., Shiota K., Noda K., Yagi S., Ogawa T., Takahashi M. Molecular cloning of cDNA for rat ovarian 20α-hydroxysteroid dehydrogenase (HSD1) Biochem. J. 1994;299:561–567. doi: 10.1042/bj2990561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alliston T. N., Maiyar A. C., Buse P., Firestone G. L., Richards J. S. Follicle stimulating hormone-regulated expression of serum/glucocorticoid-inducible kinase in rat ovarian granulosa cells: a functional role for the Sp1 family in promoter activity. Mol. Endocrinol. 1997;11:1934–1949. doi: 10.1210/mend.11.13.0033. [DOI] [PubMed] [Google Scholar]
- 14.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 15.Jackson S. P., MacDonald J. J., Lees-Miller S., Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 16.Jackson S. P., Tjian R. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell. 1988;55:125–133. doi: 10.1016/0092-8674(88)90015-3. [DOI] [PubMed] [Google Scholar]
- 17.Braun H., Koop R., Ertmer A., Nacht S., Suske G. Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res. 2001;29:4994–5000. doi: 10.1093/nar/29.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes T. R., Tengku-Muhammad T. S., Irvine S. A., Ramji D. P. A novel role of Sp1 and Sp3 in the interferon-γ-mediated suppression of macrophage lipoprotein lipase gene transcription. J. Biol. Chem. 2002;277:11097–11106. doi: 10.1074/jbc.M106774200. [DOI] [PubMed] [Google Scholar]
- 19.Ihn H., Trojanowska M. Sp3 is a transcriptional activator of the human α2(I) collagen gene. Nucleic Acids Res. 1997;25:3712–3717. doi: 10.1093/nar/25.18.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagen G., Müller S., Beato M., Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]