Abstract
We have previously reported, from the nematode worm Caenor-habditis elegans, three genes (gly-12, gly-13 and gly-14) encoding enzymically active UDP-N-acetyl-D-glucosamine:α-3-D-mannoside β1,2-N-acetylglucosaminyltransferase I (GnT I), an enzyme essential for hybrid, paucimannose and complex N-glycan synthesis. We now describe a worm with null mutations in all three GnT I genes, gly-14 (III);gly-12 gly-13 (X) (III and X refer to the chromosome number). The triple-knock-out (TKO) worms have a normal phenotype, although they do not express GnT I activity and do not synthesize 31 paucimannose, complex and fucosylated oligomannose N-glycans present in the wild-type worm. The TKO worm has increased amounts of non-fucosylated oligomannose N-glycan structures, a finding consistent with the site of GnT I action. Five fucosylated oligomannose N-glycan structures were observed in TKO, but not wild-type, worms, indicating the presence of unusual GnT I-independent fucosyltransferases. It is concluded that wild-type C. elegans makes a large number of GnT I-dependent N-glycans that are not essential for normal worm development under laboratory conditions. The TKO worm may be more susceptible to mutations in other genes, thereby providing an approach for the identification of genes that interact with GnT I.
Keywords: Caenorhabditis elegans, N-acetylglucosaminyltransferase I, null mutations, mass spectrometry (MS), N-glycan biosynthesis, paucimannose N-glycans
Abbreviations: Gn or GlcNAc, N-acetyl-D-glucosamine; GnT I, UDP-N-acetyl-D-glucosamine:α-3-D-mannoside β1,2-N-acetylglucosaminyltransferase I (EC 2.4.1.101); F1, first filial generation (etc.); Hex, hexose; L4, fourth larval stage; M or Man, D-mannose; M5Gn2, [Manα1,6(Manα1,3)Manα1,6]-(Manα1,3)Manβ1,4GlcNAcβ1,4GlcNAc; MALDI-TOF, matrix-assisted laser-desorption–ionization time-of-flight; NGM, nematode growth medium; PA, pyridylamine; TKO, triple knock-out
INTRODUCTION
There is considerable evidence that most membrane-bound cell-surface proteins are glycosylated and that a major function of protein-bound glycans is the mediation of cell–cell and cell– environment interactions [1–3]. Caenorhabditis elegans is a nematode worm that has been used extensively as a model organism to study animal development and behaviour [4–7]. We are interested in the role in C. elegans development of a particular class of asparagine-linked glycans (N-glycans) dependent on the prior action of UDP-GlcNAc (N-acetyl-D-glucosamine):α-3-D-mannoside β1,2-N-acetylglucosaminyltransferase I (GnT I, EC 2.4.1.101). GnT I is a Golgi-resident enzyme which transfers a GlcNAc residue in β1,2 linkage to the Manα1,3Manβ-terminus of [Manα1,6(Manα1,3)Manα1,6](Manα1,3)Manβ1,4-GlcNAcβ1,4GlcNAc-Asn-protein (Man5GlcNAc2-Asn-protein), thereby initiating the synthesis of hybrid, paucimannose and complex N-glycans [8] (Scheme 1). Chinese-hamster ovary cells and other cultured cell lines [9–12] with null mutations in GnT I grow and function normally. In contrast, GnT I-null mouse embryos [13,14] die at 9.5–10.5 days after fertilization, and humans [15–17] and mice [18,19] with a null mutation in UDP-GlcNAc:α-6-D-mannoside β1,2-N-acetylglucosaminyltransferase II (an enzyme downstream of GnT I) suffer severe multi-systemic developmental abnormalities. The marked discrepancy between the effects of defective synthesis of GnT I-dependent N-glycans in cultured cells and in the intact organism indicates that these N-glycans play minor roles in the day-to-day housekeeping functions of the single cell, but are essential for normal morphogenesis in mammals and possibly in most metazoans. GnT I appeared in evolution at approximately the same time as metazoans, and GnT I-dependent N-glycans are not found in protozoans. The N-glycan core, Manα1,6(Manα1,3)Manβ1,4Glc-NAcβ1,4GlcNAc-Asn, on the other hand, is an ancient structure essential for viability of protozoa, metazoa and cultured cells [20–23].
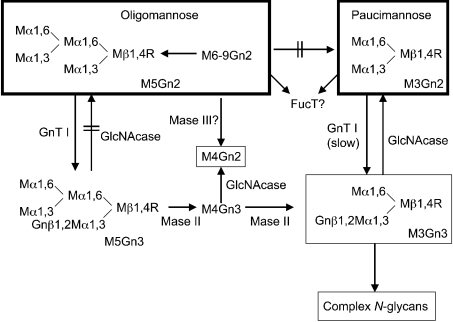
Scheme 1. N-glycan synthesis in C. elegans.
The Scheme shows the conversion of oligomannose N-glycans (M5–9Gn2) into hybrid (M3–5Gn3) and paucimannose (M3–4Gn2) N-glycans. Abbreviations: FucT, fucosyltransferase; R=GlcNAcβ1,4GlcNAc-Asn-protein. Compounds detected by MS in wild-type worms are framed by rectangles; the thicker the lines of the rectangle, the more abundant are the respective structures in C. elegans. Other compounds detected by MS are not shown in the Scheme. GnT I adds a GlcNAc residue in β1,2 linkage to the Manα1,3-arm of M5Gn2 to form the hybrid N-glycan M5Gn3. Two mannose residues are removed from M5Gn3 by the action of GnT I-dependent α3,6-mannosidase II (Mase II) to form the truncated hybrid N-glycans M4Gn3 and M3Gn3. There appears to be some GnT I-independent α3,6-mannosidase activity (Mase III) [47] in C. elegans, but the enzyme is involved only in making M4Gn2 and not M3Gn2 (the GnT I-null worm makes some M4Gn2 but not M3Gn2; Table 2). The inability by several groups to detect M5Gn3 and M4Gn3 by MS is probably due to the rapid action of Mase II. C. elegans accumulates paucimannose N-glycans M3Gn2 and M4Gn2 due to the action of a specific β-N-acetylglucosaminidase (GlcNAcase; [26]); this enzyme cannot act efficiently on M5Gn3. Fucosylation by fucosyltransferase is discussed in the text.
We have shown that C. elegans has three genes expressing enzymically active GnT I (gly-12, gly-13 and gly-14) [24]; GLY-13 is the major GnT I in normal worms [25,26]. We have previously reported worm lines with single null mutations in the gly-12, gly-13 and gly-14 genes and a double null mutation in the gly-12 and gly-14 genes (gly-14;gly-12) [27]. Only the gly-13 null worm showed an abnormal phenotype, but we have shown (S. Zhu and H. Schachter, unpublished work) that the gly-13 allele described in our previous study has an additional mutation that is responsible for this abnormality. We now describe a triple-knock-out (TKO) worm, gly-14 (III);gly-12 gly13 (X), with null mutations in all three GnT I genes, the gly-12(id47) and gly-14(id48) alleles previously reported and a new allele, namely gly-13(ok712). Although the TKO worms show a normal phenotype with several standard tests, worm extracts do not express GnT I activity and have a markedly abnormal N-glycan pattern. We conclude that N-glycans dependent on GnT I are not essential for normal development of wild-type C. elegans, at least under laboratory conditions. Survival of the TKO worm may be compromised if the worm is exposed to environmental stress or if one or more other genes are non-functional. The TKO worm may therefore provide a valuable tool for identifying genes that require an active GnT I for optimum function. A preliminary report on these findings has already appeared [28].
EXPERIMENTAL
General techniques and strains
General techniques for the handling of nematodes were as previously described [4,7]. Standard molecular-biology procedures were used [29,30]. Oligonucleotides were synthesized on an automated DNA synthesizer and purified by the cartridge method (Hospital for Sick Children Biotechnology Center, Toronto, Canada). DNA was sequenced in both directions by the double-strand dideoxy method [31]. C. elegans strains N2 (non-clumping) wild-type (Bristol), dpy-6(e14) and unc-5(e53) are maintained in our laboratory. Strain RB871 gly-13(ok712) (X) was obtained from the C. elegans Gene Knockout Project at the Oklahoma Medical Research Foundation, Oklahoma City, OK, U.S.A. AS270 gly-12(id47) (X) and AS271 gly-14(id48) (III) were described previously [27]. AS338 dpy-6 gly-13, AS339 gly-12 dpy-6 gly-13 and AS341 gly-14;gly-12 gly-13 are described below. Worms were grown on NGM (nematode growth medium) agar plates (3 g of NaCl, 2.5 g of peptone and 17 g of agar dissolved in 975 ml of water) [32] seeded with Escherichia coli strain OP50 (a leaky uracil-requiring strain) at 20 °C [4,33].
Single-worm PCR
Single-worm PCR was used to detect deletions. Worms were singled and allowed to lay eggs for 1–2 days before being picked for single-worm PCR. A single worm was put into 0.004 ml of buffer A [10 mM Tris/HCl (pH 8.0)/50 mM KCl/2.5 mM MgCl2/0.45% Igepal CO-630/0.45% Tween-20/0.01% gelatin] containing freshly added proteinase K to a final concentration of 0.2 mg/ml and incubated at 60 °C for 1 h. Proteinase K was inactivated at 83 °C for 15 min. Each gene required three PCR primers for analysis (Table 1 and Figure 1 below). Deletions in gly-13 and gly-14 were analysed in a single tube. PCR was carried out in 25 μl reaction volumes with Platinum Taq DNA Polymerase (1 unit; Gibco BRL), 5 μl of DNA template, 2 μl of 10×PCR buffer [10×PCR buffer is 0.2 M Tris/HCl (pH 8.4)/0.5 M KCl], 0.6 μl of 50 mM MgCl2, 0.4 μl of 10 mM dNTP, 0.4 μl of each gene-specific primer (10 μM). PCR conditions were: 95 °C for 2 min, followed by 40 cycles at 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s. PCR products were analysed by electrophoresis in a 2.0%-agarose gel in TBE buffer [0.089 M Tris/borate (pH 8.3)/0.025 M disodium EDTA] and visualized under UV light.
Table 1. PCR primers used for detection of GnT I deletions.
Three PCR primers are required to establish a deletion: a common (com) primer which pairs with a deletion (del) primer to identify the deletion or with a wild-type primer (wild) to identify the non-mutated allele. The positions of the primers are shown in Figure 1. F and R refer to forward or reverse directions of the open reading frame.
| Primer name | Primer sequence | Position of primer (nt) relative to ATG at 1 |
|---|---|---|
| gly-12Fdel | 5′-GCG GAA CAA CTA CCC AGC-3′ | 643–660 |
| gly-12Fwild | 5′-CGT ACC GAA ATC CAT CAC A-3′ | 2350–2367 |
| gly-12Rcom | 5′-ACC AGA GTC TTG ACC ACA-3′ | 2679–2662 |
| gly-13Fcom | 5′-TTC AAA ACG GGA ATC TGG AG-3′ | −586 to −567 |
| gly-13Rwild | 5′-TCC CGT AGT TTA TGC AGT AT-3′ | −267 to −286 |
| gly-13Rdel | 5′-ACC ACT CAC AGG CAA CAA AT-3′ | 1796–1777 |
| gly-14Fcom | 5′-CGT GCT CAT CTA CTT TTC AT-3′ | 32–51 |
| gly-14Rwild | 5′-AGC TTT TCC AAA ACT CCT CCT AC-3′ | 291–268 |
| gly-14Rdel | 5′-CGT AAC AAA ATG CGC AGA AT-3′ | 1937–1918 |
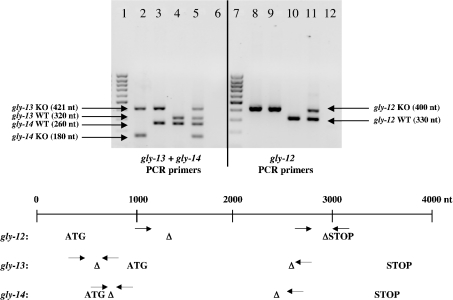
Figure 1. PCR analysis of the TKO mutant.
The scheme in the lower part of the Figure shows the PCR primers used to detect deletions (Table 1). There are three primers for every gene. Two primers (‘com’ and ‘del’; Table 1) bracket the deletion, while the third primer (‘wild’; Table 1) is within the deletion. The ‘com’ and ‘del’ PCR primers are separated by about 2000 nt using wild-type genomic DNA as template; only the smaller ‘com-wild’ PCR products (260–330 nt, WT; upper part of the Figure) are formed with wild-type DNA as template because amplification of the larger fragment cannot compete with the smaller fragment. The mutant DNA templates result in PCR products ranging from 180 to 421 nt (KO; upper part of the figure) due to the ‘com-del’ primer pairs. The upper part of the Figure shows the results of PCR analysis. Lanes: 1 and 7, 100 bp markers; 2 and 8, gly-14;gly-12 gly-13 worms; 3 and 9, gly-12 gly-13 worms; 4 and 10, N2 wild-type worms; 5 and 11, gly-14/+;gly-12 gly-13/++ heterozygotes; 6 and 12, controls (no worms added). Lanes 1–6, gly-13 and gly-14 PCR primers; lanes 7–12, gly-12 PCR primers. The analysis shows that the gly-14;gly-12 gly-13 worm is homozygous for all three deletions. Δ indicates start and stop positions of deletions. Short horizontal arrows indicate PCR primer positions. The ATG and STOP codons indicate the start and end of the open reading frame.
Production of mutant worms
Crosses were carried out by standard methods [7]. Male worms were obtained by the heat-shock method. L4 (fourth-stage) larvae were incubated at 30 °C for 6 h followed by selfing at 20 °C. The progeny contained about 2–5% male worms. Crosses were carried out by mating five or six males and one or two hermaphrodites for 24 h at 20 °C.
AS338 dpy-6 gly-13 (X) worms
The RB871 gly-13(ok712) worms we received had not been back-crossed. A dpy-6 gly-13 worm was made to facilitate this (dpy refers to the dumpy mutation). Male gly-13 worms were crossed with dpy-6(e14) hermaphrodites. Non-dumpy F1 (first filial generation) dpy-6+/+gly-13 (X) and dumpy F2, F3 and F4 hermaphrodites were singled. Single-worm PCR for the gly-13 deletion identified 24 recombinant homozygous dpy-6 gly-13 hermaphrodites in the F4 worms. N2 males were crossed with the dpy-6 gly-13 hermaphrodites, and non-dumpy F1 dpy-6 gly-13 heterozygotes were singled. Dumpy F2 hermaphrodites were picked out and crossed with N2 males. The back-crossing procedure was carried out five times to produce AS338 dpy-6(e14) gly-13(ok712) (X) worms.
AS339 gly-12 dpy-6 gly-13 (X) worms
Male gly-12 worms were crossed with dpy-6 gly-13 hermaphrodites. Non-dumpy F1+dpy-6 gly-13/gly-12++ (X) and dumpy F2 and F3 hermaphrodites were singled. Single-worm PCR for the gly-12 and gly-13 deletions identified three recombinant homozygous gly-12 dpy-6 gly-13 hermaphrodites in the F3 worms.
AS341 gly-14;gly-12 gly-13 worms
The dpy-6 gene was first removed from gly-12 dpy-6 gly-13 worms by recombination. Male gly-12 worms were crossed with gly-12 dpy-6 gly-13 hermaphrodites. Non-dumpy F1 gly-12 dpy-6 gly-13/gly-12++ (X) and non-dumpy F2, F3, F4 and F5 hermaphrodites were singled. Single-worm PCR for the gly-13 deletion identified 24 recombinant homozygous gly-13 hermaphrodites in the F5 worms. Of these 24 worms, ten produced no dumpy F6 progeny. Four non-dumpy F6 progeny from these ten F5 worms and eight non-dumpy F7 progeny from the F6 worms were singled. Single-worm PCR for the gly-12 and gly-13 deletions proved that all four F6 worms and all eight F7 worms were recombinant non-dumpy homozygous gly-12 gly-13 hermaphrodites.
Male gly-14 worms were crossed with gly-12 gly-13 hermaphrodites. The crossed F1 gly-14/+ (III);gly-12 gly-13/++ (X) hermaphrodites and F2, F3 and F4 hermaphrodites were singled. Single-worm PCR for all three deletions identified ten homozygous gly-14;gly-12 gly-13 hermaphrodites.
Phenotype analysis of the TKO worm
Brood size
A total of 20 L4 larvae were cultured singly on fresh plates and incubated at 20 °C overnight. Hermaphrodites were transferred to fresh plates to prevent overcrowding until egg laying ceased (3–4 days). The progeny at the L3 and L4 stages were counted.
Life span
About 15–20 adults were allowed to lay eggs for 1–2 h and were then removed (zero time). After the progeny reached the L4 stage, they were transferred daily to fresh plates during the egg-laying period and weekly thereafter. Animals were scored dead when they no longer moved or twitched when prodded or pushed several times with a platinum wire.
Defecation interval
Hermaphrodites which had reached adulthood ≈24 h before testing were monitored under the dissecting microscope. Defecation cycle length was defined as the duration between the posterior muscular contraction of one defecation and that of the next defecation. Ten animals were scored. For each animal, five defecation cycles were measured with a stopwatch and the average was calculated for each animal.
Osmotic avoidance test
Young adults were washed twice with M9 buffer (6 g of Na2HPO4, 3 g of KH2PO4, 5 g of NaCl, 0.25 g of MgSO4·7H2O/litre). About 0.01 ml of worm suspension containing over 100 worms was transferred to the centre of a 4 M NaCl ring which had soaked for less than 3 min. The excess M9 buffer was absorbed with tissue paper. After 15 or 20 min, animals crossing the ring were scored as non-avoiders.
Levamisole-resistance test
Young adults were washed twice with M9 buffer. About 0.2 ml of OP50-strain Escherichia coli culture containing 0.05 ml of 25 mM levamisole (Sigma) was spread in a circle on NGM plates. About 0.01 ml of worm suspension containing over 100 worms was transferred to the centre of the bacterial ring. Worms that became paralysed were scored as sensitive to levamisole.
Dauer formation and recovery
Mutant and wild-type N2 animals on plate culture were starved for 2 weeks. Dauer larvae (a developmentally arrested dispersal stage that may be formed under conditions of starvation or overcrowding) were examined under the dissecting microscope and were collected and incubated in 1% (w/v) SDS at room temperature for 30 min. After incubation, a large volume of M9 buffer was added and the suspension was centrifuged at 1500 g for 5 min. Worms were washed one more time with M9 buffer, and placed on fresh plates seeded with E. coli OP50. On the next day, the presence of L4 animals on the plates indicated recovery from the Dauer stage.
Male mating efficiency
TKO male worms were obtained by incubating hermaphrodites at 30 °C for 6 h, followed by selfing at 20 °C. The progeny contained about 2–5% male worms. TKO males (five or six worms) and unc-5 hermaphrodites (two worms) were allowed to mate for 24 h at 20 °C. The presence of non-Unc (Unc is derived from ‘unco-ordinated’) worms was scored as crossed progeny. Successful mating was further confirmed by single-worm PCR for the presence of the three GnT I deletions in the progeny.
Post-embryonic development
Ten hermaphrodites were allowed to lay eggs for 2 h and then removed. About 100 progeny were monitored daily until maturity. Animals were scored as mature adults when the vulva could be observed.
Microsome preparation
Worms from 20 10-cm-diameter agar plates were harvested and bacteria were removed by washing (three times) with M9 buffer. Worms were resuspended in 2 ml of buffer A (see above) containing proteinase inhibitors (Mini EDTA-Free Proteinase Inhibitor Cocktail Tablets; Roche Diagnostics GmbH, Mannheim, Germany) and stored at −80 °C. The frozen worms were thawed and lysed by sonication on ice as required [24]. All subsequent procedures were carried out at 4 °C. The lysate was centrifuged at 1500 g for 10 min and the supernatant was centrifuged at 126000 g for 1 h. The resulting microsomal pellet was resuspended in 0.25 ml of buffer B [25 mM Mes (pH 7.0)/1% Triton X-100 containing proteinase inhibitors] and homogenized with a hand-held Potter–Elvejhem glass homogenizer. The suspension was incubated on ice for 1 h, centrifuged at 20000 g for 20 min, and the supernatant was used for GnT I assays as soon as possible after preparation.
GnT I assays
Man5GlcNAc2-pyridylamine (M5Gn2-PA) was kindly donated by Dr Erika Staudacher (Institut für Chemie, Universität für Bodenkultur, Vienna, Austria). This substrate was used instead of the more convenient Manα1,6(Manα1,3)Manβ1-O-octyl substrate because GLY-13 acts only on Man5GlcNAc2 substrates [25,26]. The enzyme incubation contained, in a total volume of 0.02 ml, 0.017 ml of worm microsome extract, 0.25 mM M5Gn2-PA, 0.5 mM UDP-[3H]GlcNAc (New England Nuclear) diluted with non-radioactive UDP-GlcNAc (Sigma) to a specific radioactivity of 94000 d.p.m./nmol, 10 mM MnCl2, 5 mM AMP, 75 mM Mes, pH 7.0, 0.2 M GlcNAc, 1.0% Triton X-100 and 0.05 mg of BSA. Incubations were carried out for 1 h at 26 °C. The reaction was stopped by adding 0.5 ml of distilled water. Products were purified by the SepPak C18 cartridge method [34,35]. Protein concentration was determined by the bicinchoninic acid assay method (Pierce Biotechnology). Control assays for endogenous acceptors were carried out with no added acceptor substrate; these values were subtracted from the standard incubation values. Product formation was proportional to time of incubation and volume of enzyme under the conditions of the standard enzyme assay.
Sequencing of gly-12, gly-13 and gly-14 deletions
Single-worm PCR was performed to amplify the deletion bands of the gly-12, gly-13 and gly-14 genes as described above. The PCR products were purified by electrophoresis in 1.0% agarose gels and extracted using the Qiagen (Mississauga, Ont., Canada) Gel Extraction Kit. Direct sequencing of the PCR products was done by the DNA Sequencing Center at the Hospital for Sick Children, Toronto, Canada.
MS analysis of worm extracts
Mixed stage C. elegans cultures sufficient to yield 10–20 mg of protein were suspended in 1.5 ml of a lysis buffer [35 mM Tris/8 M urea/4% (w/v) CHAPS/65 mM dithiothreitol, pH 8.0], and homogenized using a Barocycler™ NEP2017 (Boston Biomedica Inc., Bridgewater, MA, U.S.A.) sample preparation system. The pressure-cycle treatment, consisting of five cycles of 20 s at 207 MPa (30000 lbf/in2), was performed at room temperature. Protein extracts were centrifuged at 10000 g for 15 min and the protein content of the supernatant was determined using the Bradford assay [36]. Protein in the supernatant was precipitated with 15% (w/v) trichloroacetic acid on ice for 1 h. The protein pellet was washed with 2 ml of acetone (three times) and 1 ml of chloroform/methanol/water (10:10:1, by vol.) (three times), dried under nitrogen and stored at −20 °C.
The protein pellet was vacuum-dried over P2O5 for 48 h immediately prior to hydrazinolysis with 0.4 ml of anhydrous hydrazine (Sigma) for 6 h at 100 °C. After cooling, hydrazine was removed under a nitrogen stream. The released glycans were re-N-acetylated with acetic anhydride (0.1 ml) in saturated NaHCO3 (0.2 ml), de-salted with a cation-exchange resin and purified on microcrystalline cellulose. Glycans were reduced with NaBH4 (0.2 ml of 15 mg/ml NaBH4 in 10 mM NaOH) at room temperature overnight. Borate was removed by adding two drops of acetic acid on ice, followed by co-evaporation with 3 ml of ethanol, 3 ml of 1% acetic acid in methanol (twice), and 3 ml of toluene (three times). The reduced glycans were desalted and subjected to MALDI-TOF (matrix-assisted laser-desorption–ionization time-of-flight) MS analysis. The glycans were also permethylated and analysed by MALDI-TOF [37] for confirmation.
RESULTS AND DISCUSSION
gly-14;gly-12 gly-13 is a triple-null mutant worm
PCR analysis of the gly-14;gly-12 gly-13 worm showed homozygosity for the gly-12, gly-13 and gly-14 mutations (Figure 1). The deleted genomic DNA regions (nucleotide numbering relative to the ATG start codon at 1) are 990 to 2597 (1608 nt) for gly-12, −309 to 1653 (1962 nt) and 2267 to 2343 (77 nt) for gly-13, and 60 to 1806 (1747 nt) for gly-14 (Figure 1). These are null deletions as shown by the absence of GnT I enzyme activity with M5Gn2-PA acceptor substrate; N2 and gly-14;gly-12 gly-13 worm microsomes showed enzyme activities of 6.9 and <0.004 nmol/h per mg of protein respectively (averages of duplicate determinations).
Phenotype analysis of the TKO worm
The TKO worms have a normal shape, show no obvious morphological defects, move well, show neither the Unc nor Egl (from ‘egg-laying defect’) phenotypes, and respond normally to touch. The TKO worms were very similar to the wild-type N2 worms in a series of commonly used phenotypic parameters (average values are given for wild-type and null worm respectively): brood size (270±17, 273±21), life span in days (12.5, 13.1), defecation interval in s/cycle (45.1±2.0, 44.9±2.0), osmotic avoidance at 4 M NaCl (>99% of both normal and mutant worms avoided salt), resistance to levamisole (all worms were paralysed at 25 mM levamisole), Dauer formation and recovery, and male mating efficiency. The post-embryonic development of over 100 embryos from each worm line was monitored for 2 days from eggs to the L4 stage and no differences from normal were observed. Differential-interference-contrast microscopy of the living TKO animals was carried out (results not shown). The only phenotype difference observed between wild-type and null worms was the occasional presence of a dead embryo in the mutant.
MS analysis of extracts from wild-type and TKO worms
Several groups have used MS and other methods to determine the N-glycan structures in extracts of C. elegans [25,38–45]. A recent review [46] summarizes the occasionally conflicting data. Our data on N2 wild-type worms (Table 2) agree with the published information.
Table 2. MS analysis of N-glycans in the TKO worm.
The Table shows ratios of peak heights of mutant C. elegans N-glycans relative to peak heights of wild-type N-glycans after MALDI-TOF MS analysis. Normal structures which are absent in the mutant worms (ratio=0/X) are shown in bold type. Structures seen in the mutant but not in the wild-type worm (ratio=Y/0) are shown in italics. The hexose residues in Hex2–Hex9 are probably mannose residues, but the presence of some galactose cannot be ruled out [45,46]; Hex10 is presumably Glc1Man9. Abbreviations: F, L-fucose; Me, O-methyl. Hex3GlcNAc3 and Hex3GlcNAc4 probably represent truncated mono-antennary and bi-antennary complex N-glycans respectively.
| Ratio of peak height | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| N-glycan | Hex2 | Hex3 | Hex4 | Hex5 | Hex6 | Hex7 | Hex8 | Hex9 | Hex10 |
| GlcNAc2 | 0/100 | 10/34 | 100/41 | 30/20 | 24/16 | 12/12 | 6/14 | 1/1 | |
| GlcNAc3 | 0/18 | ||||||||
| GlcNAc4 | 0/5 | ||||||||
| GlcNAc2F1 | 0/38 | 4/46 | 5/18 | 12/0 | 10/0 | 4/0 | 3/0 | ||
| GlcNAc2F2 | 0/23 | 0/31 | 0/48 | 2/36 | 2/17 | 2/1 | 2/0 | ||
| GlcNAc2F3 | 0/14 | 0/36 | 0/40 | 0/27 | 0/10 | 0/1 | |||
| GlcNAc2F4 | 0/10 | 0/10 | 0/7 | 0/2 | |||||
| GlcNAc2F1Me | 0/16 | 0/5 | |||||||
| GlcNAc2F2Me | 0/10 | 0/20 | 0/7 | ||||||
| GlcNAc2F3Me | 0/16 | 0/18 | 0/16 | 0/8 | 0/1 | ||||
| GlcNAc2F4Me | 0/18 | 0/11 | 0/10 | 0/3 | |||||
The differences between TKO and wild-type worms are dramatic (Table 2): 31 N-glycans seen in wild-type worms are not made by the mutant (bold type), five N-glycans not seen in wild-type worms are made by the mutant (italics) and there are several structures which show either decreased or increased amounts of N-glycan in mutant relative to wild-type worms. Although most of the hexoses detected by MS are probably mannose residues, small amounts of galactose are also present in C. elegans [45,46]; the data in Table 2 are therefore given as hexose residues. The results of our MS analyses of wild-type N2 and TKO worm extracts are summarized as follows (Table 2).
(i) Oligomannose Hex5–9GlcNAc2 N-glycans are present in relatively large amounts in the N2 worms. There are significant increases in Hex5–7GlcNAc2 in the GnT I null worm consistent with the positions of oligomannose structures upstream of GnT I in the synthetic pathway (Scheme 1).
(ii) Fucosylated oligomannose Hex5GlcNAc2Fuc1–4, Hex6-GlcNAc2Fuc2–4 and Hex7GlcNAc2Fuc3,4 are present in moderate to large amounts in wild-type worms. These structures are either absent or greatly reduced in the GnT I null worm. Hex5GlcNAc2, Hex6GlcNAc2 and Hex7GlcNAc2 are upstream of GnT I (Scheme 1). If these N-glycans are the substrates for the fucosyltransferases responsible for the above structures, GnT I-dependent N-glycans must somehow be required for fucosyltransferase activity.
There is evidence from other laboratories [46] for the existence in C. elegans of one or more families of unusual fucose-rich Hex3–7GlcNAc2-Asn-X N-glycans in which fucose is attached not only to the asparagine-linked core GlcNAc but also to peripheral residues such as mannose or galactose. Further study of the TKO worm will determine whether any of these structures are present in the GnT I-dependent fucosylated oligomannose N-glycans identified by our data.
(iii) Fucosylated oligomannose Hex6–9GlcNAc2Fuc1 and Hex8-GlcNAc2Fuc2 are observed in the GnT I null worm, but not the wild-type worm. This is an unexpected observation. The results suggest that the respective fucosyltransferases are inhibited by GnT I-dependent N-glycans by some as yet unknown mechanism.
(iv) Paucimannose Hex3,4GlcNAc2 are present in large amounts in the wild-type worm, but are absent or greatly reduced in the GnT I null worm. We have shown that the synthesis of paucimannose N-glycans by C. elegans requires the prior actions of GnT I, α3,6-mannosidase II and a specific membrane-bound β-N-acetylglucosaminidase (Scheme 1; [26]). The results in Table 2 add further support to this observation. There may be some GnT I-independent α3,6-mannosidase activity [47] in C. elegans, since the GnT I-null worm makes a small amount of Hex4GlcNAc2 (Scheme 1 and Table 2).
(v) Fucosylated paucimannose Hex3GlcNAc2Fuc1–3 and Hex4-GlcNAc2Fuc2–4 are present in large amounts in the wild-type worm but are absent in the GnT I null worm. There is a large decrease in the amount of Hex4GlcNAc2Fuc1. These findings can be explained by a lack of substrate for the respective fucosyltransferases, owing to the dependence of paucimannose N-glycan synthesis on prior GnT I action (Scheme 1; [26]). Alternatively, the core fucosyltransferase may require prior GnT I action, as previously shown for vertebrates, insects and plants [48–51].
(vi) Hybrid N-glycan Hex3GlcNAc3 (Scheme 1) is present in relatively small amounts in wild-type worms, but absent in GnT I null worms. This is consistent with the pathway shown in Scheme 1 in which prior GnT I action is essential for hybrid N-glycan synthesis.
(vii) Bi-antennary complex N-glycan Hex3GlcNAc4 is present in very small amounts in wild-type worms, but absent in GnT I null worms. This structure requires prior GnT I action [8].
(viii) Other laboratories have reported both fucose and mannose with O-methyl groups in C. elegans [40,41]. We have also observed that wild-type worms have relatively small amounts of O-methylated fucose residues, Hex4,5GlcNAc2(methyl)Fuc1, Hex3–5GlcNAc2(methyl)Fuc2, Hex3–7GlcNAc2(methyl)Fuc3 and Hex4–7GlcNAc2(methyl)Fuc4 (Table 2). These N-glycans are all absent in the GnT I null worm, probably due to the lack of fucosylated substrates for the methylation enzymes.
Conclusions
GnT I null mouse embryos die at day 10.5 [13,14] whereas, in marked contrast, we have shown that a similar GnT I null mutation in C. elegans results in a normal phenotype. This discrepancy illustrates the dangers of extrapolating conclusions from one species to another. It is concluded that wild-type C. elegans makes a large number of GnT I-dependent N-glycans that are not essential for normal worm development under laboratory conditions. However, the complexity of the machinery for the synthesis of GnT I-dependent N-glycans in worms suggests that these N-glycans have important functions. Such functions may become evident if the TKO worm is subjected to environmental stress or to defects in one or more other genes. If this hypothesis is correct, the TKO worm may provide a valuable tool for identifying genes that require an active GnT I for optimum function.
Acknowledgments
H. S. and A. M. S. were supported by grants from the Canadian Institutes of Health Research and the Canadian Protein Engineering Network of Centres of Excellence (PENCE). H. S. was also supported by the Mizutani Foundation for Glycoscience. A. H. and V. N. R. were supported in part by National Institutes of Health (NIH) grant no. P20 RR16459 (V. N. R.) from the BRIN (Biomedical Research Infrastructure Networks) Program of the National Center for Research Resources; the content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Dr W. Brent Derry, Department of Medical Genetics and Microbiology, Hospital for Sick Children, Toronto, Canada, for differential-interference-contrast microscopy of the worms.
References
- 1.Fukuda M. Cell surface carbohydrates: cell type-specific expression. Mol. Cell. Glycobiol. 2000;30:1–61. [Google Scholar]
- 2.Lowe J. B. Glycosylation in the control of selectin counter-receptor structure and function. Immunol. Rev. 2002;186:19–36. doi: 10.1034/j.1600-065x.2002.18603.x. [DOI] [PubMed] [Google Scholar]
- 3.Feizi T. Carbohydrate-mediated recognition systems in innate immunity. Immunol Rev. 2000;173:79–88. doi: 10.1034/j.1600-065x.2000.917310.x. [DOI] [PubMed] [Google Scholar]
- 4.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R. C. elegans II. Cold Spring Harbor Monogr. Ser. 1997;33 [Google Scholar]
- 6.Wood W. B. Plainview, NY: Cold Spring Harbor Laboratory Press; 1988. The nematode Caenorhabditis elegans. [Google Scholar]
- 7.Epstein H. F., Shakes D. C. Caenorhabditis elegans: modern biological analysis of an organism. Methods Cell Biol. 1995;48 [Google Scholar]
- 8.Schachter H. The “yellow brick road” to branched complex N-glycans. Glycobiology. 1991;1:453–461. doi: 10.1093/glycob/1.5.453. [DOI] [PubMed] [Google Scholar]
- 9.Stanley P., Raju T. S., Bhaumik M. CHO cells provide access to novel N-glycans and developmentally regulated glycosyltransferases. Glycobiology. 1996;6:695–699. doi: 10.1093/glycob/6.7.695. [DOI] [PubMed] [Google Scholar]
- 10.Gottlieb C., Baenziger J., Kornfeld S. Deficient uridine diphosphate-N-acetylglucosamine:glycoprotein N-acetylglucosaminyltransferase activity in a clone of Chinese hamster ovary cells with altered surface glycoproteins. J. Biol. Chem. 1975;250:3303–3309. [PubMed] [Google Scholar]
- 11.Stanley P., Narasimhan S., Siminovitch L., Schachter H. Chinese hamster ovary cells selected for resistance to the cytotoxicity of phytohemagglutinin are deficient in a UDP-N-acetylglucosamine:glycoprotein N-acetylglucosaminyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 1975;72:3323–3327. doi: 10.1073/pnas.72.9.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vischer P., Hughes R. C. Glycosyl transferases of baby-hamster-kidney (BHK) cells and ricin-resistant mutants. N-glycan biosynthesis. Eur. J. Biochem. 1981;117:275–284. doi: 10.1111/j.1432-1033.1981.tb06334.x. [DOI] [PubMed] [Google Scholar]
- 13.Ioffe E., Stanley P. Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc. Natl. Acad. Sci. U.S.A. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzler M., Gertz A., Sarkar M., Schachter H., Schrader J. W., Marth J. D. Complex asparagine-linked oligosaccharides are required for morphogenic events during post-implantation development. EMBO J. 1994;13:2056–2065. doi: 10.1002/j.1460-2075.1994.tb06480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charuk J. H. M., Tan J., Bernardini M., Haddad S., Reithmeier R. A. F., Jaeken J., Schachter H. Carbohydrate-deficient glycoprotein syndrome type II – An autosomal recessive N-acetylglucosaminyltransferase II deficiency different from typical hereditary erythroblastic multinuclearity, with a positive acidified-serum lysis test (HEMPAS) Eur. J. Biochem. 1995;230:797–805. doi: 10.1111/j.1432-1033.1995.0797h.x. [DOI] [PubMed] [Google Scholar]
- 16.Jaeken J., Schachter H., Carchon H., De Cock P., Coddeville B., Spik G. Carbohydrate deficient glycoprotein syndrome type II: A deficiency in Golgi localised N-acetylglucosaminyltransferase II. Arch. Dis. Child. 1994;71:123–127. doi: 10.1136/adc.71.2.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan J., Dunn J., Jaeken J., Schachter H. Mutations in the MGAT2 gene controlling complex N-glycan synthesis cause carbohydrate-deficient glycoprotein syndrome type II, an autosomal recessive disease with defective brain development. Am. J. Hum. Genet. 1996;59:810–817. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Schachter H., Marth J. D. Mice with a homozygous deletion of the Mgat2 gene encoding UDP-N-acetylglucosamine:α6-D-mannoside β1,2-N-acetylglucosaminyltransferase II: a model for congenital disorder of glycosylation type IIa. Biochim. Biophys. Acta. 2002;1573:301–311. doi: 10.1016/s0304-4165(02)00397-5. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y., Tan J., Sutton-Smith M., Ditto D., Panico M., Campbell R. M., Varki N. M., Long J. M., Jaeken J., Levinson S. R., et al. Modeling human congenital disorder of glycosylation type IIa in the mouse: conservation of asparagine-linked glycan-dependent functions in mammalian physiology and insights into disease pathogenesis. Glycobiology. 2001;11:1051–1070. doi: 10.1093/glycob/11.12.1051. [DOI] [PubMed] [Google Scholar]
- 20.Finnie J. W. Effect of tunicamycin on hepatocytes in vitro. J. Comp. Pathol. 2001;125:318–321. doi: 10.1053/jcpa.2001.0510. [DOI] [PubMed] [Google Scholar]
- 21.Finnie J. W., O'Shea J. D. Acute hepatotoxicity with resultant pulmonary and cerebral embolism in guinea pigs given tunicamycin. Pathology. 1989;21:194–199. doi: 10.3109/00313028909061058. [DOI] [PubMed] [Google Scholar]
- 22.Jago M. V., Culvenor C. C. Tunicamycin and corynetoxin poisoning in sheep. Aust. Vet. J. 1987;64:232–235. doi: 10.1111/j.1751-0813.1987.tb09689.x. [DOI] [PubMed] [Google Scholar]
- 23.Stewart P. L., Hooper P. T., Colegate S. M., Edgar J. A., Raisbeck M. F. Toxic effects of a single parenteral dose of tunicamycin in late stage pregnancy in rats. Vet. Hum. Toxicol. 2002;44:211–215. [PubMed] [Google Scholar]
- 24.Chen S. H., Zhou S. H., Sarkar M., Spence A. M., Schachter H. Expression of three Caenorhabditis elegans N-acetylglucosaminyltransferase I genes during development. J. Biol. Chem. 1999;274:288–297. doi: 10.1074/jbc.274.1.288. [DOI] [PubMed] [Google Scholar]
- 25.Schachter H., Chen S., Zhang W., Spence A. M., Zhu S., Callahan J. W., Mahuran D. J., Fan X., Bagshaw R. D., She Y. M., et al. Functional post-translational proteomics approach to study the role of N-glycans in the development of Caenorhabditis elegans. Biochem. Soc. Symp. 2002;69:1–21. doi: 10.1042/bss0690001. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W., Cao P., Chen S., Spence A. M., Zhu S., Staudacher E., Schachter H. Synthesis of paucimannose N-glycans by Caenorhabditis elegans requires prior actions of UDP-GlcNAc:α-3-D-mannoside β1,2-N-acetylglucosaminyltransferase I, α-3,6-mannosidase II and a specific membrane-bound β-N-acetylglucosaminidase. Biochem. J. 2003;372:53–64. doi: 10.1042/BJ20021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S., Spence A. M., Schachter H. Isolation of null alleles of the Caenorhabditis elegans gly-12, gly-13 and gly-14 genes, all of which encode UDP-GlcNAc:α-3-D-mannoside β1,2-N-acetylglucosaminyltransferase I activity. Biochimie. 2003;85:391–401. doi: 10.1016/s0300-9084(03)00009-9. [DOI] [PubMed] [Google Scholar]
- 28.Zhu S., Sarkar M., Boulianne G., Spence A. M., Schachter H. Caenorhabditis elegans and Drosophila melanogaster lines with defects in the expression of UDP-GlcNAc:α-3-D-mannoside β-1,2-N-acetylglucosaminyltransferase I. Glycobiology. 2003;13:894. [Google Scholar]
- 29.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. New York: John Wiley and Sons; 1993. Current Protocols in Molecular Biology. [Google Scholar]
- 30.Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 31.Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Church D. L., Guan K. L., Lambie E. J. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development. 1995;121:2525–2535. doi: 10.1242/dev.121.8.2525. [DOI] [PubMed] [Google Scholar]
- 33.Lewis J. A., Fleming J. T. Basic culture methods. Methods Cell Biol. 1995;48:3–29. [PubMed] [Google Scholar]
- 34.Palcic M. M., Heerze L. D., Pierce M., Hindsgaul O. The use of hydrophobic synthetic glycosides as acceptors in glycosyltransferase assays. Glycoconjugate J. 1988;5:49–63. [Google Scholar]
- 35.Sarkar M. Expression of recombinant rabbit UDP-GlcNAc:α3-D-mannoside β-1,2-N-acetylglucosaminyltransferase I catalytic domain in Sf9 insect cells. Glycoconjugate J. 1994;11:204–209. doi: 10.1007/BF00731219. [DOI] [PubMed] [Google Scholar]
- 36.Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye-binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 37.Ciucanu I., Kerek F. A simple and rapid method for the permethylation of carbohydrates. Carbohydrate Res. 1984;131:209–217. [Google Scholar]
- 38.Guerardel Y., Balanzino L., Maes E., Leroy Y., Coddeville B., Oriol R., Strecker G. The nematode Caenorhabditis elegans synthesizes unusual O-linked glycans: identification of glucose-substituted mucin-type O-glycans and short chondroitin-like oligosaccharides. Biochem. J. 2001;357:167–182. doi: 10.1042/0264-6021:3570167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosa J. C., Schachter H., Reinhold B., Reinhold V. N. The glycome of Caenorhabditis elegans, a pathway to function. Glycobiology. 2001;11:884–885. [Google Scholar]
- 40.Altmann F., Fabini G., Ahorn H., Wilson I. B. Genetic model organisms in the study of N-glycans. Biochimie. 2001;83:703–712. doi: 10.1016/s0300-9084(01)01297-4. [DOI] [PubMed] [Google Scholar]
- 41.Haslam S. M., Gems D., Morris H. R., Dell A. The glycomes of Caenorhabditis elegans and other model organisms. Biochem. Soc. Symp. 2002;69:117–134. [PubMed] [Google Scholar]
- 42.Natsuka S., Adachi J., Kawaguchi M., Nakakita S.-I., Hase S., Ichikawa A., Ikura K. Structure analysis of n-linked glycans in Caenorhabditis elegans. J. Biochem. (Tokyo) 2002;131:807–813. doi: 10.1093/oxfordjournals.jbchem.a003169. [DOI] [PubMed] [Google Scholar]
- 43.Cipollo J. F., Costello C. E., Hirschberg C. B. The fine structure of Caenorhabditis elegans N-glycans. J. Biol. Chem. 2002;277:49143–49157. doi: 10.1074/jbc.M208020200. [DOI] [PubMed] [Google Scholar]
- 44.Hirabayashi J., Hayama K., Kaji H., Isobe T., Kasai K. Affinity capturing and gene assignment of soluble glycoproteins produced by the nematode Caenorhabditis elegans. J. Biochem. (Tokyo) 2002;132:103–114. doi: 10.1093/oxfordjournals.jbchem.a003186. [DOI] [PubMed] [Google Scholar]
- 45.Hanneman A., Reinhold V. Abundant and unusual N-linked glycans from the eukaryote C. elegans. Glycobiology. 2003;13:899–900. [Google Scholar]
- 46.Haslam S. M., Dell A. Hallmarks of Caenorhabditis elegans N-glycosylation: complexity and controversy. Biochimie. 2003;85:25–32. doi: 10.1016/s0300-9084(03)00041-5. [DOI] [PubMed] [Google Scholar]
- 47.Chui D., Oh-Eda M., Liao Y. F., Panneerselvam K., Lal A., Marek K. W., Freeze H. H., Moremen K. W., Fukuda M. N., Marth J. D. α-Mannosidase-II deficiency results in dyserythropoiesis and unveils an alternate pathway in oligosaccharide biosynthesis. Cell. 1997;90:157–167. doi: 10.1016/s0092-8674(00)80322-0. [DOI] [PubMed] [Google Scholar]
- 48.Wilson J. R., Williams D., Schachter H. The control of glycoprotein synthesis. I. N-acetylglucosamine linkage to mannose residue as a signal for the attachment of L-fucose to the asparagine-linked N-acetylglucosamine residue of glycopeptide from α1-acid glycoprotein. Biochem. Biophys. Res. Commun. 1976;72:909–916. doi: 10.1016/s0006-291x(76)80218-5. [DOI] [PubMed] [Google Scholar]
- 49.Staudacher E., Dalik T., Wawra P., Altmann F., Marz L. Functional purification and characterization of a GDP-fucose:β-N-acetylglucosamine (Fuc to Asn linked GlcNAc) α1,3-fucosyltransferase from mung beans. Glycoconjugate J. 1995;12:780–786. doi: 10.1007/BF00731239. [DOI] [PubMed] [Google Scholar]
- 50.Leiter H., Mucha J., Staudacher E., Grimm R., Glössl J., Altmann F. Purification, cDNA cloning, and expression of GDP-L-Fuc:Asn-linked GlcNAc α1,3-fucosyltransferase from mung beans. J. Biol. Chem. 1999;274:21830–21839. doi: 10.1074/jbc.274.31.21830. [DOI] [PubMed] [Google Scholar]
- 51.März L., Altmann F., Staudacher E., Kubelka V. Protein glycosylation in insects. In: Montreuil J., Vliegenthart J. F. G., Schachter H., editors. Glycoproteins. vol. 29a. Amsterdam: Elsevier; 1995. pp. 543–563. [Google Scholar]