Abstract
POP (prolyl oligopeptidase) specifically hydrolyses a number of small proline-containing peptides at the carboxy end of the proline residue and POP inhibitors have been shown to have cognition-enhancing properties. It has been noted that certain functional groups at the P1 site of the inhibitor, which correspond to the substrate residue on the N-terminal side of the bond to be cleaved, increase the inhibitory potency. However, detailed mechanistic and kinetic analysis of the inhibition has not been studied. In the present study, we examined the effect of different functional groups at the P1 site of the parent inhibitor isophthalic acid bis-(L-prolylpyrrolidine) amide on the binding kinetics to POP. Addition of CHO, CN or COCH2OH groups to the P1 site increased the inhibitory potency by two orders of magnitude (Ki=11.8–0.1 nM) and caused a clear slow-binding inhibition. The inhibitor containing a CHO group had the lowest association rate constant, kon=(2.43±0.12)×105 M−1·s−1, whereas the inhibitor with a CN group exhibited the fastest binding, kon=(12.0±0.08)×105 M−1·s−1. In addition, the dissociation rate was found to be crucially dependent on the type of the functional group. Compounds with COCH2OH and CHO groups had much longer half-lives of dissociation (over 5 h) compared with the compound with the CN group (25 min), although the Ki values of the compounds were relatively similar. A possibility to optimize the duration of inhibition by changing the functional group at the P1 site is important when planning therapeutically useful POP inhibitors.
Keywords: enzyme kinetics, prolyl oligopeptidase, slow- and tight-binding inhibition, Z-prolylprolinal
Abbreviations: DTT, dithiothreitol; POP, prolyl oligopeptidase; Z-Gly-Pro-AMC, N-benzyloxycarbonylglycylprolyl-7-amino-4-methylcoumarin
INTRODUCTION
POP (prolyl oligopeptidase; EC 3.4.21.26) is a large enzyme (80 kDa), which belongs to a POP family of serine proteases, unrelated to trypsin, subtilisin and carboxypeptidase Y [1]. The POP family includes POP, dipeptidyl peptidase IV (EC 3.4.14.5), oligopeptidase B (EC 3.4.21.83) and acylaminoacyl peptidase (EC 3.4.19.1), and this enzyme family is of ancient origin [2]. POP preferentially hydrolyses small peptides at the carboxy side of a proline residue. Although POP is predominantly a cytosolic enzyme, a membrane-bound form has also been characterized [3].
Several POP substrates, such as substance P, vasopressin, neurotensin and thyroliberin, are claimed to be involved in learning and memory [4]. In addition, low levels of substance P are commonly found in the brains of patients suffering from Alzheimer's disease, and administration of substance P has been reported to block β-amyloid-induced neurotoxicity [5]. It was recently reported that POP gene transcription was decreased when rats were exposed to an enriched environment and the transcription was increased manyfold in hypothalamus and cortex from aged rats [6,7]. Therefore it has been postulated that centrally acting POP inhibitors might be beneficial in patients with cognitive disturbances. Indeed, POP inhibitors have been shown to reverse scopolamine-induced amnesia in rats and to improve cognition in old rats and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-treated monkeys [8–10]. In addition, POP inhibitors have been reported to prevent β-amyloid deposition in a neuroblastoma cell line and in senescence-accelerated mouse [11,12].
The overall principles of catalysis and inhibition of the serine protease POP are reasonably well documented, and several very potent POP inhibitors have been developed [13]. However, the relationship between the structure of the inhibitor and the rate of the reaction is still scarcely known. This would be useful information, since it could provide the means to regulate the duration of enzyme inhibition when using POP inhibitors as a therapeutic agent.
Previously, one POP inhibitor, Z-prolylprolinal (see Figure 1), has been reported to act in a slow-binding manner [14]. This compound contains a CHO group at the P1 site and has been shown to form a hemiacetal adduct with the active-site serine residue, mimicking the tetrahedral transition state of the enzyme-catalysed reaction [15]. In addition to CHO group, also CN and COCH2OH groups at the P1 site have been reported to increase the affinity of isophthalic acid bis-(L-prolylpyrrolidine) amide, probably by interacting with the active-site serine residue [16]; however, no details are available of their binding kinetics and the importance of different functional groups. Slow-binding inhibition is a phenomenon in which inhibition occurs relatively slowly and not at diffusion-controlled rates. Analysis of slow-binding inhibition provides insight into the binding mode of the inhibitors and also allows the calculation of enzyme–inhibitor association and dissociation constants [17].
Figure 1. Z-prolyl-prolinal and the inhibitors used in the study.
In the present study, we performed a detailed kinetic analysis of isophthalic acid bis-(L-prolylpyrrolidine) amide containing different 2(S) substituents at the pyrrolidine ring at the P1 site (see Figure 1). Indeed, all of the three functional groups were shown to lead to a slow-binding inhibition mechanism and this allowed us to determine the association and dissociation rates of the inhibition. Measurements indicated that both of these kinetic parameters are crucially dependent on the characteristics of the functional group, which can be used to optimize the duration of action of POP inhibitors.
EXPERIMENTAL
Reagents
Z-Gly-Pro-AMC (N-benzyloxycarbonyl-glycyl-prolyl-7-amino-4-methylcoumarin) was obtained from Bachem AG (Bubendorf, Switzerland). Na2HPO4 and KH2PO4 were from Merck (Darmstadt, Germany), methanol from Labscan (Dublin, Ireland) and DMSO from Riedel-de Haën (Seelze, Germany).
Compounds
The four inhibitors that were used in the experimental study are presented in Figure 1. The studied inhibitors were synthesized at the University of Kuopio [16]. Stock solutions of 0.1 M were made in DMSO; further dilutions were made in 0.1 M sodium/potassium phosphate buffer (pH 7.0).
Expression and purification of porcine POP
Expression and purification of porcine POP were performed as reported previously, and the active enzyme concentration was determined using the tight-binding POP inhibitor JTP-4819 [18]. Purified porcine POP was used as the enzyme in all measurements.
Determination of association and dissociation rate constants
Slow-binding inhibition measurements were performed at 23 °C in 0.1 M sodium/potassium phosphate buffer (pH 7.0), containing 0.1 mM DTT (dithiothreitol) at an enzyme concentration of 0.14 nM. From the two commercially available POP substrates, Z-Gly-Pro-AMC was used, since it does not exhibit substrate inhibition [18], which could lead to erroneous estimation of the association and dissociation constants. Z-Gly-Pro-AMC was used at 10 μM, which is 0.5 Km. Owing to its poor solubility, Z-Gly-Pro-AMC was dissolved and diluted in methanol. The final methanol concentration in the reaction mixture was 1%, which inhibited POP by <10%. The reactions were initiated by the addition of the enzyme, and the progress curves of AMC liberation were monitored every 1 min for 60 min using a Bio-Tek FL500 fluorescence plate reader at excitation and emission wavelengths of 360 and 460 nm respectively. Five or six inhibitor concentrations were used for each inhibitor. Blank values (substrate without enzyme) were subtracted from data and the progress curves were fitted to the rate equation of slow-binding inhibition [17]
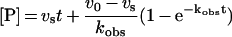 |
(1) |
using GraphPad Prism version 3.02 software (GraphPad Software, San Diego, CA, U.S.A.).In the above equation, [P] is the product concentration, v0 and vs are the initial and final steady-state reaction rates, t the time and kobs the apparent first-order rate constant for determining the final steady-state velocity.
The apparent association and dissociation constants were then calculated as described in the Results section.
Recovery of enzyme activity
The dissociation rate constants were also determined directly by measuring the recovery of enzyme activity after dilution of the enzyme–inhibitor complex. In this assay, 5 nM of the inhibitor was incubated with 5 nM of the enzyme for 1 h at 23 °C. After incubation, the mixture was diluted 200-fold into the assay buffer containing 100 μM Z-Gly-Pro-AMC and the reaction was monitored every 1 min for 100 min. The data were fitted to eqn (1), in which kobs corresponds to the dissociation rate constant.
Determination of Ki values
The Ki value for the inhibitor with R=H was calculated using eqn (2) and the results from the progress-curve experiments:
 |
(2) |
Ki (app) is an apparent inhibition constant that was converted to a real Ki value according to:
 |
(3) |
where [S] is the substrate concentration (10 μM) and Km the Henry–Michaelis–Menten constant for Z-Gly-Pro-AMC (20 μM). The Km value was determined as reported previously [18]. Since the compound with R=H did not show any time-dependent inhibition, the v0 value was calculated from the control reaction in the absence of an inhibitor.
The Ki values for compounds with R=CHO, R=COCH2OH and R=CN could not be determined by the same method as for the compound with R=H, because they were shown to be tight-binding inhibitors of POP (their Ki values are of the same order of magnitude as the enzyme concentration in the reaction mixture). Instead, their slow dissociation from the enzyme allowed direct determination of the real Ki value. The enzyme (0.57 nM) was incubated for 2 h in the presence of various concentrations of inhibitors at 23 °C. The reaction was started with Z-Gly-Pro-AMC (final concentration 10 μM) and the reaction was monitored every 1 min for 10–15 min. Over that time scale, the product formation was linear, indicating that the inhibitor did not dissociate markedly from the enzyme. The Ki values were calculated using the Morrison equation, which takes the tight-binding inhibition into account [19]:
 |
(4) |
where v0 and vi are reaction velocities in the absence and presence of the inhibitor (I) respectively, Ki is the inhibition constant of the inhibitor and E the active enzyme concentration in the reaction medium (0.57 nM). Since the inhibitor did not dissociate from the enzyme during the measurement, competition of binding between substrate and inhibitor did not occur and, hence, the calculated Ki value is the real dissociation constant of the inhibitor [20].
Inhibitor stability
The possible degradation of inhibitors by POP was tested with the parent inhibitor (compound with R=H). The compound (5 μM) was incubated with 10 nM enzyme for 0 or 24 h in 20 mM sodium/potassium phosphate buffer (pH 7.0) at 23 °C. The incubations were stopped by filtration with Amicon Ultra-4 centrifugal filter device with 30 kDa-cut-off membrane. Samples (50 μl) were injected into a reversed-phase HPLC system consisting of Shimadzu LC-10AD pump, SPD-10A UV-detector, SIL-9A autoinjector and Supelco Supelcosil LC-18-DB column (25 cm×4.6 mm; inner diameter, 5 μm). The mobile phase was 25% acetonitrile, 75% 20 mM sodium/potassium phosphate buffer (pH 7.0) and the detection wavelength and flow rate were 215 nm and 0.5 ml/min respectively. The HPLC method exhibited excellent linearity and repeatability.
RESULTS
Slow-binding inhibition
As shown in Figures 2(A) and 2(B), POP inhibition by the compound with R=CHO showed a clear time- and concentration-dependent approach to steady state, whereas the steady-state rate of substrate hydrolysis was reached instantaneously. Similar slow-binding inhibition was also observed for compounds with R=CN and R=COCH2OH. In contrast, the time course of inhibition by the compound with R=H was linear, indicating rapid binding.
Figure 2. Slow-binding inhibition of POP.
(A) Progress curves for the hydrolysis of Z-Gly-Pro-AMC in the presence of various concentrations of compound with R=CHO, monitored in 0.1 M sodium/potassium phosphate buffer (pH 7.0), containing 0.1 mM DTT at 23 °C. (B) Region of the initial velocity of the progress curve. (C) kobs as a function of inhibitor concentration for the compounds with R=CHO, R=CN and R=COCH2OH. Results are expressed as means±S.E.M. for 3–5 independent experiments.
In principle, there are two ways in which slow-binding inhibition can occur: the direct binding of inhibitor (Scheme 1a) or the isomerization of an initial enzyme–inhibitor complex (Scheme 1b) [17]. In the direct-binding model, the binding of the inhibitor to the enzyme active site is either a slow process (low-association constant, kon) or both konI and koff are low. In the latter case, the low association and dissociation rates would lead to inhibition of slow binding, even though kon was of a similar magnitude to that seen with diffusion-controlled interaction. In the isomerization model described in Scheme 1(b), the inhibitor first binds rapidly to the enzyme (complex EI formation), which then undergoes a slow conformational change to form the inactive complex EI*.
Scheme 1. Kinetic schemes for slow-binding inhibition.
(a) Binding of the inhibitor to the enzyme active site is a single-step process, whereas in the two-step scheme (b), the inhibitor first binds rapidly to the enzyme (formation of complex EI), which then undergoes a slow conformational change to form the inactive complex EI*.
The association and dissociation constants of a slow-binding inhibitor can be determined by plotting the apparent first-order rate constant kobs against inhibitor concentration. Depending on the inhibition mechanism, the plot is a straight line (for the direct-binding model; Scheme 1a) or a hyperbola (for the isomerization model; Scheme 1b). Figure 2(C) shows that the plot of kobs versus [I] is linear for all of the studied compounds, which indicates that the binding of the inhibitors to the enzyme active site is a single-step process. Therefore the data could be fitted to:
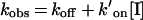 |
(5) |
where koff and k′on are the dissociation and association rate constants respectively. k′on was determined from the slope of the plot and then corrected for substrate competition using:
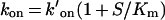 |
(6) |
The koff values of the inhibitors were so low that the interceptions of the y-axis in eqn (5) did not provide a very accurate estimate of the dissociation rate constant. Instead, the koff values were calculated with the help of Ki value according to:
 |
(7) |
Ki values were calculated using the Morrison equation (eqn 4). A representative plot for the compound with R=CHO is shown in Figure 3.
Figure 3. Measurement of Ki for the compound with R=CHO.
The enzyme (0.57 nM) was incubated for 2 h in the presence of various concentrations of the compound with R=CHO, after which the reaction was started with Z-Gly-Pro-AMC. vi/v0 was fitted with the Morrison equation (eqn 4). Results are expressed as means±S.E.M. for three independent experiments.
The kinetic parameters were determined similarly for the compounds with R=CN and R=CH2OH groups. For the compound with R=H, only the Ki value was determined, since the inhibitor did not exhibit slow-binding inhibition kinetics. The results of the kinetic measurements are presented in Table 1.
Table 1. Kinetic parameters for POP inhibitors.
The inhibition kinetic assays were conducted with Z-Gly-Pro-AMC as a substrate in 0.1 M sodium/potassium phosphate buffer (pH 7.0), containing 0.1 mM DTT at 23 °C. Results are expressed means±S.E.M. for 3–5 measurements; ND, not detected.
| Compound | Ki (nM) | Kon×105 (M−1·s−1) | koff×10−5 (s−1) | EI half-life |
|---|---|---|---|---|
| R=H | 11.8±0.5 | ND | ND | ND |
| R=CN | 0.39±0.03 | (12.0±0.08) | (46.7±0.46) | 25±3 min |
| R=CHO | 0.15±0.01 | (2.43±0.12) | (3.56±0.34) | 5.4±0.5 h |
| R=COCH2OH | 0.079±0.010 | (4.66±0.23) | (3.68±0.50) | 5.2±0.7 h |
It can be seen that the presence of CHO, CN and COCH2OH groups greatly increases the inhibitory potency of the compound, as previously noted in IC50 determinations [16]. The Ki values of these compounds are in the sub-nanomolar range, in contrast with the Ki value of 11.8 nM for the compound having R=H. The inhibitor containing the COCH2OH group is the most potent inhibitor of POP, having a Ki value of 0.079±0.010 nM. All these compounds also exhibited slow-binding inhibition kinetics. The lowest association rate constant was measured for the compound with R=CHO; the difference in association constants between the inhibitors with R=CHO and R=CN was 5-fold. The compounds with R=COCH2OH and R=CHO had equal, very low dissociation rate constants, which correspond to half-lives of more than 5 h for dissociation. In contrast, for the compound with R=CN, the half-life was only 25 min for dissociation. Thus, even though the difference in Ki values was only 2-fold between the inhibitors with R=CN and R=CHO, the half-life for dissociation was 13 times longer for the compound with R=CHO. To visualize the considerable differences in the kinetic parameters between the compounds, the association and dissociation half-lives are presented in Figure 4. The association half-life was calculated for the inhibitor concentration of 10 nM. The three compounds can be categorized as follows: (1) R=CN, fast association, fast dissociation; (2) R=CHO, slow association, slow dissociation; and (3) R=COCH2OH, intermediate association rate, slow dissociation.
Figure 4. Association (black bars, left y-axis) and dissociation (white bars, right y-axis) half-lives of slow-binding POP inhibitors at a concentration of 10 nM.
Results are means±S.E.M. for 3–5 independent measurements.
It is important to mention that the inhibitors are not cleaved by POP, as incubation with 5 μM compound with R=H during 24 h did not decrease the amount of the inhibitor as measured by HPLC (results not shown). The degradation of compounds with R=CN, R=CHO and R=COCH2OH is also unlikely, since they contain only the same peptide bonds similar to the tested compound.
The koff values were also determined by enzyme dilution experiments (Figure 5) for the compounds with R=CN, R=CHO and R=COCH2OH. The recovery of activity was so fast for the inhibitor with R=H that the dissociation rate could not be measured. In these assays, the inhibitor concentrations used were >10Ki before dilution and <35Ki (app) after dilution. The 95% confidence intervals for the half-lives of the dissociations were 29–36 min for the compound with R=CN, 3.9–10.0 h for the compound with R=CHO and 2.9–5.3 h for the compound with R=COCH2OH. These experimental values agree with the calculated dissociation rates, further supporting the direct-binding model of the inhibition (Scheme 1a).
Figure 5. Recovery of enzyme activity after the dilution of the enzyme–inhibitor complex.
Inhibitor and enzyme (5 nM) were incubated for 1 h at 23 °C, after which the complex was diluted 200-fold into the assay buffer containing 100 μM Z-Gly-Pro-AMC. The liberation of AMC was monitored every 1 min for 100 min.
The kinetic parameters of the compound with R=CHO can be compared with those of the previously reported slow-binding inhibitor, Z-prolylprolinal, because they share the same functional formyl group [14]. The association and dissociation constants were found to be almost equal (2.4×105 versus 1.6×105 M−1· s−1 and 3.6×10−5 s−1 versus 4×10−5 s−1) for these inhibitors, although their P3 and P4 sites are quite different. Hence, it seems that the functional CHO group is the principal factor determining the inhibition kinetics of those compounds.
DISCUSSION
We have shown that the addition of a CHO, CN or COCH2 group to the P1 site of isophthalic acid bis-(L-prolylpyrrolidine) amide decreases the IC50 value strongly [16]. However, as the IC50 value is an overall result of the association and dissociation rates of the inhibition and it is also affected by the enzyme and substrate concentrations used in the measurement, it does not provide any insight into the reaction mechanism or kinetic behaviour. In the present study, we have analysed the inhibition reaction with these potent inhibitors in detail and found that the inhibitors achieve their high potency in different ways.
The kinetic measurements indicated that the addition of a CHO, CN or COCH2OH group to the P1 site of the inhibitor leads to a slow-binding inhibition of POP. Inhibition with each of the studied inhibitor was a single-step process, as can be deduced from the linear relationship in the plot of kobs versus [I]. This inhibition mechanism is described in Scheme 1(a). However, distinguishing between the direct binding and the isomerization mechanisms is not straightforward. If the k2/k1 ratio in the isomerization model is much greater than the k4/k3 and the measurements are made using an inhibitor concentration lower than k2/k1, the plot of kobs versus [I] would also be linear for an inhibitor that obeys this mechanism (Scheme 1b) [19]. Irrespective of that complication, we suggest that the slow-binding inhibitors studied here follow the direct-binding model for two reasons. First, the progress curves of the compound with a CHO group (Figures 2A and 2B) show that the initial velocity, v0, does not decrease even at the highest inhibitor concentration (20 nM), when compared with the uninhibited reaction velocity. This concentration is higher than the Ki value of the compound with R=H, which was seen to bind rapidly. Secondly, if the slow-binding compounds were obeying the two-step mechanism, the core of the inhibitor should first bind rapidly to the active site (corresponding to the binding of the compound with R=H) and form a complex, as was observed with the compound having R=H. In the second step, the enzyme–inhibitor complex would undergo a slow conformational change and form a tighter complex EI* due to the CHO group. However, this first complex would have a dissociation constant of 11.8 nM (Ki for compound R=H) and, thus, the highest measured inhibitor concentration of 20 nM should cause a marked decrease in the initial velocity of the reaction. Since that was not the case (see Figures 2A and 2B), we conclude that the inhibition of POP by these slow-binding inhibitors is really a single-step process. The single-step reaction is also supported by the fact that the calculated and directly measured dissociation constants were equal.
Although addition of each of the groups, CHO, CN and COCH2OH, led to highly potent POP inhibitors and revealed single-step slow-binding inhibition mechanism, there were considerable differences in the inhibition rates between the compounds. The inhibitor with R=CN was found to have the highest association constant, followed by the compounds with R =COCH2OH and R=CHO. It has been shown that the Ser-554 at the enzyme active site attacks the CHO carbon atom of the inhibitor, resulting in a covalent hemiacetal adduct [15]. Similarly, it is probable that the CN and COCH2OH groups interact with the same Ser-554 residue and form an imino ether and a hemiketal adduct respectively, and these covalent adducts increase the affinity of the inhibitors [16]. The rate of the hemiacetal formation is considerably slower than the formation of the imino ether and the hemiketal adducts, demonstrated by the observation that the association rate of the inhibitor with R=CHO is 2- and 5-fold lower than the association rates of the inhibitors with R=COCH2OH and R=CN respectively. It seems that the formation of these adducts is a relatively slow process and mainly responsible for the slow-binding inhibition.
In addition to the differences in the association rates, differences in the dissociation rates between these slow-binding inhibitors were also found. Namely, the inhibitor with R=CN had a half-life of 25 min for dissociation, in contrast with the inhibitors with R=CHO and R=COCH2OH, which dissociated much more slowly from the enzyme, having the half-life of over 5 h. The differences in the dissociation rates often affect the Ki values of the inhibitors since Ki=koff/kon. However, the Ki values of the inhibitors studied here do not predict the dissociation rates. For example, the difference in Ki values is only 2-fold between the inhibitors with R=CN and R=CHO, even though the half-life for dissociation is 13 times higher for the compound with R=CHO. This is because the association rate of the compound with R=CN is much higher than that of the compound with R=CHO. The findings reported in the present study are of relevance, as POP inhibitors are under consideration for therapeutic use in the treatment of cognitive deficits associated with aging and neurodegenerative diseases. The slow dissociation would be predicted to result in a long duration of action and, consequently, inhibition of POP could persist even after the drug has been cleared from the plasma, as was recently shown with the anti-Alzheimer drug rivastigmine [21]. Here, we have demonstrated that the duration of action can be adjusted by changing the functional group at the P1 site of the inhibitor. This information can be used to develop potent short- or long-acting POP inhibitors.
In summary, in the present study, we have demonstrated that (i) the addition of COCH2OH, CN or CHO group at the P1 site of isophthalic acid bis-(L-prolylpyrrolidine) amide leads to slow-binding inhibition of POP and the inhibition appears to be a single-step process; and (ii) although the Ki values of the inhibitors are relatively similar, both the association and dissociation rates are crucially dependent on the type of the functional group: fast association, fast dissociation with R=CN; slow association, slow dissociation with R=CHO; intermediate association rate, slow dissociation with R=COCH2OH. These findings will be valuable when developing POP inhibitors for therapeutic use.
Acknowledgments
We thank Mrs Päivi Sutinen and Mrs Jaana Leskinen for their excellent technical assistance and we are grateful to Dr Ewen MacDonald for linguistic advice. This work was supported in part by the National Technology Agency of Finland (Tekes) and Finncovery Ltd.
References
- 1.Rennex D., Hemmings B. A., Hofsteenge J., Stone S. R. cDNA cloning of porcine brain prolyl endopeptidase and identification of the active-site seryl residue. Biochemistry. 1991;30:2195–2203. doi: 10.1021/bi00222a025. [DOI] [PubMed] [Google Scholar]
- 2.Venäläinen J. I., Juvonen R. O., Männistö P. T. Evolutionary relationships of the prolyl oligopeptidase family enzymes. Eur. J. Biochem. 2004;271:2705–2715. doi: 10.1111/j.1432-1033.2004.04199.x. [DOI] [PubMed] [Google Scholar]
- 3.O'Leary R. M., Gallagher S. P., O'Connor B. Purification and characterization of a novel membrane-bound form of prolyl endopeptidase from bovine brain. Int. J. Biochem. Cell. Biol. 1996;28:441–449. doi: 10.1016/1357-2725(95)00154-9. [DOI] [PubMed] [Google Scholar]
- 4.Huston J. P., Hasenohrl R. U. The role of neuropeptides in learning: focus on the neurokinin substance P. Behav. Brain Res. 1995;66:117–127. doi: 10.1016/0166-4328(94)00132-y. [DOI] [PubMed] [Google Scholar]
- 5.Kowall N. W., Beal M. F., Busciglio J., Duffy L. K., Yankner B. A. An in vivo model for the neurodegenerative effects of beta amyloid and protection by substance P. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7247–7251. doi: 10.1073/pnas.88.16.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rampon C., Jiang C. H., Dong H., Tang Y. P., Lockhart D. J., Schultz P. G., Tsien J. Z., Hu Y. Effects of environmental enrichment on gene expression in the brain. Proc. Natl. Acad. Sci. U.S.A. 2000;97:12880–12884. doi: 10.1073/pnas.97.23.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang C. H., Tsien J. Z., Schultz P. G., Hu Y. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider J. S., Giardiniere M., Morain P. Effects of the prolyl endopeptidase inhibitor S 17092 on cognitive deficits in chronic low dose MPTP-treated monkeys. Neuropsychopharmacology. 2002;26:176–182. doi: 10.1016/S0893-133X(01)00307-4. [DOI] [PubMed] [Google Scholar]
- 9.Atack J. R., Suman-Chauhan N., Dawson G., Kulagowski J. J. In vitro and in vivo inhibition of prolyl endopeptidase. Eur. J. Pharmacol. 1991;205:157–163. doi: 10.1016/0014-2999(91)90814-7. [DOI] [PubMed] [Google Scholar]
- 10.Marighetto A., Touzani K., Etchamendy N., Torrea C. C., De Nanteuil G., Guez D., Jaffard R., Morain P. Further evidence for a dissociation between different forms of mnemonic expressions in a mouse model of age-related cognitive decline: effects of tacrine and S 17092, a novel prolyl endopeptidase inhibitor. Learn. Mem. 2000;7:159–169. doi: 10.1101/lm.7.3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinoda M., Toide K., Ohsawa I., Kohsaka S. Specific inhibitor for prolyl endopeptidase suppresses the generation of amyloid β protein in NG108-15 cells. Biochem. Biophys. Res. Commun. 1997;235:641–645. doi: 10.1006/bbrc.1997.6730. [DOI] [PubMed] [Google Scholar]
- 12.Kato A., Fukunari A., Sakai Y., Nakajima T. Prevention of amyloid-like deposition by a selective prolyl endopeptidase inhibitor, Y-29794, in senescence-accelerated mouse. J. Pharmacol. Exp. Ther. 1997;283:328–335. [PubMed] [Google Scholar]
- 13.De Nanteuil G., Portevin B., Lepagnol J. Prolyl endopeptidase inhibitors: a new class of memory enhancing drugs. Drugs Future. 1998;23:167–179. [Google Scholar]
- 14.Bakker A. V., Jung S., Spencer R. W., Vinick F. J., Faraci W. S. Slow tight-binding inhibition of prolyl endopeptidase by benzyloxycarbonyl-prolyl-prolinal. Biochem. J. 1990;271:559–562. doi: 10.1042/bj2710559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fülöp V., Böcskei Z., Polgár L. Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis. Cell (Cambridge, Mass.) 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 16.Wallén E. A. A., Christiaans J. A. M., Forsberg M. M., Venäläinen J. I., Männistö P. T., Gynther J. Dicarboxylic acid bis(L-prolyl-pyrrolidine) amides as prolyl oligopeptidase inhibitors. J. Med. Chem. 2002;45:4581–4584. doi: 10.1021/jm020966g. [DOI] [PubMed] [Google Scholar]
- 17.Morrison J. F., Walsh C. T. The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 18.Venäläinen J. I., Juvonen R. O., Forsberg M. M., Garcia-Horsman A., Poso A., Wallén E. A. A., Gynther J., Männistö P. T. Substrate-dependent, non-hyperbolic kinetics of pig brain prolyl oligopeptidase and its tight binding inhibition by JTP-4819. Biochem. Pharmacol. 2002;64:463–471. doi: 10.1016/s0006-2952(02)01184-x. [DOI] [PubMed] [Google Scholar]
- 19.Copeland R. A. A Practical Introduction to Structure. 2nd edn. Wiley, New York: Mechanism and Data Analysis; 2000. Enzymes. [Google Scholar]
- 20.Gutheil W. G., Bachovchin W. W. Separation of L-Pro-DL-boroPro into its component diastereomers and kinetic analysis of their inhibition of dipeptidyl peptidase IV. A new method for the analysis of slow, tight-binding inhibition. Biochemistry. 1993;32:8723–8731. doi: 10.1021/bi00085a001. [DOI] [PubMed] [Google Scholar]
- 21.Bar-On P., Millard C. B., Harel M., Dvir H., Enz A., Sussman J. L., Silman I. Kinetic and structural studies on the interaction of cholinesterases with the anti-Alzheimer drug rivastigmine. Biochemistry. 2002;41:3555–3564. doi: 10.1021/bi020016x. [DOI] [PubMed] [Google Scholar]








