Abstract
C4-specific (photosynthetic) NADP+-dependent malic enzyme (NADP+-ME) has evolved from C3-malic enzymes and represents a unique and specialized form, as indicated by its particular kinetic and regulatory properties. In the present paper, we have characterized maize (Zea mays L.) photosynthetic NADP+-ME mutants in which conserved basic residues (lysine and arginine) were changed by site-directed mutagenesis. Kinetic characterization and oxaloacetate partition ratio of the NADP+-ME K255I (Lys-255→Ile) mutant suggest that the mutated lysine residue is implicated in catalysis and substrate binding. Moreover, this residue could be acting as a base, accepting a proton in the malate oxidation step. At the same time, further characterization of the NADP+-ME R237L mutant indicates that Arg-237 is also a candidate for such role. These results suggest that both residues may play ‘back-up’ roles as proton acceptors. On the other hand, Lys-435 and/or Lys-436 are implicated in the coenzyme specificity (NADP+ versus NAD+) of maize NADP+-ME by interacting with the 2′-phosphate group of the ribose ring. This is indicated by both the catalytic efficiency with NADP+ or NAD+, as well as by the reciprocal inhibition constants of the competitive inhibitors 2′-AMP and 5′-AMP, obtained when comparing the double mutant K435/6L (Lys-435/436→Ile) with wild-type NADP+-ME. The results obtained in the present work indicate that the role of basic residues in maize photosynthetic NADP+-ME differs significantly with respect to its role in non-plant MEs, for which crystal structures have been resolved. Such differences are discussed on the basis of a predicted three-dimensional model of the enzyme.
Keywords: maize (Zea mays), malic enzyme, NADP+-dependent malic enzyme, NADP+ specificity, photosynthesis, site-directed mutagenesis
Abbreviations: IF, intrinsic fluorescence; NAD+-ME, NAD+-dependent malic enzyme; NADP+-ME, NADP+-dependent malic enzyme; rH, oxaloacetate partitioning ratio; K255I, Lys-255→Ile; K435/6L, Lys-435/436→Ile
INTRODUCTION
Malic enzymes (MEs) catalyse the reversible oxidative decarboxylation of L-malate to yield pyruvate, CO2 and NAD(P)H in the presence of a bivalent metal ion [1]. The enzyme is found in most living organisms, from bacteria to animals and plants. In eukaryotic organisms, different ME isoforms are found in the cytosol, mitochondria and/or chloroplasts. MEs are involved in different metabolic pathways depending on the tissue and subcellular localization. Crystal structures of human mitochondrial NAD(P)+-dependent ME [2], pigeon cytosolic NADP+-ME (NADP+-dependent ME) [3] and the parasitic nematode Ascaris suum mitochondrial NAD+-ME (NAD+-dependent ME) [4] have indicated that ME is a new class of oxidative decarboxylase, with a distinct backbone structure. More recently, the crystal structures of ME bound to different substrates have been reported [5,6], giving new insights into the structure and catalytic mechanism of this key enzyme.
In plants, different isoforms of NADP+-ME are involved in a wide range of metabolic pathways [7,8]. The photosynthetic C4-NADP+-ME, which is involved in the CO2 concentrating mechanism that increases the photosynthetic yield of some C4 plants, is compartmentalized in bundle-sheath chloroplasts. This C4-specific NADP+-ME represents a unique and specialized form of ME, as indicated by its particular kinetic and regulatory properties [7,8]. We have recently expressed the maize (Zea mays L.) photosynthetic and non-photosynthetic NADP+-MEs in a prokaryotic system. These enzymes present intriguing differences with regards to MEs from non-plant sources [9,9a].
In the present work, site-directed mutagenesis was performed to investigate the role of conserved basic residues (Lys-255, Arg-237 and Lys-435/436) in maize photosynthetic (C4) NADP+-ME. Alanine-scanning site-directed mutagenesis carried out on all conserved lysine residues of pigeon liver NADP+-ME have shown only two residues to be involved in catalysis and coenzyme binding [10]. In NAD+-ME from A. suum, Lys-199 was identified as the proton donor in the NAD+-ME enzyme reaction based on site-directed mutagenesis experiments [11]. More recently, after the crystallization of human NAD(P)+-ME complexed with its substrates, Lys-182 (analogous to Lys-199 in NAD+-ME from A. suum) was suggested to be the general base for the oxidation step [5]. The results obtained in the present work regarding the conserved lysine (and arginine) residues of maize C4-NADP+-ME, give new insights on the role played by these residues in this particular NADP+-ME.
EXPERIMENTAL
Materials
NADP+, NADPH, NAD+, L-malate, oxaloacetate, Tris and entero-kinase were purchased from Sigma (St. Louis, MO, U.S.A.). DNA-modifying enzymes were purchased from Promega (Madison, WI, U.S.A.). All other reagents were of analytical grade.
Site-directed mutagenesis of C4-NADP+-ME and expression and purification of wild-type and mutated NADP+-ME
Site-directed mutagenesis of maize mature NADP+-ME was carried out according to the procedure described by Mikaelian and Sergeant [12]. The primers that introduced mutations in the wild-type protein were as follows: K255I (Lys-255→Ile), 5′-CCTGTAGGCATACTTGCTCTATAC-3′; K435/6L, 5′-GCGCCCAAGGTGCCGCGAATGGCTG-3′; and R237L, 5′-ACTGATGGTGAGCTAATCTTGGGA-3′ [9]. The mutated positions are emboldened and underlined in the oligonucleotide sequence. These mutagenic primers were used in combination with the following primers in order to obtain the complete sequence of NADP+-ME: 5EMMut, 5′-TAGCATGCCATGGCGATG-3′; 3EMMut, 5′-CAACGCTCGAGGCACTACC-3′; site 2, 5′-ATTACGATTCTGCAATACCAGTTCCAGC-3′; site 5, 5′-ATTCATACTAGGAGGAGTTGACCCATCAG-3′.
PCR was carried out using a mixture of Taq and Pfu polymerases (10:1) in order to minimize possible errors introduced by Taq polymerase. The resulting mutated sequences obtained by PCR were analysed by electrophoresis on agarose gels, cloned into pGEMT-Easy vector (Promega) and digested with restriction enzymes upstream and downstream of the mutated sites. The resulted fragments were separated by agarose gels, purified by QIAprep columns (Qiagen) and ligated to pET32-ME, which contains the mature sequence of wild-type photosynthetic NADP+-ME [9], previously digested with the same set of restriction enzymes. The introduced sequences in the mutated plasmids were sequenced to verify the introduction of the corresponding mutations and that no errors were added due to PCR and subcloning procedures. Escherichia coli BL21(DE3) cells were then transformed with the pET32 plasmid containing the mutated NADP+-Mes, and the different proteins were expressed and purified as described previously for the wild-type NADP+-ME [9], except that the Affi-Gel Blue column was omitted in some cases due to the lack of or low binding properties of the mutated enzymes.
Protein concentration was determined by the method of Sedmak and Grossberg [13] using BSA as molecular-mass standard.
Estimation of kinetic parameters of wild-type and mutated NADP+-ME
NADP+-ME activity was determined spectrophotometrically at 30 °C by monitoring NADPH production at 340 nm. The standard reaction mixture contained 50 mM Tris/HCl, pH 8.0, 10 mM MgCl2, 0.5 mM NADP+ and 4 mM L-malate. The reaction was started by the addition of L-malate. One unit is defined as the amount of enzyme that catalyses the formation of 1 μmol of NADPH/min under the specified conditions.
Initial-velocity studies were performed by varying the concentration of one of the substrates around its Km while keeping the other substrate concentrations at saturating levels. All kinetic parameters were calculated at least in triplicate and subjected to non-linear regression. Free concentrations of all substrates were calculated as described by Grover et al. [14].
Malic enzyme activity in the presence of NAD+ was measured in a reaction mixture, keeping free Mg2+ concentration constant while varying NAD+ concentration (as described in [15]). In this case, the reaction medium contained 50 mM Tris/HCl, pH 7.0, 10 mM MgCl2 and 4 mM L-malate. Initial activity was measured at different NAD+ concentrations by adding NAD+/MgCl2 (1:0.34) solution, adjusted to pH 7.0, to the assay mixture. This was not necessary when testing NADP+, since NADP+-ME activity is saturated by low concentrations of this substrate.
Oxaloacetate partitioning of wild-type and mutated NADP+-ME
The partitioning ratio of oxaloacetate was performed according to the method of Karsten and Cook [16]. Partitioning reaction mixtures contained 100 mM Hepes, pH 7.0, 0.8 mM NADPH, 1 mM oxaloacetate, 20 mM MgCl2 and 1–5 units of wild-type or mutated NADP+-ME. Typically partitioning was allowed to proceed for 5 min, over which the reaction rates were linear. The disappearance of oxaloacetate was monitored at 282 nm, while the formation of malate was monitored by the decrease in absorbance at 340 nm associated with the disappearance of NADPH. After subtracting the background control rate, the partitioning ratio (rH), expressed as pyruvate/malate, was calculated as follows:
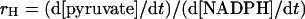 |
where d[pyruvate]/dt=(d[oxaloacetate]/dt)−(d[NADPH/dt).
Inhibition studies
Initial-velocity studies of inhibition were performed by varying the inhibitor concentration (2′-AMP or 5′-AMP) around their inhibition constants (Ki). The reaction was started by the addition of malate (4 mM) in the presence of a fixed NADP+ (0.02–2 mM) and different 2′-AMP or 5′-AMP concentrations in the assay medium, while keeping the cation concentration at saturating levels (10 mM MgCl2). The dissociation constants of the inhibitors (Ki) were calculated at least in triplicate and adjusted to the equation for a linear competitive-type inhibitor by non-linear regression:
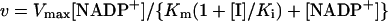 |
where Vmax is the maximum NADP+-ME activity in the absence of inhibitor and [I] is the concentration of 2′-AMP or 5′-AMP.
CD and fluorescence measurements
CD spectra were produced with a Jasco J-810 spectropolarimeter using a 1.0-cm pathlength cell and averaging five repetitive scans between 250 and 200 nm. Typically, 50 μg of the wild-type or mutated NADP+-ME in phosphate buffer (20 mM sodium phosphate, pH 8.0, and 5 mM MgCl2) were used for each assay.
IF (intrinsic fluorescence) spectra of the recombinant wild-type and mutated NADP+-ME were analysed with a PerkinElmer 650-40 fluorescence spectrophotometer set at 20 °C, using an excitation wavelength of 295 nm and slit widths of 5 nm. Typically, 5 μg of the wild-type or mutated NADP+-ME in phosphate buffer were used for each assay. All spectra were corrected for buffer absorption and the Raman spectra of water.
RESULTS
Purification and structural characterization of NADP+-ME mutants
Lys-255 and Lys-435/436 in maize photosynthetic NADP+-ME were replaced by isoleucine (K255I) or leucine (double mutant K435/6L) respectively. The mutated enzymes were successfully expressed in E. coli BL21(DE3) and purified to homogeneity using the same procedure described previously for the mature wild-type NADP+-ME [9].
To determine whether the introduced mutations resulted in a loss of overall structural integrity, CD and IF spectra were measured for all mutant proteins and compared with the one obtained for the wild-type NADP+-ME. In all cases, the CD and IF spectra were superimposable after corrections were made for protein concentration (results not shown). In this way, the changes in structure, if any, should be limited to the active site without generating a severe loss of protein structure.
Kinetic characterization of K255I and K435/6L NADP+-ME mutants
The apparent kinetic parameters of the recombinant wild-type photosynthetic NADP+-ME and the mutants K255I and K435/6L were measured (Table 1).
Table 1. Kinetic parameters of the recombinant wild-type NADP+-ME and site-directed mutants.
The indicated kinetic parameters are the means±S.E.M. for at least five separate determinations. The kinetic parameters for the mutant R237L were described previously [9], but they are included for comparison. N.A., no activity could be measured.
| Parameter | NADP+-ME… | Wild-type | K255I | K435/6L | R237L |
|---|---|---|---|---|---|
| kcat NADP (s−1) | 201.3±6.4 | 1.10±0.05 | 181.1±4.7 | 0.38±0.02 | |
| Km NADP+ (μM) | 8.0±0.3 | 123.0±5.2 | 73.0±1.9 | 290.0±6.9 | |
| Km malate (mM) | 0.23±0.01 | 2.60±0.12 | 0.31±0.02 | 2.90±0.13 | |
| kcat/Km NADP+ | 25.2 | 0.029 | 2.48 | 0.001 | |
| kcat NAD (s−1) | 13.5±0.6 | N.A. | 18.4±0.7 | N.A. | |
| Km NAD+ (mM) | 8.10±0.37 | N.A. | 1.9±0.09 | N.A. | |
| kcat/Km NAD+ | 1.7 | N.A. | 9.9 | N.A. | |
| Km NAD+/Km NADP+ | 987 | N.A. | 26 | N.A. |
The K255I mutant showed a marked decrease in kcat NADP (nearly 200-fold) with respect to the wild-type, whereas the Km values for NADP+ and malate were increased by 15- and more than 10-fold (Table 1). On the other hand, the double mutant K435/6L was not significantly affected in its catalytic capacity (the kcat NADP was almost the same as that of the wild-type). The Km for malate was also practically unaffected, whereas the Km for NADP+ increased 9-fold (Table 1). Table 1 shows also the kinetic parameters of the R237L mutant [9]; which was further characterized in the present work.
When using NAD+ as cofactor instead of NADP+, the mutants K255I and R237L presented no measurable activity, which might be caused by the low specific activity of these mutants. Instead, the K435/6L mutant presented a slightly increase in the kcat,NAD, whereas the Km for NAD+ decreased 4-fold with respect to the wild-type (Table 1). In this case, the catalytic efficiency with NAD+ presented an increase of nearly 6-fold.
Oxaloacetate partitioning studies
Partitioning of oxaloacetate in the E·NADPH·Mg·oxaloacetate complex toward either the E·NADPH·Mg·enolpyruvate or E·NADP+·Mg·malate complexes were measured for the wild-type NADP+-ME, K255I, K435/6L and R237L mutants (Table 2).
Table 2. Pyruvate/malate partitioning ratios for oxaloacetate partitioning in wild-type and K255I, K435/6L and R237L NADP+-ME mutants.
Partitioning ratios are the means±S.E.M. for at least three separate determinations for each case.
| Enzyme | rH | rH mutant/wild-type |
|---|---|---|
| Wild-type | 2.9±0.048 | 1.0 |
| K255I | 29.1±1.13 | 10.0 |
| K435/6L | 3.2±0.12 | 1.1 |
| R237L | 317.4±3.8 | 109.5 |
The partition ratio (formation rate of pyruvate/formation rate of malate) yielded a value of 2.9 for the wild-type, in agreement with the ratio of 2.3 measured for the human mitochondrial NAD+-ME [17], but significantly greater from the 0.4 value described for the NAD+-ME from A. suum [11].
The partition ratios of oxaloacetate for the K255I mutant was 10-fold higher, whereas for the R237L variant it was more than 100-fold higher, in both cases compared with the wild-type form (Table 2). Thus these mutations preferentially decrease the rate of oxaloacetate reduction with respect to its decarboxylation. On the other hand, the mutation of K435/6L does not substantially alter the rates of hydride transfer and decarboxylation relative to each other (Table 2).
Inhibition of K435/6L by 2′-AMP or 5′-AMP
The dissociation constant of 2′-AMP and 5′-AMP were calculated for the K435/6L NADP+-ME mutant and compared with the value obtained for the wild-type enzyme (Table 3). Both compounds behaved as linear competitive-type inhibitors with respect to NADP+ in both the wild-type and K435/6L mutant NADP+-ME. Nevertheless, mutation at positions K435/6 presented an opposite effect on the dissociation constants of 2′-AMP and 5′-AMP with respect to the wild-type NADP+-ME; that is, whereas the Ki for 2′-AMP increased more than 6 times, the Ki for 5′-AMP decreased 1.7 times.
Table 3. Inhibition constants for 2′-AMP and 5′-AMP with respect to NADP+ in wild-type NADP+-ME and K435/6L mutant.
The values presented are the means±S.E.M. for at least three separate determinations.
| Ki (μM) | ||
|---|---|---|
| Enzyme | 2′-AMP | 5′-AMP |
| Wild-type | 104.5±1.2 | 439.2±6.2 |
| K435/6L | 687.6±12.3 | 255.5±6.1 |
| Wild-type/K435/6L | 0.15 | 1.7 |
Three-dimensional model of maize C4-NADP+-ME
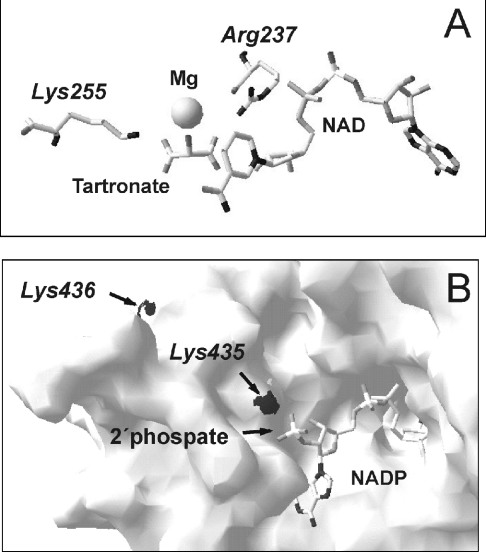
We have previously analysed [9] the three-dimensional model of the maize photosynthetic NADP+-ME based on the crystallographic data for the human mitochondrial NAD(P)+-ME (1QR6, Protein Data Bank; http://www.rcsb.org/pdb/). However, substrate binding at the active site shifts the enzyme from an open to a closed form, leading to an active-site closure and modifying the interaction of some residues with the substrates [5,18]. In this way, we obtained a new three-dimensional model of maize C4-NADP+-ME (predicted by the protein structure homology-modelling server, SWISS-MODEL; http://expasy.ch/swissmod/SWISSMODEL.html), but now using as a template the structure of the human mitochondrial NAD(P)+-ME complexed with its substrates (we specifically used the structure 1EFL from the Protein Data Bank, where tartronate was used as an analogue of malate). The model obtained show significant differences with respect to the three-dimensional model previously obtained [9], suggesting also a modification of the active site after substrate binding, as is the case for the human NAD(P)-ME. In this way, this model was used for discussing the role of the residues Lys-255 and Arg-237, which were mutated in the present work. Figure 1(A) shows the position of these residues with respect to the substrates in the context of this new model.
Figure 1. Three-dimensional model view of the maize chloroplastic C4-NADP+-ME corresponding to the active site of the enzyme.
(A) The predicted model was obtained using human mitochondrial NAD(P)+-ME complexed with their substrates as templates. The position of Lys-255 and Arg-237 relative to tartronate (analogue of malate) is shown. (B) The predicted model was obtained using pigeon liver NADP+-ME in order to analyse the position of the 2′-phosphate in NADP+ relative to Lys-435 and Lys-436. The Figures were created by using Swiss Protein Data Bank Viewer version 3.7.
On the other hand, a three-dimensional model based on pigeon liver NADP+-ME (1GQ2, Protein Data Bank) was constructed in order to discuss the role of Lys-435/6 of maize NADP+-ME (Figure 1B), due to the fact that human NAD+-ME lacks lysine residues at these positions (Figure 2), probably because of its higher affinity for NAD+ instead of NADP+ (see the Discussion).
Figure 2. Sequence alignment around Lys-435/6 of maize photosynthetic NADP+-ME.
Maize NADP+-ME sequences (C4 and non-C4) were compared with the sequences of the three NAD(P)+-MEs that have been crystallized. This comparison was performed using Clustal W (1.81). :, conserved substitution are observed; ., semi-conserved substitutions are observed.
DISCUSSION
Role of Lys-255 and Arg-237 in maize photosynthetic NADP+-ME
Malic enzyme catalyses a three-step acid-base reaction from malate into pyruvate: dehydrogenation (oxidation) of malate into oxaloacetate, decarboxylation of oxaloacetate into enolpyruvate, and tautomerization of enolpyruvate into pyruvate [19]. First, L-malate is converted into oxaloacetate in a reaction facilitated by a general base that accepts a proton from the 2-hydroxy group of L-malate. In the second step, oxaloacetate is decarboxylated into enolpyruvate and a general acid transfers a proton to the oxygen in the enolpyruvate molecule. Finally, enolpyruvate is tautomerized into pyruvate.
In the present paper, the change of Lys-255 to isoleucine in maize C4-NADP+-ME produced a significant decrease in the catalytic activity (nearly 200-fold, Table 1). The affinities for the substrates are also affected (Table 1), rendering an enzyme with 868-fold decrease in the catalytic efficiency with NADP+. The corresponding residue in pigeon liver NADP+-ME (Lys-162) and A. suum NAD+-ME (Lys-199) have been mutated, and showed decreases of 235- and 105-fold in kcat respectively [10,11]. At the same time, the affinity for the substrates was not affected in either case, indicating an additional role for Lys-255 in maize C4-NADP+-ME. The oxaloacetate partitioning rate of the K255I mutant increased 10-fold with respect to the wild-type (Table 2), which indicates that this mutation favours the decarboxylation reaction over the reverse-hydride transfer. Thus Lys-255 would have a more critical role in the reaction previous to the formation of oxaloacetate, that is, in the reductive hydrogenation.
Maize NADP+-ME mutant R237L shows also a significant decrease in kcat (530-fold), which is even more pronounced than for the K255I mutant (Table 1). The oxaloacetate partitioning of this mutant shows more than 100-fold increase with respect to the wild-type NADP+-ME (Table 2), which indicates a serious impairment in the reductive hydrogenation reaction due to the mutation of Arg-237. Previous chemical modification of maize NADP+-ME has suggested an essential role for arginine residues [21].
On the basis of kinetic and mutagenesis studies on A. suum NAD+-ME [11,20], subsequently assessed by crystallographic studies [6], Asp-279 has been proposed to be the general base for the reaction; whereas Lys-199 [corresponding to human NAD(P)+-ME Lys-183 and maize C4-NADP+-ME Lys-255] has been proposed to be the general acid. Nevertheless, crystallographic studies of human NAD(P)+-ME [5] have pointed out that the corresponding residue of Asp-279 is not correctly positioned to remove the proton from the C-2 in malate. They proposed that Lys-183 is the most likely candidate to act as the general base in the dehydrogenation step.
The results obtained in the present work, more specifically the oxaloacetate partitioning results (Table 2), clearly indicate that Lys-255 is not the general acid for the reaction, as suggested for the corresponding residue in A. suum NAD+-ME [6]. Our findings are more in accordance with the role of a general base, as proposed for the corresponding Lys-183 of human NAD(P)+-ME [5]. Nevertheless, based on the results obtained for the maize C4-NADP+-ME R237L mutant, Arg-237 is also a candidate to play such role in this particular enzyme. However, when considering the possibility of Lys-255 or Arg-237 acting as the general base of the malic enzyme reaction, the relatively small decrease in kcat observed for both K255I and R237L mutants (Table 1) is somewhat intriguing, since mutation of the general base for the reaction is expected to yield a more significant decrease in the kcat value. Nonetheless, this fact could be explained if we consider the existence of residues playing a ‘back-up’ role as proton acceptors. In this way, it is possible that the existence of residues Lys-255 and Arg-237, both with the ability to act as the general base, may explain the small change in kcat for the K255I and R237L mutants rather than a total knock-out in the kcat value.
To further evaluate this possibility, we analysed the position of both Lys-255 and Arg-237 relative to the substrate tartronate (used as analogue of malate) in the predicted three-dimensional model of maize C4-NADP+-ME (Figure 1A). This Figure clearly shows that Lys-255 is correctly located to serve as the general base for the reaction, being the likely candidate to remove a proton from malate. However, Arg-237 is also very close to the substrate (we calculated that Lys-255 is approx. 2 Å (0.2 nm) closer than Arg-237 from the OH at position 2 in tartronate). In this way, it is possible that when the positive charge of Lys-255 is mutated (as the case of K255I) the enzyme can alter its conformation in a way that approximates Arg-237 to malate, allowing this residue to play the role of the general base of the reaction, explaining the relative small decrease in kcat for the K255I mutant.
Alternatively, we cannot completely rule out the possibility that Lys-255 and/or Arg-237 may play essential roles in positioning malate for the reduction step, impairing the dehydrogenation step of the catalytic mechanism, but not participating as the general base for the reaction.
Role of Lys-435 and Lys-436 in maize photosynthetic NADP+-ME
Lys-435 of maize C4-NADP+-ME is highly conserved in NADP+-MEs, but not in NAD+-MEs. Figure 2 shows a sequence alignment around this residue, in comparison with maize non-photosynthetic NADP+-ME and other MEs whose crystal structures have been resolved. Moreover, two contiguous lysine residues corresponding to Lys-435 and Lys-436 of maize C4-NADP+-ME are highly conserved in plant NADP+-MEs. This fact is valid for all the NADP+-MEs (18 in total) used to construct the phylogenetic tree in Drincovich et al. [8], with the exception of Mesembryanthemun crystallinun NADP+-ME, where the second lysine residue is replaced by a leucine; and plastidic Ricinus communis and Lycopersicon esculentum NADP+-ME, where a conservative replacement is observed (lysine with arginine). Since the goal of the present work was to investigate the role of the positive charge in this position, we decided to mutate both Lys-435 and Lys-436 of maize C4-NADP+-ME.
The results obtained suggest that the replacement of these two residues by isoleucine has no significant effect on the kcat (Table 1) nor the oxaloacetate partitioning ratio (Table 2), indicating that these residues are not involved in the catalytic process. Nevertheless, this mutation clearly decreases the affinity for NADP+, while increasing that for NAD+ (Table 1). In addition, the Ki for 2′-AMP significantly increases in the mutant, whereas the Ki for 5′-AMP decreases (Table 3). Both inhibitors are competitive with respect to NADP+ in the wild-type as well as in the mutated NADP+-ME, which suggests that they interact with the active site of the enzyme, as was recently confirmed for the human NAD(P)+-ME using different nucleotides [22]. These observations suggest that the lysine residue at position 435 and/or 436 favour(s) the interaction of the enzyme with adenosine derivatives containing a phosphate group on the 2′ position of the ribose ring. In addition, the fact that the affinity for NAD+ and 5′-AMP was increased in the mutant indicates that the lysine residue(s) has a negative effect on the interaction with adenosine derivatives without a phosphate group on the 2′ position. Mutation of Lys-340 in pigeon liver NADP+-ME (corresponding to maize Lys-435) also produced a significant increase in the Km for NADP+ [10]. Moreover, the crystal structure of pigeon liver NADP+-ME, suggests the possibility of an interaction between Lys-340 and the 2′-phosphate of NADP+ [3]. A three-dimensional model of maize C4-NADP+-ME (Figure 1B) indicates that this is also the case for this enzyme, and suggests that the residue implicated in such interaction is Lys-435 and not Lys-436. Nevertheless, it is worth mentioning that pigeon liver NADP+-ME is not inhibited by nucleotides such as ATP, GTP or ADP, an observation attributed to the lack of the 2′-phosphate bound to ribose in these compounds [22]. This is not the case for the maize C4-NADP+-ME, which is inhibited by several nucleotide derivatives (present study and [23]), independent from the presence of a 2′-phosphate group in the ribose ring. In this way, several factors may be acting to determine the binding affinity of nucleotides to the active site of the different MEs.
Concluding remarks
The amino acid residues sequences of human mitochondrial NAD(P)+-ME, pigeon cytosolic NADP+-ME, A. suum NAD+-ME and maize chloroplastic C4-NADP+-ME are highly conserved (between 44 and 50% identity). Nevertheless, several kinetic and structural differences among these MEs are found (e.g. dual specificity for NAD+/NADP+, co-operative effect in malate binding, regulation by fumarate and ATP, inhibition by malate and tetrameric interface). In addition, the results obtained in the present work suggest further differences, such as the interaction of lysine residues with the substrates, the potential existence of alternative residues acting as general bases, as well as the interaction of the enzyme with nucleotides. Crystallographic studies of maize C4-NADP+-ME will help to elucidate the basis for these particular variations.
Acknowledgments
This research was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (PICT 1-03397 and 1-11604, Argentina), Fundación Antorchas (Project N °13887/1 and N °14116/24, Argentina) and CONICET (PIP 3029). M. F. D. and C. S. A. are members of the Researcher Career of CONICET and E. D. is a fellow of the same institution. We thank Dr A. Vila (IBR, Rosario, Argentina) for the use of the CD instrument and Dr Claudio F. Pairoba for carefully reading the manuscript and for his suggestions.
References
- 1.Chang G.-G., Tong L. Structure and function of malic enzyme, a new class of oxidative decarboxylases. Biochemistry. 2003;42:12721–12733. doi: 10.1021/bi035251+. [DOI] [PubMed] [Google Scholar]
- 2.Xu Y., Bhargava G., Wu H., Loeber G., Tong L. Crystal structure of human mitochondrial NAD(P)-dependent malic enzyme: a new class of oxidative decarboxylases. Structure. 1999;7:877–889. [PubMed] [Google Scholar]
- 3.Yang Z., Zhang H., Hung H.-H., Kuo C.-C., Tsai L.-C., Yuan H. S., Chou W.-Y., Chang G.-G., Tong L. Structural studies of the pigeon liver cytosolic NADP-dependent malic enzyme. Protein Sci. 2002;11:332–341. doi: 10.1110/ps.38002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman D. E., Jagannatha Rao G. S., Goldsmith E. J., Cook P. F., Harris B. G. Crystal structure of the malic enzyme from Ascaris suum complexed with nicotinamide adenine dinucleotide at 2.3 Å resolution. Biochemistry. 2002;41:6928–6938. doi: 10.1021/bi0255120. [DOI] [PubMed] [Google Scholar]
- 5.Tao X., Yang Z., Tong L. Crystal structures of substrates complexed of malic enzyme and insights into the catalytic mechanism. Structure. 2003;11:1141–1150. doi: 10.1016/s0969-2126(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 6.Jagannatha Rao G. S., Coleman D. E., Karsten W. E., Cook P. F. Crystallographic studies on Ascaris suum NAD-malic enzyme bound to reduced cofactor and identification of an effector site. J. Biol. Chem. 2003;278:38051–38058. doi: 10.1074/jbc.M305145200. [DOI] [PubMed] [Google Scholar]
- 7.Edwards G. E., Andreo C. S. NADP-malic enzyme from plants. Phytochemistry. 1992;31:1845–1857. doi: 10.1016/0031-9422(92)80322-6. [DOI] [PubMed] [Google Scholar]
- 8.Drincovich M. F., Casati P., Andreo C. S. NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 2001;290:1–6. doi: 10.1016/s0014-5793(00)02331-0. [DOI] [PubMed] [Google Scholar]
- 9.Detarsio E., Gerrard Wheeler M., Campos Bermúdez M. A., Andreo C. S., Drincovich M. F. Maize C4 NADP-malic enzyme: Expression in E. coli and characterization of site-directed mutants at the putative nucleotide binding sites. J. Biol. Chem. 2003;278:13757–13764. doi: 10.1074/jbc.M212530200. [DOI] [PubMed] [Google Scholar]
- 9a.Saigo M., Bologna F. P., Maurino V. G., Detarsio E., Andreo C. S., Drincovich M. F. Maize recombinant non-C4 NADP-malic enzyme: a novel dimeric malic enzyme with high specific activity. Plant Mol. Biol. 2004 doi: 10.1007/s11103-004-0472-z. in the press. [DOI] [PubMed] [Google Scholar]
- 10.Kuo C.-C., Tsai L.-C., Chin T.-Y., Chang G.-G., Chou W.-Y. Lysine residues 162 and 340 are involved in the catalysis and coenzyme binding of NADP-dependent malic enzyme from pigeon. Biochem. Biophys. Res. Commun. 2000;270:821–825. doi: 10.1006/bbrc.2000.2502. [DOI] [PubMed] [Google Scholar]
- 11.Liu D., Karsten W. E., Cook P. F. Lysine 199 is the general acid in the NAD-malic enzyme reaction. Biochemistry. 2000;39:11955–11960. doi: 10.1021/bi000790p. [DOI] [PubMed] [Google Scholar]
- 12.Mikaelian I., Sergeant A. A general and fast method to generate multiple site directed mutagenesis. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sedmak J. J., Grossberg S. E. A rapid, sensitive, and versatile assay for protein using Coomassie brillant blue G250. Anal. Biochem. 1977;79:544–552. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- 14.Grover S. D., Canellas P. F., Wedding R. T. Purification of NAD malic enzyme from potato and investigation of some physiological and kinetic properties. Arch. Biochem. Biophys. 1981;209:396–407. doi: 10.1016/0003-9861(81)90297-6. [DOI] [PubMed] [Google Scholar]
- 15.Ashton A. R. NADP-malic enzyme from C4 plant Flaveria bidentis: Nucleotide substrate specificity. Arch. Biochem. Biophys. 1997;345:251–258. doi: 10.1006/abbi.1997.0260. [DOI] [PubMed] [Google Scholar]
- 16.Karsten W. C., Cook P. F. Stepwise versus concerted oxidative decarboxylation catalyzed by malic enzyme: a reinvestigation. Biochemistry. 1994;33:2096–2103. doi: 10.1021/bi00174a016. [DOI] [PubMed] [Google Scholar]
- 17.Rishavy M. A., Yang Z., Tong L., Cleland W. W. Determination of the mechanism of human malic enzyme with natural and alternate dinucleotides by isotope effects. Arch. Biochem. Biophys. 2001;396:43–48. doi: 10.1006/abbi.2001.2598. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z., Floyd D. L., Loeber G., Tong L. Structure of a closed form of human malic enzyme and implications for catalytic mechanism. Nat. Struct. Biol. 2000;7:251–257. doi: 10.1038/73378. [DOI] [PubMed] [Google Scholar]
- 19.Cleland W. W. Mechanism of enzymatic oxidative decarboxylation. Acc. Chem. Res. 1999;32:862–868. [Google Scholar]
- 20.Karsten W. C., Chooback L., Liu D., Hwang C.-C., Lynch C., Cook P. F. Mapping the active site topography of the NAD-malic enzyme via alanine-scaning site-directed mutagenesis. Biochemistry. 1999;38:10527–10532. doi: 10.1021/bi9906165. [DOI] [PubMed] [Google Scholar]
- 21.Rao S. R., Kamath B. G., Bhagwat A. S. Chemical modification of the functional arginine residue(s) of malic enzyme from Zea mays. Phytochemistry. 1991;30:431–435. [Google Scholar]
- 22.Yang Z., Lanks C. W., Tong L. Molecular mechanism for the regulation of human mitochondrial NAD(P)-dependent malic enzyme by ATP and fumarate. Structure. 2002;10:951–960. doi: 10.1016/s0969-2126(02)00788-8. [DOI] [PubMed] [Google Scholar]
- 23.Spampinato C. P., Paneth P., O'Leary M. H., Andreo C. S. Analogues of NADP as inhibitors and coenzymes for NADP-malic enzyme from maize. Photosynth. Res. 1991;28:69–76. doi: 10.1007/BF00033716. [DOI] [PubMed] [Google Scholar]