Abstract
Background
Klebsiella pneumoniae (KP) is the second most prevalent Gram-negative bacterium causing bloodstream infections (BSIs). In recent years, the management of BSIs caused by KP has become increasingly complex due to the emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP). Although numerous studies have explored the risk factors for the development of CRKP-BSIs, the mortality of patients with KP-BSIs, and the molecular epidemiological characteristics of CRKP, the variability in data across different populations, countries, and hospitals has led to inconsistent conclusions. In this single-center retrospective observational study, we utilized logistic regression analyses to identify independent risk factors for CRKP-BSIs and factors associated with mortality in KP-BSI patients. Furthermore, a risk factor-based prediction model was developed. CRKP isolates underwent whole-genome sequencing (WGS), followed by an evaluation of microbiological characteristics, including antimicrobial resistance and virulence genes, as well as epidemiological characteristics and phylogenetic analysis.
Results
Our study included a total of 134 patients with KP-BSIs, comprising 50 individuals infected with CRKP and 84 with carbapenem-susceptible Klebsiella pneumoniae (CSKP). The independent risk factors for CRKP-BSIs were identified as gastric catheterization (OR = 9.143; CI = 1.357–61.618; P = 0.023), prior ICU hospitalization (OR = 4.642; CI = 1.312–16.422; P = 0.017), and detection of CRKP in non-blood sites (OR = 8.112; CI = 2.130-30.894; P = 0.002). Multivariate analysis revealed that microbiologic eradication after 6 days (OR = 3.569; CI = 1.119–11.387; P = 0.032), high Pitt bacteremia score (OR = 1.609; CI = 1.226–2.111; P = 0.001), and inappropriate empirical treatment after BSIs (OR = 6.756; CI = 1.922–23.753; P = 0.003) were independent risk factors for the 28-day mortality in KP-BSIs. The prediction model confirmed that microbiologic eradication after 6.5 days and a Pitt bacteremia score of 4.5 or higher were significant predictors of the 28-day mortality. Bioinformatics analysis identified ST11 as the predominant CRKP sequence type, with blaKPC−2 as the most prevalent gene variant. CRKP stains carried multiple plasmid-mediated resistance genes along with some virulence genes. Phylogenetic analysis indicated the presence of nosocomial transmission of ST11 CRKP within the ICU.
Conclusions
The analysis of risk factors for developing CRKP-BSIs and the association between KP-BSIs and 28-day mortality, along with the development of a risk factor-based prediction model and the characterization of CRKP strains, enhances clinicians’ understanding of the pathogens responsible for BSIs. This understanding may help in the timely administration of antibiotic therapy for patients with suspected KP-BSIs, potentially improving outcomes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03465-4.
Keywords: Carbapenem-resistant Klebsiella pneumoniae, Risk factors, Bloodstream infections, Virulence genes, Nosocomial transmission
Background
Bloodstream infections (BSIs) pose a critical public health threat globally, leading to high mortality rates among at-risk populations and placing substantial economic burdens on healthcare systems. Klebsiella pneumoniae (KP), a Gram-negative pathogen, is notably implicated in a substantial proportion of BSIs. Research indicates that KP is the second most prevalent Gram-negative bacterium causing BSIs [1]. Carbapenems are the recommended first-line treatment for multidrug-resistant KP-BSIs [2, 3]. However, the management of KP-induced BSIs has been complicated by the emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP). Data from the China Antimicrobial Surveillance Network (CHINET) show a significant rise in resistance to imipenem (from 3 to 24.8%) and meropenem (from 2.9 to 26%) from 2005 to 2023 (http://www.chinets.com/).
Although colistin, fosfomycin, tigecycline, and aminoglycosides have been considered effective in treating CRKP infections, the associated mortality rates for BSIs remain alarmingly high, consistently resulting in fatal outcomes [4, 5]. The reported mortality rates for CRKP-BSIs vary widely, ranging from 42–81% [6–11]. The restricted range of therapeutic options presents a significant challenge for clinicians, exacerbating the issue of antibiotic resistance and posing a serious threat to public health, as emphasized in reports from the Centers for Disease Control and Prevention (CDC) [12].
The risk factors associated with the development of CRKP-BSIs and mortality among patients with KP-BSIs vary across different studies [13–15], despite previous research indicating a global rise in KP-BSI cases [1, 16–18]. Appropriate antibiotic therapy and timely infection control measures are crucial for improving outcomes [19]. Previous studies have linked carbapenem resistance with an increased risk of death [20]. However, it remains unclear whether the increased mortality in patients infected with KP is due to inadequate therapy or to carbapenem resistance. Thus, the epidemiological characteristics, risk factors associated with development of CRKP-BSIs, mortality risk factors for KP-BSIs, and pathogenic characteristics still require further investigation and verification in clinical practice.
This single-center retrospective observational study was primarily designed to evaluate the risk factors for developing CRKP-BSIs and those associated with the mortality of KP-BSIs, and to establish a risk factor-based prediction model. Additionally, the study assessed the bacterial characteristics of CRKP strains, including antimicrobial resistance and virulence genes, as well as their epidemiological characteristics and phylogenetic relationships. This research has the potential to increase clinician awareness and significantly influence the control of KP transmission and improve patient prognosis.
Methods
Study design and participants
This single-center retrospective observational study was conducted at the 980th Hospital of the PLA Joint Logistical Support Force, a 1500-bed medical center in Shijiazhuang, Hebei province, North China. The study included all adult patients (≥ 18 years) diagnosed with KP-BSIs between January 1, 2016, and December 31, 2020. Exclusion criteria were (i) incomplete medical records; (ii) patients diagnosed with polymicrobial BSIs. Initially, patients were devided into carbapenem-susceptible Klebsiella pneumoniae (CSKP) and CRKP groups to identify risk factors for CRKP-BSIs. Subsequently, KP-BSI patients were divided into survival and non-survival groups to analyze the risk factors associated with the 28-day mortality. Lastly, the microbiological characteristics of CRKP were examined through whole-genome sequencing (WGS).
Data collection and definition
Demographic and clinical data were retrieved from the hospital’s electronic medical records system, encompassing detailed such as age, gender, underlying diseases, history of intensive care unit (ICU) stay in our hospital, use of invasive procedure and/or devices within 30 days before the onset of BSIs, antibiotic exposure, CRKP colonization, length of hospitalization before BSIs, and both empirical and definitive antimicrobial therapy. Outcomes recorded included all-cause mortality rates at 7, 14, and 28 days. The severity of bacteremia was assessed using the Pitt bacteremia score [21]. The underlying conditions were evaluated using the Charlson comorbidity index (CCI) [22].
KP-BSIs were defined as having at least one positive blood culture and accompanying signs and symptoms of bacteremia [23]. The BSIs onset was marked by the date of the first positive blood culture collection. CRKP identified in specimens other than blood were considered indicative of CRKP colonization. Antibiotic exposure was defined as the administration of antibiotics for more than 48 h within the 30 days preceding BSIs or prior to discharge. Microbiologic eradication was defined as the absence of KP in blood cultures during follow-up. Empirical antimicrobial therapy involved the prescription of antibiotics prior to obtaining bacterial identification and antimicrobial susceptibility results. Definitive antimicrobial therapy referred to the prescription of antibiotics based on the antimicrobial susceptibility results. Appropriate anti-infection therapy was defined as the administration of at least one antimicrobial agent to which KP was susceptible in vitro, for a duration of at least 48 h.
Bacterial identification and antimicrobial susceptibility testing (AST)
Blood cultures were performed using the BACT/ALERT 3D system (BioMerieux, Lyon, France). Species identification and antimicrobial susceptibility testing (AST) were conducted using the MA120 automatic microbial identification system (Zhuhai Meihua Medical Technology Co., Ltd, Zhuhai, People’s Republic of China). The antimicrobial agents tested included: piperacillin/tazobactam (TZP), amoxicillin-clavulanate (AMC), cefazolin (CFZ), cefoxitin (FOX), cefuroxime (CXM), ceftazidime (CAZ), cefotaxime (CTX), cefepime (FEP), imipenem (IPM), meropenem (MPN), minocycline (MIN), tobramycin (TOB), amikacin (AMK), gentamicin (GEN), ciprofloxacin (CIP), levofloxacin (LEV), cefoperazone/sulbactam (SCF), trimethoprim/sulfamethoxazole (STX), chloramphenicol (C), aztreonam (ATM), polymyxin B (PB), and tigecycline (TG). Escherichia coli ATCC 25,922 and Pseudomonas aeruginosa ATCC 27,853 served as quality control strains for AST. The minimum inhibitory concentrations (MICs) were interpreted according to the Clinical and Laboratory Standards Institute guidelines (CLSI-M100-S29). Carbapenem resistance was defined as an MIC of ≥ 4ug/ml for meropenem or imipenem [24]. KP isolates resistant to any carbapenem (imipenem, meropenem, ertapenem, or doripenem) or producing carbapenemase were classified as CRKP. The criteria and recommendations from the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2020) were applied specifically for tigecycline [25].
Whole-genome sequencing (WGS) and analysis
Genomic DNA was extracted from the CRKP strain using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The purified DNA underwent WGS on the Illumina HiSeq platform (Illumina, San Diego, California). Sequence reads were assembled using SOAP denovo software (version 2.04) [26]. Antimicrobial resistance genes were identified by querying the Comprehensive Antibiotic Resistance Database (CARD) [27]. Virulence-associated genes were detected using the VFDB database (https://www.mgc.ac.cn/VFs/main.htm). The whole genome data were uploaded to the MLST 2.0 database for further analysis. Multilocus sequence typing (MLST) was conducted using MLST software (http://github.com/tseemann/mlst), with sequence type (ST) determination based on seven conserved housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB).
Phylogenetic analysis
The KP strain HS11286 (GenBank NO.016845) served as the reference for comparison in our analysis. Core-genome single nucleotide polymorphisms (cgSNPs) for all strains were identified using Snippy software. Recombinogenic regions were excluded using Gubbins v2.3.4 to ensure accuracy in phylogenetic inference. Subsequently, a phylogenetic tree was constructed using RAxML-NG software (v1.1.0), employing a maximum likelihood (ML) approach.
Statistical analysis
Continuous variables were analyzed based on their distribution: normally distributed data were presented as means ± standard deviations (SDs), while non-normally distributed data were summarized as medians and interquartile ranges (IQR). Categorical variables were presented as frequencies and percentages. The Chi-square test or Fisher’s exact test were employed to compare the resistance rates of the tested antibiotic across different groups.
A logistic regression model was utilized to calculate odds ratios (ORs), 95% confidence intervals (95%CI) and P-values for the univariate analysis of risk factors associated with CRKP-BSIs, as well as factors related to mortality in patients with KP-BSIs. Variables with a P-value < 0.05 in the univariate analysis were included in multivariable logistic regression analysis to evaluate independent risk factors for CRKP-BSIs and for the 28-day mortality of KP-BSIs. A 28-day survival analysis for patients with BSIs was constructed using the Kaplan-Meier method, considering a P-value < 0.05 as statistically significant. The risk factor-based prediction model was assessed using the receiver operating characteristic (ROC) curve. All statistical analyses were performed using SPSS version 20.0 (SPSS Inc., Chicago, IL, USA).
Results
Demographic and clinical characteristics of patients with KP-BSIs
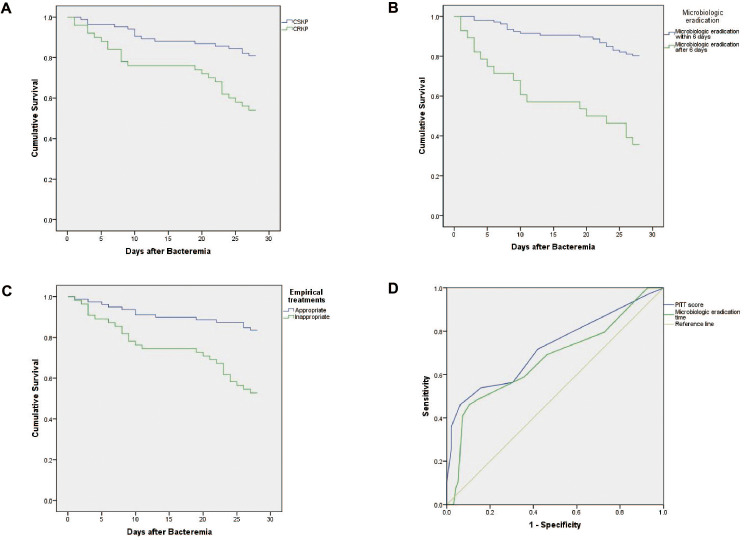
Between January 1, 2016, and December 31, 2020, a total of 152 patients experienced at least one episode of KP-BSIs in our hospital. Eighteen patients were excluded due to being under 18 years of age or having polymicrobial BSIs, leaving 134 patients for inclusion in the study: 50 in the CRKP-BSI group (CRKP group) and 84 in the CSKP-BSI group (CSKP group). The clinical and demographic characteristics of these 134 patients are outlined in Table S1. The 7-day and 28-day mortality rates were significantly higher in the CRKP group compared to the CSKP group [7-day mortality: 16% (8/50) vs. 3.6% (3/84), P = 0.019; 28-day mortality: 46% (23/50) vs. 19% (16/84), P = 0.001]. The 14-day mortality rates were 24% (12/50) in the CRKP group and 11.9% (10/84) in the CSKP group, with no significant difference observed between the groups (P = 0.068). Kaplan-Meier survival curve analysis (Fig. 1A) further demonstrated significantly lower survival probabilities for patients infected with CRKP compared to those infected with CSKP (Log-rank test, P = 0.001).
Fig. 1.
(A) Kaplan-Meier survival curve analysis of the 28-day mortality of patients with CSKP and CRKP bloodstream infections (Log-rank test, P = 0.001). (B) Kaplan-Meier survival curve analysis of the 28-day mortality of patients with KP-BSIs eradicated bacterium within 6 days and after 6 days (Log-rank test, P < 0.001). (C) Kaplan-Meier survival curve analysis of the 28-day mortality of patients with KP-BSIs empirical treated appropriately and inappropriately (Log-rank test, P < 0.001). (D) Construction of the risk factor-based prediction model by receiver operating characteristic (ROC) curve. Microbiologic eradication after 6.5 days and a Pitt bacteremia score of 4.5 or higher were with an AUROC of 0.670 (95%CI = 0.562–0.778, P = 0.002) and 0.723 (95%CI = 0.619–0.827, P < 0.001), respectively. Abbreviations: CSKP, carbapenem-susceptible K.pneumoniae; CRKP, carbapenem-resistant K.pneumoniae; KP-BSIs, Klebsiella pneumoniae bloodstream infections
Risk factors for the development of CKKP-BSIs
Univariate analyses identified several factors significantly associated with the development of CRKP-BSIs, as detailed in Table 1. These factors include a long hospital stay before BSIs, prior ICU hospitalization, and invasive procedure and/or devices. Prior exposure to carbapenems, cephalosporins, glycopeptide, antifungals, and β-lactam/β-lactamase inhibitor combinations were also significant risk factors. Additionally, a higher Pitt bacteremia score and detection of CRKP sites other than blood were associated with the development of CRKP-BSIs. The analysis also indicated that patients with cardiovascular diseases and hypoproteinemia were more susceptible to CRKP-BSIs.
Table 1.
Univariate and multivariate analysis of risk factors for the development of CRKP-BSIs
| Variables | CRKP (n = 50) | CSKP (n = 84) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |||
| Demographic | ||||||
| Gender Male, n(%) | 35 (70) | 51 (60.7) | 1.510 (0.716–3.186) | 0.280 | ||
| Age (median, IQR) | 75 (61, 87) | 65 (53, 77) | 1.027 (1.004–1.050) | 0.021 | ||
| Hospital stay before BSIs, days (median, IQR) | 16 (6, 31) | 2 (1, 12) | 1.045 (1.019–1.071) | 0.001 | ||
| Underlying diseases | ||||||
| Hepatobiliary and pancreatic diseases | 15 (30) | 37 (44) | 0.544 (0.259–1.144) | 0.109 | ||
| Kidney diseases | 17 (34) | 20 (23.8) | 1.648 (0.762–3.564) | 0.204 | ||
| Chronic lung diseases | 12 (24) | 10 (11.9) | 2.337 (0.926–5.898) | 0.072 | ||
| Diabetes mellitus | 18 (36) | 34 (40.5) | 0.827 (0.401–1.705) | 0.607 | ||
| Cardiovascular diseases | 33 (66) | 27 (32.1) | 4.098 (1.949–8.615) | < 0.0001 | ||
| Hypoproteinemia | 40 (80) | 50 (59.5) | 2.720 (1.200-6.167) | 0.017 | ||
| CCI score (median, IQR) | 5 (3, 6) | 5 (3, 6) | 0.966 (0.825–1.130) | 0.664 | ||
| Invasive procedures | ||||||
| Mechanical ventilation | 31 (62) | 6 (7.1) | 21.211 (7.743–58.106) | < 0.0001 | ||
| Urinary catheterization | 40 (80) | 26 (31) | 8.923 (3.879–20.528) | < 0.0001 | ||
| Gastric catheterization | 33 (66) | 8 (9.5) | 18.441 (7.244–46.945) | < 0.0001 | 9.143 (1.357–61.618) | 0.023 |
| Central venous catheterization | 31 (62) | 15 (17.9) | 7.505 (3.377–16.681) | < 0.0001 | ||
| Medical history | ||||||
| IUC stay (previous 1 month) | 30 (60) | 16 (19) | 6.375 (2.907–13.981) | < 0.0001 | 4.642 (1.312–16.422) | 0.017 |
| Prior surgery (previous 1 month) | 15 (30) | 20 (23.8) | 1.371 (0.625–3.010) | 0.431 | ||
| Antibiotics use within 30 days before BSIs | ||||||
| Carbapenems | 25 (50) | 16 (19) | 4.250(1.954–9.245) | < 0.0001 | ||
| Cephalosporins | 35 (70) | 25 (29.8) | 5.507(2.563–11.829) | < 0.0001 | ||
| Quinolones | 17 (34) | 17 (20.2) | 2.030 (0.921–4.478) | 0.079 | ||
| Glycopeptide | 20 (40) | 10 (11.9) | 4.933(2.068–11.771) | < 0.0001 | ||
| Antifungals | 11 (22) | 4 (4.8) | 5.641(1.688–18.855) | 0.005 | ||
| β-lactam/β-lactamase inhibitor combinations | 31 (62) | 27 (32.1) | 3.444(1.657–7.162) | 0.001 | ||
| Severity of illness at the time of BSIs | ||||||
| Pitt bacteremia score (median, IQR) | 3 (1, 5) | 1 (1, 3) | 1.434 (1.192–1.726) | < 0.0001 | ||
| CRKP detection in non-blood sites | 42 (84) | 23 (27.4) | 13.924 (5.687–34.088) | < 0.0001 | 8.112 (2.130-30.894) | 0.002 |
Data are expressed as n (%) or median (IQR)
Subsequent multivariate logistic regression analysis identified several independent risk factors for CRKP-BSIs: gastric catheterization (OR = 9.143; CI = 1.357–61.618; P = 0.023), prior ICU hospitalization (OR = 4.642; CI = 1.312–16.422; P = 0.017), and detection of CRKP in non-blood sites (OR = 8.112; CI = 2.130-30.894; P = 0.002).
Risk factors for 28-day mortality in patients with KP-BSIs
Patients with KP-BSIs were categorized into survival and non-survival groups based on 28-day outcomes. Univariate analysis (Table 2) identified several factors associated with an increased risk of 28-day mortality in patients with KP-BSIs. These factors include a long hospital stay before BSIs, microbiologic eradication after 6 days, prior ICU hospitalization, invasive procedure and/or devices, high Pitt bacteremia score, inappropriate empirical treatment after BSIs, and CRKP infection.
Table 2.
Univariate and multivariate analysis of risk factors for 28-day mortality in patients with KP-BSIs
| Variables | No survivors(n = 39) | Survivors (n = 95) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | |||
| Demographic | ||||||
| Gender Male, n(%) | 28 (71.8) | 58 (61.1) | 1.624 (0.722–3.651) | 0.241 | ||
| Age (Years, mean ± SD) | 68.6 ± 15.5 | 67.2 ± 17 | 1.004 (0.982–1.027) | 0.695 | ||
| Hospital stay before BSIs, days (median, IQR) | 17 (7, 33) | 2 (1, 13) | 1.040 (1.017–1.064) | 0.001 | ||
| Microbiologic eradication after 6 days | 18 (46.2) | 10 (10.5) | 7.286 (2.937–18.076) | < 0.0001 | 3.569 (1.119–11.387) | 0.032 |
| Underlying diseases | ||||||
| Hepatobiliary and pancreatic diseases | 9 (23.1) | 43 (45.3) | 0.363 (0.155–0.847) | 0.019 | ||
| Kidney diseases | 12 (30.8) | 25 (26.3) | 1.244 (0.549–2.823) | 0.601 | ||
| Chronic lung diseases | 10 (25.6) | 12 (12.6) | 2.385 (0.932–6.104) | 0.070 | ||
| Diabetes mellitus | 12 (30.8) | 40 (42.1) | 0.611 (0.277–1.350) | 0.223 | ||
| Cardiovascular diseases | 21 (53.8) | 39 (41.1) | 1.675 (0.791–3.549) | 0.178 | ||
| Hypoproteinemia | 27 (69.2) | 63 (66.3) | 1.143 (0.512–2.549) | 0.744 | ||
| CCI score (median, IQR) | 5 (3.5, 6) | 4.5 (3, 6) | 0.986 (0.834–1.165) | 0.866 | ||
| Invasive procedures | ||||||
| Mechanical ventilation | 18 (46.2) | 19 (20.0) | 3.429 (1.532–7.674) | 0.003 | ||
| Urinary catheterization | 25 (64.1) | 41 (43.2) | 2.352 (1.089–5.080) | 0.029 | ||
| Gastric catheterization | 19 (48.7) | 22 (23.2) | 3.152 (1.433–6.934) | 0.004 | ||
| Central venous catheterization | 21 (53.8) | 25 (26.3) | 3.267 (1.501–7.110) | 0.003 | ||
| Medical history | ||||||
| IUC stay (previous 1 month) | 19 (48.7) | 27 (28.4) | 2.393 (1.108–5.168) | 0.026 | ||
| Prior surgery (previous 1 month) | 14 (35.9) | 21 (22.1) | 1.973 (0.874–4.454) | 0.102 | ||
| Severity of illness at the time of BSIs | ||||||
| Pitt bacteremia score (median, IQR) | 4 (1, 7) | 1 (1, 3) | 1.583 (1.296–1.933) | < 0.0001 | 1.609 (1.226–2.111) | 0.001 |
| Treatment | ||||||
| Inappropriate empirical treatment after BSIs | 26 (66.7) | 29 (30.5) | 4.552 (2.053–10.091) | < 0.0001 | 6.756 (1.992–23.753) | 0.003 |
| Inappropriate definitive treatment after BSIs | 3 (7.7) | 6 (6.3) | 1.236 (0.293–5.212) | 0.773 | ||
| CRKP infection, n(%) | 23 (59) | 27 (28.4) | 3.620 (1.662–7.885) | 0.001 | ||
Data are expressed as n (%) or median (IQR)
Abbreviations: CCI, Charlson comorbidity index; CRKP, carbapenem-resistant K.pneumoniae; BSIs, bloodstream infections; OR, odds ratio
Further, multivariate logistic regression analysis highlighted independent factors associated with a higher risk of mortality from KP-BSIs: microbiologic eradication after 6 days (OR = 3.569; CI = 1.119–11.387; P = 0.032), high Pitt bacteremia score (OR = 1.609; CI = 1.226–2.111; P = 0.001), and inappropriate empirical treatment after BSIs (OR = 6.756; CI = 1.922–23.753; P = 0.003).
Survival curve analysis confirmed the finding that microbiologic eradication after 6 days was associated with a higher 28-day mortality rate in patients with KP-BSIs (Fig. 1B). Additionally, the analysis revealed that the 28-day mortality rate was significantly higher among patients who received inappropriate empirical treatment compared to those who received appropriate empirical treatment (Fig. 1C).
Further analysis using the ROC curve indicated that microbiologic eradication after 6.5 days and a Pitt bacteremia score of 4.5 or higher were effective predictive factors for the mortality in patients with KP-BSIs. The area under the ROC curve (AUROC) for these predictors was 0.670 (95% CI = 0.562–0.778, P = 0.002) for microbiologic eradication and 0.723 (95% CI = 0.619–0.827, P < 0.0001) for the Pitt bacteremia score, indicating substantial predictive value (Fig. 1D and Table S2).
Antimicrobial susceptibility profiles
The results of AST for KP isolates are detailed in Table S3 and Table S6. The CRKP group exhibited high resistance rates to key carbapenems, with 88% (44/50) of isolates resistant to imipenem and 96% (48/50) resistant to meropenem. Notably, 84% (42/50) of CRKP isolates were resistant to both imipenem and meropenem. CRKP isolates demonstrated significantly higher resistance rates across all tested antimicrobial drugs compared to CSKP isolates. The resistance rates of CRKP strains to antibiotics were 44% (22/50) for tobramycin, 40% (20/50) for amikacin, 38% (19/50) for minocycline, and 24% (12/50) for tigecycline. All CRKP isolates had MICs ≤ 1ug/mL for polymyxin B.
Bacterial characteristics of CRKP strains
The antimicrobial resistance genes of CRKP strains are detailed in Fig. 2 and Table S4. The predominant carbapenemase genes detected were blakpc−2 in 47 isolates (94%) and blaKPC−12 in 1 isolate (2%). Notably, other carbapenemase genes such as blaNDM, blaVIM, blaIMP and blaOXA−48 were not detected in any of the isolates. Two isolates, S12 and S70, displayed carbapenem resistance despite lacking detectable carbapenemase genes.
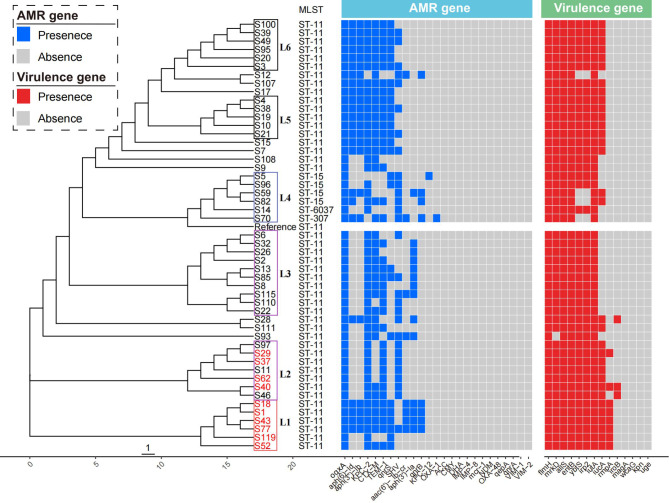
Fig. 2.
Phylogenetic analysis, MLST, virulence genes, and resistance genes of 50 CRKP strains. Strains in the cluster L1 and L2 from the ICU were color-coded on the tree (red). Blue and red represented the antimicrobial resistance genes and virulence genes, respectively. Colored blocks represented the presence of genes, and grey blocks represented the absence
Most strains (98%, 49/50) harbored β-lactamase gene, including blaCTX−M, blaTEM, blaOXA1, and blaSHV. Among these, blaCTX−M was found in several types across 39 CRKP strains, including blaCTX−M−3, blaCTX−M−65, blaCTX−M−14, blaCTX−M−82, blaCTX−M−55, and blaCTX−M−15. The Class A extended-spectrum β-lactamases (ESBLs) gene blaTEM−1 was identified in 39 isolates. The Oxacillin-hydrolyzing (OXA)-type Class D β-lactamase encoding gene blaOXA1 was specifically observed in isolate S70.
Moreover, 29 out of 50 isolates carried the chromosomally encoded antimicrobial resistance gene blaSHV. Plasmid-mediated quinolone resistance genes, including qnrS, qnrA, qnrB, oqxA, and aac(6’)-Ib-cr, were detected in all isolates. Additionally, aminoglycoside-related resistance genes aph(3’)-Ia, aph(3’’)-Ib, and aph(6)-Id were identified in 32 out of 50 isolates.
We conducted an investigation into 14 representative virulence genes within these clinical strains, which included flmH, allS, entB, mrkD, iutA, ybtS, iucA, rmpA, irp2, iroB, magA, wcaG, kpn, and uge. The presence and distribution of these virulence genes in CRKP strains are depicted in Fig. 2 and Table S5. Among the analyzed strains, all were found to carry flmH, allS, entB, iutA. The gene mrkD was highly prevalent, detected in 98% (49/50) of the strains, followed closely by ybtS and irp2, each present in 92% (46/50) of the strains. The gene iucA was found in 64% (32/50) of the strains. Notably, rmpA and iroB were less common, identified in only 16% (8/50) and 6% (3/50) of the strains, respectively. The genes magA, wcaG, kpn, and uge were not detected in any of the isolates.
Molecular epidemiological analysis identified the most prevalent ST among the CRKP isolates as ST11, which accounted for 88% (44/50) of the cases. This was followed by ST15 with 8% (4/50), and ST6037 and ST307, each representing 2% with 1 isolate. These findings are visualized in Fig. 2. Notably, all eight isolates carrying rmpA virulence gene belonged to ST11. The 50 CRKP isolates were categorized into six major clusters (L1, L2, L3, L4, L5, and L6), with isolates from the ICU predominantly found in clusters L1 and L2. Phylogenetic analysis further revealed that all 6 isolates within the L1 cluster were from the ICU, with all strains carrying the rmpA virulence genes. This pattern indicates a likely nosocomial transmission within the ICU setting.
Discussion
In this retrospective study, we identified several independent risk factors for CRKP-BSIs, including gastric catheterization, prior ICU hospitalization, and detection of CRKP in non-blood sites. These risk factors are consistent with those outlined in previous studies [28, 29]. Gastric catheterization has been shown to increase the risk of CRKP-BSIs, possibly due to mucosal barrier injury in the nasopharvnx caused by this invasive procedure [28, 30, 31]. Previous research has demonstrated a significant association between ICU admission and the development of CRKP-BSIs [29, 32]. The ICU environment, along with the extensive use of medical equipment, creates a potential for increased infection and transmission of resistant bacteria. The finding from our study confirm this, as we observed nosocomial transmission of CRKP within the ICU, emphasizing the role of the environment and equipment as reservoirs for these pathogens. Additionally, patients admitted to ICU are more likely to undergo invasive procedures and receive extensive antibiotic therapy due to their serious illness, making them more susceptible to CRKP-BSIs. Consistent with prior studies [31, 33], our findings indicate that the presence of CRKP in sites other than blood is associated with an increased risk of CRKP-BSIs. This suggests that colonization or infection at these sites may serve as reservoirs for subsequent BSIs. Inappropriate use of antibiotics has been linked to the rise of resistant strains [34, 35], which can lead to BSIs. These infections may manifest as primary bacteremia or as secondary bacteremia stemming from other infections, such as pneumonia and urinary tract infections.
In examining the independent risk factors for the mortality of KP-BSIs, findings can vary significantly depending on the population, country, and hospital settings studied. Our retrospective study identified inappropriate empirical treatment and host-associated factors, notably the Pitt bacteremia score and microbiologic eradication time, as key predictors of the 28-day mortality for KP-BSIs. The 28-day mortality rate in our study was 29.1%. The Pitt bacteremia score, a well-documented predictor of mortality in previous research [36, 37], was found to be significantly associated with mortality in our cohort. Specifically, a Pitt bacteremia score > 4 has been linked to higher mortality rates [38]. Our data supported this, showing that a Pitt bacteremia score of 4.5 or higher was particularly predictive of the 28-day mortality, highlighting the severity of the patients’ condition as a critical factor in their outcomes. The timing of microbiologic eradication also emerged as a crucial factor. Consistent with other studies [14, 39], we found that patients achieving microbiologic eradication within 6 days had significantly better survival rates. Conversely, eradication times after 6 days were independently associated with increased mortality, and times after 6.5 days were identified as good predictive factors for mortality. This suggests the importance of rapid and effective microbiologic eradication, which depends on factors such as the patient’s immune function, the severity of the illness, underlying diseases, accurate and prompt microbiological diagnosis, and appropriate antibiotic therapy. Appropriate antibiotic treatment has been shown to improve outcomes significantly [19, 40]. However, empirical therapy with non-active antibiotics may lead to unfavourable outcomes [41, 42]. Our analysis highlighted that patients receiving inappropriate empirical therapy had a higher 28-day mortality compared to those receiving appropriate treatment. Interestingly, we found no association between inappropriate definitive therapy and mortality, suggesting that the initial response to empirical therapy plays a more critical role in patient survival. Regarding CRKP, while our study confirmed its association with the death of KP-BSI patients, it did not emerge as an independent risk factor for mortality. This finding contrasts with other studies [30, 43], which have identified CRKP as an independent risk factor for mortality in patients with KP-BSIs. This discrepancy highlights the complex interplay of host, microbial, and therapeutic factors in determining outcomes for patients with KP-BSIs.
In addition to analyzing clinical characteristics of KP-BSIs, our study also delved into the microbiological characteristics of CRKP strains. This analysis aimed to assess the mechanisms of antimicrobial resistance and investigate the potential for nosocomial transmission during the study period. Most CRKP strains carried at least one ESBLs-producing gene associated with nosocomial infectious outbreak, suggesting the potential for evolution and transmission of resistance genes among clinical strains. A previous study reported resistance rates of 49% for amikacin and 64.7% for polymyxinB [44]. According to another study, tigecycline exerted the highest susceptibility rate (100%), followed by colistin (96.8%), and amikacin (87.1%) [45]. Results of our study demonstrated that the resistance rate for amikacin reached to 40%, and the resistance rates for tigecycline were over 20%. Although the high resistance rates of CRKP strains to tested antibiotics, all strains were susceptible to polymyxinB. These findings provide valuable guidance for selecting appropriate empirical treatments prior to obtaining AST results. The predominant mechanism of carbapenem resistance in CRKP strains is carbapenemase production. Commonly acquired genes that encode carbapenemase production include K.pneumoniae carbapenemases (KPC), New Delhi β-lactamases (NDM), oxacillinase 48-like (OXA-48), verona integron-encoded metallo-β-lactamase (VIM). Consistent with prior research [44, 46], our data also indicated that KPC-type carbapenemases were the main molecular mechanism of carbapenem resistance among the CRKP-BSIs. Despite the occurrence of nosocomial transmission of CRKP in the ICU, CRKP strains producing NDM were not detected, which contrasts with previous studies where NDM-1 was a significant mechanism during nosocomial spread [45]. The molecular epidemiologic features of CRKP vary across different countries and regions. In our hospital, KPC-2-producing ST11 KP strains were widely disseminated, aligning with findings from a nationwide surveillance of clinical CRKP strains in China [47–50]. However, this is distinct from the United States, where a predominance of ST258 has been observed [51]. Additionally, ST307, first described in Europe in 2008 [52, 53], has been detected globally, including a report of its spread in pediatric patients from Shenzhen Children’s Hospital in China, co-producing CTX-M, SHV and KPC [54]. In our study, we identified only one ST307 strain co-harboring blaCTX−M−15, blaOXA1, blaTEM, blaSHV, with no evidence of nosocomial transmission. Furthermore, a strain of ST6037 carrying blaKPC and ESBL encoding gene blaSHV, was detected for the first time in our hospital. The pathogenic potential of the ST6037 strain requires additional comprehensive investigation to be fully understood. Two specific strains, S12 and S70, demonstrated resistance to carbapenems in vitro despite not possessing carbapenemase genes. This resistance might be due to alternative mechanisms, such as reduced or altered pore proteins, efflux pump that excrete carbapenems from bacteria cells, changes in the structure of the penicillin-binding protein (PBP) [55], among other factors. Ceftazidime-avibactam (CAZ-AVI), effective against both KPC and OXA-48 carbapenemases, is often used to treat BSIs caused by CRKP that do not carry metalloenzyme [56]. For NDM-producing CRKP, a combination therapy involving ATM with CAZ-AVI can be considered [57]. Therefore, understanding these epidemiological and resistance characteristics is crucial for clinicians when selecting empirical therapeutic strategies for patients with CRKP-BSIs. Additionally, identifying both the genotypic and phenotypic characteristics of antibiotic resistance is also essential for rational and targeted antimicrobial therapy.
In our study, all strains that exhibited resistance to quinolones carried plasmid-mediated quinolone resistance (PMQR) genes, marking a significant finding as it contrasts with previous research where only the PMQR genes qnrS and aac(6’)-Ib-cr were detected in a mere 2.0% CRKP strains [44]. This indicates a broader prevalence of quinolone resistance genes among our CRKP isolates. We also noted a high level of resistance to aminoglycosides, with over 60% of the strains harboring aminoglycoside-related resistance genes that were plasmid-mediated. This trend toward increased aminoglycoside resistance could be linked to the mobility of the resistance genes. Polymyxins, including colistin and polymyxin B, are considered last-resort antibiotics for treating infections caused by carbapenem-resistant enterobacteriaceae (CRE), despite their toxicity [58]. In our study, all CRKP strains exhibited low MICs (≤ 1ug/mL) for polymyxin B. However, the plasmid-mediated colistin resistant gene mcr-7 was detected in three strains (S12, S85, and S107). These results highlight the urgent need for vigilant monitoring of antimicrobial resistance and the implementation of stringent infection control practices to inhibit the spread of resistant strains. The detection of high levels of resistance to commonly used antibiotics and the presence of last-resort antibiotic resistance genes should prompt a reevaluation of current therapeutic strategies and encourage efforts to prevent the horizontal transfer of resistance genes by plasmids.
A variety of hypervirulence-associated genes (e.g., rmpA, iroB, iucA, iutA, wcaG, fimH, mrkD, and allS), the hypermucoviscous phenotype, and capsule serotypes (K1 and K2) are link to hypermucoviscous K.pneumoniae (HvKP). The rmpA/rmpA2 genes are known to encode proteins that enhance capsule production, which contributes to the hypermucoviscosity phenotype. The genes iucA/iutA and iroB facilitate the biosynthesis of the siderophores aerobactin and salmochelin, respectively [59]. Moreover, fimH, mrkD, and allS play roles in bacterial colonization and fimbriae adhesion. Zeng et al. reported the absence of allS and iroB in CRKP isolates [44]. In contrast, Wu et al. observed that 81.3% (13/16) of CRKP isolates harbored iutA [60]. Our finding, however, contradicts these earlier studies; all isolates carried iutA and allS, and only 6% (3/50) carried iroB. The definition of HvKP remains unclear to date. Zeng et al. characterized HvKP as a strain that tests positive for both the string test and the rmpA gene concurrently. Additionally, iucA is considered a significant virulence determinant for HvKP [61]. In this retrospective study, we analyzed the homology of 50 CRKP strains and identified a nosocomial transmission of CRKP in the ICU. Integrated epidemiological and genomic data revealed that the dissemination in the ICU were caused by ST11 CRKP carring KPC-2 carbapenemases. Notably, 6 strains positive for the string test also carried the fimH, mrkD, allS, rmpA and iucA virulence genes. Among the patients infected with CRKP strains that possessed the virulence coding genes rmpA and iucA, 5 succumbed in the hospital. The role of hypervirulence in patient mortality warrants further investigation, necessitating additional experiments such as the Galleria mellonella infection model, virulent plasmid analysis, capsule serotype identification, and neutrophil killing assay.
Considering the risk factors for CRKP-BSIs, the factors influencing mortality in KP-BSIs, and the potential for nosocomial transmission of CRKP, it is essential to implement a multi-faceted approach. This should include: (1) An antimicrobial stewardship program to ensure the appropriate use of antibiotics; (2) Enhanced bacterial surveillance systems to prevent the further spread of drug-resistant bacteria; (3) Laboratory identification of carbapenem resistance genotypes to guide definitive therapy; (4) Comprehensive infection prevention and control measures, such as safe patient care practices, hand hygiene, strict adherence to aseptic technique, thorough environment cleaning and sterilization, especially in ICUs, and the isolation of colonized or infected patients.
Conclusions
Our study identified independent risk factors for CRKP-BSIs and factors associated with mortality in KP-BSIs patients through univariate and multivariate logistic regression analyses. Moreover, we developed a risk factor-based prediction model that could assist clinicians in administering prompt and effective therapy to patients suspected of having KP-BSIs, thereby improving patient outcomes. Additionally, our analysis of the primary mechanisms of carbapenem resistance offers valuable insights for hospital infection control and the clinical management of CRKP-BSIs. The widespread presence of ST11 CRKP strains with multiple plasmid-mediated resistance and hypervirulence genes underscores the need for effective control strategies to prevent nosocomial transmission.
Limitations
Our retrospective study has several notable limitations. Firstly, the study was limited to a single center and a relatively small patient cohort, which restricted our ability to perform a comprehensive multivariate analysis and may have introduced bias. Secondly, the limited number of cases prevented us from analyzing independent risk factors for mortality in patients with CRKP-BSIs. Lastly, although we evaluated virulence genes in CRKP strains, the study did not examine virulence genes in CSKP strains or investigate the impact of these genes on the mortality associated with KP-BSIs.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- CCI
Charlson comorbidity index
- CRKP
carbapenem-resistant K. pneumoniae
- BSIs
bloodstream infections
- OR
odds ratio
Author contributions
All authors contributed to this work. All authors have read and agreed to the published version of the manuscript. Shu Yu and Yan Cheng designed the study. Qi Cheng, Rong Zhang, Jie-ying Gao, Wei Li, Fu-kun Wang, Zheng-xin He, Qing-qing Sun, and Han-bing Meng performed the experiments and interpreted the data. Yan Cheng and Qi Cheng wrote the first draft of the paper. Yan Cheng and Shu Yu reviewed and approved the final report.
Funding
This research was funded by the Key Research of the 980th Hospital (No.FYJHZD-05).
Data availability
Data on the system exactly matches the statement provided in the manuscript. The sequencing data presented in the study are deposited in the National Library of Medicine repository (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1106931), accession number PRJNA1106931.
Declarations
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics statement
This study was approved by the Ethics Committee of 980th Hospital of the PLA Joint Logistical Support Force (No.2023-KY-198).
Author contribution statement
All authors contributed to this work. All authors have read and agreed to the published version of the manuscript. SY and YC designed the study. QC, RZ, J-y G, WL, F-k W, Z-x H, Q-q S, and H-b M performed the experiments and interpreted the data. YC and QC wrote the first draft of the paper. YC and SY reviewed and approved the final report.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yan Cheng and Qi Cheng contributed equally to this work.
References
- 1.Diekema DJ, Hsueh PR, Mendes RE, Pfaller MA, Rolston KV, Sader HS, Jones RN. The Microbiology of Bloodstream infection: 20-Year trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother 2019, 63(7). [DOI] [PMC free article] [PubMed]
- 2.Livermore DM. The impact of carbapenemases on antimicrobial development and therapy. Curr Opin Investig Drugs. 2002;3(2):218–24. [PubMed] [Google Scholar]
- 3.Logan LK, Weinstein RA. The epidemiology of Carbapenem-Resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl1):S28–36. 10.1093/infdis/jiw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rafailidis PI, Falagas ME. Options for treating carbapenem-resistant Enterobacteriaceae. Curr Opin Infect Dis. 2014;27(6):479–83. 10.1097/QCO.0000000000000109 [DOI] [PubMed] [Google Scholar]
- 5.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. 10.1016/S1473-3099(17)30489-9 [DOI] [PubMed] [Google Scholar]
- 6.Xu L, Sun X, Ma X. Systematic review and meta-analysis of mortality of patients infected with carbapenem-resistant Klebsiella pneumoniae. Ann Clin Microbiol Antimicrob. 2017;16(1):18. 10.1186/s12941-017-0191-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohler PP, Volling C, Green K, Uleryk EM, Shah PS, McGeer A. Carbapenem Resistance, initial antibiotic therapy, and Mortality in Klebsiella pneumoniae Bacteremia: a systematic review and Meta-analysis. Infect Control Hosp Epidemiol. 2017;38(11):1319–28. 10.1017/ice.2017.197 [DOI] [PubMed] [Google Scholar]
- 8.Falcone M, Bassetti M, Tiseo G, Giordano C, Nencini E, Russo A, Graziano E, Tagliaferri E, Leonildi A, Barnini S, et al. Time to appropriate antibiotic therapy is a predictor of outcome in patients with bloodstream infection caused by KPC-producing Klebsiella pneumoniae. Crit Care. 2020;24(1):29. 10.1186/s13054-020-2742-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balkan II, Alkan M, Aygun G, Kuskucu M, Ankarali H, Karagoz A, Sen S, Arsu HY, Bicer M, Kaya SY, et al. Colistin resistance increases 28-day mortality in bloodstream infections due to carbapenem-resistant Klebsiella pneumoniae. Eur J Clin Microbiol Infect Dis. 2021;40(10):2161–70. 10.1007/s10096-020-04124-y [DOI] [PubMed] [Google Scholar]
- 10.Meng H, Han L, Niu M, Xu L, Xu M, An Q, Lu J. Risk factors for mortality and outcomes in hematological malignancy patients with carbapenem-resistant Klebsiella pneumoniae bloodstream infections. Infect Drug Resist. 2022;15:4241–51. 10.2147/IDR.S374904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Yang Z, Sun L, Wang Z, Sun L, Xu J, Zeng L, Sun T. Clinical Observation and Prognostic Analysis of patients with Klebsiella pneumoniae bloodstream infection. Front Cell Infect Microbiol. 2020;10:577244. 10.3389/fcimb.2020.577244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Simmonds A, Greenman M, Sullivan SB, Tanner JP, Sowash MG, Whittier S, Uhlemann AC. Population structure of Klebsiella pneumoniae causing bloodstream infections at a New York City Tertiary Care Hospital: diversification of Multidrug-Resistant isolates. J Clin Microbiol. 2015;53(7):2060–7. 10.1128/JCM.03455-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang P, Wang J, Hu H, Zhang S, Wei J, Yang Q, Qu T. Clinical characteristics and risk factors for bloodstream infection due to Carbapenem-resistant Klebsiella pneumoniae in patients with hematologic malignancies. Infect Drug Resist. 2020;13:3233–42. 10.2147/IDR.S272217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu KS, Tong YS, Lee MT, Lin HY, Lu MC. Risk factors of 30-Day all-cause mortality in patients with carbapenem-resistant Klebsiella pneumoniae bloodstream infection. J Pers Med 2021, 11(7). [DOI] [PMC free article] [PubMed]
- 15.Cienfuegos-Gallet AV, Ocampo de Los Rios AM, Sierra Viana P, Ramirez Brinez F, Restrepo Castro C, Roncancio Villamil G, Del Corral Londono H, Jimenez JN. Risk factors and survival of patients infected with carbapenem-resistant Klebsiella pneumoniae in a KPC endemic setting: a case-control and cohort study. BMC Infect Dis. 2019;19(1):830. 10.1186/s12879-019-4461-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543(7643):15. 10.1038/nature.2017.21550 [DOI] [PubMed] [Google Scholar]
- 17.Potter RF, D’Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat. 2016;29:30–46. 10.1016/j.drup.2016.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pepin J, Yared N, Alarie I, Lanthier L, Vanasse A, Tessier P, Deveau J, Chagnon MN, Comeau R, Cotton P, et al. Klebsiella pneumoniae bacteraemia in a region of Canada. Clin Microbiol Infect. 2010;16(2):141–6. 10.1111/j.1469-0691.2009.02912.x [DOI] [PubMed] [Google Scholar]
- 19.Cano A, Gutierrez-Gutierrez B, Machuca I, Gracia-Ahufinger I, Perez-Nadales E, Causse M, Caston JJ, Guzman-Puche J, Torre-Gimenez J, Kindelan L, et al. Risks of infection and mortality among patients colonized with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: validation of scores and proposal for management. Clin Infect Dis. 2018;66(8):1204–10. 10.1093/cid/cix991 [DOI] [PubMed] [Google Scholar]
- 20.Soontaros S, Leelakanok N. Association between carbapenem-resistant Enterobacteriaceae and death: a systematic review and meta-analysis. Am J Infect Control. 2019;47(10):1200–12. 10.1016/j.ajic.2019.03.020 [DOI] [PubMed] [Google Scholar]
- 21.Rhee JY, Kwon KT, Ki HK, Shin SY, Jung DS, Chung DR, Ha BC, Peck KR, Song JH. Scoring systems for prediction of mortality in patients with intensive care unit-acquired sepsis: a comparison of the Pitt bacteremia score and the Acute Physiology and Chronic Health evaluation II scoring systems. Shock. 2009;31(2):146–50. 10.1097/SHK.0b013e318182f98f [DOI] [PubMed] [Google Scholar]
- 22.Charlson ME, Pompei P, Ales KL, MacKenzie CR. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–83. 10.1016/0021-9681(87)90171-8 [DOI] [PubMed] [Google Scholar]
- 23.Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, Raad II, Rolston KV, Young JA, Wingard JR, et al. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis. 2011;52(4):427–31. 10.1093/cid/ciq147 [DOI] [PubMed] [Google Scholar]
- 24.Institute CaLS. Performance standards for antimicrobial susceptibility testing CLSI document M100-S29 2021.
- 25.Testing TECAS. Breakpoint tables for interpretation of MICs and zone diameters. version 100 2020.
- 26.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–72. 10.1101/gr.097261.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao T, Zhu Y, Zhang S, Wang Y, Shen P, Zhou Y, Yu X, Xiao Y. A retrospective analysis of risk factors and outcomes of Carbapenem-resistant Klebsiella pneumoniae Bacteremia in Nontransplant patients. J Infect Dis. 2020;221(Suppl 2):S174–83. 10.1093/infdis/jiz559 [DOI] [PubMed] [Google Scholar]
- 29.Giannella M, Trecarichi EM, De Rosa FG, Del Bono V, Bassetti M, Lewis RE, Losito AR, Corcione S, Saffioti C, Bartoletti M, et al. Risk factors for carbapenem-resistant Klebsiella pneumoniae bloodstream infection among rectal carriers: a prospective observational multicentre study. Clin Microbiol Infect. 2014;20(12):1357–62. 10.1111/1469-0691.12747 [DOI] [PubMed] [Google Scholar]
- 30.Chang H, Wei J, Zhou W, Yan X, Cao X, Zuo L, Chen S, Yao K, Huang R, Chen Y, et al. Risk factors and mortality for patients with bloodstream infections of Klebsiella pneumoniae during 2014–2018: clinical impact of carbapenem resistance in a large tertiary hospital of China. J Infect Public Health. 2020;13(5):784–90. 10.1016/j.jiph.2019.11.014 [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Li J, Hu T, Hu J, Song N, Zhang Y, Chen Y. Five-year change of prevalence and risk factors for infection and mortality of carbapenem-resistant Klebsiella pneumoniae bloodstream infection in a tertiary hospital in North China. Antimicrob Resist Infect Control. 2020;9(1):79. 10.1186/s13756-020-00728-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian L, Tan R, Chen Y, Sun J, Liu J, Qu H, Wang X. Epidemiology of Klebsiella pneumoniae bloodstream infections in a teaching hospital: factors related to the carbapenem resistance and patient mortality. Antimicrob Resist Infect Control. 2016;5:48. 10.1186/s13756-016-0145-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin RM, Cao J, Brisse S, Passet V, Wu W, Zhao L, Malani PN, Rao K, Bachman MA. Molecular Epidemiology of Colonizing and Infecting isolates of Klebsiella pneumoniae. mSphere 2016, 1(5). [DOI] [PMC free article] [PubMed]
- 34.Drlica K, Zhao X. Mutant selection window hypothesis updated. Clin Infect Dis. 2007;44(5):681–8. 10.1086/511642 [DOI] [PubMed] [Google Scholar]
- 35.McGowan JE Jr. Antimicrobial resistance in hospital organisms and its relation to antibiotic use. Rev Infect Dis. 1983;5(6):1033–48. 10.1093/clinids/5.6.1033 [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez-Gutierrez B, Salamanca E, de Cueto M, Hsueh PR, Viale P, Pano-Pardo JR, Venditti M, Tumbarello M, Daikos G, Pintado V, et al. A predictive model of mortality in patients with bloodstream infections due to carbapenemase-producing Enterobacteriaceae. Mayo Clin Proc. 2016;91(10):1362–71. 10.1016/j.mayocp.2016.06.024 [DOI] [PubMed] [Google Scholar]
- 37.Pan H, Lou Y, Zeng L, Wang L, Zhang J, Yu W, Qiu Y. Infections caused by carbapenemase-producing Klebsiella pneumoniae: microbiological characteristics and risk factors. Microb Drug Resist. 2019;25(2):287–96. 10.1089/mdr.2018.0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee NY, Tsai CS, Syue LS, Chen PL, Li CW, Li MC, Ko WC. Treatment outcome of Bacteremia due to Non-carbapenemase-producing Carbapenem-resistant Klebsiella pneumoniae Bacteremia: role of Carbapenem Combination Therapy. Clin Ther. 2020;42(3):e33–44. 10.1016/j.clinthera.2020.01.004 [DOI] [PubMed] [Google Scholar]
- 39.Nguyen M, Eschenauer GA, Bryan M, O’Neil K, Furuya EY, Della-Latta P, Kubin CJ. Carbapenem-resistant Klebsiella pneumoniae bacteremia: factors correlated with clinical and microbiologic outcomes. Diagn Microbiol Infect Dis. 2010;67(2):180–4. 10.1016/j.diagmicrobio.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 40.Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, Prekates A, Themeli-Digalaki K, Tsakris A. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin Microbiol Infect. 2011;17(12):1798–803. 10.1111/j.1469-0691.2011.03514.x [DOI] [PubMed] [Google Scholar]
- 41.Akturk H, Sutcu M, Somer A, Aydin D, Cihan R, Ozdemir A, Coban A, Ince Z, Citak A, Salman N. Carbapenem-resistant Klebsiella pneumoniae colonization in pediatric and neonatal intensive care units: risk factors for progression to infection. Braz J Infect Dis. 2016;20(2):134–40. 10.1016/j.bjid.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satlin MJ, Cohen N, Ma KC, Gedrimaite Z, Soave R, Askin G, Chen L, Kreiswirth BN, Walsh TJ, Seo SK. Bacteremia due to carbapenem-resistant Enterobacteriaceae in neutropenic patients with hematologic malignancies. J Infect. 2016;73(4):336–45. 10.1016/j.jinf.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Guo LY, Song WQ, Wang Y, Dong F, Liu G. Risk factors for carbapenem-resistant K. pneumoniae bloodstream infection and predictors of mortality in Chinese paediatric patients. BMC Infect Dis. 2018;18(1):248. 10.1186/s12879-018-3160-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng L, Yang C, Zhang J, Hu K, Zou J, Li J, Wang J, Huang W, Yin L, Zhang X. An outbreak of Carbapenem-resistant Klebsiella pneumoniae in an Intensive Care Unit of a major Teaching Hospital in Chongqing, China. Front Cell Infect Microbiol. 2021;11:656070. 10.3389/fcimb.2021.656070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J, Li Q, Li X, Kang J, Song Y, Song J, Yin D, Duan J. Successful control of the first carbapenem-resistant Klebsiella pneumoniae outbreak in a Chinese hospital 2017–2019. Antimicrob Resist Infect Control. 2020;9(1):91. 10.1186/s13756-020-00757-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Yang Y, Chen G, Lin M, Chen Y, He R, Galvao KN, El-Gawad El-Sayed Ahmed MA, Roberts AP, Wu Y, et al. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg Microbes Infect. 2021;10(1):700–9. 10.1080/22221751.2021.1906163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, Li Y, Liao K, Chen S. Nationwide Surveillance of Clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. 10.1016/j.ebiom.2017.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–12. 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 49.Jia X, Li C, Chen F, Li X, Jia P, Zhu Y, Sun T, Hu F, Jiang X, Yu Y, et al. Genomic epidemiology study of Klebsiella pneumoniae causing bloodstream infections in China. Clin Transl Med. 2021;11(11):e624. 10.1002/ctm2.624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, Zhang Y, Jin L, Chen H, Wang Z, et al. Phenotypic and genotypic characterization of Carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE Study in China (2012–2016). Clin Infect Dis. 2018;67(suppl2):S196–205. 10.1093/cid/ciy660 [DOI] [PubMed] [Google Scholar]
- 51.Giani T, Pini B, Arena F, Conte V, Bracco S, Migliavacca R, Participants A-CS, Pantosti A, Pagani L, Luzzaro F et al. Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro Surveill 2013, 18(22). [PubMed]
- 52.Heiden SE, Hubner NO, Bohnert JA, Heidecke CD, Kramer A, Balau V, Gierer W, Schaefer S, Eckmanns T, Gatermann S, et al. A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med. 2020;12(1):113. 10.1186/s13073-020-00814-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peirano G, Chen L, Kreiswirth BN, Pitout JDD. Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob Agents Chemother 2020, 64(10). [DOI] [PMC free article] [PubMed]
- 54.Patil S, Chen H, Guo C, Zhang X, Ren PG, Francisco NM, Wen F. Emergence of Klebsiella pneumoniae ST307 co-producing CTX-M with SHV and KPC from Paediatric patients at Shenzhen Children’s Hospital, China. Infect Drug Resist. 2021;14:3581–8. 10.2147/IDR.S324018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma J, Song X, Li M, Yu Z, Cheng W, Yu Z, Zhang W, Zhang Y, Shen A, Sun H, et al. Global spread of carbapenem-resistant Enterobacteriaceae: epidemiological features, resistance mechanisms, detection and therapy. Microbiol Res. 2023;266:127249. 10.1016/j.micres.2022.127249 [DOI] [PubMed] [Google Scholar]
- 56.Tumbarello M, Raffaelli F, Giannella M, Mantengoli E, Mularoni A, Venditti M, De Rosa FG, Sarmati L, Bassetti M, Brindicci G, et al. Ceftazidime-Avibactam Use for Klebsiella pneumoniae carbapenemase-producing K. pneumoniae infections: a Retrospective Observational Multicenter Study. Clin Infect Dis. 2021;73(9):1664–76. 10.1093/cid/ciab176 [DOI] [PubMed] [Google Scholar]
- 57.Falcone M, Daikos GL, Tiseo G, Bassoulis D, Giordano C, Galfo V, Leonildi A, Tagliaferri E, Barnini S, Sani S, et al. Efficacy of Ceftazidime-Avibactam Plus Aztreonam in patients with bloodstream infections caused by Metallo-beta-lactamase-producing enterobacterales. Clin Infect Dis. 2021;72(11):1871–8. 10.1093/cid/ciaa586 [DOI] [PubMed] [Google Scholar]
- 58.Macesic N, Nelson B, McConville TH, Giddins MJ, Green DA, Stump S, Gomez-Simmonds A, Annavajhala MK, Uhlemann AC. Emergence of Polymyxin Resistance in Clinical Klebsiella pneumoniae through Diverse Genetic adaptations: a genomic, retrospective cohort study. Clin Infect Dis. 2020;70(10):2084–91. 10.1093/cid/ciz623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 2019, 32(3). [DOI] [PMC free article] [PubMed]
- 60.Wu X, Shi Q, Shen S, Huang C, Wu H. Clinical and bacterial characteristics of Klebsiella pneumoniae affecting 30-Day mortality in patients with bloodstream infection. Front Cell Infect Microbiol. 2021;11:688989. 10.3389/fcimb.2021.688989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Zhao C, Wang Q, Wang X, Chen H, Li H, Zhang F, Li S, Wang R, Wang H. High prevalence of Hypervirulent Klebsiella pneumoniae infection in China: Geographic distribution, clinical characteristics, and Antimicrobial Resistance. Antimicrob Agents Chemother. 2016;60(10):6115–20. 10.1128/AAC.01127-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data on the system exactly matches the statement provided in the manuscript. The sequencing data presented in the study are deposited in the National Library of Medicine repository (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1106931), accession number PRJNA1106931.