Abstract
In March 2003, a novel coronavirus was isolated from patients exhibiting atypical pneumonia, and was subsequently proven to be the causative agent of the disease now referred to as SARS (severe acute respiratory syndrome). The complete genome of the SARS-CoV (SARS coronavirus) has since been sequenced. The SARS-CoV nucleocapsid (SARS-CoV N) protein shares little homology with other members of the coronavirus family. In the present paper, we show that SARS-CoV N is capable of inducing apoptosis of COS-1 monkey kidney cells in the absence of growth factors by down-regulating ERK (extracellular-signal-regulated kinase), up-regulating JNK (c-Jun N-terminal kinase) and p38 MAPK (mitogen-activated protein kinase) pathways, and affecting their downstream effectors. SARS-CoV N expression also down-regulated phospho-Akt and Bcl-2 levels, and activated caspases 3 and 7. However, apoptosis was independent of the p53 and Fas signalling pathways. Furthermore, activation of the p38 MAPK pathway was found to induce actin reorganization in cells devoid of growth factors. At the cytoskeletal level, SARS-CoV N down-regulated FAK (focal adhesion kinase) activity and also down-regulated fibronectin expression. This is the first report showing the ability of the N protein of SARS-CoV to induce apoptosis and actin reorganization in mammalian cells under stressed conditions.
Keywords: actin reorganization, caspase, cell signalling, growth factor, programmed cell death, SARS coronavirus nucleocapsid
Abbreviations: AP1, activator protein 1; DMEM, Dulbecco's modified Eagle's medium; ERK, extracellular-signal-regulated kinase; FAK, focal adhesion kinase; HA, haemagglutinin; HSP27, heat-shock protein 27; JNK, c-Jun N-terminal kinase; MAPK, mitogen-activated protein kinase; N, nucleocapsid; ORF, open reading frame; p-, phospho-; PI3K, phosphoinositide 3-kinase; SARS, severe acute respiratory syndrome; SARS-CoV, SARS coronavirus; TUNEL, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling; Z-VAD-FMK, benzyloxycarbonyl-Val-Ala-DL-Asp-fluoromethylketone
INTRODUCTION
The causative viral agent of SARS (severe acute respiratory syndrome) caused severe outbreaks of atypical pneumonia in different regions of the world in 2003 [1]. The virus is a newly identified member of the coronavirus family [2,3]. The SARS virus (SARS-CoV) is an enveloped, positive-sense RNA virus with a genome comprising approx. 30000 nucleotides predicted to encode 13–15 ORFs (open reading frames) [4]. Sequence comparison with ORFs of other known coronaviruses shows a similar pattern of gene organization with the non-structural genes clustered at the 5′ end, and the structural genes placed at the 3′ end of the genome, resembling most closely the group II coronaviruses [3,4]. The high viral virulence resulting in a significant mortality rate of infected patients has created widespread scientific interest to understand the mechanisms of pathogenicity of this virus.
The SARS-CoV nucleocapsid (N) protein is a predicted phosphoprotein of 46 kDa, which self-associates to form a dimer [5] and shares homology with other members of the coronavirus family. However, unique to it are a short serine-rich stretch, and a putative bipartite nuclear localization signal, thus suggesting its involvement in other important functions besides capsid assembly during the viral life cycle. The N protein has also been reported to activate the AP1 (activator protein 1) signal transduction pathway [6].
Recently, the SARS virus has been shown to induce apoptosis in cell culture [7]. In the present paper, we describe a possible role for the N protein in virus-induced apoptosis. Experiments with COS-1 cells expressing the N protein induced significant programmed cell death, particularly in the absence of growth factors. Data on the pro-apoptotic nature of the N protein revealed down-regulation of ERK (extracellular-signal-regulated kinase), but up-regulation of JNK (c-Jun N-terminal kinase) and p38 MAPK (mitogen-activated protein kinase) activities. These observations are in agreement with their corresponding downstream effectors as well. Furthermore, activation of the p38 MAPK cascade was found to induce actin reorganization. N expression was also found to down-regulate the levels of phospho (p)-Akt and Bcl-2, and activate caspases 3 and 7, leading to apoptosis. This is the first report describing the ability of the N protein of SARS-CoV to induce apoptosis in mammalian cells.
MATERIALS AND METHODS
Plasmids and Reagents
The N region (28120–29385) of the SARS-CoV genome (GenBank® accession number NC_004718) was PCR-amplified and cloned into pCR-XL-TOPO. The clone was confirmed by sequencing, and further subcloned into BamHI and ApaI sites of pcDNA3.1 to generate a C-terminal myc-tagged N gene. For generating an N-terminal HA (haemagglutinin)-tagged clone, the N gene in pGADT7 was digested with BglII and subcloned into the pSGI vector. Both clones were verified by restriction mapping and sequencing. Antibodies against HA, Myc (9E10), fibronectin, p-FAK (focal adhesion kinase), integrin αV, Bcl-2, p-Akt, calnexin, Bcl-XL, p-ERK, total ERK, p-Jun, c-Jun, p-JNK, mouse (Texas-Red-conjugated), p53, Fas, Fas ligand and Bax were purchased from Santa Cruz Biotechnology. Antibodies against p38 MAPK, p-p38 MAPK, p-MAPKAP-K2 (MAPK-activated protein kinase 2), p-HSP27 (heat-shock protein 27), caspase 3 and cleaved caspase 7 were obtained from Cell Signaling Technology. Rhodamine-conjugated phalloidin, cytochalasin D, Z-VAD-FMK (benzyloxycarbonyl-Val-Ala-DL-Asp-fluoromethylketone) and SB203580 were purchased from Sigma Chemicals.
Cell culture and transfection
COS-1 and HuH7 cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with penicillin, streptomycin and 10% foetal bovine serum. Cells were transfected with Lipofectin® reagent (Invitrogen) according to the manufacturer's instructions. At 24 h post-transfection, cells were maintained for a further 24 h either in the absence of serum (for growth factor deprivation) or in the presence of serum. Mock-transfected cells were transfected with the respective empty vectors. Cytochalasin D was added 1 h before harvesting the cells. Z-VAD-FMK and SB203580 were added 12 h before harvesting the cells.
Metabolic labelling and immunoprecipitation
At 40 h post-transfection, cells were starved for 1 h in cysteine/methionine-deficient medium, and were then labelled with 100 μCi of [35S]cysteine/[35S]methionine Promix for 4 h. After labelling, cells were washed once in PBS, and lysed in RIPA buffer (150 mM NaCl, 1% Nonidet P40, 0.5% deoxycholate, 0.1% SDS and 50 mM Tris/HCl, pH 8.0) with protease inhibitor cocktail. For immunoprecipitation, an equal amount of protein was incubated with 1 μg of the corresponding antibody overnight, followed by incubation with 100 μl of 10% Protein A–Sepharose suspension for 1 h. Beads were washed four times in RIPA buffer, and proteins were eluted by boiling samples in 2×SDS dye.
Western blotting
For Western immunoblotting of total cell lysates, samples were lysed directly in 1×SDS lysis buffer, and subjected to SDS/PAGE. Immunoprecipitated samples were also resolved by SDS/PAGE. The samples were subsequently transferred on to nitrocellulose membranes, incubated with respective antibodies, and developed by the ECL® (enhanced chemiluminescence) detection method according to the manufacturer's specifications (Amersham Biosciences). Gels shown are representative of three independent sets of experiments. Results were quantified and normalized values were calculated using the NIH (National Institutes of Health) Image version 1.32 program. Histograms show means±S.E.M. for three independent sets of experiments.
TUNEL (terminal deoxynucleotidyl transferase-mediated dUTP nick-end labelling) assay
Cell death was detected using an in vivo cell-death-detection TUNEL assay kit (Roche Biochemicals), according to the manufacturer's instructions. Cell death was quantified by counting TUNEL-positive cells from a total sample size of 500 cells in each set of experiments.
Immunofluorescence assay
Cells were washed once in PBS and fixed in 2% formaldehyde for 10 min. The cells were subsequently blocked with 5% normal goat serum in PBS in the presence of 0.1% saponin for 1 h. The cells were then incubated with anti-HA antibody for 1 h, followed by rhodamine-conjugated phalloidin and FITC-conjugated anti-mouse IgG antibody for 1 h, and washed three times in PBS. Following this, the cells were mounted on slides with antifade reagent (Bio-Rad Laboratories). Photographs were taken using a Nikon TE 2000U immunoflourescence microscope at ×100 magnification.
RESULTS AND DISCUSSION
To understand the role of the N protein of SARS-CoV in a mammalian cell environment, we cloned the N gene into two different eukaryotic expression vectors. In order to avoid blockage of the N- or C-terminal regions of the SARS-CoV N protein leading to functional impairment due to the fusion tag, we cloned N into pcDNA3.1 with a C-terminal myc tag, and into pSGI with an N-terminal HA tag. Both constructs were transfected into COS-1 cells, and immunoprecipitated with corresponding antibodies after labelling the protein with [35S]cysteine/[35S]methionine Promix. Both constructs expressed the N protein migrating at approx. 48 kDa (Figure 1A, lanes 2 and 4). In COS-1 cells, the pSGI construct led to stronger expression of the N protein than the pcDNA3.1 N construct. This may be attributed to the higher efficiency of the SV40 (simian virus 40) promoter in COS-1 cells. To exclude any functional variation arising from the positional effect of the myc and HA tags with the overexpression of N, the levels of p-ERK, p-Akt, Bcl-2 and p-FAK were checked using both the transfected clones and found to be identical in all cases (results not shown).
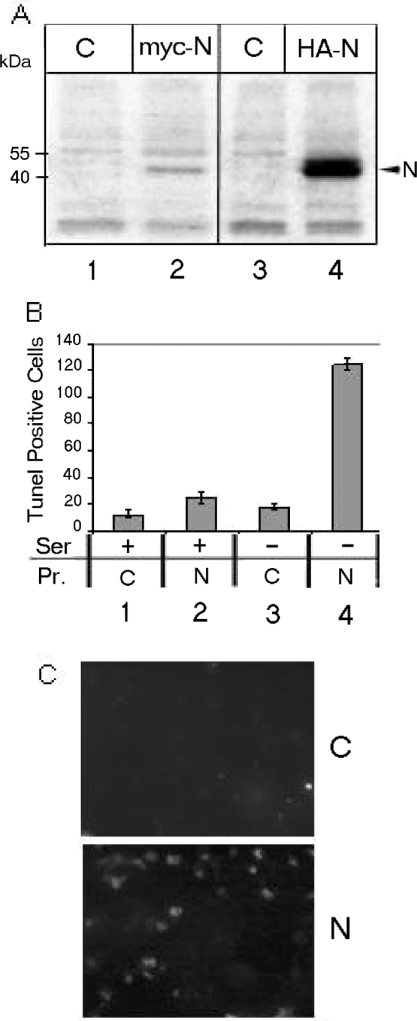
Figure 1. SARS Co-V N induces cell death.
(A) Expression of N protein was analysed by immunoprecipitating mock-transfected (C) or pcDNA3.1 myc-tagged N- (myc-N) or pSGI HA-tagged N- (HA-N) transfected cell lysates with corresponding antibodies. Lanes 2 and 4 show N expression, while lanes 1 and 3 are respective controls. (B) Results of TUNEL assay. Lanes 1 and 2 respectively show mock- (C) or N-transfected (N) cells in presence of serum (Ser), while lanes 3 and 4 show the same cells in the absence of serum. TUNEL-positive cells were scored from a total of 500 cells viewed from each of the three sets of experiments. The histogram shows the means±S.E.M. for three independent sets of experiments. The P values are 0.17 for lanes 1 and 2, and 0.02 for lanes 3 and 4. TUNEL-positive cells in the absence of serum constituted approx. 30% of the total cell count. Pr., protein. (C) A representative field of TUNEL-positive cells as observed under the microscope.
Interestingly, cells transfected with SARS-CoV N showed significant cell death when cultured under conditions of stress induced by serum starvation. In order to quantify cell death under these conditions, we conducted a TUNEL assay. As shown in Figure 1(B), serum starvation induced approx. 30% cell death in the presence of N (lane 4). However, mock-transfected cells (transfected with the empty vector alone) did not show significant cell death in the absence of serum (lane 3), nor did N-transfected cells in the presence of serum (lane 2). Moreover, human hepatoma cells (HuH7) maintained under similar conditions did not show significant cell death (results not shown). The relatively moderate percentage of death of COS-1 cells observed in our experiments may be attributed to the limitations of the transient transfection method. However, the results were reproducible in repeated experiments. Figure 1(C) shows a representative field of TUNEL-positive N-expressing cells in the absence of serum.
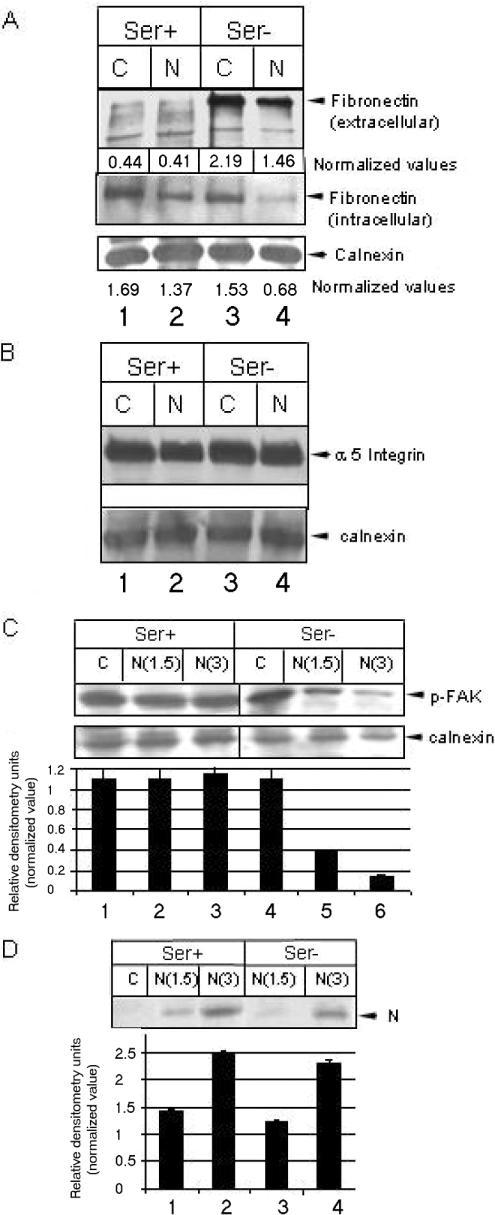
Given that N-induced cell death was observed at significant levels only in the absence of growth factors, and since it is known that binding of ligands to integrin receptors maintains the major survival pathway through PI3K (phosphoinositide 3-kinase) and MAPK pathways when growth-factor-mediated signalling is absent [8], we thus postulated that N may be interfering with the integrin signalling pathway. Experiments were designed to monitor the expression levels of the integrin receptor and ligand in N-expressing cells. We checked the level of one of the most abundant ligands, fibronectin. N expression clearly down-regulated the level of extracellular fibronectin in the absence of serum (Figure 2A, upper panel). However, we were unable to observe a significant difference in the level of fibronectin in the presence of serum, since only a small quantity of protein could be detected by immunoprecipitation. This may be due to lower stability of fibronectin protein in the presence of serum factors. Next, we investigated whether N-mediated down-regulation of extracellular fibronectin level was due to suppression of fibronectin gene expression. For this, we restricted protein transport beyond the endoplasmic reticulum by treatment with brefeldin A, and immunoprecipitated the cell lysate with anti-fibronectin antibody, which showed the total amount of nascent fibronectin protein produced by the cells. As expected, fibronectin levels were found to be lower in N-expressing cells irrespective of the presence or absence of serum (Figure 2A, lower panel), suggesting that N down-regulated fibronectin gene expression. Subsequently, we analysed the levels of the fibronectin receptor (integrin αV), but there was no change in the level of the latter (Figure 2B). We then assayed for the activity of FAK, which is a major intermediate factor that co-ordinates signal communication between integrins and downstream effectors such as MAPKs [9]. The level of phosphorylated FAK was significantly down-regulated in N-expressing cells in the absence of growth factors (Figure 2C). This result supported further our hypothesis that N-induced cell death was due to interference with the integrin signalling pathway, and that serum factors inhibited this phenomenon dominantly by maintaining the activity of FAK through alternative pathways. Figure 2(D) shows the expression pattern of N protein in the presence and absence of serum. Two different concentrations of DNA (1.5 μg and 3 μg) were used in all the Western blot experiments to rule out any possible experimental artifact.
Figure 2. N down-regulates fibronectin expression.
(A) Mock-transfected (C) or N-transfected (N) cells were maintained in the presence (Ser+) or absence (Ser−) of serum. For extracellular fibronectin level (upper panel), growth medium was immunoprecipitated using the respective antibody. For intracellular fibronectin level, cells were treated for 2 h with brefeldin A (10 μg/ml), lysed in RIPA buffer, immunoprecipitated using anti-fibronectin antibody, resolved by SDS/6% PAGE and immunoblotted. Values were normalized with reference to a non-specific band (calnexin) in the same gel (for extracellular fibronectin or to the level of calnexin for intracellular fibronectin). (B) Cells grown under similar conditions as above were lysed in SDS buffer, resolved by SDS/7% PAGE and immunoblotted using anti-(integrin αV) or anti-calnexin antibody (loading control). (C) Level of p-FAK (Tyr397). The lower gel shows calnexin level as a loading control. N(1.5) and N(3) represents samples transfected with 1.5 μg and 3 μg DNA respectively. Mock samples were transfected with 3 μg of empty vector DNA. The histogram shows the means±S.E.M. for three independent sets of experiments. (D) N expression as assessed by immunoprecipitation with anti-Myc antibody in the presence (lanes 1, 2 and 3) or absence of (lanes 4 and 5) serum respectively, with different concentrations of DNA. The histogram shows the means±S.E.M. for three independent sets of experiments.
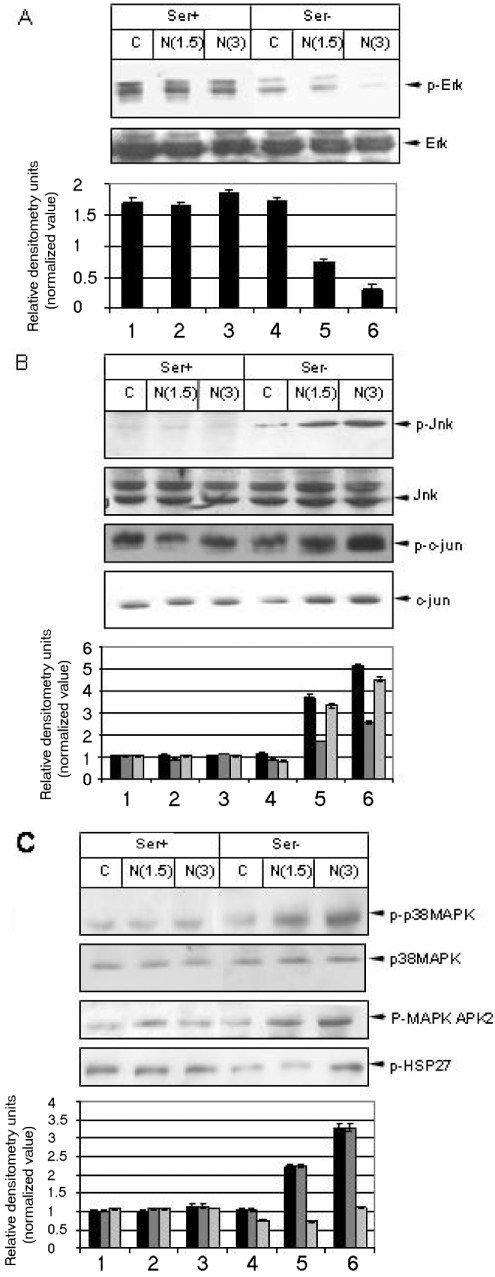
Next, we checked the activity of different MAPKs, since these are the major downstream effectors of the integrin signalling cascade. Interestingly, N expression modulated all three major MAPK signalling pathways. In the absence of serum, N expression down-regulated the activity of p-ERK (Figure 3A, lanes 4–6). Furthermore, N expression up-regulated the activity of stress-activated protein kinases, namely the JNK and p38 MAPK pathways. The activity was significantly high in the absence of serum (Figure 3B, top panel, lanes 5 and 6). The up-regulation of JNK activity was reflected further by the elevated p-Jun level (Figure 3B, third panel, lanes 5 and 6). Moreover, the level of c-Jun also increased in the absence of serum (Figure 3B, fourth panel, lanes 5 and 6). These data are congruent with a recent report [6], which demonstrated that AP1 was up-regulated in the presence of N. In addition, phosphorylation of downstream substrates of the p38 MAPK pathway were also found to be up-regulated (Figure 3C, third and fourth panels, lanes 5 and 6).
Figure 3. N protein expression modulates cellular MAPK activity.
(A) Mock-transfected or Myc–N clone-transfected cells were maintained in serum (Ser+) for 48 h (lanes 1–3) or serum-starved for 24 h (Ser−, lanes 4–6); harvested with SDS lysis buffer, resolved by SDS/12% PAGE, and immunoblotted using corresponding antibodies. The Figure shows the levels of p-ERK and total ERK. The histogram shows means±S.E.M. for three independent sets of experiments. (B) Levels of p-JNK, p-Jun and total c-Jun level. Calnexin was used to ensure equal loading. The histogram shows the means±S.E.M. for three independent sets of experiments. Black bars represent p-JNK, dark grey bars represent p-c-Jun and light grey bars represent c-Jun. (C) The level of p-p38 MAPK, p-MAPKAPK2 (MAPK-activated protein kinase 2) and p-HSP27. Total p38 MAPK level served as the loading control for p-p38 MAPK. The histogram shows the means±S.E.M. for three independent sets of experiments. Black bars represent p-p38 MAPK, dark grey bars represent p-MAPKAPK2 and light grey bars represent p-HSP27.
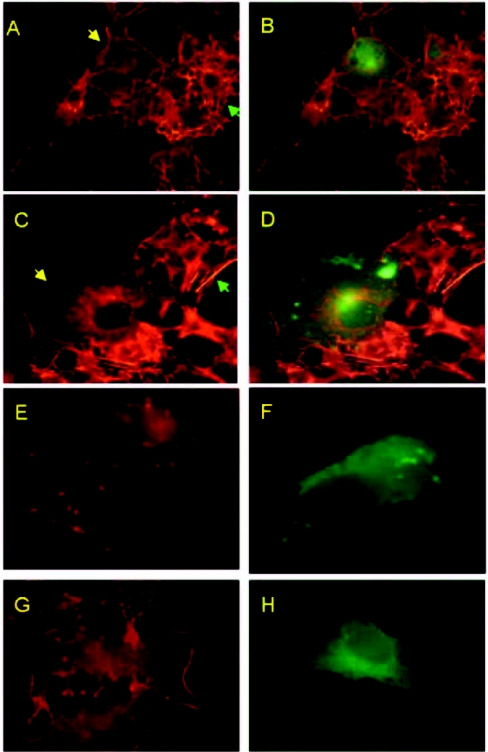
Since HSP27 phosphorylation was up-regulated, we questioned whether actin organization is affected in N-expressing cells. To address this, we used rhodamine-conjugated phalloidin to analyse actin distribution in the presence of N. In the presence of serum, N expression had no visible effects on actin distribution (Figure 4A, compare N-protein-transfected cells with non-transfected cells). Figure 4(C) shows actin distribution in N-expressing cells in the absence of growth factors, i.e. actin filaments concentrated around the nuclear membrane forming a ring-like structure. In contrast, actin is distributed extensively throughout the cytoplasm of non-transfected cells. To prove that the ring-like structure was indeed actin, we employed the specific actin-depolymerizing agent, cytochalasin D, which effectively blocked actin ring formation at a concentration as low as 5 nM (Figure 4E). Furthermore, SB203580 was used to verify whether actin reorganization was dependent on the p38 MAPK pathway. As shown in Figure 4(G), SB203580 inhibited actin redistribution in N-transfected cells. Figures 4(B), 4(D), 4(F) and 4(H) show N expression in the respective cells (superimposition of FITC-stained image on rhodamine-stained image). Thus we conclude that N-induced activation of the p38 MAPK pathway in the absence of growth factors leads to actin reorganization.
Figure 4. Actin reorganization induced by the SARS Co-V N protein.
Immunofluorescence study using rhodamine-conjugated phalloidin and HA-tagged N. (A) Actin distribution in the presence of serum. The yellow arrow shows an N-transfected cell, and the green arrow shows a non-transfected cell. (B) The same field for N expression stained with FITC-conjugated anti-mouse antibody (merged image). (C) Actin distribution in the absence of serum. Arrows as in (A). (E) Actin redistribution in the presence of cytochalasin D (5 nM). (G) Effect of SB203580 (10 μg/ml). (D, F and H) Superimposition of FITC-stained images on rhodamine-stained images showing N expression in the absence of serum, cytochalasin D or SB203580 respectively.
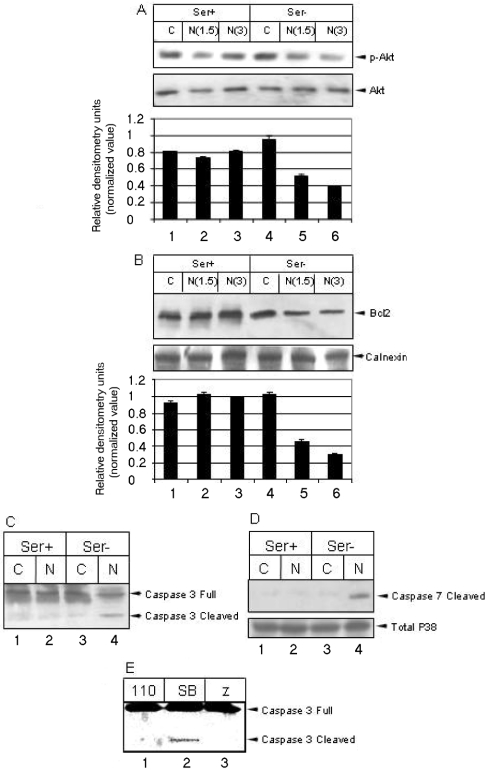
Furthermore, we investigated the activity of Akt, a major intermediate of the PI3K pathway, which is the predominant survival pathway in a cell. Phosphorylation of Akt at Thr308 by PDK (phosphoinositide-dependent kinase) is a key event for activating downstream targets of Akt [10]. The level of p-Akt was found to be significantly down-regulated in the absence of serum (Figure 5A, lanes 4–6). This observation was confirmed further using an expression construct of p110α (pCDNA3 PI3K caax 110 HA; Dr A. S. Baur, University of Erlangen, Erlangen, Germany) subunit of PI3K, which has been shown to act as a constitutively active mutant of PI3K. Expression of this construct together with N rescued the cells from a death phenotype as exhibited by inhibition of caspase 3 activation (Figure 5E, lane 1). We then analysed the levels of Bcl-2 expression in the presence of N. Immunoprecipitation of Bcl-2 showed strong down-regulation in the absence of serum (Figure 5B, lanes 4–6). However, the level of Bcl-XL was not affected in N-expressing cells, irrespective of the presence or absence of growth factors (results not shown).
Figure 5. N expression down-regulates pro-survival factors and induces apoptosis in the absence of growth factors.
(A) The level of p-Akt (Thr308) in the presence (Ser+) and absence (Ser−) of serum. The lower gel shows the level of total Akt under the same conditions. The histogram shows the means±S.E.M. for three independent sets of experiments. (B) Bcl-2 was immunoprecipitated from mock (C) or N-transfected (N) cell lysates in the presence (Ser+) or absence (Ser−) of serum respectively, and was detected by Western blotting. The calnexin level is shown as loading control in the bottom gel. The histogram shows the means±S.E.M. for three independent sets of experiments. (C) The activation of caspases 3 and 7 in N-expressing cells (lane 4). Anti-(caspase 3) antibody detects both the precursor and cleavage product. (D) Anti-(caspase 7) antibody is specific for cleaved product only. (E) Western blot of caspase 3 in serum-deprived N-expressing cells in the presence of overexpressed p110α subunit of PI3K (110, lane 1), 10 μg/ml SB203580 (SB, lane 2) or 100 μM Z-VAD-FMK (Z, lane 3).
Finally, we analysed the status of two major downstream caspases in the presence and absence of serum. Caspase 3 activation yields a 17 kDa product, which was present in N-expressing cells only in the absence of growth factors. This activation was detected using an antibody that detects both the precursor and cleaved product (Figure 5C, lane 4). Caspase 7 activation produces a major product of 20 kDa. Like caspase 3, caspase 7 was also activated only in the absence of growth factors in N-expressing cells. This was detected by an antibody specific for the cleaved caspase 7 only (Figure 5D, lane 4). Activation of caspase 3 was confirmed further by Z-VAD-FMK, which blocks its activation (Figure 5E, lane 3). However, blocking the p38 MAPK pathway by SB203580 was unable to inhibit caspase 3 activation. Thus we propose that p38 MAPK-dependent actin reorganization and caspase activation are probably two independent functional properties of N. In addition, N-mediated apoptosis was found to be independent of Fas, or the Fas-ligand-mediated pathway, and did not involve modulation of p53 and Bax (results not shown). From these experiments, we conclude that the N protein of SARS-CoV induces apoptosis and actin reorganization by interfering with the integrin signalling pathway during cellular stress induced by the absence of growth factors.
Reports strongly suggest that the cytokine storm at the early stages of SARS in most patients contributes to the progression of SIRS (systemic inflammatory response syndrome). The principal histopathologic lesions of the atypical pneumonia of SARS are serous, fibrinous and haemorrhagic inflammation in most pulmonary alveoli, alveolar thickening with interstitial mononuclear inflammatory infiltration, diffuse alveolar damage, desquamation of pneumocytes, formation of hyaline membrane and multinucleated pneumocytes, with capillary engorgement and microthrombosis. SARS patients also display lymphopenia, as well as abnormal structures of lymph nodes and spleen including haemorrhagic necrosis, reduced numbers of lymphocytes, depletion of lymph node follicles and atrophy of splenic nodules. Pneumocytes represent the primary target of infection. Severe and similar damage to the pulmonary, lymphohaematopoietic, hepatic and other tissues infected by SARS-CoV is responsible for the clinical features of SARS and may lead to the death of patients [11,12]. The SARS N protein has been shown to be highly immunogenic. There is predominant antibody response in patients against N [13]. This process may in turn trigger cytokine production which may actually induce apoptosis of the host cells. Our results also show that the N protein of SARS is capable of inducing actin reorganization. Hence the focal sites of infection with an altered cytokine milieu and cytoskeletal reorganization may be able to alter the duration of interaction between the host cells engaged in combating infection, the lowering of which would lead to an unsuccessful immune initiation event and persistent viral presence and replication.
Although our studies clearly demonstrate that N expression leads to programmed cell death, the proximal initiator of the death programme remains to be identified. Moreover, the observed cell death we observed was tissue-type-specific. We could not detect significant cell death in human hepatoma cells which are of epithelial cell lineage. However, we cannot rule out the possibility that the observed stress tolerance of HuH7 cells is a specific property of that particular cell line rather than tissue specificity, since the mechanism of transformation of COS-1 cells and Huh7 cells are different and hence may have resulted from deregulation of different regulatory networks in these two cell lines, leading to an altered end response. Identification of the differentially regulated factors in these two cell lines during N expression might provide a clue in addressing problems associated with viral infection. Also, identification of the upstream effectors of apoptosis in COS-1 cells will reveal the mechanisms responsible for triggering cell death. Future work in these directions will help us evaluate the specific role of N protein in apoptosis induction and classify it as a host response or a response due to the cytopathic effect of N expression. Research along these lines holds great promise for designing therapeutics to alleviate tissue injury during the early stages of SARS viral infection.
Acknowledgments
We thank Dr A.S. Baur for kindly providing the pCDNA3 PI3K caax p110 HA expression construct. We gratefully acknowledge the assistance of Dr Chetan Chitnis for use of the fluorescence microscopy facility. We thank Anup, Suchi, Ravinder Kumar and Wee Ming for technical assistance. M.S. is a CSIR (Council of Scientific and Industrial Research) senior research fellow. This work was supported by internal funds from ICGEB (International Centre for Genetic Engineering and Biotechnology) and NUS (National University of Singapore), and a Department of Biotechnology research grant to S.K.L. The ASM (American Society for Microbiology)-UNESCO (United Nations Educational Scientific and Cultural Organization) Visiting Resource Person fellowship to S.K.L. for the years 2002 and 2003 is gratefully acknowledged.
References
- 1.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H. R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R. A., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 2.Ksiazek T. G., Erdman D., Goldsmith C. S., Zaki S. R., Peret T., Emery S., Tong S., Urbani C., Comer J. A., Lim W., et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 3.Rota P. A., Oberste M. S., Monroe S. S., Nix W. A., Campagnoli R., Icenogle J. P., Penaranda S., Bankamp B., Maher K., Chen M. H., et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Marra M. A., Jones S. J., Astell C. R., Holt R. A., Brooks-Wilson A., Butterfield Y. S., Khattra J., Asano J. K., Barber S. A., Chan S. Y., et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 5.Surjit M., Liu B., Kumar P., Chow V. T. K., Lal S. K. The nucleocapsid protein of the SARS coronavirus is capable of self-association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 2004;317:1030–1036. doi: 10.1016/j.bbrc.2004.03.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He R., Leeson A., Andonov A., Li Y., Bastien N., Cao J., Osiowy C., Dobie F., Cutts T., Ballantine M., Li X. Activation of AP-1 signal transduction pathway by SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 2003;311:870–876. doi: 10.1016/j.bbrc.2003.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H., Xiao G., Zhang J., Hu Y., Yuan F., Cole D. K., Zheng C., Gao G. F. SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol. 2004;73:323–331. doi: 10.1002/jmv.20094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aplin A. F., Howe A., Alahari S. K., Juliano R. L. Signal transduction and signal modulation by cell adhesion receptors: the role of integrins, cadherins, immunoglobulin–cell adhesion molecules, and selectins. Pharmacol. Rev. 1998;5:197–263. [PubMed] [Google Scholar]
- 9.Schratt G., Philippar U., Berger J., Schwarz H., Heidenreich O., Nordheim A. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 2002;156:737–750. doi: 10.1083/jcb.200106008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta S. R., Brunet A., Greenberg M. E. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 11.Lang Z. W., Zhang L. J., Zhang S. J., Meng X., Li J. Q., Song C. Z., Sun L., Zhou Y. S., Dwyer D. E. A clinicopathological study of three cases of severe acute respiratory syndrome (SARS) Pathology. 2003;35:526–531. doi: 10.1080/00313020310001619118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tse G. M., To K. F., Chan P. K., Lo A. W., Ng K. C., Wu A., Lee N., Wong H. C., Mak S. M., Chan K. F., et al. Pulmonary pathological features in coronavirus associated severe acute respiratory syndrome (SARS) J. Clin. Pathol. 2004;57:260–265. doi: 10.1136/jcp.2003.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung D. T., Tam F. C., Ma C. H., Chan P. K., Cheung J. L., Niu H., Tam J. S., Lim P. L. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190:379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]