Abstract
Research on metabolism of nucleotides and their derivatives has gained increasing interest in the recent past. This includes de novo synthesis, analysis of salvage pathways, breakdown and transport of nucleotides, nucleosides and nucleobases. To perform a further step towards the analysis of nucleoside transport in Arabidopsis, we incubated leaf discs with various radioactively labelled nucleosides. Leaf cells imported labelled nucleosides and incorporated these compounds into RNA, but not into DNA. Furthermore, we report on the biochemical properties of three so far uncharacterized members of the Arabidopsis ENT (equilibrative nucleoside transporter) family (AtENT4, AtENT6 and AtENT7). After heterologous expression in yeast, all three proteins exhibited broad substrate specificity and transported the purine nucleosides adenosine and guanosine, as well as the pyrimidine nucleosides cytidine and uridine. The apparent Km values were in the range 3–94 μM, and transport was inhibited most strongly by deoxynucleosides, and to a smaller extent by nucleobases. Typical inhibitors of mammalian ENT proteins, such as dilazep and NBMPR (nitrobenzylmercaptopurine ribonucleoside, also known as nitrobenzylthioinosine) surprisingly exerted almost no effect on Arabidopsis ENT proteins. Transport mediated by the AtENT isoforms differed in pH-dependency, e.g. AtENT7 was not affected by changes in pH, AtENT3, 4 and 6 exhibited a less pronounced pH-dependency, and AtENT1 activity was clearly pH-dependent. Using a GFP (green fluorescent protein)-fusion protein transiently expressed in tobacco leaf protoplasts, a localization of AtENT6 in the plant plasma membrane has been revealed.
Keywords: Arabidopsis thaliana, equilibrative nucleoside transporter (ENT), salvage pathway, transport
Abbreviations: AtENT, equilibrative nucleoside transporter from Arabidopsis thaliana; AttDT, dicarbonic acid transporter from Arabidopsis thaliana; CCCP, carbonyl cyanide m-chlorophenylhydrazone; CNT, concentrative nucleoside transporter; ENT, equilibrative nucleoside transporter; GFP, green fluorescent protein; LeSUT, plasma-membrane-localized sucrose transporter from tomato (Lycopersicon esculentum); NBMPR, nitrobenzylmercaptopurine ribonucleoside (also known as nitrobenzylthioinosine); WT, wild-type
INTRODUCTION
Nucleotides are building blocks of DNA and RNA, represent major energy carriers and are also core elements of cofactors such as NAD, NADP, S-adenosylmethionine or coenzyme A. During de novo nucleotide biosynthesis, nucleosides do not occur as intermediates, but these metabolites appear during nucleotide turnover. This nucleotide turnover is catalysed in so called ‘salvage pathways’ that are known to occur in animals and plants. Via salvage pathways, cells preserve the energy and carbon inherent in the breakdown products of DNA and RNA, namely nucleosides. De novo synthesis of purine nucleotides, for example, requires the hydrolysis of five nucleotides, whereas the reaction of adenosine kinase, phosphorylating adenosine to AMP, only requires one ATP [1]. In plants, the salvage pathway involved in adenylate recycling is the best studied, although enzymes for the recovery of other nucleosides also exist [1,2]. In contrast with enzymic reactions involved in nucleoside salvage in plants, the transport of corresponding nucleosides is still poorly characterized. In general, nucleoside transport proteins can be divided into CNT (concentrative nucleoside transporter) and ENT (equilibrative nucleoside transporter) types [3,4]. The CNT family exhibits 12–13 predicted transmembrane domains and catalyses either a Na+- or H+-energized co-transport of nucleosides against a concentration gradient. CNT proteins have been identified in a number of bacterial species and in eukaryotes such as Caenorhabditis elegans, Drosophila melanogaster and mammals [5], but not in plants.
Members of the ENT family of nucleoside transporters typically exhibit 11 predicted transmembrane domains and catalyse transport energized by an existing nucleoside concentration gradient. So far, more than 40 members of the ENT protein family have been identified in eukaryotic cells, and it is supposed that they are evolutionarily related to prokaryotic nucleoside transporters [6].
Some protozoan nucleoside transporters are structurally closely related to ENT proteins, but surprisingly catalyse a concentrative proton-coupled nucleoside co-transport [7,8]. In this respect, the first plant nucleoside transporter characterized on the molecular level, ENT1 from Arabidopsis thaliana, behaves similarly, since this protein catalyses a proton-coupled adenosine uptake [9]. The Arabidopsis genome harbours eight isoforms of ENT-type proteins in total, and so far only two isoforms, namely AtENT1 and AtENT3, have been characterized on both the molecular and functional levels [9,10]. The aims of the present study were to deepen our insight into nucleoside metabolism in Arabidopsis and to characterize some of the remaining ENT members. The observation that various disturbances in plant nucleoside metabolism induce dramatically negative effects on both development and metabolism [11,12] clearly emphasizes that we have to increase our knowledge on plant nucleoside metabolism, which includes the corresponding transport proteins.
EXPERIMENTAL
Uptake experiment with leaf discs
Arabidopsis thaliana, ecotype Columbia, was used throughout the present study. To measure the uptake of radiolabelled nucleosides into Arabidopsis leaves, discs (7 mm diameter) were cut from fully developed leaves. A total of 100 leaf discs were incubated in 20 ml of 5 mM Mes/KOH (pH 5.5) supplemented with 5 μM of the indicated nucleoside (185 MBq/mmol; Moravek Biochemicals, CA, U.S.A.). Leaf discs were constantly agitated in Petri dishes. At the given time points, 500 μl of the incubation medium was withdrawn and counted for radioactivity. After 24 h, the incubation was stopped and the leaf discs were washed three times in ice-cold incubation buffer, dried and frozen in liquid nitrogen. To extract soluble components, RNA and DNA, leaf material was homogenized by grinding in liquid nitrogen and 100 mg aliquots were transferred into 1.5 ml reaction tubes. The subsequent extraction was essentially as given in Ashihara and Nobusawa [13].
Strains and media
Plasmids were propagated in Escherichia coli cells (XL1Blue; Stratagene, Heidelberg, Germany) grown in YT medium (0.8% peptone, 0.5% yeast extract and 0.25% NaCl) with or without ampicillin (50 mg/l) and tetracycline (2.5 mg/l). Plasmids harbouring AtENT3, 4, 6, 7 or 8 genes were transformed into ΔFUI1 Saccharomyces cerevisiae yeast cells (W303; Mat α; ura3-1; his3-11; leu2-3_112; trp1Δ2; ade2-1; can1-100; YBL042c 11,1902::kanMX4) obtained from EUROSCARF [European Saccharomyces cerevisiae Archive for Functional Analysis (Institut für Mikrobiologie, Johann Wolfgang Goethe-Universität, Frankfurt am Main, Germany)] applying the method of Ito et al. [14]. Cells were grown on minimal medium containing 0.67% yeast nitrogen base (Remel, distributed by Ceratogene Biosciences, Augsburg, Germany) and supplements as required to maintain auxotropic selection.
cDNA cloning
The genomic sequences available from the Arabidopsis Genome Initiative [15] were used to design primers for the amplification of AtENTs, so that the amplification products covered the complete coding regions. The primers contained restriction sites for BamHI and XhoI for subsequent cloning into the pYES2 vector (Invitrogen, Groningen, The Netherlands) for subsequent expression in yeast cells. The following primers were used: AtENT3: forward, 5′-GTTCTTTGGATCCCAATGGCGGATAGATATG-3′ and reverse, 5′-TGAACCACTGTTCTCGAGTGATTAAAACTC-3′; AtENT4: forward, 5′-GAGGGATTCATAACTATTTAAAAAATGGCG-3′ and reverse, 5′-TAATTGCACAAAATCTCGAGAACTCAG-3′; AtENT6: forward, 5′-GTTCTTTGGATCCCTCTGTAACAATTTCG-3′ and reverse, 5′-GAAACACTGGTTCTCGAGTGATTAAAACTC-3′; AtENT7: forward, 5′-GATTTAGGGGATCCGTGATCGCCTGATGAC-3′ and reverse, 5′-AAAGACTCGAGTTATATGATCTCTAAAACG-3′; AtENT8: forward, 5′-AAAACTCGAGTTACAATGGTTGATGAG-3′ and reverse, 5′-TAAGGTCTAGATGAGCCAGAGCCAACC-3′.
The GFP (green fluorescent protein)-fusion construct was prepared by amplification of the complete coding region of AtENT6 using primers: forward, 5′-ATGTTCTTTTGATCTCTCTAGAACAATTTC-3′ and reverse, 5′-GATTAACTCGAGAAAGGCATTCTTCTTACC-3′ with Pfu polymerase. The primers contained XbaI and XhoI restriction sites for insertion into pGFP2 [16]. The resulting plasmid was sequenced to verify ‘in-frame’ cloning of AtENT6 and GFP.
Transport experiments
To initiate expression, cells inoculated in minimal medium containing 2% glucose and 1% raffinose as the carbon source were harvested by centrifugation at 4300 g for 10 min and transferred to induction-medium containing 2% galactose and 1% raffinose. Cells were grown for at least 6 h to allow for induction and then harvested at a D600 of 0.5–1.5, washed twice with 25 mM NaH2PO4 buffer medium (pH 6.0), and resuspended in the same buffer medium resulting in a final D600 of 10. For experiments with differing pH in the incubation medium, cells were resuspended in 5 mM NaH2PO4 buffer medium (pH 6.0). To initiate uptake, 100 μl of cell suspension was added to 100 μl of incubation medium containing the 3H-labelled transport substrate in 100 mM NaPO4 buffer medium at the given pH. For all other experiments, cells were added to incubation medium with 25 mM NaH2PO4 buffer medium (pH 6.0). Cells were incubated for the indicated time intervals at 30 °C and subsequently filtered on membrane filters (Supor® membrane disc filters, 0.45 μm pore size; Pall Gelman Laboratory, Ann Arbor, MI, U.S.A.) previously set under vacuum. The radioactivity on the filters was quantified by liquid-scintillation counting in a Tricarb 2500 TR scintillation counter (Packard, Dreieich, Germany).
Transient expression of GFP-fusion constructs
Protoplasts were prepared from sterile-grown tobacco (Nicotiana tabacum cv. W38) plants as given in Wendt et al. [17]. Protoplasts were transformed with column-purified plasmid DNA (30 μg per 0.5×106 cells). After 18 h of incubation in the dark at room temperature (20 °C), protoplasts were checked for green fluorescence using the Zeiss laser-scanning system LSM510 (Carl Zeiss, Jena, Germany). GFP was excited at 488 nm, and emission was detected by a photomultiplier through a 505–530 nm band-pass filter using an Achroplan 40×/0.75W objective. Vacuoles from fluorescing protoplasts were prepared as given in Frangne et al. [18] and checked for green fluorescence using a light microscope (Axioskop, Carl Zeiss, Oberkochen, Germany) equipped with filter set 09.
RESULTS
Uptake of 3H-labelled nucleosides into Arabidopsis leaf discs
To take a further step to understand transport and metabolism of nucleosides in Arabidopsis we incubated leaf discs in medium containing tritiated adenosine, guanosine, uridine or cytidine respectively. All nucleosides tested were provided at a concentration of 5 μM, which is in the range of the apparent Km values of nucleoside transporters analysed so far.
Within the first hours of incubation, a linear uptake of all four nucleosides tested could be observed, and initial rates were 2.2 nmol·(g of fresh weight)−1·h−1 (Figure 1A). Uptake of guanosine equilibrated after about 4 h, whereas uptake rates of the other three nucleosides remained constant.
Figure 1. Uptake of 3H-labelled nucleosides into Arabidopsis leaf discs and separation of incorporation products.
(A) Time course of uptake [in nmol·(g of fresh weight)−1·h−1 (nmol/gFW/h)] of 3H-labelled adenosine (•), guanosine (▪), uridine (▴) or cytidine (♦). Leaf discs were incubated for the given time in incubation medium containing 5 μM nucleoside. (B) Separation of the soluble fraction, RNA and DNA after incubating leaf discs under the conditions given in (A) for 24 h. The key to the shading is given beneath the ‘soluble’ group of bars. Results are means±S.E.M. for three independent experiments.
At the end of the incubation, soluble components, RNA and DNA were extracted. Most of the radioactivity in leaf discs incubated with labelled adenosine was found in soluble components and the RNA fraction (40% and 56% respectively), whereas only 2% was recovered in DNA (Figure 1B). The molecular nature of the residual radioactivity (less than 2%) has not been identified (Figure 1B). Partitioning of radioactive label in samples supplemented with [3H]cytidine was similar to samples incubated with labelled adenosine (Figure 1B). [3H]Uridine was mostly present in the soluble fraction, whereas 82% of the [3H]guanosine was found in the RNA fraction (Figure 1B).
Nucleoside transport into yeast cells heterologously synthesizing AtENT3, 4, 6, 7 and 8
The Arabidopsis genome harbours eight genes encoding putative members of the ENT family. However, only two of these proteins, namely AtENT1 and AtENT3, have been analysed so far at the functional level [9,10]. To characterize the biochemical properties of some of the remaining isoforms, we amplified full-length cDNAs of AtENT 3, 4, 6, 7 and 8 from first-strand cDNA, prepared from a mixture of root, leaf, stem, flower and silique tissues. The gene-specific primers used were designed according to information taken from the Arabidopsis Genome Initiative [15], and the full-length transporter cDNAs were subsequently cloned into the vector pYES2 and sequenced.
The sequence information obtained were identical with data published previously by others under accession numbers AF426400, AtENT3; AF426401, AtENT4, AF426402, AtENT6; AF426403, AtENT7 and AY187030, AtENT8 [10]. AtENT1 had previously been characterized by us [9], AtENT2 and 5 could not be amplified yet, which is possibly due to the low expression level of corresponding genes in the Arabidopsis tissues used to prepare the first-strand cDNA. AtENT3 had already been initially analysed [10], but was also included into this work here to enlarge the knowledge on the biochemical properties of this nucleoside transporter isoform. The transport properties of Arabidopsis ENT-type proteins have been characterized using the yeast mutant ΔFUI1 lacking the sole yeast plasma membrane-located nucleoside transporter FUI1 [19]. FUI1 is highly specific for uridine transport, and uridine is thought to represent a general permeant accepted by most nucleoside transporters analysed so far. Therefore use of the ΔFUI1 mutant should allow us to analyse the Arabidopsis ENT proteins that mediate uridine transport.
AtENT1 from Arabidopsis exhibits a broad substrate spectrum as revealed by competition studies [9]. To check whether AtENT3, 4, 6, 7 and 8 transporters are also able to transport purine and pyrimidine nucleosides, we performed growth studies in the presence of either adenosine (a purine nucleoside) or fluorouridine (a toxic pyrimidine nucleoside analogue). Figure 2 presents the growth pattern of WT (wild-type) (W303)-, ΔFUI1-, or ΔFUI1 cells expressing AtENT3, 4, 6, 7 and 8 on various media. Plate A contained complete YPD medium; plate B contained a synthetic minimal medium lacking uracil, but containing adenine; plate C lacked uracil and adenine, but contained adenosine; and plate D lacked uracil, but contained toxic fluorouridine.
Figure 2. Growth analysis of yeast cells expressing AtENTs.
Growth of S. cerevisiae yeast cells harbouring expression plasmids containing AtENT3, 4, 6, 7 and 8, as indicated or controls (W303; ΔFui1: W303, ΔFui1 transformed with empty pYES vector). (A) Growth on complete medium (YPD). (B) Growth on synthetic minimal medium lacking uracil and containing galactose (inducing conditions). (C) Growth on synthetic minimal medium lacking uracil and adenine, but containing adenosine (150 μM) and galactose. (D) Growth on synthetic minimal medium lacking uracil, but containing fluorouridine (1 mM) and galactose. Cells were grown for 2–4 days at 30 °C.
All cells tested were able to grow on a YPD medium (Figure 2A), whereas WT cells were not able to grow on medium lacking uracil (Figure 2B). With the exception of AtENT8, all other Arabidopsis ENT proteins, AtENT3, 4, 6, and 7, were able to complement the ade2 mutation on an adenosine-containing medium (Figure 2C), clearly indicating the competence for purine transport. AtENT8-expressing cells and the ΔFUI1 mutant grew in the presence of the toxic compound fluorouridine (Figure 2D), indicating that AtENT3, 4, 6, and 7 additionally mediate pyrimidine transport.
Following these indirect uptake studies, we next analysed nucleoside transport in more detail. For a correct characterization of biochemical transport properties, it is required that apparent Km and Vmax values are taken within the linear phase of import. Therefore we first checked that nucleoside uptake catalysed by each of the heterologously synthesized Arabidopsis ENT proteins occurred linearly with time. Nucleoside uptake catalysed by all transporters tested was linear with time at saturating nucleoside concentrations for at least 5 min (results not shown).
Exemplified in Figures 3(A) and 3(B), the Michaelis–Menten kinetics of adenosine or uridine uptake catalysed by the AtENT6 transporter are shown. The deduced Hanes plot (inset of Figures 3A and 3B) indicates apparent Km values for adenosine and uridine of 3.0 and 6.4 μM respectively. The maximal rates of transport were 0.25 nmol/107 cells per h for adenosine, and 0.83 nmol/107 cells per h for uridine.
Figure 3. Substrate-dependence of nucleoside uptake into yeast cells expressing AtENT6.
(A) Uptake of 3H-labelled adenosine. Cells were incubated for 2 min at the indicated substrate concentration. The affinity constant was calculated to be 3.0 μM on the basis of the Hanes analysis (inset). (B) Uptake of [3H]uridine. Cells were incubated for 2 min at the indicated substrate concentration. The affinity constant was calculated to be 6.4 μM on the basis of the Hanes analysis (inset). Induction of protein expression and uptake measurements were performed as described in experimental procedures. Results are means±S.E.M. for three experiments.
Table 1 summarizes the affinity constants of transporters AtENT1, 3, 4, 6 and 7. AtENT1 exhibited high affinity for adenosine, but much lower affinities for guanosine and cytidine, as indicated by IC50 values (Table 1) [9]. AtENT3 exhibited slightly higher affinities for pyrimidine nucleosides (9.5 and 10.8 μM for uridine and cytidine respectively; Table 1) as compared with purine nucleosides (15.5 and 18 μM for adenosine and guanosine; Table 1). AtENT4 is characterized by the highest affinity for guanosine (apparent Km, 7.3 μM); for all other nucleosides, AtENT4 exhibited the lowest apparent affinities of all ENT proteins analysed, ranging from 27.8 to 49.1 μM (Table 1). AtENT6 exhibited the highest affinities for adenosine and uridine (3.0 and 6.4 μM respectively) and lower affinities for guanosine and cytidine (11.5 and 21.2 μM respectively; Table 1). Finally, AtENT7 appeared to be more specific for purine nucleosides (Kms of 9.8 μM for adenosine and 9.4 μM for guanosine; Table 1) than for pyrimidine nucleosides (Kms of 40 μM for cytidine and 13.4 μM for uridine; Table 1).
Table 1. Affinity constants of AtENT1, 3, 4, 6 and 7 for purine and pyrimidine nucleosides.
Km values were obtained by incubating yeast cells, expressing the respective proteins in 3H-labelled nucleosides for 2 min. The affinity constants were calculated according to Hanes plot. Results are means±S.E.M. for three independent experiments. n.d., not determined.
| Km (μM) | ||||
|---|---|---|---|---|
| Transporter | Adenosine | Guanosine | Cytidine | Uridine |
| AtENT1 | 3.6* | IC50 28.0* | IC50 20.0* | n.d. |
| AtENT3 | 15.5 | 18.0 | 10.8 | 9.5 |
| AtENT4 | 49.1 | 7.3 | 94.2 | 27.8 |
| AtENT6 | 3.0 | 11.5 | 21.2 | 6.4 |
| AtENT7 | 9.8 | 9.4 | 40.0 | 13.4 |
* Data taken from Möhlmann et al. [9].
To analyse whether structurally related compounds affect nucleoside transport, we performed competition experiments. For this, we quantified the uptake of [3H]adenosine given at a concentration of 25 μM (AtENT4), 1.8 μM (AtENT6) or 5 μM (AtENT7) in the absence or presence of these compounds. From the nucleobases tested, hypoxanthine inhibited AtENT4-mediated adenosine transport most efficiently (58% residual activity; Figure 4). All other nucleobases had only little or no effect on transport catalysed by AtENT4, 6 or 7 (Figure 4). From all nucleosides tested, inosine acted inhibitorily on AtENT6 and 7 (below 38% residual activity; Figure 4). Deoxycytidine, deoxyguanosine and deoxyinosine interfered strongly with adenosine transport of AtENT7, and also slightly inhibited activity of AtENT6, whereas AtENT4 was only affected by deoxyguanosine. (Figure 4). The nucleotides tested did not exert strong effects on the three AtENT proteins (Figure 4).
Figure 4. Inhibition of 3H-labelled adenosine uptake mediated by AtENT4, 6 and 7 by analogous substrates.
Adenosine was given at a concentration of 25 μM, (AtENT4), 1.8 μM (AtENT6) and 5 μM (AtENT7), and yeast cells were incubated for 2 min. Analogous substrates were provided at 10 times the concentration of adenosine, simultaneously with the 3H-labelled adenosine. Induction of protein expression and uptake measurements were performed as described in the Experimental section. Results are means±S.E.M. for three independent experiments.
Typical inhibitors of mammalian ENT proteins such as dilazep and NBMPR (nitrobenzylmercaptopurine ribonucleoside, also known as nitrobenzylthioinosine) were tested on AtENT4, 6 and 7 catalysed transport. Only extraordinarily high concentrations of dilazep (20 μM) resulted in a substantial inhibition of AtENT6 activity (17.9% residual activity; Table 2). AtENT4 and 7 were inhibited to 30% and 27% of the control rate respectively (Table 2). NBMPR strongly impaired transport mediated by AtENT4 and 6, only when supplied at the high concentration of 20 μM (17.7% and 14.5% residual activity respectively), whereas AtENT7 was less affected by NBMPR (Table 2).
Table 2. Effect of vasodilator drugs and CCCP on transport of adenosine by AtENT4, 6 and 7.
Adenosine was provided at a concentration of 25 μM (AtENT4), 1.8 μM (AtENT6) and 5 μM (AtENT7) and yeast cells were incubated for 2 min together with the indicated effectors. Results are means±S.E.M. for three independent experiments.
| Transporter (residual activity, %) | |||
|---|---|---|---|
| Inhibitor | AtENT4 | AtENT6 | AtENT7 |
| Dilazep (20 nM) | 71.5±17.0 | 76.3±3.8 | 81.7±4.3 |
| Dilazep (20 μM) | 30.0±12.0 | 17.9±1.1 | 27.5±5.6 |
| NBMPR (20 nM) | 79.3±7.0 | 86.7±6.6 | 84.0±4.8 |
| NBMPR (20 μM) | 17.7±9.0 | 14.5±1.0 | 62.3±3.9 |
| CCCP (50 μM) | 52.0±17.0 | 31.5±2.5 | 76.0±2.5 |
The protonophore CCCP (carbonyl cyanide m-chlorophenylhydrazone) was used to assess the question of whether or not nucleoside transport catalysed by ENT proteins was dependent upon an existing proton gradient. Adenosine import catalysed by AtENT4 was reduced to 52%, AtENT6 activity to 32% and AtENT7 activity only to 76% (Table 2).
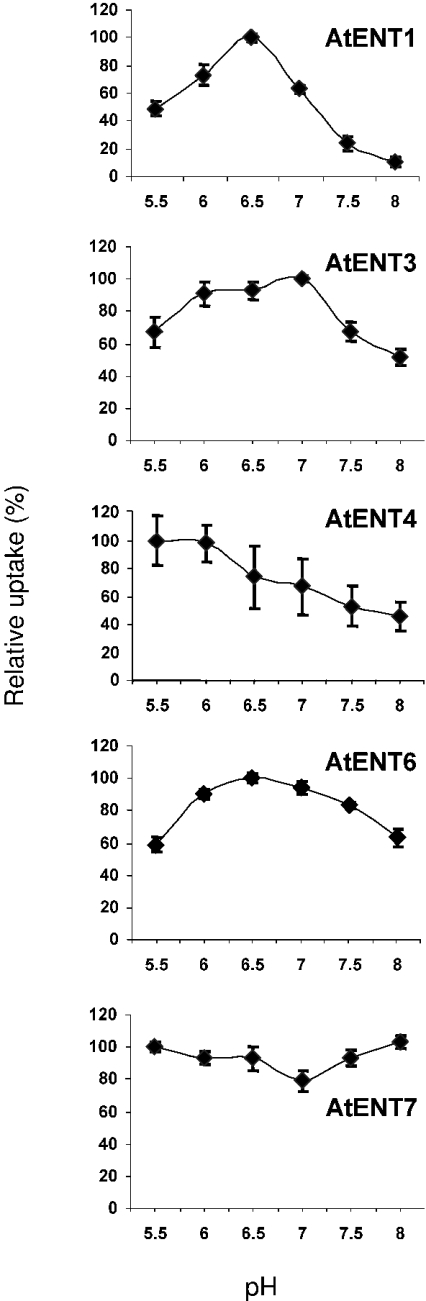
To address the pH-dependency in detail, we quantified nucleoside transport into yeast cells at different pH values. Figure 5 clearly reveals that AtENT1-dependent nucleoside import responds strongly to changing proton concentrations in the medium, exhibiting maximal transport rates at pH 6.5 and only 10% residual activity at pH 8. In contrast with this, AtENT7 activity is not influenced by the pH of the incubation medium (Figure 5). AtENT3 activity was much less inhibited by alkalization of the medium when compared with AtENT1 (Figure 5). The same holds true for AtENT4 and 6, which also exhibited a broad pH optimum (Figure 5).
Figure 5. pH-dependency of nucleoside uptake into yeast cells.
Influence of the pH of the incubation medium on the uptake of [3H]adenosine, mediated by AtENT1, 3, 4, 6 and 7. After induction, yeast cells were harvested, washed and resuspended in 5 mM NaPO4 (pH 6.0) and put on ice until uptake was initiated. The incubation medium consisted of 100 mM NaPO4 at various pH values. [3H]Adenosine was provided at a concentration of 2 μM and cells were incubated for 2 min. Results are means±S.E.M.
Subcellular localization of AtENT6
Recently it has been shown that AtENT1 and AtENT3 reside in the plant plasma membrane [10,20]. However, this does not mean that other AtENT transporters also reside in this membrane. To analyse the subcellular localization of AtENT6, we prepared an AtENT6–GFP fusion construct and transiently transformed tobacco protoplasts. As controls GFP alone, GFP-fusion constructs carrying a plasma-membrane-localized sucrose transporter from tomato (LeSUT–GFP) or a vacuolar dicarbonic acid transporter from Arabidopsis (AttDT–GFP, [21]) have been used. Figure 6 revealed cytosolically located fluorescence when GFP alone is expressed (Figure 6A). In contrast, fluorescence surrounding the protoplast appeared when LeSUT–GFP or AtENT6–GFP was expressed (Figures 6B and 6C), indicating a plasma membrane localization of AtENT6. The reported plasma membrane localization of AtENT1 was also confirmed using a corresponding GFP-fusion construct (Figure 6D).
Figure 6. Transient transformation of tobacco protoplasts with GFP-fusion constructs.
Tobacco protoplasts were prepared as given in the Experimental section and transformed using poly(ethylene glycol) (PEG). Transformed protoplasts were analysed with confocal laser scanning microscopy. (A) GFP alone. (B) GFP fused to a plasmalemma control (LeSUT). (C) GFP fused to AtENT6. (D) GFP fused to AtENT1. (E) Vacuoles prepared from AttDT–GFP-expressing protoplasts (arrows mark vacuoles, red fluorescence is emitted from protoplasts that were not lysed). (F) Vacuoles prepared from AtENT6–GFP-expressing protoplasts. (G) GFP fluorescence from vacuoles in (E). (H) GFP fluorescence from vacuoles in (F).
For a better differentiation between plasma membrane and tonoplast localization, we prepared vacuoles from protoplasts expressing GFP alone, LeSUT–GFP, AtENT6–GFP and AttDT–GFP. Vacuoles prepared from protoplasts expressing AttDT–GFP [viewed with a DAPI (4,6-diamidino-2-phenylindole) fluorescence filter set] exhibit light-blue fluorescence (Figure 6E), which is similar to vacuoles prepared from AtENT6–GFP-expressing cells (Figure 6F). However, only vacuoles from protoplasts expressing AttDT–GFP exhibited green fluorescence (Figure 6G), but not vacuoles from protoplasts expressing AtENT6–GFP (Figure 6H) or LeSUT–GFP (results not shown).
DISCUSSION
Although there has been increasing interest in metabolism of nucleotides and their derivatives in higher plants [1,2], still many of the processes are poorly understood. Until now, nucleoside transport in higher plants has only been demonstrated for a limited number of tissues, e.g. germinating petunia (Petunia hybrida) pollen [22,23], cotyledons of germinating castor bean endosperm [24] and discs of potato tubers [25].
We therefore performed an initial analysis of nucleoside transport in Arabidopsis, since, to our knowledge, this has not been done before. Arabidopsis leaf cells clearly exhibit the capacity to import purine as well as pyrimidine nucleosides at rates of about 2.2 nmol·(g of fresh weight)−1·h−1 (Figure 1A). Besides a soluble fraction, approx. 40% of the imported nucleosides appear to be incorporated into RNA, whereas conversion into DNA was negligible (Figure 1B). This indicates that nucleosides are preferentially used in anabolic reactions rather than being degraded. Incorporation of significant amounts of exogenously supplied nucleosides into RNA has also been described for cultured white spruce (Picea glauca) cells [26,27] and for potato discs [25]. Therefore, in addition to our data, we have to assume that the capacity to import nucleosides is a phenomenon widely present in plant tissues.
From eight ENT-type nucleoside transporters present in Arabidopsis, seven have been analysed for their tissue specificity by RT (reverse transcriptase)-PCR and all of those were found to be present in leaf tissues [10]. The biochemical characterization of AtENT1 revealed that this transporter is highly specific for adenosine, but also transports guanosine and cytidine [9] (results not shown), and AtENT3 exhibits an even broader substrate specificity ([10]; and Table 1). AtENT1 and AtENT3 are supposed to reside in the plasma membrane ([10,20]; and Figure 6), a localization that has now also been shown for AtENT6 (Figure 6). As AtENT1, 3 and 6 are also able to transport a range of nucleosides ([10]; and Table 1), we have to assume that plant cells use a number of nucleoside transporters simultaneously to import these metabolites. This nucleoside import may function in the salvage of nucleosides from the apoplast. The assumption of a highly active salvage pathway in leaves is consistent with low levels of nucleosides reported for this tissue [28].
The enzymes necessary for the conversion of nucleosides into ribonucleotides the direct precursors for RNA synthesis are clearly present in plant cells, although only few corresponding genes have been identified so far [1,26,27]. The low incorporation of nucleosides into DNA (Figure 1B) is most probably not due to restrictions in the enzyme activity necessary for the conversion of nucleosides in this tissue, but is most likely a result of the low cell division rate in mature leaves.
In Arabidopsis, the members of the AtENT family, comprising eight isoforms in total, catalyse the transport of nucleosides and two of them (AtENT1 and AtENT3) have already been analysed at the molecular and biochemical level [9,10]. In the present paper, we also report on the biochemical properties of AtENT4, 6 and 7, but not on AtENT2 and 5. The latter two transporters could not be amplified from cDNA at full length, possibly due to low expression, as shown for AtENT2 [10], or tightly regulated, cell-type-specific expression.
Initial growth analysis of yeast cells expressing AtENTs indicated that AtENT3, 4, 6 and 7 transport both purine and pyrimidine nucleosides, but AtENT8 does not exhibit any transport competence (Figure 2). The lack of transport activity of AtENT8 was confirmed by direct transport measurements on yeast cells (results not shown). Whether this lack of transport is due to incorrect or missing integration into the yeast plasma membrane or whether AtENT8 transports substrates other than nucleosides will be elucidated in forthcoming experiments.
Direct transport studies of the three novel members of the AtENT family (AtENT4, 6 and 7), as well as an extended analysis of AtENT3, showed that all of these four ENTs transport adenosine, guanosine, cytidine and uridine with high affinities. The apparent affinity of adenosine and uridine uptake mediated by AtENT3 had been analysed before, and was determined to be slightly higher [10]. This is probably due to the different yeast strains used.
Competition experiments revealed that adenosine uptake by AtENT4, 6 and 7 is inhibited by deoxynucleosides, inosine and only to a smaller extent by nucleobases (Figure 4). Nucleotides do not exert strong inhibitory effects on adenosine uptake by any of the three proteins. Uptake studies using 32P-labelled ATP and ADP confirmed that these metabolites are not substrates for AtENT4 and 7 (results not shown). These results indicate that other metabolites than nucleosides may also be substrates for plant nucleoside transporters. In this respect, the transport properties of AtENT6 and 7 resemble the biochemical features of AtENT1 [9]. It seems remarkable that transport facilitated by AtENT3 is also inhibited by the bases cytosine and uracil and moreover by the nucleotides ADP and ATP [10], indicating an extended substrate spectrum of this transporter.
Now that five of eight AtENTs have been analysed at the functional level, a high affinity and broad substrate specificity seem to be main features of plant nucleoside transport. This is in good agreement with our findings on Arabidopsis leaves (Figure 1), and also with import capacities of cotyledons from germinating castor beans, potato tuber discs, cultured cells of white spruce (Picea glauca) and petunia (Petunia hybrida) pollen [1,23–26]. All of these tissues are competent for uptake of multiple nucleosides and related metabolites. In this respect, plant ENT-type transporters resemble characteristics known for mammalian ENT transporters. All mammalian ENT proteins analysed so far showed a wide substrate spectrum, clearly different from the narrow substrate spectrum of most CNT proteins [7,29].
To explain the differences in substrate specificity and affinity between individual AtENTs, additional physiological studies on, for instance, knockout or RNAi (RNA interference) mutants are necessary.
The low sensitivity of AtENT4, 6 and 7 towards dilazep and NBMPR represents a similarity to AtENT1 and 3 investigated before [9,10]. Molecular studies on mammalian ENTs revealed that conserved amino acid residues are important for sensitivity against vasodilator drugs [7,30]. Comparing the protein sequences of AtENTs with those of the human homologue hENT1, Li et al. [10] identified two amino acid exchanges (position 154 of hENT1, Gly→Asp, and position 33 of hENT1, Met→Ile/Leu) in conserved domains of all AtENTs. With the evidence that AtENT4, 6 and 7 are also resistant against dilazep and NBMPR presented in the present work, these residues are most likely to be responsible for the resistance against vasolidator drugs of all five AtENTs tested so far.
Recently, a first cytokinin transporter, AtPUP1 was identified [31] belonging to the Arabidopsis family of purine transporters that is, most interestingly, able to transport adenosine in addition. Therefore we speculated that some AtENTs might function as cytokinin transporters, but no indication for transport of kinetin, zeatin or zeatin riboside was found in competition studies (results not shown).
Nucleoside transport mediated by all AtENTs tested so far is inhibited by protonophores, but to a quite different extent ([9,10]; and Table 2). Therefore we analysed the pH-dependency in more detail. The less pronounced response of adenosine transport to the protonophore CCCP by AtENT3, 4, 6 and 7 in comparison with AtENT1 is reflected by a lower dependency towards pH changes of the incubation medium (Figure 5).
The observation that AtENT1, 3, and 6 reach maximal transport rates at pH 6.5 and reduced rates at pH 5.5 might reflect a regulatory mechanism of transport activity in cells that are subjected to drastic pH changes, e.g. companion cells. However, for a physiological interpretation of this pH effect, the exact localization of the corresponding proteins in specific cell types has to be analysed with mutants expressing promoter–reporter gene fusions.
AtENT7 exhibits no pH-dependency and may transport nucleosides against a pH gradient, e.g. from the cytosol into the apoplastic space, thus functioning as an exporter. The observation that Arabidopsis possesses eight members of the ENT family, but no homologue with CNT-type transporters can now be explained, because both functions are inherent in AtENT members.
From our transient expression studies using an AtENT6–GFP fusion construct, we obtained good evidence for a plasma membrane localization (Figure 6), as already demonstrated for AtENT1 and 3. To obtain final evidence that the plasma membrane is the physiological destination, independent experiments using specific antisera will be needed. Whether or not the remaining five AtENT family members share the plasma membrane localization must be questioned. Sequence analysis of AtENT2 and AtENT5 reported by the ARAMEMNON database (http://aramemnon.botanik.uni-koeln.de/index.html) using the algorithm of the iPsort software [32] indicates a possible plastidic or mitochondrial localization respectively. Plant vacuoles, in addition, serve as a compartment to store carbohydrates, dicarbonic acids and a wide range of products of the cellular detoxification system. Degrading enzyme activities as acid hydrolases and proteases are also present in the vacuole. In purified tomato vacuoles, RNase activity as well as RNA oligonucleotides and uridine were detected [33,34]. This nucleolytic activity of the vacuole requires the presence of a transport mechanism for the export of nucleic acid breakdown products such as nucleosides. Interestingly, yeast cells possess an internal nucleoside transporter, FUN26 [19], located in the ‘post-Golgi’ compartment that is related to the vacuole. Furthermore, the human transporter hENT3 is also supposed to reside predominantly in an intracellular compartment [35], strengthening the position that AtENTs may well be present in intracellular compartments.
ATP export from Arabidopsis leaves has been reported [36], and it is thought that this ATP export is mediated by ABC (ATP-binding cassette) transporters involved in establishing resistance to xenobiotics. Furthermore, this extracellular ATP may act as a signalling agent [37]. In any case, re-import of adenosine into the cell is necessary to avoid accumulation of ATP as a result of arising concentrations of breakdown products in the extracellular space. AtENT1 is, in contrast with the other AtENTs, highly specific for adenosine, moreover it shows a strict proton-dependency and is present in all tissues. Possibly, AtENT1 is involved in one of the systems described above.
AtENT3 and AtENT6, both also residing in the plasma membrane, exhibit broader substrate specificity and are most probably involved in import of nucleosides resulting from nucleotide breakdown of damaged or dead cells. Senescence and pathogen attack are also events leading to the liberation of nucleosides due to nucleolytic breakdown of RNA and DNA and to re-import of these products into neighbouring cells.
If these assumptions hold true and if other physiological roles for nucleoside transporters can be envisaged, then this has to be analysed in future work.
Acknowledgments
We thank Inka Zschiedrich for help during protoplast transformation and Dr Christian Lohr for expert help during the confocal analysis. This work was supported by the Deutsche Forschungsgemeinschaft (Arabidopsis Functional Genomics Network, AFGN).
References
- 1.Moffatt B. A., Ashihara H. Purine and pyrimidine nucleotide synthesis and metabolism. In: Somerville C. R., Meyerowitz E. M., editors. The Arabidopsis Book. Rockville: American Society of Plant Biologists; 2002. doi/10.1199/tab.0018, http://www.aspb.org/publications/arabidopsis/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boldt R., Zrenner R. Purine and pyrimidine biosynthesis in higher plants. Physiol. Plant. 2003;117:297–304. doi: 10.1034/j.1399-3054.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- 3.Griffith D. A., Jarvis S. M. Nucleoside and nucleobase transport systems of mammalian cells. Biochim. Biophys. Acta. 1996;1286:153–181. doi: 10.1016/s0304-4157(96)00008-1. [DOI] [PubMed] [Google Scholar]
- 4.Cass C. E., Young J. D., Baldwin S. A. Recent advances in the molecular biology of nucleoside transporters of mammalian cells. Biochem. Cell. Biol. 1998;76:761–770. doi: 10.1139/bcb-76-5-761. [DOI] [PubMed] [Google Scholar]
- 5.Huang Q. Q., Yao S. Y., Ritzel M. W., Paterson A. R., Cass C. E., Young J. D. Cloning and functional expression of a complementary DNA encoding a mammalian nucleoside transport protein. J. Biol. Chem. 1994;269:17757–17760. [PubMed] [Google Scholar]
- 6.Acimovic Y., Coe I. R. Molecular evolution of the equilibrative nucleoside transporter family: identification of novel family members in prokaryotes and eukaryotes. Mol. Biol. Evol. 2002;19:2199–2210. doi: 10.1093/oxfordjournals.molbev.a004044. [DOI] [PubMed] [Google Scholar]
- 7.Hyde R. J., Cass C. E., Young J. D., Baldwin S. A. The ENT family of eukaryote nucleoside and nucleobase transporters: recent advances in the investigation of structure/function relationships and the identification of novel isoforms. Mol. Membr. Biol. 2001;18:53–63. [PubMed] [Google Scholar]
- 8.Mäser P., Sütterlin Ch., Kralli A., Kaminsky R. A nucleoside transporter from Trypanosoma brucei involved in drug resistance. Science. 1999;285:242–244. doi: 10.1126/science.285.5425.242. [DOI] [PubMed] [Google Scholar]
- 9.Möhlmann T., Mezher Z., Schwerdtfeger G., Neuhaus H. E. Characterisation of a concentrative type of adenosine transporter from Arabidopsis thaliana (ENT1,At) FEBS Lett. 2001;509:370–374. doi: 10.1016/s0014-5793(01)03195-7. [DOI] [PubMed] [Google Scholar]
- 10.Li G., Liu K., Baldwin S. A., Wang D. Equilibrative nucleoside transporters of Arabidopsis thaliana: cDNA cloning, expression pattern and analysis of transport activities. J. Biol. Chem. 2003;278:35732–35742. doi: 10.1074/jbc.M304768200. [DOI] [PubMed] [Google Scholar]
- 11.Moffatt B., Somerville C. Positive selection for male-sterile mutants of Arabidopsis lacking adenine phosphoribosyl transferase activity. Plant Physiol. 1988;86:1150–1154. doi: 10.1104/pp.86.4.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffatt B., Weretilnyk E. A. Sustaining S-adenosyl-L-methionine-dependent methyltransferase activity in plant cells. Physiol. Plant. 2001;113:435–442. [Google Scholar]
- 13.Ashihara H., Nobusawa E. Metabolic fate of [8-14C]-adenine and [8-14C]-hypoxanthine in higher plants. Z. Pflanzenphysiol. 1981;104:443–458. [Google Scholar]
- 14.Ito H., Fukuda J., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature (London) 408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 16.Kost B., Spielhofer P., Chua N. H. A GFP–mouse talin fusion protein labels plant actin filaments and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- 17.Wendt U. K., Wenderoth I., Tegeler A., von Schaewen A. Molecular characterization of a novel glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.) Plant J. 2000;23:723–733. doi: 10.1046/j.1365-313x.2000.00840.x. [DOI] [PubMed] [Google Scholar]
- 18.Frangne N., Eggmann T., Koblischke C., Weissenbock G., Martinoia E., Klein M. Flavone glucoside uptake into barley mesophyll and Arabidopsis cell culture vacuoles: energization occurs by H+-antiport and ATP-binding cassette-type mechanisms. Plant Physiol. 2002;128:726–733. doi: 10.1104/pp.010590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vickers M. F., Yao S. Y. M., Baldwin S. A., Young J. D., Cass C. E. Nucleoside transporter proteins of Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:25931–25939. doi: 10.1074/jbc.M000239200. [DOI] [PubMed] [Google Scholar]
- 20.Li J., Wang D. Cloning and in vitro expression of the cDNA encoding a putative nucleoside transporter from Arabidopsis thaliana. Plant Sci. 2000;157:23–32. doi: 10.1016/s0168-9452(00)00261-2. [DOI] [PubMed] [Google Scholar]
- 21.Emmerlich V., Linka N., Reinhold T., Hurth M. A., Traub M., Martinoia E., Neuhaus H. E. The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11122–11126. doi: 10.1073/pnas.1832002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kamboj R. K., Jackson J. F. Pyrimidine nucleoside uptake by petunia pollen. Plant Physiol. 1985;79:801–805. doi: 10.1104/pp.79.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamboj R. K., Jackson J. F. Purine nucleoside transport in petunia pollen is an active, carrier-mediated system not sensitive to nitrobenzylthioinosine and not renewed during pollen tube growth. Plant Physiol. 1987;84:688–691. doi: 10.1104/pp.84.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kombrink E., Beevers H. Transport of purine and pyrimidine bases and nucleosides from endosperm to cotyledons in germinating castor bean seedlings. Plant Physiol. 1983;73:370–376. doi: 10.1104/pp.73.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katahira R., Ashihara H. Profiles of pyridine biosynthesis, salvage and degradation in disks of potato (Solanum tuberosum L.) tubers. Planta. 2002;215:821–828. doi: 10.1007/s00425-002-0806-5. [DOI] [PubMed] [Google Scholar]
- 26.Ashihara H., Stasolla C., Loukanina N., Thorpe T. A. Purine and pyrimidine metabolism in cultured white spruce (Picea glauca) cells: metabolic fate of 14C-labeled precursors and activity of key enzymes. Physiol. Plant. 2000;108:25–33. [Google Scholar]
- 27.Ashihara H., Stasolla C., Loukanina N., Thorpe T. A. Purine metabolism during white spruce somatic embryo development: salvage of adenine, adenosine, and inosine. Plant Sci. 2001;160:647–657. doi: 10.1016/s0168-9452(00)00441-6. [DOI] [PubMed] [Google Scholar]
- 28.Wagner K. G., Backer A. I. Dynamics of nucleotides in plants studied on a cellular basis. Int. Rev. Cytol. 1992;134:1–84. [Google Scholar]
- 29.Cabrita M. A., Baldwin S. A., Young J. D., Cass C. E. Molecular biology and regulation of nucleoside and nucleobase transporter proteins in eukaryotes and prokaryotes. Biochem. Cell. Biol. 2002;80:623–638. doi: 10.1139/o02-153. [DOI] [PubMed] [Google Scholar]
- 30.Visser F., Vickers M. F., Ng A. M. L., Baldwin S. A., Young J. D., Cass C. E. Mutation of residue 33 of human equilibrative nucleoside transporters 1 and 2 alters sensitivity to inhibition of transport by dilazep and dipyridamole. J. Biol. Chem. 2001;277:395–401. doi: 10.1074/jbc.M105324200. [DOI] [PubMed] [Google Scholar]
- 31.Burkle L., Cedzich A., Dopke C., Stransky H., Okumoto S., Gillissen B., Kuhn C., Frommer W. B. Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 2003;34:13–26. doi: 10.1046/j.1365-313x.2003.01700.x. [DOI] [PubMed] [Google Scholar]
- 32.Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18:298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- 33.Leinhos V., Krauss G. J., Glund K. Evidence that a part of cellular uridine of a tomato (Lycopersicon esculentum) cell suspension culture is located in the vacuole. Plant Sci. 1986;47:15–20. [Google Scholar]
- 34.Abel S., Blume B., Glund K. Evidence for RNA-oligonucleotides in plant vacuoles isolated from cultured tomato cells. Plant Physiol. 1990;94:1163–1171. doi: 10.1104/pp.94.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldwin S. A., Beal P. R., Yao S. Y., King A. E., Cass C. E., Young J. C. The equilibrative nucleoside transporter family, SLC29. Pflugers Arch. 2004;447:735–743. doi: 10.1007/s00424-003-1103-2. [DOI] [PubMed] [Google Scholar]
- 36.Thomas C., Rajagopal A., Windsor B., Dudler R., Lloyd A., Roux S. J. A role for ectophosphatase in xenobiotic resistance. Plant Cell. 2000;12:519–534. doi: 10.1105/tpc.12.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demidchik V., Nichols C., Oliynyk M., Dark A., Glover B. J., Davies J. M. Is ATP a signaling agent in plants? Plant Physiol. 2003;133:456–461. doi: 10.1104/pp.103.024091. [DOI] [PMC free article] [PubMed] [Google Scholar]