Abstract
EEN (extra eleven nineteen), also known as EA2 (endophilin A2), a fusion partner of the MLL (mixed-lineage leukaemia) gene in human acute leukaemia, is a member of the endophilin A family, involved in the formation of endocytic vesicles. We present evidence to show that EEN/EA2 is localized predominantly in nuclei of various cell lines of haemopoietic, fibroblast and epithelial origin, in contrast with its reported cytoplasmic localization in neurons and osteoclasts, and that EEN/EA2 exhibits nucleocytoplasmic shuttling. During the cell cycle, EEN/EA2 shows dynamic localization: it is perichromosomal in prometaphase, co-localizes with the bipolar spindle in metaphase and anaphase and redistributes to the midzone and midbody in telophase. This pattern of distribution coincides with changes in protein levels of EEN/EA2, with the highest levels being observed in G2/M-phase. Our results suggest that distinct subcellular localization of the endophilin A family members probably underpins their diverse cellular functions and indicates a role for EEN/EA2 in the cell cycle.
Keywords: cell cycle, extra eleven nineteen (EEN), endophilin, leukaemia, mitosis, nucleocytoplasmic shuttling
Abbreviations: CHO cells, Chinese-hamster ovary cells; EA2, endophilin A2; EEN, extra eleven nineteen; EGF, epidermal growth factor; EGFP, enhanced green fluorescent protein; FBS, foetal bovine serum; GST, glutathione S-transferase; LMB, leptomycin B; MEM, minimal essential medium; MLL, mixed-lineage leukaemia; NLS, nuclear localization signal; SH3 domain, Src homology 3 domain
INTRODUCTION
EEN (extra eleven nineteen), also known as EA2 (endophilin A2), was originally identified as a fusion partner of MLL (mixed-lineage leukaemia) translocation in childhood leukaemia [1]. EEN/EA2 is the founding member of the EEN/EA SH3 domain (Src homology 3 domain)-containing protein family, which shares approx. 70% identity with each other [2]. EA2 is widely expressed when compared with the brain-specific expression of EEN-B1/EA1 and the brain, thymus and testis expression pattern of EEN-B2/EA3 [2,3]. The biological function of the endophilin family is best known from studies on EEN-B1/EA1, which plays an essential role in the formation of endocytic vesicles at synaptic nerve terminals [4]. Through its SH3 domain, EA1 interacts with the endocytic proteins dynamin, amphiphysin and synaptojanin. In addition, it possesses a lysophosphatidic acid acyltransferase activity, which is essential for its role in shaping membrane lipid for vesicle fission [3,5,6]. This is confirmed by in vivo studies in which Drosophila mutants deficient in endophilin caused the depletion of synaptic vesicle in nerve terminals [7,8].
There is growing evidence linking the endophilin A family to non-endocytic functions. On the basis of yeast two-hybrid studies, non-endocytic proteins known to interact with the SH3 domain of EA1 include the β1-adrenergic receptor, metalloprotease disintegrins, such as transmembrane glycoproteins associated with cell adhesion and growth factor signalling and rat germinal centre kinase-like kinases, a modulator of c-Jun intracellular signalling as well as Cbl-interacting protein of 85 kDa, which is involved in the down-regulation of c-Met and EGF (epidermal growth factor) receptors [9–13]. EA2 has been shown to interact with Moloney-murine-leukaemia virus Gag protein and to modulate virion production [14]. Another member, EA3, has been shown to co-localize and interact with huntingtin protein in patients suffering from Huntington's disease and promotes the formation of insoluble polyglutamine-containing aggregates [15]. We have recently identified a novel EA2-binding partner, EBP (EEN-binding protein), which possesses inhibitory effects on Ras signalling and on cellular transformation induced by Ras [16].
To explore further the role of EEN/EA2 in cells, we have raised a specific polyclonal antibody to investigate the expression and localization of endogenous EEN/EA2 both in interphase cells of diverse cellular lineages and during various phases of the cell cycle. We have also studied the protein levels of EA2 in various phases of the cell cycle as well as the effects of transfection studies using deletion mutants of EEN/EA2. Our results show that endogenous EEN/EA2 shows predominantly nuclear localization in interphase cells of the haemopoietic, fibroblast and epithelial lineage, in contrast with the cytoplasmic localization in neurons and osteoclasts, and is also distributed to different subcellular compartments in the mitotic cycle. Taken together, our results suggest that EEN/EA2 can show diverse subcellular localizations in different cell types and, in addition to its involvement in endocytosis, may play a role in the cell cycle.
MATERIALS AND METHODS
Cell lines and cultures
Human leukaemia cell lines U937 and HL60 were cultured at 37 °C in RPMI 1640, supplemented with 10% (v/v) FBS (foetal bovine serum), streptomycin (100 units/ml) and penicillin (100 μg/ml; Gibco, Invitrogen). HeLa and HEK-293 cells (human embryonic kidney 293 cells) were cultured in Dulbecco's modified Eagle's medium containing 10% FBS, COS7 cells in MEM (minimum essential medium Eagle) containing 10% FBS, CHO cells (Chinese-hamster ovary cells) in Ham's F-12 medium containing 10% FBS and NIH3T3 cells in Dulbecco's modified Eagle's medium containing 10% (v/v) calf serum. The neuroblastoma cell line (A.T.C.C. CRL-2266, SH-SY5Y) was cultured at 37 °C in 44% (v/v) MEM and 44% (v/v) F-12 supplemented with 10% FBS, streptomycin, penicillin, 1 mM sodium pyruvate (Gibco, Invitrogen) and 2 mM L-glutamine (Sigma).
Plasmid constructs
EEN/EAs cDNA in pBluescript vector used for subcloning and PCR amplification was prepared as described in [1]. Constructs for GST (glutathione S-transferase)–EEN/EAs were generated as described in [2]. To generate untagged and C-terminal myc-tagged EEN/EA2 constructs, cDNAs for EEN/EA2 and mutant without the stop codon were subcloned in BamHI and EcoRI sites of pcDNA-3.1 myc-His vector (Invitrogen). To generate N-terminal FLAG-tagged EEN/EA2, cDNA was subcloned into the EcoRI and EcoRV sites of pFLAG-CMV2 (where CMV stands for cytomegalovirus; Sigma). EEN/EA2ΔNLS (where NLS stands for nuclear localization signal) was generated by recombinant PCR with the mutagenic primers (forward primer: 5′-CGCCTGGACTTTGACTACGCAGCTGCGGCGCAGGGCAAG; reverse primer: 5′-GTAGTCAAAGTCCAGGCGGCGGCCCTCCAGTGCAGCCAGGTGGTG; the mutated residues are underlined). The preparation of EGFP (enhanced green fluorescent protein) fusion constructs and primer sequences used are available on request. All plasmid constructs were confirmed by DNA sequencing and the expression of the protein was confirmed by immunoblotting with Ab2 or antibody to the respective epitope (N. Cheung and L. C. Chan, unpublished work).
Antibodies
Rabbit polyclonal antibody (Ab2) was raised against a peptide antigen corresponding to amino acids 162–182 of EEN/EA2 by Sigma-Genosys (Haverhill, Cambridgeshire, U.K.). The specificity of the antibody was first verified by dot-blotting with the peptide antigen (N. Cheung and L. C. Chan, unpublished work) and Western blotting on recombinant GST fusion proteins and cell line extracts. Mouse monoclonal antibodies to α-tubulin (B-5-1-2) and FLAG epitope (M5) were purchased from Sigma. Goat anti-lamin B (sc-6217), goat anti-β-actin (sc-1616), goat anti-B23 (sc-6013), rabbit anti-myc (sc-789), rabbit anti-cyclin A (sc-751) and mouse anti-cyclin B (sc-245) antibodies were purchased from Santa Cruz Biotechnology.
Western blotting
Total cell extracts were prepared by lysing cells for 30 min on ice in lysis buffer containing 50 mM Tris (pH 6.8), 150 mM NaCl, 50 mM sodium glycerophosphate, 10 mM NaF, 1 mM sodium orthovanadate, complete protease inhibitor (Roche) and 1% Nonidet P40. The extracts were cleared by centrifugation for 30 min at 17000 g and then boiled in SDS sample buffer for 10 min. Total cell lysate (20–30 μg) was resolved by SDS/PAGE (10% gel), transferred on to an Immobilon-P membrane (Millipore) and the signal was visualized by ECL® (Amersham Biosciences). Protein molecular-mass standards and horseradish peroxidase-conjugated secondary antibodies were purchased from MBI Fermentas (Flamborough, ON, Canada) and Jackson Immunoresearch Laboratories (West Grove, PA, U.S.A.) respectively. For the competition of antibody binding, antiserum was preincubated with 50 μg of peptide antigen at room temperature (22–24 °C) for 1 h.
Immunofluorescence
For the leukaemia cell lines, exponentially growing cells were cytospun on to a slide using Shandon Cytospin 2. Cells were fixed in 4% (w/v) paraformaldehyde in PBS for 20 min and then permeabilized with 0.5% Triton X-100 in PBS for 10 min. Cultured cells were grown on poly-L-lysine-coated glass coverslips in a 35 mm disc and fixed and processed as mentioned above, with the exception of α-tubulin staining in which cells were fixed in −20 °C methanol for 5 min and then permeabilized in 0.1% Triton X-100 for 5 min. Cells were blocked either with 5% (v/v) normal donkey serum or normal goat serum for 30 min, incubated with primary antibody at 4 °C overnight and fluorochrome-conjugated secondary antibody for 1 h at room temperature. Depleted antiserum was prepared by preclearing with 200 μg of bead-coupled GST–EEN/EA2 protein at room temperature for 1 h. FITC and Texas Red-conjugated secondary antibodies were purchased from Jackson Immunoresearch Laboratories. Cells were counterstained with 4,6-diamidino-2-phenylindole (2 μg/ml; Calbiochem, La Jolla, CA, U.S.A.) before they were mounted in Vectashield antifade mountant (H1000; Vector Laboratories, Burlingame, CA, U.S.A.). All images were captured using a fluorescent microscope equipped with charge-coupled-device camera (Leica, Wetzlar, Germany), except that an MRC1024 confocal laser scanning microscope (Bio-Rad) was used for the co-localization of EEN/EA2 and B23, and an inverted fluorescence microscope equipped with digital camera (Nikon) was used for taking the phase-contrast image. All images were processed using Adobe Photoshop.
Subcellular fractionation
Cytoplasmic and nuclear fractions were prepared by hypo-osmotic cell lysis as described in [12]. Nuclear (50 μg) and cytoplasmic (100 μg) proteins were separated by SDS/PAGE (10% gel), transferred on to an Immobilon-P membrane and immunoblotted with Ab2, anti-lamin B and anti-β-actin antibodies.
Transfection and LMB (leptomycin B) treatment
Transfection of cells by calcium phosphate precipitation was performed as described in [18]. Semiconfluent cells in a 100 mm plate were transfected with 20 μg of plasmid DNA for 12–16 h and harvested after 24 h. Cells seeded on poly-L-lysine-coated coverslips were transfected using Fugene 6 reagent according to the manufacturer's instructions (Roche). Transfected cells were treated with LMB (10 ng/ml; Sigma) or mock-treated with methanol carrier for 6 h before fixation.
Cell synchronization and flow cytometry
HeLa cells were arrested in the S- and M-phases by 4 mM hydroxyurea and 100 ng/ml nocodazole respectively for 20 h. EEN/EA2 protein levels in the treated and untreated cells were determined by densitometry from three individual experiments. For double-thymidine blocks, HeLa cells were treated with 2 mM thymidine (Calbiochem) for 16 h, washed with PBS three times and released in fresh medium for 9 h. Cells were again treated with 2 mM thymidine for 16 h. The block was released by washing out thymidine and replenishing with fresh medium. Cells were harvested at various time points after the release and subjected to FACS analysis and Western blotting with Ab2, cyclin B1, cyclin A and β-actin antibodies. For cell-cycle profiling, cells were harvested by trypsinization and washed twice in ice-cold PBS with 1% calf serum. Cells were fixed in 80% (v/v) cold ethanol and then stained with TE buffer (50 mM Tris/HCl, pH 7.4/5 mM EDTA) containing 40 μg/ml propidium iodide and 40 μg/ml RNase A at 37 °C for 30 min. FACS analysis was performed using EPICS-XL-MCL (Beckman Coulter, Fullerton, CA, U.S.A.).
Intercellular bridge assay
HeLa and HEK-293 cells were transfected for 24–48 h with EGFP–C1 vector and constructs for EGFP–EEN/EA2, EGFP–EEN/EA2 (1–260), EGFP–EEN/EA2 (1–179), EGFP–EEN/EA2 (1–156) and EGFP–EEN/EA2 (180–368) and then immunostained for microtubules with anti-α-tubulin antibody. The number of intercellular bridges in 200–400 transfected cells was determined in four independent experiments for each cell type.
RESULTS
Characterization of the specificity of a polyclonal antibody (Ab2) raised against an internal peptide of EEN/EA2
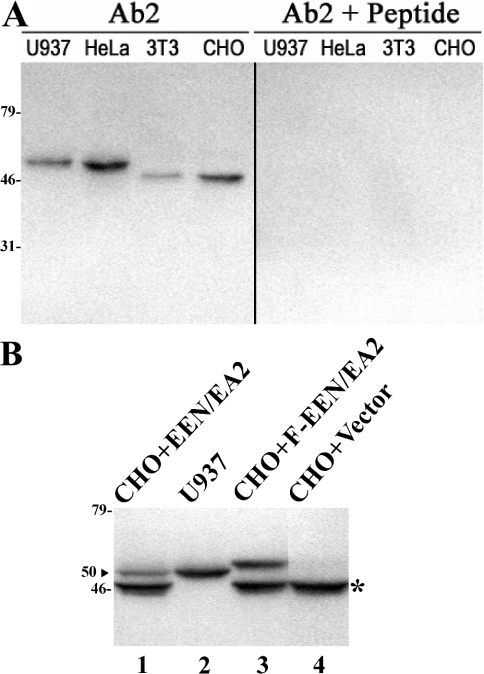
First, we immunoblotted fusion proteins GST–EEN/EA2 to verify the specificity of Ab2, a polyclonal antibody raised against an internal peptide of EEN/EA2. Using Western blots prepared from cell extracts from a variety of human cell lines of different cell types including HL60 (granulocytic), U937 (monocytic), Raji (B-lymphoid), MOLM1 (myeloid), HeLa (cervical cancer) and HEK-293 (embryonic kidney), only a polypeptide of approx. 50 kDa was detected (N. Cheung and L. C. Chan, unpublished work). Since the reported mass of EA2 is 45 kDa in rodent tissues [19], we further validated the antibody specificity of Ab2 by Western blotting of the total cell extract from two rodent cell lines (NIH3T3 and CHO) and two human cell lines (U937 and HeLa). In agreement with the published results, Ab2 can detect the 45 kDa EA2 in both NIH3T3 and CHO cells; competition with the peptide antigen eliminated the specific signal of all cell lines (Figure 1A). Furthermore, we confirmed that the 50 kDa protein in human cell lines is the authentic EEN/EA2 by probing Western blots prepared from untagged as well as FLAG-tagged human EEN/EA2, independently expressed in CHO cells. As shown in Figure 1(B), in addition to the endogenous 45 kDa protein (lanes 1, 3 and 4), Ab2 could detect an additional 50 kDa polypeptide in CHO cells expressing untagged human EEN/EA2 (lane 1), which co-migrated with the 50 kDa protein in U937 cells (lane 2) as well as FLAG-tagged EEN/EA2 with a lower mobility (lane 3). From the above experiments, we conclude that the 50 kDa protein detected by Ab2 represents EEN/EA2.
Figure 1. Characterization of the polyclonal antibody Ab2.
(A) Detection of EEN/EA2 protein in human cell lines U937 and HeLa and rodent cell lines NIH3T3 and CHO. The specific signal was completely eliminated after competition of the antiserum with the antigen peptide (right panel). (B) Immunoblot analysis of cell extracts from U937 cells (lane 2) and rodent CHO cells transfected with untagged (CHO+EEN/EA2; lane 1), FLAG-tagged human EEN/EA2 (CHO+F-EEN/EA2; lane 3) or empty vector (CHO+Vector; lane 4) by Ab2 antibody. Endogenous EEN/EA2 protein from CHO cells is indicated by an asterisk.
Localization of EEN/EA2 in different cell types
We next examined the localization of endogenous EEN/EA2 in various cell types by immunofluorescence staining. In contrast with the reported cytoplasmic localization of EEN/EA2 proteins in neurons and osteoclasts [19,20], we observed mainly nuclear staining but occasionally observed large nuclear dots suggestive of nucleolus localization in U937 cells, HL60 leukaemia cells, monkey kidney COS7 cells and NIH3T3 cells. As expected, cytoplasmic staining was observed in the neuroblastoma cell line SH-SY5Y (Figure 2A). In the control experiment, antiserum precleared with GST–EEN/EA2 as well as the preimmune serum gave only background signal (Figure 2A; N. Cheung and L. C. Chan, unpublished work). To validate further the nuclear localization of EEN/EA2 in non-neuronal cells, subcellular fractionation was performed using four cell lines, NIH3T3, CHO, U937 and HeLa, followed by immunoblotting. As shown in Figure 2(B), EEN/EA2 was detected in the nuclear fraction but not in the cytoplasmic fraction, which is in line with the results from immunofluorescence. The integrity of the subcellular fractionation was validated by lamin B, which is a component of the nuclear lamina and is solely present in the nuclear fraction, whereas the loading of cytoplasmic protein was justified by immunodetection with anti-β-actin antibody. Using confocal microscopy, we showed that EEN/EA2 is co-localized with nucleolar protein B23 in HL60 and U937 leukaemia cells (Figure 2C; N. Cheung and L. C. Chan, unpublished work).
Figure 2. Nuclear localization of EEN/EA2.
(A) Immunofluorescence staining of non-neuronal HL60, U937, COS7 and NIH3T3 cells and the neuroblastoma cell line SH-SY5Y with Ab2 antibody. In the control staining, the depleted antiserum as well as the preimmune serum (N. Cheung and L. C. Chan, unpublished work) gave only a low background signal. Scale bar, 5 μM. (B) Immunoblot analysis of the nuclear (N) and cytoplasmic (C) fractions of NIH3T3, CHO, U937 and HeLa cells. Nuclear and cytoplasmic fractions were prepared from hypo-osmotic cell lysis and then subjected to immunoblot analysis. The loading of cytoplasmic protein was confirmed by β-actin antibody. (C) Confocal analysis of HL60 cells co-labelled with Ab2 antibody (FITC) and anti-B23 antibody (Texas Red). EEN/EA2 co-localized with nucleolus marker B23 mainly at the core but not at the periphery region.
Localization of EEN/EA2 in different phases of the mitotic cycle of leukaemia cells
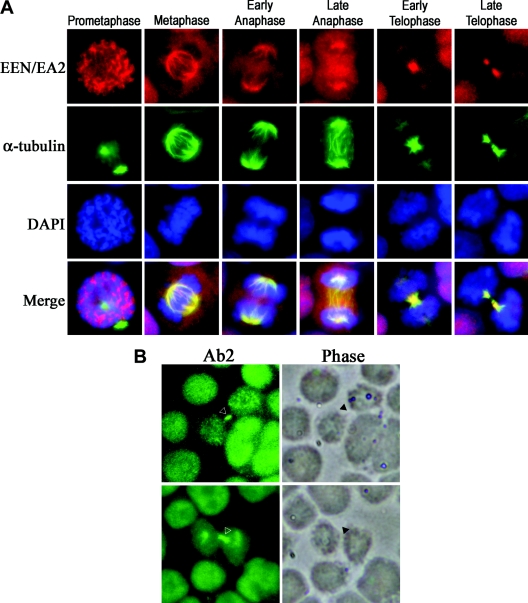
Based on its nuclear localization, we next studied the pattern of EEN/EA2 staining during mitosis of both U937 and HL60 leukaemia cells with Ab2 and anti-α-tubulin antibodies to visualize the microtubules. As shown in Figure 3(A), EEN/EA2 is predominantly nuclear, with a weak cytoplasmic signal during interphase and is present in the perichromosomal region during prometaphase and mainly localized to the mitotic spindle during metaphase. EEN/EA2 is localized to the spindle throughout the anaphase and concentrated in the midzone in late anaphase. In telophase, EEN/EA2 is redistributed to the midbody, the cytoplasmic bridge linking the two daughter cells (Figure 3A). To validate further the localization of EEN/EA2 during cytokinesis, immunodetection of EEN/EA2 consistently demonstrated its co-localization with the intercellular bridge as revealed by phase-contrast microscopy (Figure 3B).
Figure 3. Subcellular localization of EEN/EA2 during mitosis.
(A) Dynamic localization of EEN/EA2 during mitosis of U937 leukaemia cells. Immunofluorescence analysis of EEN/EA2 localization during mitosis of U937 cells by labelling with Ab2 antibody (Texas Red), anti-α-tubulin antibody (FITC) for microtubules and 4,6-diamidino-2-phenylindole (DAPI) for DNA. (B) Immunodetection of EEN/EA2 protein revealed its co-localization with the intercellular bridges of HL60 leukaemia cells shown by phase contrast (indicated by arrowheads).
The dynamic localization of EEN/EA2 during mitosis allowed us to examine protein levels during cell cycle. After release of HeLa cells from the double-thymidine block, the amount of EEN/EA2 protein at different times was analysed by immunoblotting using Ab2, cyclin B1, cyclin A and β-actin (Figure 4A). EEN/EA2 protein levels increased from a baseline level in G1 cells (0 h) throughout the S-phase (4 h) and attained the highest level in G2/M-phase (8–9 h). As cells exited mitosis (10–11 h), as indicated by an increase in G1 population and a decrease in G2/M population, the level of EEN/EA2 decreased slightly. Identical protein loading was confirmed by β-actin immunoblot, and both cyclin A and cyclin B1 levels, used as a control for cell-cycle synchronization, showed the typical accumulation of protein in G2/M-phase and degradation after mitotic exit. Quantification of EEN/EA2 protein level from asynchronized HeLa cells and cells arrested in S-phase by hydroxyurea and M-phase by nocodazole revealed that the protein level was reduced in S-phase by 56% and increased in M-phase by 84% relative to the asynchronized cells (Figure 4B).
Figure 4. EEN/EA2 protein level varies during cell cycle.
(A) EEN/EA2 protein levels in various phases of the cell cycle of synchronized HeLa cells after their release from the double-thymidine block. FACS analysis revealed the progression of diploid (2N) cells from G1-phase (0 h) to S-phase (4–6 h) and G2/M-phase (8–10 h), followed by mitotic exit (11–12 h) (upper panel). Accumulation of cyclin A and cyclin B1 protein in S-phase (4–6 h) and G2/M-phase (8–10 h) respectively validated the synchronization. Equal loading of protein was confirmed by probing with β-actin antibody. (B) Immunoblot analysis of total cell lysate prepared from asynchronized HeLa cells (AS), cells arrested in S-phase by hydroxyurea (HU) and nocodazole-blocked mitotic cells (Noco). The ratio of EEN/EA2 protein expression levels was determined by densitometry from three individual experiments.
Nucleocytoplasmic shuttling of exogenously expressed EEN/EA2
Although endogenous EEN/EA2 is predominantly nuclear, we noted strong cytoplasmic localization of transfected untagged, N-terminal FLAG- and C-terminal myc-tagged exogenous EEN/EA2 in cultured cell lines (Figure 5A; N. Cheung and L. C. Chan, unpublished work). Since subcellular localization of proteins by nucleocytoplasmic shuttling is a common phenomenon, we explored the contribution of nucleocytoplasmic shuttling to the localization of exogenously transfected EEN/EA2 [21,22]. When HeLa cells were transfected with a construct encoding FLAG-tagged EEN/EA2 in the presence of LMB, a fungal toxin commonly used to block the CRM1-dependent nuclear export pathway, it was noted that FLAG-tagged EEN/EA2 accumulated in the nucleus of treated but not untreated cells (Figure 5B) [23]. This suggests that, under normal conditions, EEN/EA2 is shuttled between the nucleus and cytoplasm. The steady-state cytoplasmic localization in cells not treated with LMB with transfected EEN/EA2 may be attributed to CRM1-dependent nuclear export. However, the consensus leucine-rich nuclear export signal (LX2–3LX2LXL) that mediated interaction with CRM1 exportin was not identified in EEN/EA2 [24].
Figure 5. Nucleocytoplasmic shuttling of EEN/EA2.
(A) COS7 cells were transfected with wild-type untagged EEN/EA2 and immunostained with Ab2 antibody. Exogenous wild-type EEN/EA2 accumulated predominantly in the cytoplasm, suggesting that cytoplasmic localization was not attributed to epitope tagging. (B) Nucleocytoplasmic shuttling of EEN/EA2 in HeLa cells transfected with N-terminal FLAG-tagged EEN/EA2, immunodetected with anti-FLAG antibody and then probed with FITC-conjugated secondary antibody. (C) Schematic representation of the structural domains of EEN/EA2 and the various deletion mutants. The Lys-Arg-rich motif (KR1; amino acids 159–177) harbours the predicted bipartite basic NLS (amino acids 159–174). The coiled-coil region (amino acids 180–260) is linked to the C-terminal SH3 domain (amino acids 303–368) through the variable domain (amino acids 261–302), which is not well conserved in the endophilin family members. (D) Subcellular localization of EGFP–EEN/EA2 and its deletion mutants in HeLa cells. EGFP–EEN/EA2 (amino acids 1–179) also demonstrated midbody localization (arrowhead). Nuclear localization of the N-terminal region (amino acids 1–156) of EEN/EA2 suggested that the predicted bipartite basic NLS (amino acids 159–174) was dispensable for nuclear import. (E) Subcellular localization of FLAG-tagged NLS mutant of EEN/EA2 (ΔNLS). This indicated the dispensable nature of the predicted NLS on the regulation of nuclear import.
Identification of domains of EEN/EA2 mediating sublocalization
To delineate the domains governing the subcellular localization of EEN/EA2, HeLa cells were transiently transfected with the wild-type and the various N-terminal (amino acids 134–368, 180–368 and 210–368) and C-terminal (amino acids 1–260, 1–236, 1–179 and 1–156) deletion mutants tagged with EGFP (Figures 5C and 5D). As shown in Figure 5(D), C-terminal deletion mutants showed accumulation of the protein in the nucleus with the minimal deletion of SH3 domain (amino acids 304–368) as the common deleted region. In contrast, the three N-terminal deletion mutants (amino acids 134–368, 180–368 and 210–368) showed cytoplasmic localization. The result indicated that the nuclear export is regulated by the C-terminal SH3 domain but not by the N-terminal motif. Although a putative bipartite basic NLS (amino acids 159–174, KKLEGRRLDFDYKKKR) was identified by PSORT database search, entrapment of NLS mutant (amino acids 159–174, AALEGRRLDFDYAAAA) in the nucleus by LMB as well as in the C-terminal deletion mutant (amino acids 1–156) indicates that it is dispensable for nuclear localization (Figure 5E).
Recruitment of the C-terminal deletion mutant EEN/EA2 (1–179) to the midbody matrix inhibited midbody abscission
In our analysis of the localization of EEN/EA2 deletion mutants, we observed one of the C-terminal deletion mutants, i.e. EEN/EA2 (amino acids 1–179), to be concentrated to the midbody of HeLa (Figure 5D). Using anti-tubulin antibody, we showed that EEN/EA2 (amino acids 1–179) is present in the midbody matrix, a centrally located zone devoid of microtubule staining and comprising mostly uncharacterized amorphous material (Figure 6A) [25]. We also observed that 23% of the cells transfected with EEN/EA2 (amino acids 1–179) showed persistent intercellular bridges in comparison with 4–6% of such cells in the EGFP control, wild-type as well as N-terminal deletion mutant EEN/EA2 (amino acids 180–368) and other C-terminal deletion mutants (amino acids 1–260 and 1–156; Figure 6B). Since there was no increase in binucleated cells, which is a hallmark of cytokinesis failure, this suggests the ‘bridged cell phenotype’ in cells expressing EEN/EA2 (amino acids 1–179) is probably due to impairment of the intercellular bridge breakdown, i.e. a defect in midbody abscission rather than furrow ingression and central spindle formation. Although matrix localization was also detected in the scanty intercellular bridges of cells expressing wild-type and EEN/EA2 (amino acids 1–156), the absence of increased bridged cell formation indicated that matrix localization was dispensable for the inhibitory effect (Figure 6A). Intercellular bridge assay performed in HEK-293 cells also yielded a similar observation (N. Cheung and L. C. Chan, unpublished work).
Figure 6. Deletion mutant EGFP–EEN/EA2 (amino acids 1–179) was strongly associated with the midbody matrix and inhibited midbody abscission.
(A) HeLa cells were transfected with EGFP fusion constructs and then labelled with anti-α-tubulin antibody to stain for the microtubule. EGFP–EEN/EA2 (amino acids 1–179) was robustly recruited to the matrix of the midbody, a centrally located zone devoid of microtubule staining (arrowhead). Matrix localization of wild-type EEN/EA2 (WT) and EEN/EA2 (amino acids 1–156) was also detected in the scanty bridged cells (arrowhead). (B) Intercellular bridge assay. HeLa cells were transfected with EGFP vector, EGFP–EEN/EA2 and various EGFP–EEN/EA2 deletion mutants for 24–48 h and the number of transfected cells still linked by the intercellular bridge, as visualized by α-tubulin staining, was determined.
We also noted a minor population of cells expressing EEN/EA2 (amino acids 1–179) localized to the midbody without any intercellular bridge (results not shown). This suggests that either the possibility of an alternative pathway was utilized to overcome the midbody abscission defect or intercellular bridges can break simply due to cells migrating away from each other after division [26].
DISCUSSION
Subcellular localization of EEN/EA2
At present, most of the studies on the EEN/EA family have described their endocytic functions in neuronal cells. However, the distinct tissue distributions and diverse interacting partners of EEN/EA family members identified so far suggest that each member may have related but diverse functional roles in various cellular pathways.
To investigate the role of EEN/EA2 in non-neuronal cells, we have raised a polyclonal antibody Ab2 against an internal peptide and confirmed using immunoblotting analysis that EEN/EA2 is the only member of the EEN/EA family expressed that is recognized by Ab2 in a variety of human and rodent cell lines. Immunofluorescence as well as subfractionation analysis revealed a predominant nuclear localization of EEN/EA2 in several human leukaemia cell lines, HeLa cells, mouse fibroblasts and monkey kidney epithelial cells. This pattern of nuclear staining in haemopoietic, fibroblast and epithelial cells differs from the cytoplasmic localization in neurons and osteoclasts, suggesting that the subcellular localization of EEN/EA2 may be cell-context-dependent [19,20]. We also show that there is nuclear retention of the mutant with the minimal deletion of C-terminal SH3 domain, which suggests a coupling of the SH3 domain with the nuclear export pathway either directly or indirectly through adaptor protein interaction. Although a predicted bipartite basic NLS (amino acids 159–174) was identified in EEN/EA2, it was dispensable for nuclear import since nuclear entrapment was observed in the NLS mutant. Using LMB, which inhibits CRM1-dependent nuclear export, we were able to demonstrate nucleocytoplasmic shuttling of EEN/EA2. Based on this, we propose that the steadystate cytoplasmic distribution of exogenous EEN/EA2 in transfected cells can be attributed to CRM1-dependent nuclear export. It is recognized that regulated subcellular localization through the differential control of nucleocytoplasmic shuttling is a physiological process employed by diverse cellular signalling pathways [17]. This is also consistent with recent reports on two endocytic proteins eps15 and epsin, both of which were also found to shuttle in and out of the nucleus [26].
EEN/EA2 and cell cycle
We have presented results to show the remarkable dynamic localization of EEN/EA2 within cells in different phases of the mitotic cell cycle, i.e. perichromosomal in prometaphase, co-localized with the bipolar spindle in metaphase and anaphase and redistributed to the midzone and midbody in telophase. This together with EEN/EA2 protein levels reaching a peak in G2/M-phase suggests a possible function of EEN/EA2 related to mitosis.
Of particular interest is the localization of endogenous EEN/EA2 at late mitosis in the midzone and midbody of leukaemia cells, a pattern which resembles the spatial and temporal distribution of proteins involved in cytokinesis, including cytoskeletal proteins (actomyosin complex, microtubule), the kinesin-like motor proteins [MKLP (mitotic kinesin-like protein)/CHO1], vesicle trafficking proteins (Rab6KIFL, syntaxin) and protein kinases (citron kinase, aurora-B and Polo-like kinases) (reviewed in [27–31]). We are intrigued by the observation that N-terminal EEN/EA2 (amino acids 1–179) is concentrated in the midbody and leads to an increase in intercellular bridge formation, an event related to failure of midbody abscission. Although other fusion partners of MLL translocation in human acute leukaemia, such as septin 9 [MSF (MLL septin-like fusion), AF17q25] and septin 5 (CDCrel1), have putative roles in cytokinesis and vesicle trafficking respectively [32–36], it is premature to link EEN/EA2 with a role in cytokinesis on the basis of our findings, since N-terminal EEN/EA2 (amino acids 1–179) lacks the SH3 domain and will, therefore, be unable to interfere with endogenous EEN/EA2. In addition, we did not observe any effect on cytokinesis with the overexpression of wild-type protein. This suggests that the effect on cytokinesis with N-terminal EEN/EA2 may reflect improper folding of the N-terminus or mimicking of another protein.
It is intriguing that the effect on midbody abscission was seen only in the C-terminal deletion mutant EEN/EA2 (amino acids 1–179) but not in the N-terminal deletion mutant (amino acids 180–368) as well as in two other C-terminal deletion mutants (amino acids 1–260 and 1–156). It is possible that this is due to the lack of acyltransferase activity in the N-terminal deletion mutant (amino acids 180–368) and impairment of its activity in the C-terminal deletion mutant (amino acids 1–156) without the Lys-Arg-rich motif (KR1; amino acids 159–177). The failure of EEN/EA2 (amino acids 1–260) to inhibit midbody abscission would also suggest that the retention of the coiled-coil domain (amino acids 180–260) is essential and competent for the completion of midbody abscission, perhaps through the co-operative interaction with other coiled-coil proteins or dimerization with endogenous protein.
Taken together, we propose that EEN/EA2 has several functions, which can be cell-context-dependent as well as temporally and spatially regulated. Our studies indicate a nuclear localization pattern of EEN/EA2 with dynamic distribution in the cell cycle, in contrast with its cytoplasmic localization in neurons and osteoclasts. Further studies on the functions of EEN/EA2, including the use of siRNA, should provide important insight into the role of EEN/EA2 in diverse physiological processes.
Acknowledgments
We thank the members of Hematology Research Laboratory (Department of Pathology, University of Hong Kong) for technical support and also thank P. Jackson and M. Cleary (Department of Pathology, Stanford University) and P. Donovan (Kimmel Cancer Center, Thomas Jefferson University) for helpful discussions. We gratefully acknowledge the support given to this project by the Research Grants Council of the Hong Kong Special Administrative Region, People's Republic of China (project no. HKU7252/99M). C.W.S. is supported by a special fellowship and D.-Y.J. by a scholarship from the Leukemia and Lymphoma Society.
References
- 1.So C. W., Caldas C., Liu M. M., Chen S. J., Huang Q. H., Guo L. J., Sham M. H., Wiedemann L. M., Chan L. C. EEN encodes for a membrane of a new family of proteins containing an Src homology 3 domain and is the third gene located on chromosome 19p13 that fuses to MLL1 in human leukemia. Proc. Natl. Acad. Sci. U.S.A. 1997;94:2563–2568. doi: 10.1073/pnas.94.6.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.So C. W., Sham M. H., Chew S. L., Cheung N., So C. K., Chung S. K., Caldas C., Wiedemann L. M., Chan L. C. Expression and protein-binding studies of the EEN gene family, new interacting partners for dynamin, synaptojanin and huntingtin protein. Biochem. J. 2000;348:447–458. [PMC free article] [PubMed] [Google Scholar]
- 3.Ringstad N., Nemoto Y., De Camilli P. The SH3p4/SH3p8/SH3p13 protein family: binding partner for synaptojanin and dynamin via a Grb2-like Src homology SH3 domain. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8569–8574. doi: 10.1073/pnas.94.16.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huttner W. B., Schmidt A. Lipids, lipid modification and lipid–protein interaction in membrane budding and fission-insight from the roles of endophilin A1 and synaptophysin in synaptic vesicle endocytosis. Curr. Opin. Neurobiol. 2000;10:543–551. doi: 10.1016/s0959-4388(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 5.So C. W., So C. K., Cheung N., Chew S. L., Sham M. H., Chan L. C. The interaction between EEN and Abi-1, two MLL fusion partners, and synaptojanin and dynamin: implications for leukaemogenesis. Leukemia. 2000;14:594–601. doi: 10.1038/sj.leu.2401692. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt A. A., Wolde M., Thiele C., Fest W., Kratzin H., Podtelejnikov A. V., Witke W., Huttner W. B., Söling H.-D. Endophilin I mediates synaptic vesicle formation by transfer of arachidonate to lysophosphatidic acid. Nature (London) 1999;401:133–141. doi: 10.1038/43613. [DOI] [PubMed] [Google Scholar]
- 7.Guichet A., Wucherpfennig T., Dudu V., Etter S., Wilsh-Bräuniger M., Hellwig A., González-Gaitan M., Huttner W. B., Schmidt A. A. Essential role of endophilin A in synaptic vesicle budding at the Drosophila neuromuscular junction. EMBO J. 2002;21:1661–1672. doi: 10.1093/emboj/21.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verstreken P., Kjaerulff O., Lloyd T. E., Atkinson R., Zhou Y., Meinertzhagen I. A., Bellen H. J. Endophilin mutations block clathrin-mediated endocytosis but not neurotransmitter release. Cell (Cambridge, Mass.) 2002;109:101–112. doi: 10.1016/s0092-8674(02)00688-8. [DOI] [PubMed] [Google Scholar]
- 9.Tang Y., Hu L. A., Miller W. E., Ringstad N., Hall R. A., Pitcher J. A., DeCamilli P., Lefkowitz R. J. Identification of the endophilins (SH3p4/p8/p13) as novel binding partners for the β1-adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12559–12564. doi: 10.1073/pnas.96.22.12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard L., Nelson K. K., Maciewicz R. A., Blobel C. P. Interaction of the metalloprotease disintegrins MDC9 and MDC15 with two SH3 domain-containing proteins, endophilin I and SH3PX1. J. Biol. Chem. 1999;274:31693–31699. doi: 10.1074/jbc.274.44.31693. [DOI] [PubMed] [Google Scholar]
- 11.Ramjaun A. R., Angers A., Legendre-Guillemin V., Tong X. K., McPherson P. S. Endophilin regulates JNK activation through its interaction with the germinal center kinase-like kinase. J. Biol. Chem. 2001;276:28913–28919. doi: 10.1074/jbc.M103198200. [DOI] [PubMed] [Google Scholar]
- 12.Petrelli A., Gilestro G. F., Lanzardo S., Comoglio P. M., Migone N., Giordano S. The endophilin-CIN85-cbl complex mediates ligand-dependent downregulation of c-Met. Nature (London) 2002;416:187–190. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- 13.Soubeyran P., Kowanetz K., Szymkiewicz I., Langdon W. Y., Dikic I. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptor. Nature (London) 2002;416:183–187. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- 14.Wang M. Q., Kim W., Gao G., Torrey T. A., Morse H. C., III, De Camilli P., Goff S. P. Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production. J. Biol. 2003;3:4. doi: 10.1186/1475-4924-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sittler A., Walter S., Wedemeyer N., Hasenbank R., Scherzinger F., Eickhoff H., Bates G. P., Lehrach H., Wanker E. E. SH3GL3 associates with the Huntingtin exon 1 protein and promotes the formation of polygln-containing protein aggregates. Mol. Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 16.Yam J. W. P., Jin D. Y., So C. W., Chan L. C. Identification and characterization of EBP, a novel EEN binding protein that inhibits ras signaling and is recruited into the nucleus by the MLL-EEN fusion protein. Blood. 2004;103:1445–1453. doi: 10.1182/blood-2003-07-2452. [DOI] [PubMed] [Google Scholar]
- 17.Sternsdorf T., Jensen K., Zuchner D., Will H. Cellular localization, expression, and structure of the nuclear dot protein 52. J. Cell Biol. 1997;138:435–448. doi: 10.1083/jcb.138.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Struhl K. New York: John Wiley & Sons; 1994. Current Protocols in Molecular Biology. [Google Scholar]
- 19.Ringstad N., Nemoto Y., De Camilli P. Differential expression of endophilin 1 and 2 dimers at nervous system synapses. J. Biol. Chem. 2001;276:40424–40430. doi: 10.1074/jbc.M106338200. [DOI] [PubMed] [Google Scholar]
- 20.Ochoa G., Slepnev V. I., Neff L., Ringstad N., Takei K., Daniell L., Kim W., Cao H., McNiven M., Baron R., et al. A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 2000;150:377–389. doi: 10.1083/jcb.150.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mattaj I. W., Engimeier L. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 22.Kaffman A., O'shea E. K. Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda M., Asano S., Nakamura T., Adachi M., Yoshia M., Yanafida M., Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 24.Bogerd H. P., Fridell R. A., Benson E. R., Hua J., Cullen B. R. Protein sequence requirement for function of the human T-cell leukemia virus type 1 rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullins M. J., McIntosh R. J. Isolation and initial characterization of the mammalian midbody. J. Cell Biol. 1982;94:654–661. doi: 10.1083/jcb.94.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barton K., Taylor L. D. Traction forces of cytokinesis measured with optically modified elastic substrata. Nature (London) 1997;385:450–454. doi: 10.1038/385450a0. [DOI] [PubMed] [Google Scholar]
- 27.Vecchi M., Polo S., Poupon V., van de Loo J.-W., Benmerah A., Di Fiore P. P. Nucleocytoplasmic shuttling of endocytic proteins. J. Cell Biol. 2001;153:1511–1517. doi: 10.1083/jcb.153.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pines J., Rieder C. L. Re-staging mitosis: a contemporary view of mitotic progression. Nat. Cell Biol. 2001;3:E3–E6. doi: 10.1038/35050676. [DOI] [PubMed] [Google Scholar]
- 29.Straight A. F., Field C. M. Microtubules, membrane and cytokinesis. Curr. Biol. 2000;10:R760–R770. doi: 10.1016/s0960-9822(00)00746-6. [DOI] [PubMed] [Google Scholar]
- 30.Glotzer M. Animal cell cytokinesis. Annu. Rev. Cell Dev. Biol. 2001;17:351–386. doi: 10.1146/annurev.cellbio.17.1.351. [DOI] [PubMed] [Google Scholar]
- 31.Guertin D. A., Trautmann S., McCollum D. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Surka M. C., Tsang C. W., Trimble W. S. The mammalian septin MSF localizes with microtubules is required for completion of cytokinesis. Mol. Biol. Cell. 2002;13:3532–3545. doi: 10.1091/mbc.E02-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osaka M., Rowley J. D., Zeleznik-Le N. J. MSF (MLL septin-like fusion), a fusion partner gene of MLL, in a therapy-related acute myeloid leukemia with a t(11;17)(q23;q25) Proc. Natl. Acad. Sci. U.S.A. 1999;96:6428–6433. doi: 10.1073/pnas.96.11.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taki T., Ohnishi H., Shinohara K., Sako M., Bessho F., Yanagisawa M., Hayashi Y. AF15q25, a putative septin family gene, fuses the MLL gene in acute myeloid leukemia with t(11;17)(q23;25) Cancer Res. 1999;59:4261–4265. [PubMed] [Google Scholar]
- 35.Megonigal M. D., Rappaport E. F., Jones D. H., Williams T. M., Lovett B. D., Kelly K. M., Lerou P. H., Moulton T., Budarf M. L., Felix C. A. t(11;22)(q23;q11.2) in acute myeloid leukemia of infant twins fuses MLL with hCDCrel1, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes. Proc. Natl. Acad. Sci. U.S.A. 1998;95:6413–6418. doi: 10.1073/pnas.95.11.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beites C. L., Xie H., Bowser R., Trimble W. S. The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 1999;2:434–439. doi: 10.1038/8100. [DOI] [PubMed] [Google Scholar]