Abstract
Inducible mineralization of TSA (4-toluenesulphonate) by Comamonas testosteroni T-2 is initiated by a secondary transport system, followed by oxygenation and oxidation by TsaMBCD to 4-sulphobenzoate under the regulation of TsaR and TsaQ. Evidence is presented for a novel, presumably two-component transport system (TsaST). It is proposed that TsaT, an outer-membrane porin, formed an anion-selective channel that works in co-operation with the putative secondary transporter, TsaS, located in the inner membrane. tsaT was identified as a 1017-bp ORF (open reading frame) on plasmid pTSA upstream of the TSA-catabolic genes in the tsa operon. Expression of tsaT was regulated by TsaR, the transcriptional activator of the tsa regulon. The presence of tsaT was concomitant with the presence of the tsa operon in different TSA-degrading isolates. tsaT was expressed in Escherichia coli and was detected in the outer membrane. A 22-amino-acid leader peptide was identified. Purified protein reconstituted in lipid bilayer membranes formed anion-selective channels with a single-channel conductance of 3.5 nS in 1 M KCl. Downstream of tsaT, a constitutively expressed 720-bp ORF (tsaS) was identified. tsaS coded for a hydrophobic protein predicted to have six transmembrane helices and which is most likely localized in the cytoplasmic membrane. tsaS is adjacent to tsaT, but showed a different transcriptional profile.
Keywords: bacterial transport, Comamonas testosteroni, innermembrane component, porin-component, 4-toluenesulphonate uptake
Abbreviations: ABC, ATP-binding cassette; IPTG, isopropyl β-D-thiogalactoside; PC, diphytanoyl phosphatidylcholine; PSB, 4-sulphobenzoate; RT, reverse transcription; TSA, 4-toluenesulphonate
INTRODUCTION
Organosulphonates are the salts of strong acids, have widespread natural and xenobiotic origin, and a common trait of a charged sulphonate group, which is regarded as Nature's way to prevent a compound (coenzyme M) from crossing a biological membrane [1]. Nonetheless, organosulphonates are widely utilized by bacteria as sources of carbon or sulphur for growth, and ABC (ATP-binding-cassette) transporters and tripartite ATP-independent transporters are known in these pathways [2–5]. Neither of these systems is associated with catabolism of TSA (4-toluenesulphonate) (see below).
The outer membrane of Gram-negative bacteria forms a hydrophobic barrier that prevents access of water-soluble substrates to enzymes located in the periplasm, just as the cytoplasmic membrane prevents access to the cytoplasm. At least two different proteins, one in each membrane, are thus required for the overall transport process.
The outer membrane contains several classes of proteins that are responsible for either its molecular-sieving properties, i.e. porins, or for the active uptake of nutrients, i.e. receptors (e.g. [6,7]). The latter form single β-barrel cylinders [8,9]. Most porin channels sieve on the basis of molecular mass, which means that they are less specific (e.g. [6]). Some porin channels (specific porins) contain binding sites for substrates such as carbohydrates or nucleosides, which represents an advantage for solute transport in dilute media (e.g. [10,11]). A third class of porins consists of the TolC family, which link an outer-membrane channel with an inner-membrane export system and thus bridge the periplasmic space [12,13]. The primary structure of many porins is known: the amino acid composition is similar to that of water-soluble proteins, so the arrangement of the amino acid sequence in secondary and tertiary structure is responsible for the function of the transmembrane channels (e.g. [14]). The biodiversity of transport systems is such that some 360 different families are already known ([15]; see also the Transport Protein Database at http://tcdb.ucsd.edu/tcdb/).
Our understanding of the entry of TSA into the cell is largely limited to a physiological description of a secondary transport system [16]. The degradation of TSA in Gram-negative Comamonas testosteroni T-2 involves oxygenation and oxidation of the methyl sidechain to PSB (4-sulphobenzoate), and the same enzymes convert the analogous 4-toluenecarboxylate into terephthalate [17,18]. These enzymes, TsaMBCD, are encoded in the tsa operon (Figure 1), which is part of the plasmid-encoded TSA transposon, Tntsa, whose presence suffices for transport of TSA and its conversion into PSB (Figure 1) [19–21]. The regulator of the inducible TsaMBCD enzymes is the LysR-type TsaR, which, with a second regulator, the IclR-type TsaQ (Figure 1), is also involved in the regulation of transport [22,23].
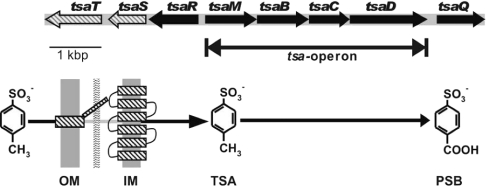
Figure 1. Organization of the relevant genes and a sketch of the function of the gene products involved in uptake and initial degradation of TSA in C. testosteroni T-2.
TsaT is the porin component that works in co-operation with TsaS, which is the putative secondary transport component located in the cytoplasmic membrane. TsaR is a LysR-type regulator which is involved in the regulation of tsaT and the tsa operon [23]. TsaQ is an IclR-type regulator involved in the expression of TsaT [22]. Intracellular TSA is metabolized by a mono-oxygenase system (oxygenase TsaM and reductase TsaB) and two dehydrogenases (TsaC and TsaD) to give PSB [19]. OM, outer membrane; IM, inner membrane.
In the present paper, we report that the last two genes to be recognized on Tntsa, tsaST, encode membrane-bound proteins found in the inner and outer membranes respectively, which appear to represent a membrane transport system that has no analogy in the Transport Protein Database.
EXPERIMENTAL
Material
Sodium [U-ring-14C]TSA, originally from Sigma, was kindly made available by Professor B. Poolman (University of Groningen, The Netherlands).
Analytical methods
Proteins were separated by SDS/PAGE in 12% separatory gels [24,25], and were subsequently stained with Coomassie Brilliant Blue G-250 [26]. Immunological detection of TsaTHis after SDS/PAGE was carried out by Western blotting following standard protocols [27]. Monoclonal antibodies against a His4 epitope were provided by Qiagen (QIAexpress Detection Kit); anti-mouse IgG coupled to alkaline phosphatase (Roche) was used as secondary antibody. Signal detection was performed with the CSPD® [disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2-(5-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate]-system (Roche). Immunological detection of expression of the tsaMBCD genes from vector pJB866-MBCD was monitored by Western blotting using antibodies against TsaB [28]; the secondary antibody and signal detection were as for TsaTHis.
Functional expression of the tsaMBCD genes from vector pJB866-MBCD was shown by assaying the activity of the multicomponent oxygenase, TsaMB. Substrate-dependent oxygen uptake of TsaMB was determined using a Clark-type oxygen electrode and TSA as substrate [29].
A Beckman LS 1801 liquid-scintillation counter was used with the Beckman Ready Value™ cocktail to quantify 14C radioactivity. N-terminal amino acid sequences were determined by Edman degradation. Protein in solution was assayed by dye binding [30]. The protein concentration in a suspension of bacteria was derived from a correction curve [23].
Bacteria, plasmids and growth conditions
C. testosteroni T-2 (DSM 6577), its pTSA−-mutant TER1 [21], C. testosteroni PSB-4 (DSM 11414) and C. testosteroniT (DSM 50244) were grown in carbon-limited mineral salts medium as described previously [28,31]. Environmental isolates (16) described elsewhere [20] were used to develop the data in Table 3. Escherichia coli DH5α [pJB866], donor of the broad-host-range vector pJB866 (GenBank® accession no. U82001), was kindly provided by J. M. Blatny and grown as described previously [32]. Host strain E. coli M15[pREP4] (Qiagen), which contains an IPTG (isopropyl β-D-thiogalactoside)-inducible expression system (pQE70), and the derived clones (Table 1), were grown in Luria–Bertani medium [33] with 25 μg/ml kanamycin and 100 μg/ml ampicillin respectively, according to the manufacturer's recommendations. Porin-deficient E. coli KS26 was grown in DYT medium (2 g/l glucose, 10 g/l yeast extract and 10 g/l tryptone) as described previously [34].
Table 3. Genetic coupling of tsaS and tsaT to genes of the tsa operon in bacteria which mineralize TSA.
n.d., not determined.
| Plasmids | Genes | |||||||
|---|---|---|---|---|---|---|---|---|
| Organism | Number | pTSA | pPSB | pT2T | TSA metabolism | tsa operon | tsaS | tsaT |
| C. testosteroni | ||||||||
| T-2 | + | − | + | + | + | + | + | |
| TER-1 | − | − | + | − | − | − | − | |
| PSB-4 | − | + | − | − | − | − | − | |
| Type strain | − | − | − | − | − | − | − | |
| Transconjugants* | ||||||||
| T-2::PSB-4 | + | + | − | + | n.d. | n.d. | n.d. | |
| T-2::Type strain | + | − | + | + | n.d. | n.d. | n.d. | |
| Environmental isolates† | ||||||||
| Group I | 9 | n.d. | n.d. | n.d. | + | + | + | + |
| Group II | 7 | n.d. | n.d. | n.d. | + | − | − | − |
Table 1. Bacteria and plasmids and their relevant characteristics (AmpR, ampicillin-resistant; TetR, tetracycline-resistant).
| Organisms and plasmids | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli DH5α | supE44, hsdR17, recA1, endA1, gyrA96, thi-1, relA1 | [51] |
| E. coli M15 | pREP4 | Qiagen |
| E. coli M70 | pQE-70, pREP4 | Qiagen |
| E. coli M70S | pQE70-S, pREP4 | Present paper |
| E. coli M70T | pQE70-T, pREP4 | Present paper |
| E. coli KS26 | MC4100 Δ(lamB) 106 Φ (ompF'-lacZ+) Hyb 16-13, Δ(ompC) 178 zei-198::Tn10, TetR | [34] |
| E. coli KS26-T | pQE70-T | Present paper |
| C. testosteroni T-2 | pTSA, pT2T, pT2L; TSA, 4-toluenecarboxylate, terephthalate, PSB, protocatechuate | [20,31] |
| C. testosteroni PSB-4 | pPSB; terephthalate, PSB, protocatechuate | [31] |
| C. testosteroni P48M | Derivative of strain PSB-4; pJB866-MBCD, pPSB; terephthalate, PSB, protocatechuate | Present paper |
| C. testosteroni P48T | Derivative of strain PSB-4; pJB866-T, pPSB; terephthalate, PSB, protocatechuate | Present paper |
| pTSA | IncP1β; Tntsa | [20] |
| pQE70 | IPTG-inducible ATG-expression vector; AmpR | Qiagen |
| pQE70-S | Introduction of restriction sites (see text) | Present paper |
| pQE70-T | Introduction of restriction sites (see text) | Present paper |
| pJB866 | Broad host range RK2 expression vector, TetR | [32] |
| pJB866-T | Introduction of restriction sites (see text) | Present paper |
| pJB866-MBCD | Introduction of restriction sites (see text) | Present paper |
DNA methodology
Plasmid DNA was prepared by the alkaline lysis protocol [33]. Restriction endonucleases and T4 DNA ligase were purchased from MBI Fermentas or New England Biolabs and used in accordance with the manufacturers' protocols. DNA was quantified fluorimetrically (DyNA Quant 200; Hoefer) according to the manufacturer's instructions. The DNA markers used were the 0.1-kb and the 1-kb ladders (MBI Fermentas) with ranges 0.1–3 kb and 0.25–10 kb respectively. DNA sequencing was performed with PCR-amplified template purified with the Qiaquick PCR Purification Kit (Qiagen). The fragment (85 ng of DNA/kb) was added to the sequencing reaction (ABI Prism Big Dye Terminator Kit; Applied Biosystems) and subjected to the following PCR cycle sequencing program: 70 s at 95 °C, then 26 cycles of 20 s at 95 °C, 30 s at 50 °C and 4 min at 60 °C. The sequence was determined by capillary electrophoresis (GATC; Konstanz, Germany).
PCR and RT (reverse transcription)-PCR
PCR was carried out in a final volume of 20–50 μl. All reactions contained 10% (v/v) DMSO. Genomic DNA (180–200 ng), prepared by cetyltrimethylammonium bromide precipitation [35], or a bacterial culture was used as template. Templates (≤5 kb) were amplified with Taq polymerase (MBI Fermentas) or (>5 kb) with the Expand Long PCR-System (Roche). The latter has a proofreading activity, and was also used for the generation of PCR products designed for cloning. Reactions were performed according to the manufacturers' recommendations. Primers used for PCR are listed in Table 2.
Table 2. PCR primers used in the present study for cloning and detection.
| PCR primer | Function | Sequences (5′→3′) |
|---|---|---|
| tsaS-Sph-N | Cloning tsaS in pQE-70 | GGGCTGGCAAGCATGCCTGTACCCCGCCAGCG |
| tsaS-Bam-C | Cloning tsaS in pQE-70 | GCCCCGCCGGGATCCCCGCCAGAGGTGCTGGG |
| tsaT-Sph-N | Cloning tsaT in pQE-70 | CTGGAGAGCATGCATAATTTCCGCCGCCGCC |
| tsaT-Bgl-C | Cloning tsaT in pQE-70 | AGTAGATCTGCGGTTGGCGGCGGCCTGG |
| tsaT-Nco-N | Cloning tsaT in pJB866; detection of tsaT | GGAGACAACCATGGATTTCCGCCGCCGCC |
| tsaT-Not-C | Cloning tsaT in pJB866; detection of tsaT; RT-PCR, first strand synthesis and reverse primer | GGCCCGGGGCGGCCGCACACTAGTGGCTTAGCGGTTGGCGGCGGCC |
| tsa-op-5′ | Cloning tsaMBCD in pJB866 | CAAGTCTGAGTCATGATCATCCGCAATTGC |
| tsa-op-3′ | Cloning tsaMBCD in pJB866 | TACTTAAGCTTAGCTCGCCCTCGGAAAAACAAGAC |
| tsaT-RT-F | RT-PCR, forward primer | GGAGACAAACATGGATTTCCGCCGCCGCC |
| tsaS-RT-F | RT-PCR, forward primer; detection of tsaS | GGCGGCCTATGCGGTGATGGGCATCACCGG |
| tsaS-RT-R1 | RT-PCR, first strand synthesis | GGAGGCGGCCACCACCATCACCACCGGCACG |
| tsaS-RT-R2 | RT-PCR, reverse primer; detection of tsaS | GGGCAATGACCATGGGGCCGGCGGTGGCG |
| tsaR-U | Detection of tsaRMBCD | CTGCAGCAGGGCAATGACCAT |
| tsaD-L | Detection of tsaRMBCD | AGCTCGCCCTCGGAAAAACAAGAC |
Total RNA of bacterial cells (4×1010 cells/preparation) was prepared with ‘RNeasy Mini Kit’ (Qiagen) and the ‘RNase-free DNase Set’ (Qiagen) following the protocols of the manufacturer. Cells for RNA preparation were harvested at mid-exponential phase (D580<0.4). For reverse transcription of RNA (0.5 μg), the First Strand cDNA Synthesis Kit (MBI Fermentas) was used. RNA was quantified photometrically at 260 nm. Amplification of cDNA by PCR was stopped in the exponential phase after 25 cycles to allow comparisons of signal intensity.
Construction of vectors
Standard protocols [33] were used to construct three vectors. Plasmid pQE70-S was generated to express a modified TsaS, TsaSHis, containing an extended C-terminus, Gly-Ser-Arg-Ser-(His)6, and an altered N-terminal sequence. Gene tsaS was amplified by PCR with primer pair tsaS-Sph-N plus tsaS-Bam-C (Table 2): the former altered the second codon of tsaS [GCT (alanine) to CCT (proline)] in order to generate the SphI site needed for ATG-cloning into pQE70, whereas the latter replaced the stop codon (TGA) by a glycine codon (GGA) to generate a BamHI site. Plasmid pQE70 was digested with SphI and BamHI, and ligation with the PCR fragment yielded vector pQE70-S. Vector pQE70-T was generated to express a modified TsaT, TsaTHis, containing an extended C-terminus, Arg-Ser-(His)6, and an additional amino acid (His2) at the N-terminus. Gene tsaT was amplified by PCR with the primer pair tsaT-Sph-N and tsaT-Bgl-C (Table 2): the former introduced an additional histidine codon (CAT) following the ATG start codon to generate a SphI site, whereas the latter replaced the authentic stop codon (TAA) with an arginine codon (AGA) to create a BglII site. Plasmid pQE70 was digested with SphI and BglII, and ligation with the PCR fragment yielded vector pQE70-T. Vector pJB866-T was generated to express an essentially unaltered TsaT. Gene tsaT was amplified by PCR using primer pair tsaT-Nco-N plus tsaT-Not-C (Table 2): the former changed the second codon of tsaT from AAT (asparagine) to GAT (aspartic acid) to create a NcoI site, whereas the latter introduced a SpeI site and a NotI site downstream of the stop codon of tsaT. Plasmid pJB866 was digested with AflIII and NotI, and ligation with the PCR fragment yielded vector pJB866-T. Vector pJB866-MBCD was generated to express genes of the tsa operon (tsaMBCD) under control of the XylR/XylS regulatory system (see [32]). The tsa operon was amplified by PCR using primer pair tsa-op-5′ plus tsa-op-3′ (Table 2): the former introduced a nucleotide exchange in tsaM [(TTC (phenylalanine) to ATC (isoleucine)] in order to generate the BspHI site required for ATG cloning, whereas the latter provided a HindIII site downstream of the stop codon of tsaD. Plasmid pJB866 was digested with AflIII and HindIII, and ligation yielded vector pJB866-MBCD. Vectors with an insert were introduced into target cells either by electroporation as detailed elsewhere [23] or by chemical transformation (E. coli M15[pREP4]) according to the supplier's instructions (QIAexpress Kit Type ATG; Qiagen).
Separation of outer-membrane proteins
E. coli M15 (pQE-70) or E. coli M70T was grown at 30 °C to early exponential phase (D580=0.8), induced with 2 mM IPTG, and incubated further to a D580 of 1.2. Cells were harvested by centrifugation at 9000 g for 10 min at 4 °C and washed twice in ice-cold 50 mM Tris/HCl, pH 7.5. Cell pellets (2 g) were resuspended in 3 ml of 20 mM Tris/HCl, pH 7.5, containing 0.5 mM EDTA (buffer A), and DNase I was added to a final concentration of 20 μg/ml. After three passages through a chilled French pressure cell at 135 MPa, PMSF was added to give a final concentration of 30 μM. Cell debris were removed by centrifugation at 9000 g for 10 min at 4 °C. Proteins of the membrane fraction were separated from soluble proteins by ultracentrifugation at 48000 rev./min for 90 min at 4 °C in a Beckman 70.1 Ti rotor. The pellet containing inner membrane, outer membrane and murein was resuspended in 30% sucrose dissolved in buffer A. Outer membranes were separated by sucrose-density-gradient centrifugation at 30000 rev./min for approx. 16 h at 8 °C in a Beckman SW40 Ti rotor using a step gradient of 30% (7 ml), 55% (2.5 ml) and 70% (1.5 ml) sucrose.
Isolation and purification of TsaT
TsaTHis was isolated from envelopes of E. coli KS26 (pQE70-T); the untransformed organism lacks the outer-membrane proteins OmpC, OmpF, and LamB [36]. Cultures were grown for approx. 3 h in 500 ml of DYT medium supplemented with kanamycin (approx. 20 μg/ml) and ampicillin (100 μg/ml) until a D600 of 0.5–0.7 was reached. The expression of TsaT was then induced by the addition of IPTG to 2 mM. The cells were grown for additional 2 h and harvested by centrifugation at 4200 g for 15 min at 4 °C. The cell pellet was washed with 10 mM Tris/HCl, pH 8.0, and cells were resuspended in 7.5 ml of the same buffer and passed three times through a chilled French pressure cell at 6.2 MPa. Undisrupted cells were removed by centrifugation at 1000 g for 15 min at 4 °C. The supernatant was then centrifuged at 100000g for 1.5 h at 4 °C. The pellet, containing the cell envelopes, was washed once with 10 mM Tris/HCl, pH 8.0. The cell pellet was then treated with lysis buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole, pH 8) supplemented with Ni-NTA (Ni2+-nitrilotriacetate)–agarose. The mixture was shaken for 30 min at room temperature (20 °C) and pelleted by centrifugation at 1000 g for 15 min at 4 °C. The pellet was washed twice with wash buffer (50 mM NaH2PO4, 300 mM NaCl and 20 mM imidazole, pH 8.0) before pure TsaTHis was separated in elution buffer (50 mM NaH2PO4, 300 mM NaCl and 250 mM imidazole, pH 8).
Lipid bilayer experiments
The methods used for the lipid bilayer experiments have been described in detail in [37]. Black lipid bilayer membranes were obtained from a 1% (w/v) solution of PC (diphytanoyl phosphatidylcholine) (Avanti Polar Lipids, Alabaster, AL, U.S.A.) in n-decane. All salts (Merck, Darmstadt, Germany) were of analytical grade. The salt solutions were used unbuffered and had a pH of approx. 6, if not otherwise indicated. The temperature was maintained at 20 °C during all experiments. Zero-current membrane potentials were measured by establishing a salt gradient across membranes containing 100–1000 channels, as described previously [38].
Determination of TSA-uptake
Uptake of 14C-labelled TSA was routinely performed by a filtration method as described previously [16]. For this purpose, 200 ml of C. testosteroni or E. coli strains was induced by IPTG or m-toluate in early exponential phase (D580=0.2 for C. testosteroni or 0.3 for E. coli), then harvested at mid-exponential phase (D580=0.35 or 0.5) by centrifugation, washed twice with ice-cold 50 mM potassium phosphate buffer, pH 7.0, and stored on ice until use.
Sequence analyses
Nucleotide or amino acid sequences were analysed using standard software (EditView from PerkinElmer and Lasergene from DNASTAR). The NCBI BLAST programs [39] were used to search for similarities of the sequences to those in databases, which included the Transport Protein Database. Secondary structures and leader peptides were predicted using the Protean software of the Lasergene package, and using different prediction servers implemented on the World Wide Web (PredictProtein, http://www.embl-heidelberg.de/predictprotein/; SignalP and TMHMM from the CBS server, http://www.cbs.dtu.dk/services/; and PSORT, http://psort.nibb.ac.jp/). We set a scanning window of 20 residues in the Kyte and Doolittle algorithm for hydrophobicity profiling. Algorithms, based on the physical principle that membrane-spanning helices will have a contiguous stretch of 19 or more hydrophobic residues, have been proven to be highly accurate [40]. The TsaT sequence was analysed in detail using the neuralnetwork-based program omp_topo_predict (http://strucbio.biologie.uni-konstanz.de/~kay/om_topo_predict2.html) [41] and the COILS program (version 2.1, http://www.ch.embnet.org/software/COILS_form.html) [42].
RESULTS
Putative transport genes, tsaST: widespread, but without a homologue in the Transport Protein Database
Preliminary experiments on the binding of the transcriptional activator TsaR to DNA in Tntsa [22,23] revealed an unexpected target, downstream of tsaR. Two contiguous ORFs (open reading frames) were found (Figure 1): tsaST, which occupied the final available coding capacity on Tntsa, so we hypothesized that they would encode the transport of TSA. The tsaS gene, 720 bp (72% G+C content), could encode a protein of 239 amino acids (25.7 kDa), 49% of which were hydrophobic. There was a gap of 19 bp between tsaR and tsaS, and of 138 bp between tsaS and tsaT. The tsaT gene, 1017 bp (66% G+C content), could encode a protein of 338 amino acids (36.5 kDa), 41% of which were hydrophobic. However, neither TsaS nor TsaT showed any similarity to proteins in Transport Protein Database. Thus if TsaST represented the minimum of two proteins needed for a transport system (see the Introduction), it seemed to be a novel system.
Our culture collection contains four relevant strains of C. testosteroni (Table 3), and 16 environmental isolates of bacteria able to mineralize TSA, so we explored using PCR the distribution of the tsaS and tsaT genes, and of the tsa operon. Only one of the four strains of C. testosteroni, T-2, was able to utilize TSA, in agreement with earlier work [20], and only in strain T-2 were the tsa operon, tsaS and tsaT detected (Table 3), associated with plasmid pTSA. The environmental isolates comprised two general groups, those involving the tsa operon and the others: only the group containing the tsa operon also contained tsaST (Table 3). The tsaST genes are not represented in the Transport Protein Database, but they seem to be representative for catabolism of TSA via TsaMBCD.
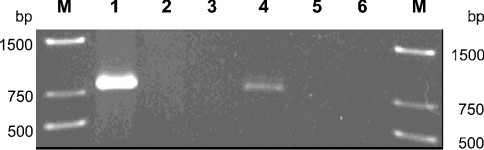
TSA uptake by C. testosteroni T-2 was induced when TSA or 4-toluenecarboxylate is the carbon source [16]. We assayed using RT-PCR the transcription of tsaT in response to different carbon sources. Expression of tsaT was detected only when the carbon source was TSA (Figure 2); undetectable transcription was the rule during growth with succinate (Figure 2), protocatechuate, terephthalate, PSB or 4-toluenecarboxylate (results not shown). This showed that TsaT was induced concomitantly with transport (and degradation) of TSA, which was consistent with the hypothesis that TsaT is part of an inducible transport system.
Figure 2. Expression of TsaT by C. testosteroni T-2 growing with different sources of carbon.
The organism was grown in 6 mM TSA-mineral-salts medium until mid-exponential phase, washed twice aseptically, and resuspended in sterile potassium phosphate buffer, pH 7.2, at 30 °C. Portions were then transferred to fresh pre-warmed medium containing the appropriate carbon source, here succinate or TSA. When early-exponential phase was reached, total RNA was isolated and tsaT mRNA was amplified by RT-PCR. Lane 1, positive control with genomic DNA as template; lane 2, negative control with genomic DNA from strain TER-1 as template; lane 3, RT-PCR with mRNA from succinate-grown cells as template; lane 4, RT-PCR with mRNA from TSA-grown cells as template; lane 5, PCR with mRNA from succinate-grown cells as template; lane 6, PCR with mRNA from TSA-grown cells as template. The other lanes (M) contain standard markers (bp).
In contrast with a TsaT induced under the regulation of TsaR and TsaQ [22], the tsaS gene was expressed constitutively. mRNA from tsaS was detected by RT-PCR when C. testosteroni T-2 utilized not only TSA and 4-toluenecarboxylate, but also PSB, terephthalate, protocatechuate and succinate, as sole sources of carbon for growth (results not shown). The regulation of this putative transport system is complex.
Cellular localization and purification of TsaT
The level of hydrophobic amino acids in TsaT, 41%, was significantly lower than in 15 typical integral inner-membrane proteins, approx. 50%. The overall hydrophobicity was low, judged by a Kyte and Doolittle analysis (results not shown), with an observed maximum value of 1.22 (maximum possible value: 4.5; [43]). A leader sequence and a cleavage site for a leader peptidase between positions 22 and 23 (ASQA-QNN) were predicted (see below). TsaT should thus be located in the periplasm or the outer membrane.
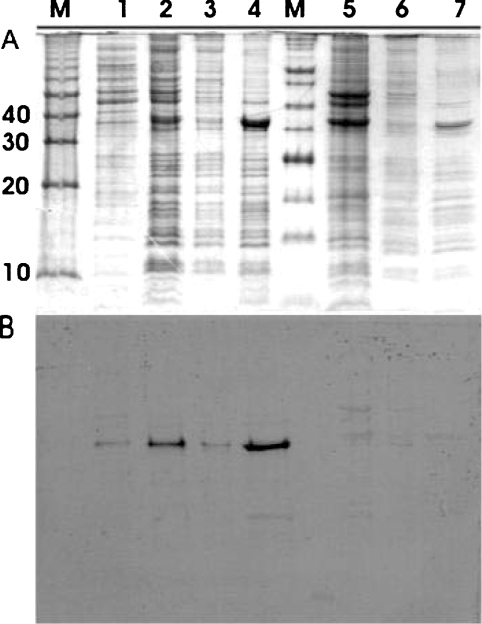
Vector pQ70-T enabled inducible expression of TsaTHis in E. coli. The soluble proteins (Figure 3A, lane 1) were clearly separated from the particulate proteins (Figure 3A, lane 2), which could be fractionated into cytoplasmic (Figure 3A, lane 3) and outer-membrane proteins (Figure 3A, lane 4). The poly-His epitope was detected in the particulate fraction (Figure 3B, lane 2), in the outer-membrane proteins only (Figure 3B, lane 4). The negative control contained no poly-His epitope (Figure 3, lanes 5–7).
Figure 3. Cellular location of TsaTHis.
TsaTHis was expressed in E. coli M15 (pQ70-T). E. coli M15 (pQE-70) was used as negative control. The soluble proteins were first separated from the membranous material, which was further separated into inner- and outer-membrane fractions. A red protein band was observed in the fraction in 55% sucrose, whereas a white band was observed in the fraction in 70% sucrose. Proteins in the different fractionations were separated by SDS/PAGE (A), and then subjected to Western hybridization to detect the His-epitope (B). Lanes M, molecular-mass standards (kDa); lanes 1 to 4, fractions from E. coli (pQ70-T); lanes 5 to 7, fractions from E. coli (pQE-70). Lane 1, supernatant protein after ultracentrifugation; lane 2, pellet after ultracentrifugation; lane 3, red-coloured band (cytoplasmic membrane); lane 4, white-coloured band (outer membrane); lane 5, combined soluble and particulate fraction; lane 6, red-coloured band (cytoplasmic membrane); lane 7, white-coloured band (outer membrane).
The protein with a poly-His epitope had a molecular mass of approx. 36 kDa (Figure 3A), which corresponded with the prediction from the derived sequence (see above), allowing for both cleavage of a leader peptide and the extension of the C-terminus with the poly-His epitope. These data confirm that tsaT encodes an outer-membrane protein, as predicted above.
TsaTHis was expressed in the porin-deficient Escherichia coli KS26 (pQE70-T) and purified by affinity chromatography. SDS/PAGE of isolated TsaTHis yielded an essentially homogeneous band at 36 kDa, as expected (see above). This sample was subjected to N-terminal amino acid sequencing. The first seven amino acids were QNNTXLL (where X is an unidentified amino acid); no contaminants were detected, which confirmed that the protein was essentially pure. This sequence was found within the derived amino acid sequence of TsaT, which confirms the prediction (see above) that the leader sequence would be cleaved between positions 22 and 23.
Transport activity of TsaT
The uptake of TSA into whole cells of C. testosteroni T-2 could be measured readily (Table 4), as observed previously [16]. The presence of TsaT in the C. testosteroni PSB-4 (pJB866-T; strain P48T) was insufficient to allow transport (Table 4). Each putative transport gene was expressed individually in E. coli, and failed to catalyse uptake of TSA. This indicates that both TsaS and TsaT are needed for transport.
Table 4. TSA-uptake and mineralization activities of organisms used in the present study.
| Activity | |||||
|---|---|---|---|---|---|
| Genotype | |||||
| Organism | tsaS | tsaT | tsaMBCD | [14C]TSA uptake (pmol/mg per min) | TSA-mineralization |
| E. coli | |||||
| M70 | − | − | − | 45.7 | − |
| M70S | + | − | − | 22.4 | − |
| M70T | − | + | − | 16.9 | − |
| C. testosteroni | |||||
| T-2 | + | + | + | 802.9 | + |
| P48M | − | − | + | 1.8 | − |
| P48T | − | + | − | 1.5 | − |
The putative channel-forming activity of TsaT was studied by the addition of small amounts of the protein to the aqueous phase bathing black lipid bilayers formed from PC. In these experiments we observed a substantial increase of the specific membrane conductance in the presence of TsaT. Approx. 2 min after addition of the protein, the membrane conductance started to rise, and increased by several orders of magnitude in approx. 20 min. Only a small further increase, as compared with the initial one, occurred after that time. The time course of the increase in conductance was similar, whether the protein was added to one or both sides of the membrane.
Single-channel analysis and selectivity of TsaT
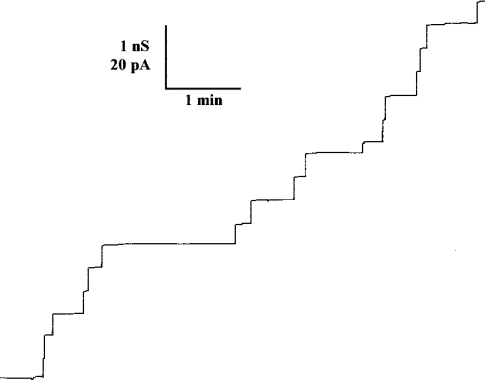
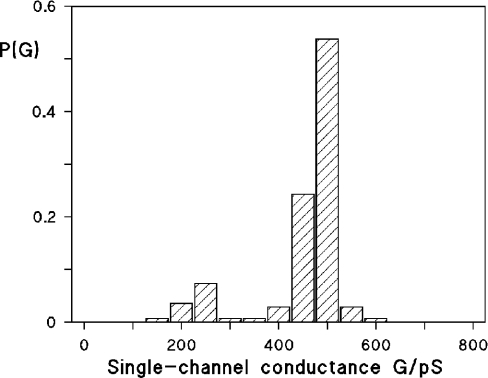
Single-channel experiments revealed that the membrane activity described above for TsaT was caused by the formation of ion-permeable channels. Figure 4 shows single-channel recordings of a PC membrane in the presence of TsaT, which was added to a black membrane at a concentration of approx. 10 ng/ml. The single-channel recording demonstrated that TsaT formed defined channels. Their single-channel conductance was about 500 pS in 0.1 M KCl. Only a minor fraction of channels with other conductance was observed (see Figure 5). It is relevant that the channels formed by TsaT had a long lifetime (mean lifetime of at least 5 min), similar to those that have been detected previously for other porins from Gram-negative bacteria [6].
Figure 4. Single-channel recording of a PC/n-decane membrane in the presence of 10 ng/ml TsaT protein added to both sides of the membrane.
The aqueous phase contained 0.1 M KCl. The applied voltage, Vm, was 20 mV; the temperature was 20 °C.
Figure 5. Histogram of all conductance steps observed with PC/n-decane membranes in the presence of 10 ng/ml TsaT protein.
P(G) is the probability that a given conductance increment G is observed in the single-channel experiments. It was calculated by dividing the number of fluctuations with a given conductance increment by the total number of conductance fluctuations. The average single-channel conductance was approx. 0.48 nS for 136 steps. The aqueous phase contained 0.1 M KCl, pH 6; the temperature was 20 °C; Vm=20 mV.
Single-channel experiments were also performed with salts other than KCl to obtain information on the size of the channels formed by TsaT and its ion-selectivity (Table 5). The replacement of potassium by the less mobile lithium ion had little influence on the conductance of the channels. The influence of the anion on the single-channel conductance in different 1 M salt solutions was substantial (Table 5), which suggests that TsaT was anion-selective. Table 5 shows also the average single-channel conductance, G, as a function of two KCl concentrations in the aqueous phase. Interestingly, the single-channel conductance was approximately a linear function of the bulk aqueous concentration. This is expected for wide water-filled channels without point charges in the vicinity of the channel [7].
Table 5. Average single-channel conductance, G, of TsaT in different salt solutions.
The membranes were formed from PC dissolved in n-decane. The aqueous solutions were unbuffered and had a pH of approx. 6. The applied voltage was 20 mV and the temperature was 20 °C. The average single-channel conductance, G, was calculated from at least 80 single events.
| Salt | Concentration (M) | G (nS) |
|---|---|---|
| LiCl | 1.0 | 2.5 |
| KCl | 0.1 | 0.48 |
| KCl | 1.0 | 3.5 |
| KCH3COO | 1.0 | 1.4 |
Zero-current membrane-potential measurements allowed the calculation of the permeability ratio Pcation/Panion in multichannel experiments. PC membranes were formed in 100 mM salt solution and concentrated TsaT was added to the aqueous phase when the membranes were in the black state. After incorporation of 100–1000 channels into a membrane, a 5-fold salt gradient was established by the addition of small amounts of concentrated KCl solution to one side of the membrane. In all experiments with TsaT, the more diluted side of the membrane became negative, which indicated preferential movement of anions through the channel. The zero-current membrane potential for KCl was approx. −16 mV (average of four experiments) on the more dilute side. Analysis of the zero-current membrane potential using the Goldman–Hodgkin–Katz equation [38] suggested that cations could also permeate through the TsaT channel because the ratio of the permeability coefficients Pcation/Panion was 0.35.
TsaS, the putative inner-membrane component of the TsaST uptake system
Sequence analysis of TsaS indicated that no leader peptide is present. Hydrophobicity plots indicated the existence of six transmembrane helices (results not shown), a structure first found in LysE from Corynebacterium glutamicum. This secondary transport system presumably uses the protonmotive force as the energy source [44], and we postulate the same energy source for TsaS. Between helices 2 and 3 (positions 106–122) of TsaS, a hydrophilic loop characteristic for inner-membrane transporters was deduced. When the sequence of this loop was searched (in April 2004) against proteins in databases (BLAST-P), no significant alignments were observed. TsaS showed significant similarity to hypothetical, inner-membrane proteins from a broad variety of organisms in the NCBI database (e.g. Bordetella parapertussis, Pseudomonas putida, Corynebacterium efficiens and Burkholderia fungorum), but not to any identified protein.
DISCUSSION
The uptake of TSA by C. testosteroni T-2, a secondary transport system with some capacity to pump against a concentration gradient [16], was suspected to be plasmid-encoded [21]. The locus was narrowed to Tntsa in genetic experiments [20,22] and now seems to be pinpointed at TsaST (Table 3). Neither gene product alone enables transport (Table 4), but the presence of Tntsa suffices for transport in a wide range of bacteria (Table 3), and there are only two candidate genes for transport on Tntsa [22]. The complex regulation of expression of the two genes (e.g. Figure 2), with two regulators for the induction of TsaT [22] contrasting with constitutive expression of TsaS, has prevented us from using heterologous expression to confirm that the presence of both proteins suffices for the transport.
TsaST appears to be a novel transport system, and it is the first system to be characterized for the catabolism of an arylsulphonate. The set of environmental isolates able to catabolize TSA includes organisms without Tntsa and tsaST (Table 3), so other transport systems are possible. The transport system for TSA in the assimilation of sulphonate sulphur involves an ABC transporter [4].
TsaT is an outer-membrane protein (Figure 3), consistent with the leader peptide, which was first predicted from the deduced primary sequence of the polypeptide, and then confirmed experimentally. Further predictions are that the mature protein comprises three domains, A (approx. 90 amino acids), which is amphipathic with short stretches of α-helices and β-sheets, B (approx. 110 amino acids), which contains two transmembrane sequences, and C (approx. 100 amino acids), which consists largely of one hydrophilic α-helix (results not shown). This protein has a high single-channel conductance (Figure 4) that is a linear function of the aqueous salt concentration. TsaT thus forms a wide water-filled channel with a conductance that is approximately twice that of the outer-membrane porins OmpF and OmpC of E. coli and other enteric bacteria [6]. The protein is thus a porin.
All general diffusion pores crystallized to date have a similar structure. The channels in a trimer are β-barrel cylinders, which contain a central constriction formed by external loop 3 (e.g. [45,46]). The cross-section of the TsaT channel, given its high conductance, is apparently larger than those of general diffusion pores of the E. coli outer membrane, which have diameters of approx. 0.8–0.9 nm [45]. Zero-current membrane-potential measurements demonstrated that the channels were selective for Cl− over K+, which indicates that there exists a positive net charge in or near the channel. These findings agree with the single-channel measurements, which revealed a more substantial effect of the mobility of the anions on the single-channel conductance. It follows that selectivity of TsaT is based on the aqueous mobility of the single anions. These data show that TsaT forms a large, general diffusion pore with a preference for anions, precisely what is required for the salt of the bulky, strong arylsulphonic acid, TSA.
The sequence of TsaT does not indicate its function as a porin, but many features typical of outer-membrane proteins are present. The leader sequence (22 amino acids) is close to the average length (25.1 amino acids) of leader peptides in Gram-negative bacteria [47]. The low cysteine content (e.g. [48]) is found (one residue). Protein sections, which span the outer membrane, are encoded by amphipathic stretches, and which are organized as antiparallel all-next-neighbour β-barrels of low hydrophobicity (e.g. [48]) are observed in section A (see above). Furthermore, the total of 338 residues encoded by tsaT is consistent with the class of general porins (e.g. [48]).
Porin TsaT shows no homology with known porins, as emphasized when an artificial neural network trained with classical porins to compare topology plots [41] detected no significant similarity. Presumably, novel features in TsaT cause this effect. Specifically, the overall hydrophobicity of TsaT is low, but two stretches long enough to span the membrane as an α-helix are detected (see above). These structures are absent from classical porin Omp32 [49].
TolC is a representative of a newly discovered class of outer membrane proteins denoted as ‘chunnels’ [12]. The α-helical barrel that spans the periplasmic space comprises 67 residues. The crystal structure of TolC revealed that these helices are organized as coiled coils [13]. TsaT also contains a long hydrophilic α-helical C-terminus, presumably fused to the C-terminal β-strand of the archetypal core porin (site of fusion F240/M241). This α-helix is 97 residues long, and is thus long enough to span the periplasmic space as an α-helix, but no coiled coils were predicted from the sequence. The hydrophilic stretches in section B of TsaT are short and might represent loops exposed to the cell surface. Porin TsaT thus contains one general feature of the TolC-like porins, but is markedly different from them.
Two-component uptake systems are well known in Gram-negative bacteria, e.g. the TonB-dependent iron siderophore transporter FhuA [8]. TolC, though involved in efflux processes [50], involves a periplasmic tunnel in co-operation with an inner-membrane component [12]. We hypothesize that TsaT comprises an anion-specific pore and a trans-periplasmic channel to the inner membrane, where TsaS completes the transport process (Figure 1).
Acknowledgments
We are grateful to B. Poolman for providing [14C]TSA, to K. Mendgen and R. Vögele for support with the radioactive uptake measurements, and to W. Boos, M. Ehrmann, W. Welte and H. Engelhardt for discussions. J.M. and T.T. were supported by a grant from the Deutsche Forschungsgemeinschaft to A.M.C. and J.R. (CO 206/4) and by funds from the University of Konstanz. E.M. was supported by a grant from the Deutsche Forschungsgemeinschaft to R.B. (Be 865/10).
References
- 1.Graham D. E., Xu H., White R. H. Identification of coenzyme M biosynthetic phosphosulfolactate synthase: a new family of sulfonate biosynthesizing enzymes. J. Biol. Chem. 2002;277:13421–13429. doi: 10.1074/jbc.M201011200. [DOI] [PubMed] [Google Scholar]
- 2.Cook A. M., Laue H., Junker F. Microbial desulfonation. FEMS Microbiol. Rev. 1999;22:399–419. doi: 10.1111/j.1574-6976.1998.tb00378.x. [DOI] [PubMed] [Google Scholar]
- 3.Kertesz M. A. Riding the sulfur cycle – metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 2000;24:135–175. doi: 10.1016/S0168-6445(99)00033-9. [DOI] [PubMed] [Google Scholar]
- 4.Kertesz M. A. Bacterial transporters for sulfate and organosulfur compounds. Res. Microbiol. 2001;152:279–290. doi: 10.1016/s0923-2508(01)01199-8. [DOI] [PubMed] [Google Scholar]
- 5.Brüggemann C., Denger K., Cook A. M., Ruff J. Enzymes and genes of taurine and isethionate dissimilation in Paracoccus denitrificans. Microbiology. 2004;150:805–816. doi: 10.1099/mic.0.26795-0. [DOI] [PubMed] [Google Scholar]
- 6.Benz R. Solute uptake through bacterial outer membrane. In: Hackenbeck R., Ghuysen J.-M., editors. Bacterial Cell Wall. Amsterdam: Elsevier; 1994. pp. 397–423. [Google Scholar]
- 7.Benz R. Porins – structure and function. In: Winkelmann G., editor. Microbial Transport Systems. Weinheim: Wiley-VCH; 2001. pp. 227–246. [Google Scholar]
- 8.Ferguson A. D., Hofmann E., Coulton J. W., Diederichs K., Welte W. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 9.Locher K. P., Rees B., Koebnik R., Mitschler A., Moulinier L., Rosenbusch J. P., Moras D. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell. 1998;95:771–778. doi: 10.1016/s0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 10.Luckey M., Nikaido H. Specificity of diffusion channels produced by lambda phage receptor protein of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1980;77:167–171. doi: 10.1073/pnas.77.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier C., Bremer E., Schmid A., Benz R. Pore-forming activity of the Tsx protein from the outer membrane of Escherichia coli: demonstration of a nucleoside-specific binding site. J. Biol. Chem. 1988;263:2493–2499. [PubMed] [Google Scholar]
- 12.Andersen C., Hughes C., Koronakis V. Chunnel vision: export and efflux through bacterial channel-tunnels. EMBO Rep. 2000;1:313–318. doi: 10.1093/embo-reports/kvd075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koronakis V., Sharff A., Koronakis E., Luisi B., Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature (London) 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- 14.Schirmer T., Keller T. A., Wang Y. F., Rosenbusch J. P. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 15.Saier M. H., Jr A functional-phylogenetic system for the classification of transport proteins. J. Cell. Biochem. 1999;32–33(Suppl.):84–94. doi: 10.1002/(sici)1097-4644(1999)75:32+<84::aid-jcb11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 16.Locher H. H., Poolman B., Cook A. M., Konings W. N. Uptake of 4-toluene sulfonate by Comamonas testosteroni T-2. J. Bacteriol. 1993;175:1075–1080. doi: 10.1128/jb.175.4.1075-1080.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Locher H. H., Leisinger T., Cook A. M. Degradation of p-toluenesulphonic acid via sidechain oxidation, desulphonation and meta ring cleavage in Pseudomonas (Comamonas) testosteroni T-2. J. Gen. Microbiol. 1989;135:1969–1978. doi: 10.1099/00221287-135-7-1969. [DOI] [PubMed] [Google Scholar]
- 18.Locher H. H., Malli C., Hooper S., Vorherr T., Leisinger T., Cook A. M. Degradation of p-toluic acid (p-toluenecarboxylic acid) and p-toluenesulphonic acid via oxygenation of the methyl sidechain is initiated by the same set of enzymes in Comamonas testosteroni T-2. J. Gen. Microbiol. 1991;137:2201–2208. [Google Scholar]
- 19.Junker F., Kiewitz R., Cook A. M. Characterization of the p-toluenesulfonate operon tsaMBCD and tsaR in Comamonas testosteroni T-2. J. Bacteriol. 1997;179:919–927. doi: 10.1128/jb.179.3.919-927.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tralau T., Cook A. M., Ruff J. Map of the IncP1β plasmid pTSA encoding the widespread genes (tsa) for p-toluenesulfonate degradation in Comamonas testosteroni T-2. Appl. Environ. Microbiol. 2001;67:1508–1516. doi: 10.1128/AEM.67.4.1508-1516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Junker F., Cook A. M. Conjugative plasmids and the degradation of arylsulfonates in Comamonas testosteroni. Appl. Environ. Microbiol. 1997;63:2403–2410. doi: 10.1128/aem.63.6.2403-2410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tralau T., Cook A. M., Ruff J. An additional regulator, TsaQ, is involved with TsaR in regulation of transport during the degradation of p-toluenesulfonate in Comamonas testosteroni T-2. Arch. Microbiol. 2003;180:319–326. doi: 10.1007/s00203-003-0594-8. [DOI] [PubMed] [Google Scholar]
- 23.Tralau T., Mampel J., Cook A. M., Ruff J. Characterization of TsaR, an oxygen-sensitive LysR-type regulator for the degradation of p-toluenesulfonate in Comamonas testosteroni T-2. Appl. Environ. Microbiol. 2003;69:2298–2305. doi: 10.1128/AEM.69.4.2298-2305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 26.Neuhoff V., Arold N., Taube D., Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9:255–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg I. M. Birkhäuser, Boston: Benchtop Techniques; 1996. Protein Analysis and Purification. [Google Scholar]
- 28.Schläfli Oppenberg H. R., Chen G., Leisinger T., Cook A. M. Regulation of the degradative pathways from 4-toluenesulphonate and 4-toluenecarboxylate to protocatechuate in Comamonas testosteroni T-2. Microbiology. 1995;141:1891–1899. doi: 10.1099/13500872-141-8-1891. [DOI] [PubMed] [Google Scholar]
- 29.Locher H. H., Leisinger T., Cook A. M. 4-Sulphobenzoate 3,4-dioxygenase: purification and properties of a desulphonative two-component enzyme system from Comamonas testosteroni T-2. Biochem. J. 1991;274:833–842. doi: 10.1042/bj2740833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Thurnheer T., Köhler T., Cook A. M., Leisinger T. Orthanilic acid and analogues as carbon sources for bacteria: growth physiology and enzymatic desulfonation. J. Gen. Microbiol. 1986;132:1215–1220. [Google Scholar]
- 32.Blatny J. M., Brautaset T., Winther-Larsen H. C., Karunakaran P., Valla S. Improved broad-host-range RK2 vectors useful for high and low regulated gene expression levels in Gram-negative bacteria. Plasmid. 1997;38:35–51. doi: 10.1006/plas.1997.1294. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J., Fritsch E. F., Maniatis T. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 34.Forst D., Schülein K., Wacker T., Kreutz W., Benz R., Welte W. Crystallization and preliminary X-ray analysis of ScrY, a specific bacterial outer membrane porin. J. Mol. Biol. 1993;229:258–262. doi: 10.1006/jmbi.1993.1028. [DOI] [PubMed] [Google Scholar]
- 35.Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. New York: John Wiley & Sons; 1987. Current Protocols in Molecular Biology. [Google Scholar]
- 36.Schülein K., Andersen C., Benz R. The deletion of 70 amino acids near the N-terminal end of the sucrose-specific porin ScrY causes its functional similarity to LamB in vivo and in vitro. Mol. Microbiol. 1995;17:757–767. doi: 10.1111/j.1365-2958.1995.mmi_17040757.x. [DOI] [PubMed] [Google Scholar]
- 37.Benz R., Janko K., Boos W., Läuger P. Formation of large, ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli. Biochim. Biophys. Acta. 1978;511:305–319. doi: 10.1016/0005-2736(78)90269-9. [DOI] [PubMed] [Google Scholar]
- 38.Benz R., Janko K., Läuger P. Ionic selectivity of pores formed by the matrix protein (porin) of Escherichia coli. Biochim. Biophys. Acta. 1979;551:238–247. doi: 10.1016/0005-2736(89)90002-3. [DOI] [PubMed] [Google Scholar]
- 39.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 41.Diederichs K., Freigang J., Umhau S., Zeth K., Breed J. Prediction by a neural network of outer membrane β-strand protein topology. Protein Sci. 1998;7:2413–2420. doi: 10.1002/pro.5560071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 43.Kyte J., Doolittle R. F. A simple method for displaying the hydrophobic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 44.Vrljic M., Garg J., Bellmann A., Wachi S., Freudl R., Malecki M. J., Sahm H., Kozina V. J., Eggeling L., Saier M. H., Jr The LysE superfamily: topology of the lysine exporter LysE of Corynebacterium glutamicum, a paradyme for a novel superfamily of transmembrane solute translocators. J. Mol. Microbiol. Biotechnol. 1999;1:327–336. [PubMed] [Google Scholar]
- 45.Cowan S. W., Schirmer T., Rummel G., Steiert M., Ghosh R., Pauptit R. A., Jansonius J. N., Rosenbusch J. P. Crystal structures explain functional properties of two E. coli porins. Nature (London) 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 46.Kreusch A., Neubuser A., Schiltz E., Weckesser J., Schulz G. E. Structure of the membrane channel porin from Rhodopseudomonas blastica at 2.0 Å resolution. Protein Sci. 1994;3:58–63. doi: 10.1002/pro.5560030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen H., Engelbrecht J., Brunak S., von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 48.Koebnik R., Locher K. P., Van Gelder P. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 49.Gerbl-Rieger S., Engelhardt H., Peters J., Kehl M., Lottspeich F., Baumeister W. Topology of the anion-selective porin Omp32 from Comamonas acidovorans. J. Struct. Biol. 1992;108:14–24. doi: 10.1016/1047-8477(92)90003-s. [DOI] [PubMed] [Google Scholar]
- 50.Sharff A., Fanutti C., Shi J., Calladine C., Luisi B. The role of the TolC family in protein transport and multidrug efflux: from stereochemical certainty to mechanistic hypothesis. Eur. J. Biochem. 2001;268:5011–5026. doi: 10.1046/j.0014-2956.2001.02442.x. [DOI] [PubMed] [Google Scholar]
- 51.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]