Abstract
SAG (salivary agglutinin), which is identical to gp-340 (glycoprotein-340) from the lung, is encoded by DMBT1 (deleted in malignant brain tumours 1). It is a member of the SRCR (scavenger receptor cysteine-rich) superfamily and contains 14 SRCR domains, 13 of which are highly similar. SAG in saliva is partially complexed with IgA, which may be necessary for bacterial binding. The goal of the present study was to characterize the binding of purified SAG to IgA. SAG binds to a variety of proteins, including serum and secretory IgA, alkaline phosphatase-conjugated IgGs originating from rabbit, goat, swine and mouse, and lactoferrin and albumin. Binding of IgA to SAG is calcium dependent and is inhibited by 0.5 M KCl, suggesting that electrostatic interactions are involved. Binding of IgA was destroyed after reduction of SAG, suggesting that the protein moiety is involved in binding. To pinpoint further the binding domain for IgA on SAG, a number of consensus-based peptides of the SRCR domains and SRCR interspersed domains were designed and synthesized. ELISA binding studies with IgA indicated that only one of the peptides tested, comprising amino acids 18–33 (QGRVEVLYRGSWGTVC) of the 109-amino-acid SRCR domain, exhibited binding to IgA. This domain is identical to the domain of SAG that is involved in binding to bacteria. Despite this similar binding site, IgA did not inhibit binding of Streptococcus mutans to SAG or peptide. These results show that the binding of IgA to SAG is specifically mediated by a peptide sequence on the SRCR domains.
Keywords: DMBT1 (deleted in malignant brain tumours 1), gp-340, IgA, salivary agglutinin, scavenger receptor
Abbreviations: CUB domain, C1r/C1s, Uegf, BMP-1 domain; gp-340, glycoprotein-340; HRP, horseradish peroxidase; SAG, salivary agglutinin; SRCR, scavenger receptor cysteine-rich; SID, SRCR interspersed domain; SP-D, surfactant protein-D
INTRODUCTION
SAG (salivary agglutinin) is identical to lung gp-340 (glycoprotein-340), a receptor for SP-D (surfactant protein-D) and SP-A, and to DMBT1 (deleted in malignant brain tumours-1), a putative tumour suppressor [1–3]. It is a member of the SRCR (scavenger receptor cysteine-rich) superfamily, a group of proteins possessing SRCR domains. Members of this group are involved in various types of ligand binding. For example, CD5 and CD6 are involved in leucocyte interactions, and CD163 scavenges haptoglobin–haemoglobin complexes [4].
SAG is composed of 13 highly similar SRCR domains which are separated by SIDs (SRCR interspersed domains), two CUB (C1r/C1s, Uegf, Bmp1) domains separated by a 14th SRCR domain, and a ZP (zona pellucida) domain [5]. SRCR domains each comprise approx. 100–110 amino acid residues, and are highly conserved across species boundaries [6]. The three-dimensional conformation of SRCR domains is stabilized by three (type A) or four (type B) disulphide bridges [7].
Lung gp-340 has been proposed as a potential opsonin receptor for SP-D, a member of the collectin family [5,8]. Collectins are pattern recognition receptors that play an important role in innate immunity by binding to specific carbohydrate structures on the surfaces of pathogenic micro-organisms [9,10]. This binding enhances the phagocytosis and killing of micro-organisms by neutrophils and alveolar macrophages. gp-340 binds to SP-D through a protein–protein interaction in a calcium-dependent manner [8].
SAG is a 300–400 kDa glycoprotein that was isolated originally by affinity adsorption of parotid saliva to Streptococcus mutans, the causative agent of dental caries [11]. It contains 25% (w/w) carbohydrate, which has been implicated in binding to S. mutans [12,13]. In addition to S. mutans, a variety of other oral and non-oral bacteria are agglutinated by SAG, such as S. oralis, S. gordonii, Staphylococcus aureus and Helicobacter pylori [3,14,15]. In addition, SAG binds to several proteins, including bovine lactoferrin, SP-D, complement C1q, mucin MUC5B and IgA [1,12,16–19]. IgA and SAG were suggested to act synergistically in saliva-induced bacterial agglutination [19].
The purpose of the present study was to identify the domains on SAG involved in binding to IgA. The interaction of SAG with IgA has been studied by stepwise degradation of SAG–IgA complexes present in saliva [11,12,19]. In the present study, the interaction of purified SAG and IgA has been studied and the effects of chemical treatment affecting its protein moiety have been tested. Furthermore, we have synthesized a series of peptides covering the complete SRCR domains and SIDs, and examined their IgA-binding properties. Only one 16-mer peptide of the SRCR domain, identified previously as a binding site for S. mutans, was found to bind IgA. Binding of IgA to SAG was inhibited by the same substances as was the binding of S. mutans to SAG, suggesting a similar type of binding.
EXPERIMENTAL
Chemicals
A monoclonal antibody against SAG (mAb143) with specificity for a reduction-sensitive protein epitope on SAG [20] was kindly provided by Dr D. Malamud (University of Pennsylvania, PA, U.S.A.). Monoclonal antibody 5E9 was raised originally by immunization with a high-molecular-mass salivary mucin (MUC5B) and is directed to a sialylated carbohydrate epitope [21]. Alkaline phosphatase-conjugated and HRP (horseradish peroxidase)-conjugated goat anti-rabbit IgG antibodies, swine anti-rabbit IgG antibodies, rabbit anti-mouse IgG antibodies (whole molecule) and HRP-conjugated avidin were obtained from DAKO (Glostrup, Denmark). HRP-conjugated goat antibodies against IgA were obtained from Sigma (St. Louis, MO, U.S.A.). Alkaline phosphatase-conjugated Fab fragments of sheep anti-mouse and sheep anti-digoxigenin antibodies were from Boehringer (Mannheim, Germany). Biotinylated UEA-1 lectin was from Sigma. BSA was from Roche Molecular Systems (Mannheim, Germany), and bovine lactoferrin was a gift from DMV-Campina (Veghel, The Netherlands). Blood group A, Lewis a and sulpho-Lewis a coupled to polyacrylamide were from Synthesome (Munich, Germany). The DNA-binding fluorescent probe SYTO-13 was from Molecular Probes (Leiden, The Netherlands).
Purification of SAG
SAG was purified by gel filtration as described previously [13]. Approx. 30 ml of parotid saliva was collected with a Lashley cup under stimulation with menthol candies. The sample was incubated on ice–water, resulting in the formation of precipitates containing SAG. After centrifugation (15 min, 5000 g, 4 °C) the pellet was resuspended in 0.1 vol. of PBS supplemented with 10 mM EDTA and applied on a Sephacryl S-400 (Amersham Biosciences, Roosendaal, The Netherlands) gel filtration column (15 mm×490 mm) run in PBS. Fractions were monitored at A280. SAG migrated in the first peak fraction.
Peptide synthesis and purification
Based on the published amino acid sequence of gp-340 [5], consensus sequences of the first 13 SRCR domains and 11 SIDs were determined using alignment software (Vector NTI; InforMax Inc., Oxford, U.K.) [15]. Nine peptides, together spanning the complete sequence, were synthesized using a Milligen 9050 peptide synthesizer (Milligen, Bedford, MA, U.S.A.).
Peptides were purified by reverse-phase HPLC on a JASCO (Tokyo, Japan) HPLC System. Peptides were dissolved in 0.1% (v/v) trifluoroacetic acid and applied on a Vydac C18 column (218TP; 1.0 cm×25 cm, 10 μm particles; Vydac, Hesperia, CA, U.S.A.) equilibrated in 0.1% trifluoroacetic acid. Elution was performed with a linear gradient from 30% to 45% (v/v) acetonitrile containing 0.1% trifluoroacetic acid in 20 min at a flow rate of 4 ml/min. The absorbance of the column effluent was monitored at 214 nm, and peak fractions were pooled, lyophilized and re-analysed by reverse-phase-HPLC and by capillary electrophoresis on a Biofocus 2000 apparatus (Bio-Rad). The authenticity of the (monomeric) peptides was confirmed by Q-TOF (quadrupole time of flight) MS on a tandem mass spectrometer (Micromass Inc., Manchester, U.K.) as described previously [22]. Despite the presence of cysteine residues, exclusively monomeric peptides were detected, indicating that peptide multimerization by disulpide-bond formation had not occurred. The purity of the peptides was at least 90%.
ELISA
To examine the binding of SAG to various proteins, microplates (Microlon; Greiner, Recklinghausen, Germany) were coated with proteins in 0.1 M sodium carbonate buffer (pH 9.6) overnight at 4 °C. All further incubations were for 1 h at 37 °C, and between each incubation microplates were washed with PBS containing 0.1% Tween 20 (PBST). After coating, microplates were incubated with parotid saliva or SAG serially diluted in PBST containing 0.5 mM CaCl2 (PBST-Ca). After washing, bound SAG was revealed with biotinylated UEA-1 lectin followed by incubation with avidin-conjugated HRP, or with antibody 143 or 5E9 followed by incubation with HRP-conjugated rabbit antibodies against mouse antibodies. These incubations were conducted in PBST supplemented with 1% (w/v) albumin. Microplates were developed at room temperature with 0.01% 3,3′,5,5′-tetramethylbenzidine (Pierce) and 0.003% H2O2 in 100 mM sodium acetate buffer, pH 5.5. The reaction was stopped with 1 M sulphuric acid.
For inhibition experiments, inhibitors were added to parotid saliva or SAG solution and subsequently serially diluted in PBST containing the same concentration of inhibitor on IgA-coated microplates. Bound SAG was revealed as described above.
To examine the binding of IgA to SAG or peptides, microplates were coated with serial dilutions of SAG or peptide solution. Thereafter, microplates were incubated with 2 μg/ml IgA in PBST-Ca. After washing, microplates were incubated with HRP-conjugated antibodies against IgA in PBST supplemented with 1% (w/v) albumin. Microplates were developed as described above.
SDS/PAGE and Western blotting
SDS/PAGE was conducted on a Pharmacia Phast System (Pharmacia-LKB, Uppsala, Sweden) according to the manufacturer's instructions. Proteins were dissolved in sample buffer containing 15 mM Tris/HCl, pH 6.8, 0.5% SDS, 2.5% glycerol and 0.05% Bromophenol Blue. For reduction, samples were incubated with 25 mM dithiothreitol (ICN Biomedicals, Aurora, OH, U.S.A.). Samples were separated on 4–15% or 7.5% (w/v) polyacrylamide gels. For Western blotting, proteins were transferred on to nitrocellulose membranes by diffusion blotting. Membranes were blocked by incubation for 1 h in TBS/Tween 20 containing 1% (w/v) gelatin. Then membranes were incubated with antibodies in TBS/Tween 20. 5-Bromo-4-chloro-3-indolyl phosphate (Roche Diagnostics) was used as substrate.
Adhesion assay
Bacterial adhesion was examined using a microplate method based on labelling of micro-organisms with a cell-permeable DNA-binding probe, as reported previously [23]. S. mutans strain Ingbritt was cultured on Todd Hewitt broth (Difco Laboratories, Detroit, MI, USA) as described previously [1]. Cells were harvested by centrifugation (3000 g at 4 °C for 10 min), washed in PBS and suspended in PBST-Ca to a density of 5×108 cells/ml. Fluotrac 600 microplates (Greiner) were coated with various amounts of either synthetic peptides or purified SAG in coating buffer (100 mM sodium carbonate, pH 9.6). After incubation at 4 °C for 16 h, plates were washed twice with PBST-Ca. Subsequently, 100 μl of a bacterial suspension (5×108 bacteria/ml) in PBST-Ca was added to each well and incubated at 37 °C for 2 h. After washing, bound bacteria were detected using 100 μl/well of 1 mM SYTO-13 solution (Molecular Probes), a cell-permeating and DNA-binding fluorescent probe. Plates were incubated in the dark at ambient temperature for 15 min and then washed three times with the same buffer. Fluorescence was measured in a Fluostar Galaxy microplate fluorescence reader (BMG Laboratories, Offenburg, Germany) at 488 nm excitation and 509 nm emission wavelengths.
RESULTS
Binding of SAG to various proteins
To investigate the presence of complexes of SAG with salivary IgA and with other proteins in parotid saliva, a sandwich ELISA was used. Microplates were coated with antibodies against IgA or secretory component. Microplates coated with antibodies against CEA-1 antigen, which is not present in parotid saliva, were used as a negative control. Microplates were then incubated with serial dilutions of parotid saliva in PBST-Ca, and bound SAG was revealed with antibody 5E9. Similar dose–response curves were obtained with the anti-IgA antibodies as with the negative control (CEA-1), suggesting non-selective binding of SAG to these antibodies. Also, the coating of microplates with albumin, initially used as a blocking agent, resulted in a good dose–response, suggesting that SAG also bound to albumin.
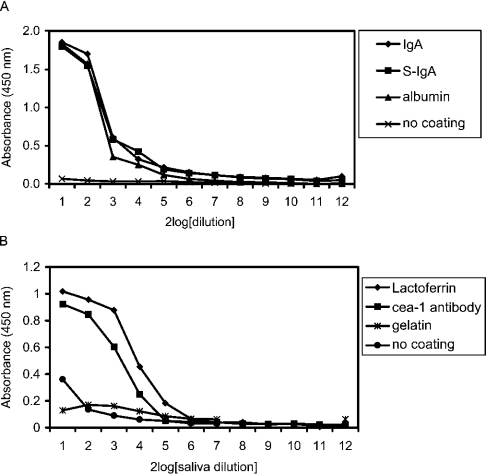
These results prompted us to study the binding of SAG to a wider variety of proteins. Microplates were coated directly with IgA, secretory IgA, human albumin, bovine lactoferrin or gelatin. SAG diluted in PBST-Ca bound to all antibodies, and also to albumin and lactoferrin, but only weakly to gelatin. In the presence of Tween 20, SAG did not bind to non-coated microplates (Figure 1).
Figure 1. Binding of SAG to various proteins.
Microplates were coated with serum IgA, secretory IgA (S-IgA) (4 μg/ml), human albumin (10 μg/ml) (A), bovine lactoferrin (1 mg/ml), rabbit IgG antibodies against CEA-1 antigen (10 μg/ml) or gelatin (1 mg/ml) (B). After coating, microplates were incubated with parotid saliva serially diluted in PBST-Ca [1:1 (v/v) starting dilution]. Bound SAG was revealed with antibody 5E9. SAG bound to antibodies, albumin and lactoferrin, but only weakly to gelatin.
Effects of Ca2+ ions
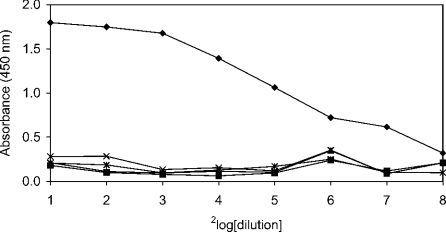
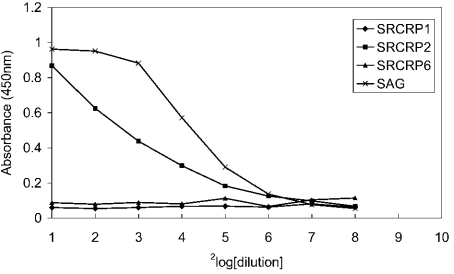
The influence of Ca2+ and other bivalent ions on the binding of SAG to IgA was tested in an ELISA. IgA-coated microplates were incubated with purified SAG in PBST containing 0.5 mM CaCl2 (with or without 5 mM EDTA), BaCl2, MgCl2 or ZnCl2 (Figure 2). It was demonstrated that Ca2+ ions were required for binding and could not be replaced by other bivalent ions, suggesting that binding of SAG to IgA was specifically dependent on Ca2+ ions.
Figure 2. Effects of bivalent ions on SAG binding to IgA.
Microplates coated with serum IgA (2 μg/ml) were incubated with serial dilutions of SAG in PBST containing different bivalent ions (0.5 mM). After washing, bound SAG was revealed with monoclonal antibody 143. SAG binding was Ca2+-dependent (♦). Binding completely disappeared in the presence of 5 mM of the calcium-chelating agent EDTA (▪). Ca2+ ions could not be replaced by other bivalent ions such as Mg2+ (▴), Ba2+ (×) or Zn2+ (✱).
Inhibition of SAG binding to IgA
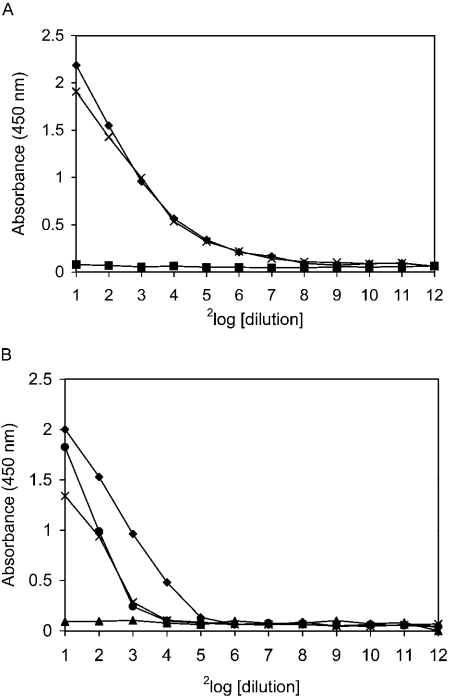
To characterize further the binding of SAG to IgA, several substances were tested for inhibition of binding. For this purpose, IgA-coated microplates were incubated with serial dilutions of parotid saliva in the presence of inhibitory substances (Figure 3). KCl at 0.5 M (Figure 3B) inhibited binding, suggesting that electrostatic interactions play a role in binding. Strikingly, amine-containing components, such as ammonium chloride (Figure 3A) and 0.1 M glycine (results not shown), also inhibited binding. In contrast, binding of SAG to IgA was not inhibited by maltose (Figure 3A), glucose, Lewis a, blood group A or sulpho-Lewis a oligosaccharides (results not shown). Negatively charged ions such as carbonate and nitrate (0.1 M) were only weakly inhibitory (Figure 3B).
Figure 3. Inhibition of SAG binding to IgA.
IgA-coated microplates (2 μg/ml) were incubated with parotid saliva serially diluted in PBST-Ca containing inhibitory substances. After washing, bound SAG was revealed with antibody 143. (A) Binding of SAG to IgA was completely inhibited by amine-containing substances such as 0.1 M NH4Cl (▪), but not by carbohydrates such as 0.1 M maltose (×), in the presence of which binding was comparable with that in the control (♦). (B) Negatively charged ions such as nitrate (•) and carbonate (×) (0.1 M) showed low or intermediate inhibition compared with the control (♦). KCl at 0.5 M (▴) completely inhibited binding.
Binding of antibodies to SAG after reduction
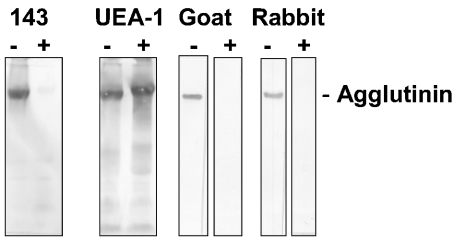
To investigate the effect of reduction of SAG on its binding to antibodies, parotid saliva was separated by SDS/PAGE, transferred on to nitrocellulose, and tested for binding of goat and rabbit IgG antibodies conjugated to alkaline phosphatase (Figure 4). SAG that was separated under non-reducing conditions bound these antibodies, but SAG that was separated under reducing conditions did not. The binding of antibody 143, directed against SAG, also disappeared after reduction. UEA-1 lectin blotting demonstrated that SAG migrated more slowly in the gel. These results suggest that reduction induced a conformational change in SAG that destroyed a conformation-dependent binding site.
Figure 4. Binding of antibodies to SAG and effect of chemical reduction.
Parotid saliva was separated by SDS/PAGE under reducing (+) or non-reducing (−) conditions, transferred on to nitrocellulose and incubated with antibody 143 against SAG, UEA-1 lectin or alkaline phosphatase-conjugated goat anti-rabbit (goat) or rabbit anti-mouse (rabbit) IgG antibodies. After reduction, the binding of antibody 143 as well as of conjugated antibodies disappeared. With UEA-1 lectin it was observed that SAG migrated more slowly in the gel, suggesting a conformational change.
Pinpointing the IgA-binding site on SAG
The SRCR domains are a major part of the protein core of SAG and their tertiary structure is stabilized by disulphide bridges. Therefore the involvement of SRCR domains in the binding of SAG to IgA was investigated. Using alignment software, the consensus sequences of the SRCR domains and SIDs were determined (Figure 5B). This resulted in a consensus sequence of the SRCR domain of 109 amino acids, and two consensus sequences for the SIDs of 20 and 22 amino acids. Seven synthetic peptides, representing loops that run in between disulphide bridges of the SRCR domain, and two peptides covering the sequences of the SIDs were synthesized (Table 1). Peptides were coated on to microplates and tested for binding of IgA. Only peptide SRCRP2, corresponding to residues 18–33 of the 109-residue SRCR domain, showed strong binding of the antibodies. The other peptides did not bind these antibodies (Figure 6).
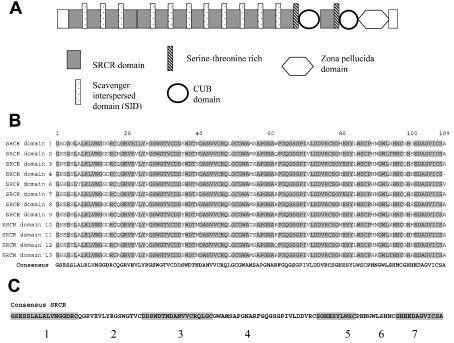
Figure 5. (A) Structure of the SAG molecule, (B) alignment of SRCR domains and (C) design of peptides.
(A) SAG contains a unique N-terminal sequence, 13 SRCR domains (dark grey) separated by SIDs (dotted boxes), a serine-proline-threonine-rich sequence (striated black), two CUB domains (circles) separated by a 14th SRCR domain, a zona pellucida domain (hexamer) and a unique sequence. (B) The first 13 highly similar SRCR domains were aligned and a consensus sequence was developed. Grey-shaded sequences are identical in all 13 SRCR domains. (C) Seven peptides were synthesized based on the consensus sequence, representing loops in the SRCR domain between disulphide bridges.
Table 1. SRCR domain and SID consensus sequence-based peptides.
SRCR peptides 1–7 (SRCRP1–7) cover the consensus sequence of 13 SRCR domains. Peptides SID20 and SID22 cover the consensus sequence of the 12 SIDs.
| Peptide | Amino acid sequence | Amino acids | Residues of SRCR domain consensus sequence |
|---|---|---|---|
| SRCRP1 | GSESSLALRLVNGGDRC | 17 | 1–17 |
| SRCRP2 | QGRVEVLYRGSWGTVC | 16 | 18–33 |
| SRCRP3 | DDSWDTNDANVVCRQLGC | 18 | 34–51 |
| SRCRP4 | GWAMSAPGNARFGQGSGPIVLDDVRC | 26 | 52–77 |
| SRCRP5 | SGHESYLWSC | 10 | 78–87 |
| SRCRP6 | PHNGWLSHNC | 10 | 88–97 |
| SRCRP7 | GHHEDAGVICSA | 12 | 98–109 |
| SID20 | SQSQPTPSPDTWPTSHASTA | 20 | |
| SID22 | AQSWSTPRPDTLPTITLPASTV | 22 |
Figure 6. Binding of IgA to SRCR and SID peptides.
Peptides were coated at serial dilutions on to microplates and tested for IgA binding. Bound IgA was revealed using HRP-conjugated antibodies against IgA. Only SRCRP2 bound IgA. None of the other peptides, of which SRCRP1 and SRCRP6 are shown as typical examples, bound SAG.
Influence of IgA on adhesion of S. mutans
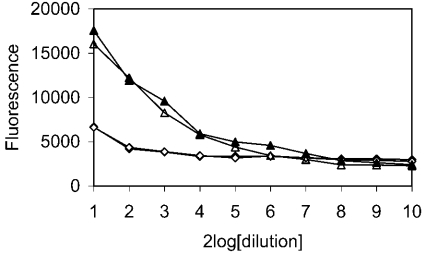
Since IgA and S. mutans have the same binding site on SAG, represented by SRCRP2, we tested whether IgA could inhibit the binding of S. mutans to SAG or SRCRP2 (Figure 7). Microplates coated with SAG or SRCRP2 peptide were incubated with IgA and tested for binding of S. mutans in an adhesion assay. The binding of S. mutans either to SAG or to peptide was not inhibited by preincubation with IgA.
Figure 7. Effect of IgA on binding of S. mutans to SAG and SRCRP2.
Microplates, coated with SAG (▴, ▵; 10 μg/ml starting concentration) or SRCRP2 (♦, ⋄; 40 μg/ml starting concentration), were incubated with PBST-Ca (▴, ♦) or with 10 μg/ml IgA (▵, ⋄) in PBST-Ca. After washing, binding of S. mutans was analysed using the fluorescent probe SYTO-13, as described in the Experimental section. The binding of S. mutans either to SAG or to SRCRP2 peptide was not inhibited by IgA. Note that the lines for SRCRP2 (♦, ⋄) overlap.
DISCUSSION
We have investigated the binding of SAG to IgA. Interaction of SAG with IgA or other proteins has been described previously [8,12,19]. In most of these studies, IgA was co-purified with SAG and the SAG–IgA complex was dissociated in further purification procedures [8,19]. In the present study, the binding of SAG to IgA was characterized further by studying the interaction between purified SAG and IgA. In addition, we demonstrated that SAG also bound to other substances, such as IgG antibodies of various species, albumin and lactoferrin.
Binding of SAG to IgA was inhibited by EDTA, suggesting the involvement of calcium ions in binding. Calcium could not be replaced by other bivalent ions (Figure 2). This suggests that calcium is part of the binding site and that the function of calcium is more than merely overcoming repulsion between negatively charged proteins. The interaction of SAG with S. mutans, SP-D or gp-120 of HIV is also calcium-dependent [1,8,11,24], suggesting some similarity in the method of binding.
As has been described for the binding of SAG to S. mutans, its binding to IgA was inhibited by amine-containing substances and high concentrations of KCl, but not by several neutral mono- or di-saccharides (Figure 3). This suggests an involvement of electrostatic interactions in binding.
The binding of SAG to IgA was destroyed by the chemical reduction of SAG (Figure 4), which suggests that the binding site is a protein epitope whose conformation is stabilized by disulphide bridges. Possible peptide-binding domains were the SRCR domains, whose tertiary structure is stabilized by four disulphide bridges. By alignment, a consensus sequence of the SRCR domains was designed that was used for the synthesis of peptides [15]. Of these peptides, only one, SRCRP2, showed affinity for IgA (Figure 6). It has also been shown that this peptide binds to S. mutans and several other bacteria [15].
Although the binding of IgA and S. mutans to SAG was mediated by the same peptide, and in both cases binding to SAG was calcium-dependent, IgA did not inhibit the binding of S. mutans to SAG or peptide SRCRP2. Possibly, the multi-ligand interaction of bacteria with SAG is much stronger than the mono-ligand interaction of IgA with SAG. This suggests that IgA does not interfere with the bacteria-binding function of SAG.
In conclusion, the SRCR domains of SAG seem to be involved in binding of this agglutinin to IgA. Binding is calcium-dependent and seems to take place according to a mechanism similar to that for the binding of S. mutans by SAG.
Acknowledgments
We thank Mrs Hella Jak and Mr Ilan Jacobs for technical assistance, and Mr Kamran Nazmi for his skilful work on peptide synthesis.
References
- 1.Ligtenberg T. J. M., Bikker F. J., Groenink J., Tornoe I., Leth-Larsen R., Veerman E. C. I., Nieuw Amerongen A. V., Holmskov U. Human salivary agglutinin binds to lung surfactant protein-D and is identical with scavenger receptor protein gp-340. Biochem. J. 2001;359:243–248. doi: 10.1042/0264-6021:3590243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mollenhauer J., Wiemann S., Scheurlen W., Korn B., Hayashi Y., Wilgenbus K. K., von Deimling A., Poustka A. DMBT1, a new member of the SRCR superfamily, on chromosome 10q25.3–26.1 is deleted in malignant brain tumours. Nat. Genet. 1997;17:32–39. doi: 10.1038/ng0997-32. [DOI] [PubMed] [Google Scholar]
- 3.Prakobphol A., Xu F., Hoang V. M., Larsson T., Bergstrom J., Johansson I., Frangsmyr L., Holmskov U., Leffler H., Nilsson C., et al. Salivary agglutinin, which binds Streptococcus mutans and Helicobacter pylori, is the lung scavenger receptor cysteine-rich protein gp-340. J. Biol. Chem. 2000;275:39860–39866. doi: 10.1074/jbc.M006928200. [DOI] [PubMed] [Google Scholar]
- 4.Kristiansen M., Graversen J. H., Jacobsen C., Sonne O., Hoffman H. J., Law S. K., Moestrup S. K. Identification of the haemoglobin scavenger receptor. Nature (London) 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 5.Holmskov U., Mollenhauer J., Madsen J., Vitved L., Gronlund J., Tornoe I., Kliem A., Reid K. B. M., Poustka A., Skjodt K. Cloning of GP-340, a putative opsonin receptor for lung surfactant protein D. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10794–10799. doi: 10.1073/pnas.96.19.10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hohenester E., Sasaki T., Timpl R. Crystal structure of a scavenger receptor cysteine-rich domain sheds light on an ancient superfamily. Nat. Struct. Biol. 1999;6:228–232. doi: 10.1038/6669. [DOI] [PubMed] [Google Scholar]
- 7.Resnick D., Pearson A., Krieger M. The SRCR superfamily: a family reminiscent of the Ig superfamily. Trends Biochem. Sci. 1994;19:5–8. doi: 10.1016/0968-0004(94)90165-1. [DOI] [PubMed] [Google Scholar]
- 8.Holmskov U., Lawson P., Teisner B., Tornoe I., Willis A. C., Morgan C., Koch C., Reid K. B. M. Isolation and characterization of a new member of the scavenger receptor superfamily, glycoprotein-340 (GP340), as a lung surfactant protein D binding molecule. J. Biol. Chem. 1997;272:13743–13749. doi: 10.1074/jbc.272.21.13743. [DOI] [PubMed] [Google Scholar]
- 9.Hartshorn K. L., White M. R., Mogues T., Ligtenberg T., Crouch E., Holmskov U. Lung and salivary scavenger receptor glycoprotein-340 contribute to the host defense against influenza A viruses. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;285:L1066–L1076. doi: 10.1152/ajplung.00057.2003. [DOI] [PubMed] [Google Scholar]
- 10.Holmskov U., Malhotra R., Sim R., Jensenius J. C. Collectins: collagenous C-type lectins of the innate immune defense system. Immunol. Today. 1994;15:67–73. doi: 10.1016/0167-5699(94)90136-8. [DOI] [PubMed] [Google Scholar]
- 11.Ericson T., Rundegren J. Characterization of a salivary agglutinin reacting with a serotype c strain of Streptococcus mutans. Eur. J. Biochem. 1983;133:255–261. doi: 10.1111/j.1432-1033.1983.tb07456.x. [DOI] [PubMed] [Google Scholar]
- 12.Oho T., Yu H., Yamashita Y., Koga T. Binding of salivary glycoprotein-secretory immunoglobulin a complex to the surface protein antigen of Streptococcus mutans. Infect. Immun. 1998;66:115–121. doi: 10.1128/iai.66.1.115-121.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ligtenberg A. J. M., Veerman E. C. I., Nieuw Amerongen A. V. A role for Lewis a antigens on salivary agglutinin in binding to Streptococcus mutans. Antonie van Leeuwenhoek. 2000;77:21–30. doi: 10.1023/a:1002054810170. [DOI] [PubMed] [Google Scholar]
- 14.Lamont R. J., Rosan B. Adherence of mutans streptococci to other oral bacteria. Infect. Immun. 1990;58:1738–1743. doi: 10.1128/iai.58.6.1738-1743.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bikker F. J., Ligtenberg A. J. M., Nazmi K., Veerman E. C. I., van't Hof W., Bolscher J. G. M., Poustka A., Nieuw Amerongen A. V., Mollenhauer J. Identification of the bacteria-binding peptide domain on salivary agglutinin (gp-340/DMBT1), a member of the scavenger receptor cysteine-rich superfamily. J. Biol. Chem. 2002;277:32109–32115. doi: 10.1074/jbc.M203788200. [DOI] [PubMed] [Google Scholar]
- 16.Mitoma M., Oho T., Shimazaki Y., Koga T. Inhibitory effect of bovine milk lactoferrin on the interaction between a streptococcal surface protein antigen and human salivary agglutinin. J. Biol. Chem. 2001;276:18060–18065. doi: 10.1074/jbc.M101459200. [DOI] [PubMed] [Google Scholar]
- 17.Boackle R. J., Connor M. H., Vesely J. High molecular weight nonimmunoglobulin salivary agglutinins (NIA) bind C1Q globular heads and have the potential to activate the first complement component. Mol. Immunol. 1993;30:309–319. doi: 10.1016/0161-5890(93)90059-k. [DOI] [PubMed] [Google Scholar]
- 18.Thornton D. J., Davies J. R., Kirkham S., Gautrey A., Khan N., Richardson P. S., Sheehan J. K. Identification of a nonmucin glycoprotein (gp-340) from a purified respiratory mucin preparation: evidence for an association involving the MUC5B mucin. Glycobiology. 2001;11:969–977. doi: 10.1093/glycob/11.11.969. [DOI] [PubMed] [Google Scholar]
- 19.Rundegren J., Arnold R. R. Differentiation and interaction of secretory immunoglobulin A and a calcium-dependent parotid agglutinin for several bacterial strains. Infect. Immun. 1987;55:288–292. doi: 10.1128/iai.55.2.288-292.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takano K., Bogert M., Malamud D., Lally E., Hand A. R. Differential distribution of salivary agglutinin and amylase in the Golgi apparatus and secretory granules of human salivary gland acinar cells. Anat. Rec. 1991;230:307–318. doi: 10.1002/ar.1092300303. [DOI] [PubMed] [Google Scholar]
- 21.Veerman E. C. I., Valentijn-Benz M., van den Keijbus P. A. M., Rathman W. M., Sheehan J. K., Nieuw Amerongen A. V. Immunochemical analysis of high-molecular-weight human salivary mucins (Mg1) using monoclonal-antibodies. Arch. Oral Biol. 1991;36:923–932. doi: 10.1016/0003-9969(91)90125-e. [DOI] [PubMed] [Google Scholar]
- 22.Nagle G. T., Jong-Brink M., Painter S. D., Li K. W. Structure, localization and potential role of a novel molluscan trypsin inhibitor in Lymnaea. Eur. J. Biochem. 2001;268:1213–1221. doi: 10.1046/j.1432-1327.2001.01972.x. [DOI] [PubMed] [Google Scholar]
- 23.Bosch J. A., Veerman E. C. I., Turkenburg M., Hartog K., Bolscher J. G. M., Nieuw Amerongen A. V. A rapid solid-phase fluorimetric assay for measuring bacterial adherence, using DNA-binding stains. J. Microbiol. Methods. 2003;53:51–56. doi: 10.1016/s0167-7012(02)00220-8. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z., Van Ryk D., Davis C., Abrams W. R., Chaiken I., Magnani J., Malamud D. Salivary agglutinin inhibits HIV type 1 infectivity through interaction with viral glycoprotein 120. AIDS Res. Hum. Retroviruses. 2003;19:201–209. doi: 10.1089/088922203763315704. [DOI] [PubMed] [Google Scholar]