Abstract
The bacterial ribosome comprises 30 S and 50 S ribonucleoprotein subunits, contains a number of binding sites for known antibiotics and is an attractive target for selection of novel antibacterial agents. On the 30 S subunit, for example, the A site (aminoacyl site) close to the 3′-end of 16 S rRNA is highly important in the decoding process. Binding by some aminoglycoside antibiotics to the A site leads to erroneous protein synthesis and is lethal for bacteria. We targeted the A site on purified 30 S ribosomal subunits from Escherichia coli with a set of overlapping, complementary OMe (2′-O-methyl) 10-mer oligoribonucleotides. An equilibrium dialysis technique was applied to measure dissociation constants of these oligonucleotides. We show that there is a single high-affinity region, spanning from A1493 to C1510 (Kd, 29–130 nM), flanked by two lower-affinity regions, within a span from U1485 to G1516 (Kd, 310–4300 nM). Unexpectedly, addition of the aminoglycoside antibiotic paromomycin (but not hygromycin B) caused a dose-dependent increase of up to 7.5-fold in the binding of the highest affinity 10-mer 1493 to 30 S subunits. Oligonucleotides containing residues complementary to A1492 and/or A1493 showed particularly marked stimulation of binding by paromomycin. The results are consistent with high-resolution structures of antibiotic binding to the A site and with greater accessibility of residues of A1492 and A1493 upon paromomycin binding. 10-mer 1493 binding is thus a probe of the conformational switch to the ‘closed’ conformation triggered by paromomycin that is implicated in the discrimination by 30 S subunits of cognate from non-cognate tRNA and the translational misreading caused by paromomycin. Finally, we show that OMe oligonucleotides targeted to the A site are moderately good inhibitors of in vitro translation and that there is a limited correlation of inhibition activity with binding strength to the A site.
Keywords: aminoglycoside, antibiotic, equilibrium dialysis, oligonucleotide, ribosome, translation inhibition
Abbreviations: A site, aminoacyl site; E site, exit site; OMe, 2′-O-methyl; PNA, peptide nucleic acid; P site, peptidyl site; TAR, trans-activation-responsive region
INTRODUCTION
Ribosomes are large ribonucleoprotein complexes that are the sites for the synthesis of cellular proteins (reviewed in [1]). Their molecular mechanisms are crucially dependent on their component rRNAs, which in prokaryotes are known as 5 S, 16 S and 23 S. The core of the interface between the two ribosomal subunits (known as 30 S and 50 S) is dominated by the rRNAs and these form the basis for the three binding sites for tRNA: A (aminoacyl), P (peptidyl) and E (exit). In addition, during translation, mRNA becomes anchored to the ribosome by binding of its Shine–Dalgarno sequence to the 3′-end of the 16 S rRNA. Furthermore, rRNAs are central to the translation process, for example in mediating the accommodation of cognate tRNA, in catalysing the formation of peptide bonds, in promoting the translocation of peptidyl and deacylated tRNA between the A site, the P site and the E site, and in catalysing the hydrolysis and release of the peptide chain from tRNA.
Interference with the function of the rRNAs results in inhibition of the translation process and leads to bacterial growth-inhibition or lethality [2]. Indeed, most inhibitors of translation exert their activity by binding to specific sites within rRNAs, rather than by binding to ribosomal proteins as had been thought until quite recently [3]. Aminoglycoside antibiotics, such as paromomycin, bind to the internal loop of helix 44 of the 16 S rRNA and give rise to an increase in the level of incorporation of incorrect amino acids [4]. Other antibiotics have been found to bind 23 S rRNA at the peptidyl transferase centre and inhibit protein synthesis, either directly by preventing peptide-bond formation (chloramphenicol and clindamycin) or indirectly by causing dissociation of peptidyl tRNA (macrolides) [5–7]. Oxazolidinone antibiotics inhibit the initiation process, probably by competing with the fMet (formylmethionine) tRNA for the binding to the peptidyl transferase centre of the 23 S rRNA [8,9], whereas tetracycline binds 16 S rRNA and inhibits decoding [10].
Since rRNA binding is a prominent feature of the activity of many effective antibacterial therapeutics, the screening of large libraries of compounds for rRNA binding, in parallel with antibacterial screening, might produce novel leads. Oligodeoxyribonucleotides and their analogues are reagents with well-known sequence-specific RNA-binding properties and which may be readily derivatized with fluorescent or radioactive labels. They have been used to probe the secondary structures of rRNAs on the surface of bacterial ribosomes [11], and as inhibitors of translation in vitro; for example, by blocking the 3′-end of 16 S rRNA [12,13] or by binding to an internal rRNA loop such as the α-sarcin loop [14,15]. However, the binding constants of oligodeoxyribonucleotides to RNA are poor, they are unstable to cellular nucleases and also form substrates for RNase H upon binding to the rRNA. A number of ribosome photochemical cross-linking studies have been carried out using oligoribonucleotides [16,17] as mRNA analogues or their OMe (2′-O-methyl) analogues [18] derivatized with photolabile groups as antisense agents. Such reagents were found to be more effective as ribosome-targeting agents than previously used oligodeoxyribonucleotide derivatives [19] due to their higher binding to RNA. OMe oligoribonucleotides have the added advantage of considerable resistance to cellular nucleases, lack of induction of RNase H, as well as good strand invasion properties on structured RNA such as the HIV TAR (trans-activation-responsive region) element [20]. We therefore supposed that OMe oligonucleotides might be potential reagents for use in drug displacement assays on known or new potential rRNA drug-binding sites.
To our knowledge, no systematic or quantitative probing of the binding of OMe oligoribonucleotides to RNA sites on the bacterial ribosome has been reported, nor have such reagents been studied in relation to the rRNA-binding of known antibiotics. To this end, we designed a novel equilibrium dialysis assay to measure accurately the affinities of such RNA-specific ligands to the native 30 S ribosomal subunit. We tested a set of OMe oligoribonucleotides complementary to the ribosomal A site RNA on the small 30 S subunit, a known antibiotic-binding site that is important in translation, and defined the boundaries of high-affinity oligonucleotide binding as being beyond those residues structurally implicated in the mRNA decoding process [21,22]. Unexpectedly, addition of the A-site-selective antibiotic paromomycin, rather than causing displacement of the oligonucleotide, markedly raised the affinity of certain OMe oligoribonucleotides that bound well to the A site. The ability to confer increased affinity correlated with the ability, or not, of the antibiotic to alter the conformation of the A site. We also found that OMe oligonucleotides targeted to the A site were able to inhibit in vitro translation.
MATERIALS AND METHODS
30 S ribosomal subunits
30 S subunits were purified essentially as described in [23]. In brief, RNase-deficient Escherichia coli strain MRE600 was grown in 2×TY medium (16 g of Bacto-tryptone, 10 g of Bacto-yeast extract, 5 g of NaCl and double-distilled water to 1 litre), harvested at a concentration of 0.5 A650 unit·ml−1, and broken open by passage through a French press at 12000 p.s.i. (1 p.s.i.=6.9 kPa). Following clarification and passage through a sucrose cushion (1.1 mM), sucrose/salt-washed ribosomes were resuspended in a low-magnesium buffer (1.1 mM Mg2+) and centrifuged through a sucrose gradient (10–30%; 28000 rev./min in a Beckman SW28 Ti rotor for 16 h at 4 °C). After harvesting gradients, fractions containing the 30 S subunit were pooled, dialysed against a high-magnesium buffer (10 mM Mg2+) and stored at −80 °C. Composite gel electrophoresis (0.5% agarose and 3% acrylamide) was used to estimate the purity of the preparation. Two 30 S bands were seen, which correspond to with and without protein S1, migrating at 500 and 600 bp of the DNA ladder.
OMe oligoribonucleotides
OMe oligomers of various length (6-, 8-, 10- and 12-mers) were designed complementary to both C- and G-rich strands of double-stranded A site RNA (E. coli) with an overlap of x−2, where x is the length of the oligomer. Random as well as mismatched oligomers were also designed. The OMe oligomers were synthesized by Dharmacon Research (Lafayette, CO, U.S.A.) or on an Applied Biosystems 394 DNA/RNA Synthesizer using OMe amidites obtained from Transgenomics (Scotland) and 32P-labelled by treatment with [γ-32P]ATP and T4 polynucleotide kinase (approx. 5 mCi/mmol; New England BioLabs, Beverly, MA, U.S.A.).
30 S–oligoribonucleotide dissociation constants by equilibrium dialysis
This technique enables an accurate measurement of the intrinsic affinity of a reversible bimolecular interaction [24], provided that the molecular masses of the two interacting ligands are different enough to allow only one of them to pass through a membrane connecting two equal-volume chambers. Since the mass of the 30 S subunit is 2.5 MDa and that of the OMe oligomer is only a few kDa, these two represent an ideal pair for such an analysis. A semi-permeable membrane between the 30 S-free and the 30 S-containing chambers enables oligomer equilibration, while retaining the 30 S subunit within a single chamber.
Two-chamber equilibrium dialysis devices were purchased from the Nest Group (Southborough, MA, U.S.A.). These consist of Teflon blocks into which two rigid cylinders of exactly 100 μl capacity are cut and in between which a cellulose ester membrane with molecular mass cut-off of 100 kDa (Spectrum Laboratories, Rancho Dominguez, CA, U.S.A.) is held rigidly. The diameter of the membrane is very small (3 mm) such that the membrane area is small compared with the total size of the chambers. The design is such that significant volume alteration between the two chambers during dialysis is prevented. This allowed complete equilibration of the oligomers within 8 h at 4 °C on a shaker, while the 30 S subunit was 100% retained in chamber B (as measured by spectrophotometry). The membrane was cut to fit the slot in the device, washed with double-distilled water to remove sodium azide and soaked in the binding buffer: 50 mM Tris/HCl (pH 7.4 at 4 °C), 10 mM MgCl2, 30 mM NH4Cl and 100 mM KCl. After membrane positioning, one of the chambers (A) was filled to capacity with the buffer containing the 32P-labelled OMe oligomer only, while B was filled to capacity with the 32P-labelled OMe oligomer plus the 30 S subunit. After overnight equilibration (16 h at 4 °C; ample for the oligomer equilibration and accompanied by only insignificant degradation of 30 S; results not shown), the amounts of radioactivity on each side of the membrane were measured by taking 20 μl aliquots from the chambers and radioactivity was determined by use of a liquid-scintillation counter (LS6500; Beckman Instruments). For each oligonucleotide, experiments were carried out in duplicate and then repeated (total of four determinations). Starting concentrations were 0.05 μM oligomer in each chamber and 1.6 μM 30 S subunit in chamber B (see the Results section).
To measure the dissociation constant (Kd) of the interaction between these two ligands, let 30 S=R, free OMe oligomer=O, and the complex between them=RO. The distribution of the radioactivity between the chambers matches exactly the distribution of the oligomer. The Kd can be calculated using the mass-action-law equation:
 |
where square brackets represent molar concentrations.
In the two (A and B) chamber setting, as a result of the 30 S–oligomer interaction in one of the chambers (e.g. B), the distribution of the oligomer in chamber B is given by:
 |
Since at equilibrium [O]B=[O]A (measured by determining the radioactivity in chamber A), then
 |
[O]totalB is calculated by measurement of the UV absorption at 260 nm of an aliquot of the [32P]oligonucleotide stock solution from chamber B and calculation of the molar concentration (from the summation of the known absorption coefficients of the individual nucleosides in the oligonucleotide). Rtotal is measured also by spectrophotometry (1 A260 unit is 69.0 pmol [23]) and then [R] is determined by [R]total−[RO].
Double-filter binding assay
A 0.45-μm-pore-size nitrocellulose membrane (PROTRAN, Schleicher and Schuell GmbH, Dassel, Germany) was used that adsorbs the 30 S subunits, but not the unbound oligomers. In addition, a positively charged nylon hybridization transfer membrane (Hybond-N+; Amersham Biosciences, Little Chalfont, Bucks., U.K.) was used to trap the unbound oligomers, which served as a method of radioactivity normalization for each slot [25]. The 30 S saturation binding curves were obtained by filtering through the double membrane (using slot blot MINIFOLDII apparatus; Schleicher and Schuell) mixtures of 30 S subunit and 32P-labelled oligomer, and obtaining autoradiographs of the membranes (storage phosphor screen autoradiography; Phosphor-Imager, Molecular Dynamics, Sunnyvale, CA, U.S.A.), followed by their analysis using ImageQuant software (Molecular Dynamics), expressed as an integrated intensity of the pixels.
A second-order transformation of the mass-action-law equation is normally used to calculate apparent dissociation constants (Kd), which assumes that dissociation of the ligands is slow [26]. In filter binding of oligonucleotides with 50 S ribosomal subunits, others have fitted their data to a two-population model to allow for non-specific binding by oligomers at high [oligomer]/[30 S] ratios [19]. We found that our binding data, where there is an observed plateau, fit best by transforming the mass-action-law equation into a quadratic function where the apparent Kd may be calculated by use of a simple in-house computer program. In calculations, we have assumed that the plateau point corresponds to 98.5% saturation of the 30 S subunit.
Competition with antibiotics
For these experiments, optimal competition stoichiometry has been used where the 30 S subunit (0.4 μM) was bound to near saturation by an oligomer. Antibiotics (paromomycin and hygromycin B) were tested in concentrations of up to 10 mM. Anisomycin (an antibiotic that acts on eukaryotic ribosomes only) and chloramphenicol (that acts on the 23 S RNA P site) were used as negative controls.
In vitro translation-inhibition assay
In vitro translation inhibition by oligonucleotides was quantified in a coupled transcription/translation assay [27], by use of E. coli S30 extract with the pBestLuc plasmid (Promega) as template. Translation reactions (10 μl) were incubated for 60 min at 37 °C, and translation was quantified by measuring the luminescence in a Victor II plate reader (PerkinElmer) after addition of 50 μl of luciferase reagent (Promega). The inhibition constant (IC50) was obtained from fitting concentration–response curves to the data of at least three independent experiments as the concentration of half-maximal inhibition.
RESULTS
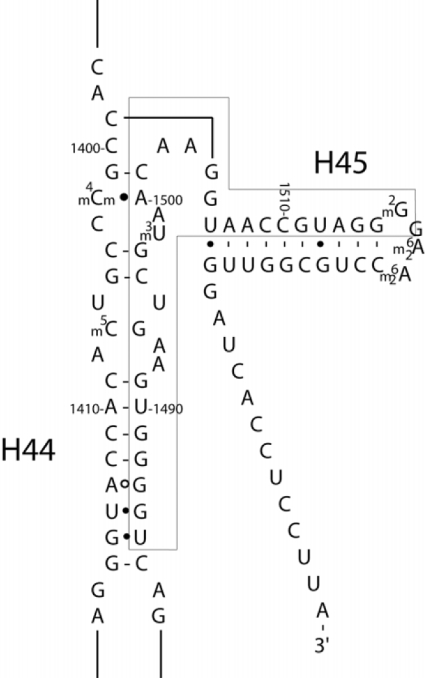
We chose the ribosomal A site (decoding site) within helix 44 of the 16 S rRNA as the model system for targeting OMe oligomers. First, in structural models, the A site is seen to be exposed on the surface of the small 30 S subunit [28]. Secondly, unpaired base residues A1492 and A1493 within the A site are crucial to the decoding process and to codon/anticodon recognition [4], the selection of tRNA by the ribosome [29], as well as to the binding of initiation factor 1 [30]. Thirdly, the A site is well known to be the site of interaction of aminoglycoside antibiotics [4,31], which can therefore provide tools for assay development and target validation. Figure 1 shows the sequence and secondary structure of the A site RNA. Surprisingly, to our knowledge, oligonucleotides have not been targeted to this site previously. In our preliminary tests, OMe oligomers targeted to the C-rich strand (residues C1399 to C1412) showed poor binding, presumably because the oligomers contained a stretch of G-residues (5′-GGGCGG-3′) that caused them to aggregate (results not shown). In contrast, oligomers targeted against the G-rich strand of the A-site RNA showed promising binding characteristics and therefore were used in binding studies.
Figure 1. Secondary structure of a section of the 3′-end of 16 S rRNA of E. coli showing the A site and the nucleotide numbers.
Residues within the box are targeted by the series of OMe oligonucleotides used in the present study (see Figure 3). Helix numbers (H44 and H45) are marked. Modifications: m4Cm, N-4-methyl-2′-O-methylC; m5C, 5-methylC; m3U, N-3-methylU; m2G, N-2-methylG; m26A, N-6-dimethylA.
Saturation filter-binding experiments
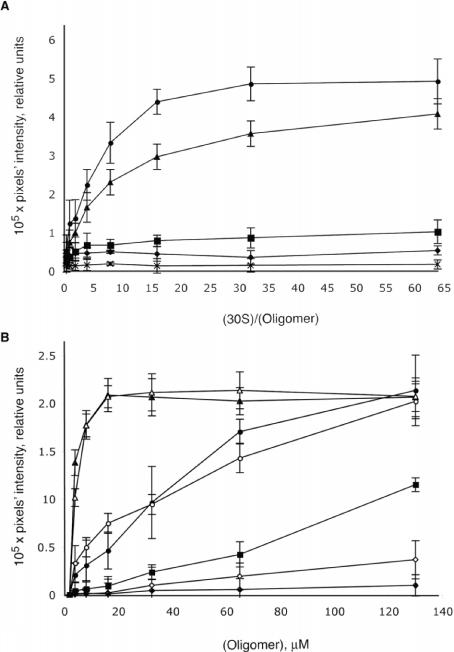
We have utilized previously a well-known technique to assess binding strengths of proteins to RNA [32,33] or oligonucleotides to RNA [20], which is the measurement of the amount of complex retention on nitrocellulose filters. This technique has been applied to binding of photolabile OMe oligonucleotides to 50 S ribosomal subunits [18,19], but not previously to 30 S subunits. We therefore assessed the binding strengths of the oligomers using the filter-binding technique. First, the OMe oligonucleotides were held in fixed concentration (5 nM) and the concentration of 30 S subunit increased. OMe oligomers of different length, each targeted such that their 3′-termini are complementary to position 1487 of 16 S RNA, were found to bind to the 30 S subunit in rank order of size as follows: 12-mer>10-mer≫8-mer>6-mer>random 10-mer (Figure 2A). In a second experiment, 10-mers were selected for a preliminary scan of the A-site region. Sequences of 10-mers are shown in Figure 3. Here, the 30 S subunit was used at a fixed concentration of 0.76 μM, while the concentration of oligomer was varied. In most cases, it was not possible to obtain plateaux for the curves within the range of the oligomer concentrations used (up to 130 μM). Only two 10-mers, 1493 and 1495, bound to 30 S subunits strongly enough (as determined by their retention on the filters) for their apparent affinities to be estimated using this experimental approach (Figure 2B). Measured apparent Kd values for these two 10-mers were approx. 330 nM. Thus filter-binding data are not sufficiently amenable to determination of accurate Kd values over a wide enough range of oligonucleotide lengths and affinities, although it may still be valuable for semi-quantitative scans.
Figure 2. Filter-binding assay.
(A) Effect of the OMe 1487 oligomer length on the binding to 30 S ribosomal subunits. All oligomers have their 3′-ends complementary to residue 1487 in 16 S RNA. Complementary 12-mer (•), 10-mer (▴), 8-mer (▪), 6-mer (♦) and a random 10-mer (×). (B) Binding curves of the OMe 10-mers 1485 (▪), 1489 (○), 1493 (▵), 1495 (▴), 1501 (•), 1505 (⋄), 1507 (♦). For details of oligomers used, see Figure 3. Results are means±S.D. for four replicate experiments.
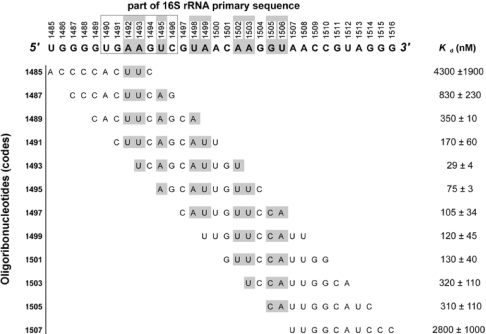
Figure 3. Scan of binding strength through use of equilibrium dialysis.
Across the top: the targeted sequence of the 16 S rRNA. Grey-shaded nucleotides are those not seen to be in hydrogen-bonded pairings according to the crystal structure of the 30 S subunit [29]. From left to right: the code number of the oligomer, its sequence, and its measured Kd (±S.D.; nM) for binding to the 30 S subunit.
Kd measurements of the binding of OMe oligoribonucleotides to the 30 S subunit by equilibrium dialysis
Consequently, we designed and applied an equilibrium-dialysis-based method to measure the binding strength (see the Materials and methods section). An advantage of this technique is that there is a true equilibrium established and that therefore a true dissociation constant (Kd) can be measured. A starting concentration of 0.05 μM oligomer in each chamber and 1.6 μM 30 S subunit in chamber B was used in these equilibrium dialysis experiments, which represents an excess of 30 S ribosomal subunits over oligomer (i.e. sub-saturating for the 30 S subunit). This is the reverse situation compared with filter-binding experiments of Figure 2(B). Oligomers of different size bound the 30 S in the same rank order as in Figure 2(A), but with different relative strengths: 12-mer=10-mer>8-mer≫6-mer≥random 10-mer (results not shown). From these preliminary studies, it was clear that 10-mers are sufficient to show strong binding to the A site.
We therefore carried out a binding scan of the A site on 30 S subunits by use of 12 overlapping 10-mers, complementary to a span from U1485 to G1516 on 16 S RNA, and calculated their Kd values (Figure 3). The Kd values varied by two orders of magnitude, from as low as 29 nM (oligomer 1493) to as high as 4300 nM (oligomer 1485), and there is a clear U-shaped pattern of affinity distribution, with a single region where 10-mers bind with high affinity flanked by lower-affinity regions. A random OMe 10-mer (3′-AUUGCAACAC-5′) showed very poor binding (Kd of 8.42 μM). Although the buffer conditions are not identical, it is of interest to note that the best 10-mer 1493 has a Kd value (29 nM) similar to that previously measured by a direct equilibrium fluorescence method for a 12-mer OMe oligonucleotide binding to a stem-loop TAR RNA in a region with apical loop, double helix and internal bulge (approx. 20 nM) [34]. This suggests that the nucleotides bound within the A site may not be dissimilar in accessibility to those found within other structured RNAs and thus not significantly hindered by the overall 30 S ribosomal subunit nucleoprotein assembly. Another interesting observation is that the measured Kd falls, with statistical significance, as the number of expected contacts increases of the oligomer with nucleotides on the targeted strand of the 16 S RNA secondary structure that are not in hydrogen-bonded pairs within helix 44, as seen in the crystal structure [29] (shaded grey in Figure 3) (r=−0.65; P<0.05). This is suggestive of the role that such unpaired nucleotides play in facilitating the binding of the oligomers to the 16 S RNA within the 30 S subunit.
Binding specificity
In addition to the non-complementary, random-sequence oligomer, which showed very poor binding (see above), we also assessed the binding affinity of partially mismatched oligomers. Either the third or the eighth nucleotide, or both, were substituted to create single- or double-mismatched oligomers. All mismatched variants bound less tightly than the correct complementary oligomers, but the relative effect of mismatches on binding affinity varied greatly. For example, whereas the position three A to C mismatch in the oligomer 1491 led to an approx. 5-fold fall in affinity, the position eight U to C mismatch led to an approx. 50-fold fall. When both mismatches were present, the fall was approx. 60-fold. In another example (oligomer 1493), whereas the position three U to C mismatch led to a 18.5-fold fall in affinity and the position eight A to C mismatch led to an 8-fold fall, the introduction of both mismatches led to only a 4.6-fold fall. The variation in magnitude of the fall may reflect contextual differences of the mismatches in that alternative base-pairings may be possible in some cases that may partially mitigate the affinity fall. Taken overall, the results are strongly consistent with correct targeting of the ribosomal A-site by the OMe oligonucleotides.
Competition of the oligomers with antibiotics for the binding to the 30 S subunit
We then tested whether the oligomers that bind to the A site of the 16 S RNA could compete with antibiotics paromomycin and hygromycin B, both known to bind the 16 S RNA A site [4,35]. In these experiments, a two-to-one ratio between oligomer and the 30 S subunit was used (0.8 μM and 0.4 μM respectively), which was a compromise between the likely optimal conditions for competition and the ability to detect the oligomer redistribution in dialysis. For example under these conditions, 75% of 30 S subunits (0.3 μM) became saturated by 1493 10-mer, and correspondingly the concentration of free oligomer falls by a significant margin (from 0.8 to 0.5 μM). Note that the apparent Kd of 10-mer 1493 under these conditions of oligonucleotide excess is 0.17 μM, somewhat higher than under the conditions of ribosome excess, which is consistent with previous reports of biphasic binding under oligonucleotide excess [19].
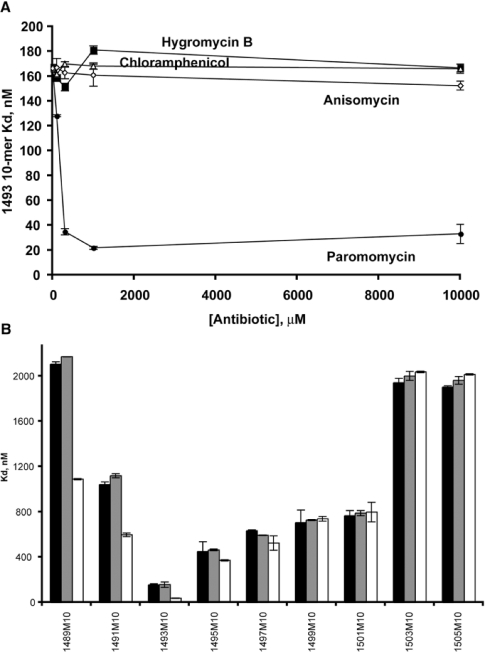
None of the antibiotics tested was found to inhibit the oligomer binding, but, unexpectedly, paromomycin addition brought about a significant (<7.5-fold) increase of the 1493 10-mer binding affinity in a dose-dependent manner (Figure 4A). The effect of increase in oligomer-binding affinity was seen also for 10-mers 1489 and 1491 (although less dramatic). Increases in affinity were seen for 1495 and 1497, but not for oligomers 1499–1505 (Figure 4B). Possible structural reasons for this enhancement are discussed below. In contrast, another A-site-specific antibiotic, hygromycin B, had no significant effect on any of the 10-mer binding affinities at any concentration. As expected, addition of neither anisomycin nor chloramphenicol, which are not known to bind to the A-site, affected the binding affinity.
Figure 4. Competition with antibiotics.
(A) Dose-dependency on the Kd values of the OMe 10-mer 1493. Paromomycin (•), hygromycin B (▪), anisomycin (⋄), chloramphenicol (▵). (B) Effects of 300 μM hygromycin B (grey bars) or paromomycin (white bars) on the Kd values of the 10-mers. Black bars, no antibiotic. Results are means±S.D. for four replicate experiments.
In vitro translation inhibition
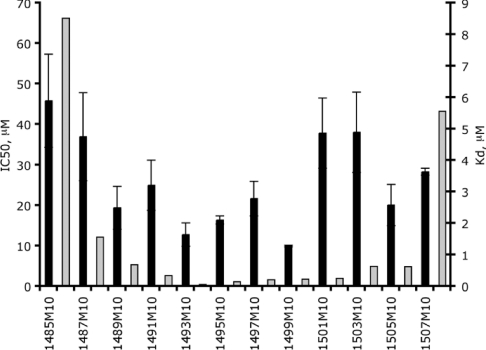
The set of 10-mer OMe oligonucleotides was also assayed for inhibition of in vitro translation in an E. coli lysate assay. All the OMe oligonucleotides showed some level of activity in the in vitro translation-inhibition assay, and there is a limited, but statistically significant, correlation (r=0.58; P<0.05) between the IC50 values obtained and the Kd values of the oligomers binding to the 30 S subunit (Figure 5). For example, the best 10-mer in binding (1493) was found to be one of the two best 10-mers in in vitro translation inhibition. Similarly, 10-mer 1485 was the weakest in both binding and in translation inhibition. A stronger correlation would not be anticipated, since in vitro translation involves numerous and complex steps, and an oligomer could potentially act at a number of levels (e.g. as an mRNA competitor or as a tRNA-binding inhibitor) in addition to any role in A-site-function inhibition. However, the best two OMe 10-mers (1493 and 1499) are as strong translation inhibitors in vitro (IC50 of approx. 10 μM) as many known antibiotics that target the small ribosomal subunit, including those used in the present study, paromomycin (IC50 of 10 μM) and hygromycin B (IC50 of 50 μM) [36].
Figure 5. In vitro translation inhibition (black bars; left y-axis) and binding strength (grey bars; right y-axis) of the oligomers.
Results are means±S.D. for four replicate experiments.
DISCUSSION
Filter binding has been the primary method used to date for assessing the binding of oligonucleotides to 50 S ribosomal subunits [18,19]. We found that where excess oligonucleotide was used (Figure 2B), some of the 10-mer oligonucleotides did not reach a plateau in the graph where the level of filter retention was titrated against a fixed concentration of 30 S subunits, particularly for weaker interactions. A second problem is the possibility of additional binding to secondary lower-affinity binding sites when excesses of oligonucleotide are used over 30 S subunits (Figure 2B). Similar non-plateau binding curves have been reported in a previous study involving the use of excess photolabelled OMe oligonucleotides binding and cross-linking to the peptidyl transferase centre of 50 S subunits [18]. Thus filter binding appears to be inadequate for measurement of dissociation constants over a wide affinity range. We also developed an assay for binding that made use of mobility shift of radiolabelled OMe oligonucleotides when bound to 30 S ribosomal subunits and when subjected to electrophoresis on composite agarose/polyacrylamide gels. However, once again consistent quantitative data over a range of oligonucleotide affinities could not be obtained (results not shown).
We therefore established an equilibrium-binding assay for the determination of dissociation constants of OMe oligonucleotides from 30 S ribosomal subunits, where the 16 S rRNA A site was used as the target model system. Unlike assays that involve filter binding or other indirect methods, the data produced give true Kd values in solution. The values are not affected by parameters such as dissociation occurring within the time taken to carry out gel-based assays or the requirement to bind to a membrane in filter-binding assays. We found that this dialysis technique gives reliable and reproducible binding data over a wide range of Kd values (nanomolar to micromolar). Furthermore, the use of an excess of ribosomal subunits over oligonucleotides ensures that the oligonucleotide is used under conditions that reflect binding only at the highest affinity site on the 30 S subunit. It should be noted that we used a fixed concentration (1600 nM) of subunits to determine Kd values. For the tightest-binding oligomers (e.g. 1493), almost all is bound to the 30 S subunit under these conditions. A more accurate Kd value may be obtained by varying the 30 S excess and by plotting the fraction bound against concentration of 30 S. The value we have obtained is therefore maximal for the tightest-binding oligomers, and binding may possibly be even a little stronger in these cases.
To our knowledge, an equilibrium-binding technique has not been applied to binding of oligonucleotides or other assayable molecules to 30 S subunits, and the technique should in principle be transferrable to 50 S subunits or even whole 70 S ribosomes. Mismatched-oligonucleotide-binding experiments were fully consistent with correct targeting of the A site. We have not been successful to date, however, in attempts to map the site of UV cross-linking of psoralen-labelled OMe 1493 to 30 S subunits by reverse transcription, probably because of difficulties in priming reverse transcription at the compact 3′-end of the 16 S RNA and the close proximity of the A site to the 3′-end (results not shown).
We chose to target 30 S subunits specifically because of the availability of high-resolution crystal structures of these subunits, not only as native structures, but also in complex with a range of antibiotics as well as tRNA and translation proteins. For example, structures are known of several different aminoglycosides bound to 30 S subunits [4,35]. It was our initial expectation that OMe oligonucleotides might compete with such antibiotics for binding to the 30 S subunit and that this might form the basis for a potential displacement assay for target-specific drug testing, which might be then transferable to other less well-established RNA targets on the ribosome in order to search for drugs that might bind to novel ribosomal targets. In contrast, the binding of OMe oligonucleotides was not reduced by the addition of excesses of aminoglycoside antibiotics, paromomycin and hygromycin B, that target the 30 S ribosomal A site. No effect of hygromycin B was seen on the binding of 10-mer oligonucleotides, but paromomycin addition gave rise to a substantial increase in the binding of 10-mers 1489, 1491 and, particularly, the most strongly interacting 10-mer 1493. Slight increases were seen for 1495 and 1497, but not for other 10-mers targeted further along the strand.
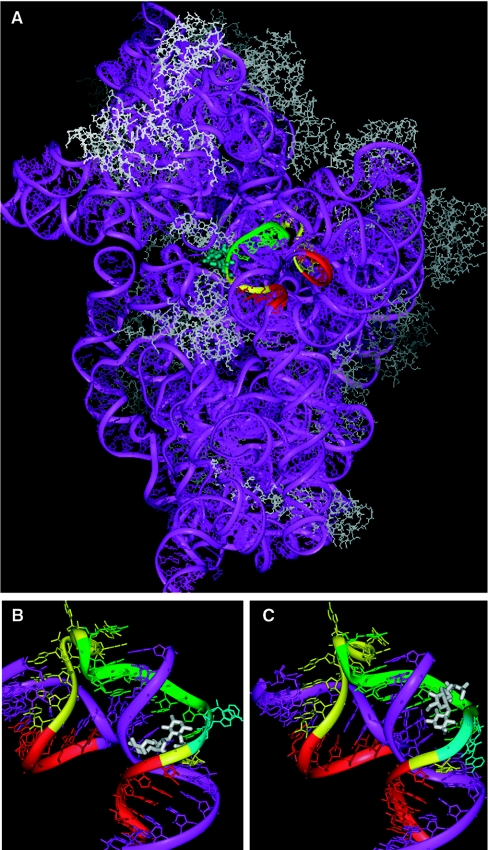
We suggest that the enhancement of binding in the case of paromomycin, and the failure to enhance in the case of hygromycin B, are a consequence of the different ways the two antibiotics interact with the A site. First, we noticed from inspection of the sequence of the 16 S RNA (Figure 1) that the three 10-mers where enhancement of binding is particularly pronounced in the presence of paromomycin include one or two U residues complementary to A residues 1492 and 1493. Secondly, the crystal structures of paromomycin complexes with 30 S ribosomes [4,35], as well as that of the antibiotic with an isolated A site RNA [37], show that the drug is inserted into the major groove of the helix and distorts the RNA structure significantly, such that the two A residues 1492 and 1493 are extruded from the RNA helix and are made available for interaction externally (Figures 6A and 6B). In a similar manner, the repositioning of residues A1492 and A1493 outside of helix 44 has been suggested as the explanation for an increase in binding affinity by a factor of 15 in the presence of paromomycin of near cognate tRNA to 30 S ribosomal subunits [29]. In the absence of the antibiotic, the conformational change is seen only when cognate tRNA binds. The stabilization of the alternative ‘closed’ conformation of the 30 S subunit is therefore required for tRNA selection and rationalizes the strong effect of paromomycin on translational fidelity [36]. The slight enhancement of binding of 10-mers 1495 and 1497, but not 1499 onwards, probably reflects a limited regional effect of the drug in distorting the RNA helix, and thus slightly enhancing accessibility, but the large binding enhancements seem to be more related to the particular residues A1492 and A1493.
Figure 6. Binding of antibiotics to the bacterial ribosomal 30 S subunit.
(A) Representation of the ribosomal 30 S subunit from Thermus thermophilus when bound by ‘near-cognate’ tRNA analogue and paromomycin {[29]; Protein Data Bank (PDB) code 1N33}, highlighting the residues (except A1493) targeted for base-pairing complementarity with the OMe 10-mer 1493 (green) and additional residues targeted by 10-mers 1491, 1495, 1497, 1499 and 1501 (yellow). Remaining residues targeted by other oligonucleotides used in the present study are in red. Residues A1492 and A1493 are shown in liquorice format in light blue. Non-targeted nucleotides are in purple. Proteins are shown in grey. The view is from the subunit interface. (B) Close-up view of the highlighted region shown in (A), with paromomycin shown in white and RNA residues complementary to targeted residues in purple. The view is rotated approx. 180° about the y-axis relative to that shown in (A). Proteins and near-cognate tRNA analogue together with non-targeted regions of ribosomal RNA are not shown. Colour coding is as in (A). (C) Analogous view of the structure of hygromycin B bound to the 30 S subunit ([35]; PDB code 1HNZ). Colour coding is as in (A) and (B).
In contrast, in the case of hygromycin B, the antibiotic binds predominantly on the surface of the major groove along the helix axis and does not induce the conformational change for residues A1492 and A1493, which instead remain stacked within the helix (Figure 6C) [35]. The lack of effect of hygromycin B on binding of OMe oligonucleotides 1489, 1491 and 1493 is therefore fully consistent with the model presented above, since no conformational change occurs in this case. Hygromycin B is considerably weaker in its ability to cause translational misreading [36], although it still has other translational inhibition activities. Therefore affinity measurement of radiolabelled OMe 10-mer 1493 oligonucleotide binding to 30 S subunits might be used perhaps as a simple and effective assay for new compounds thought to target the ribosome. Hence their ability to trigger the closed conformation, which includes the realignment of residues 1492 and 1493, might act as a predictor of a compound's likelihood to cause translational misreading.
If the model we propose is correct, then one might expect other aminoglycoside antibiotics that cause repositioning of residues A1492 and A1493 outside the RNA helix to have a similar effect to that of paromomycin. Recently the crystal structure of tobramycin bound to A site RNA was reported [38], which showed that in this case also, residues A1492 and A1493 are indeed repositioned in this way. Accordingly, we have carried out preliminary studies of the effect of tobramycin on the binding affinity of OMe 10-mer 1493 and found that the enhancement observed is of a similar magnitude to that of paromomycin (results not shown). This result strengthens the case for our proposal.
We have looked for evidence as to whether paromomycin is displaced when OMe 10-mer 1493 binds to the 30 S subunit. Preliminary NMR experiments suggest that paromomycin remains bound to the 30 S subunit when 10-mer 1493 is present, but we are unable to ascertain readily whether or not the paromomycin is still at the A site or elsewhere on the ribosome (F. Aboul-ela, unpublished work). It should be noted that aminoglycosides such as paromomycin are notoriously promiscuous in their binding to RNA and, if present in excess, can bind to a range of lower-affinity sites. Even if the paromomycin is still at the A-site, its binding mode might be significantly different. It is not clear whether all of the residues on the target RNA strand complementary to the OMe 10-mer 1493 become available for pairing. We have also not been successful to date in obtaining diffracting co-crystals of 30 S subunits in the presence of 1493 10-mer (J. Murray, personal communication). But we are clear that OMe oligonucleotides do not show competition with aminoglycoside drugs for A site binding, and therefore are not likely to be suitable reagents for general high-throughput screening of potential ribosomal-binding drugs, but instead may be very suitable as specific probes of biologically important RNA conformations induced by drug classes.
All OMe oligonucleotides showed some level of inhibitory activity in the in vitro translation assay, but those that had the highest affinities in the binding assay did show some correlation with an increased level of translation inhibition (Figure 5). Phosphodiester [14,15,39,40], methylphosphonate [13] and PNA (peptide nucleic acid) [41] oligonucleotides complementary to a range of ribosomal RNA sites have been reported previously to inhibit translation in vitro, but, to our knowledge, translation inhibition by OMe oligonucleotides has not been reported before, nor has the A site been reported as a target for oligonucleotide derivatives. The levels of inhibition activity of 10-mers 1493 and 1499 (approx. 10 μM) are similar to those of many aminoglycoside antibiotics [36].
The translation-inhibition activity of specific binding oligonucleotides is further good evidence for targeting of the A site. It should be noted, however, that penetration of oligonucleotides and their analogues into bacterial cells in culture is rather poor. For example, antibacterial activity of PNA is strong in the case of an unusually permeable E. coli strain AS19, but much weaker for wild-type strain K12 [41]. The use of cell-penetrating peptides such as PNA conjugates was shown to enhance activity in these cell lines [42]. But in general, it would be necessary to solve the bacterial-cell-penetration problem before such materials would be worth exploring as potential antibiotics. Nevertheless, significant improvements may be possible in levels of inhibition of in vitro translation by exploration of the incorporation of tighter-binding analogues that maintain an RNA-like conformation, such as LNAs (locked nucleic acids) [43,44]. This might allow even stronger binding to the A site on ribosomes or ribosomal subunits.
Acknowledgments
We thank Donna Williams for synthesis of OMe oligoribonucleotides. We also thank Venki Ramakrishnan and Andrew Carter (MRC-LMB), and Natalia Matassova and James Murray (Vernalis) who gave invaluable advice support throughout the project. We are grateful to Peter Kierstan (Vernalis) for technical support. The work was supported by an MRC Link grant award to MRC-LMB and to RiboTargets (now Vernalis).
References
- 1.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 2.Spahn C. M. T., Prescott C. D. Throwing a spanner in the works: antibiotics and the translational apparatus. J. Mol. Med. 1996;74:423–439. doi: 10.1007/BF00217518. [DOI] [PubMed] [Google Scholar]
- 3.Neu H. C., Gootz T. D. Antimicrobial chemotherapy. In: Baron S., editor. Medical Microbiology, chapter 11. Galveston: University of Texas Medical Branch; 1996. [PubMed] [Google Scholar]
- 4.Carter A. P., Clemons W. M., Brodersen D. E., Morgan-Warren R. J., Wimberley B. T., Ramakrishnan V. Functional insights from the structure of the 30 S ribosomal subunit and its interactions with antibiotics. Nature (London) 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 5.Odom O. W., Picking W. D., Tsalkova T., Hardesty B. The synthesis of polyphenylalanine on ribosomes to which erythromycin is bound. Eur. J. Biochem. 1991;198:713–722. doi: 10.1111/j.1432-1033.1991.tb16071.x. [DOI] [PubMed] [Google Scholar]
- 6.Tenson T., Lovmar M., Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. [DOI] [PubMed] [Google Scholar]
- 7.Schlunzen F., Zarivach R., Harms J., Bashan A., Tocilj A., Albrecht R., Yonath A., Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature (London) 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 8.Matassova N. B., Rodnina M. V., Endermann R., Kroll H.-P., Pleiss U., Wild H., Wintermeyer W. Ribosomal RNA is the target for oxazolidinones, a novel class of translational inhibitors. RNA. 1999;5:939–946. doi: 10.1017/s1355838299990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aoki H., Ke L., Poppe S. M., Poel T. J., Weaver E. A., Gadwood R. C., Thomas R. C., Shinabarger D. L., Ganoza M. C. Oxazolidinone antibiotics target the P site on Escherichia coli ribosomes. Antimicrob. Agents Chemother. 2002;46:1080–1085. doi: 10.1128/AAC.46.4.1080-1085.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chopra I., Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 2001;65:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill W. E., Camp D. G., Tapprich W. E., Tassanakajohn A. Probing ribosome structure and function using short oligodeoxyribonucleotides. Methods Enzymol. 1988;164:401–419. doi: 10.1016/s0076-6879(88)64057-2. [DOI] [PubMed] [Google Scholar]
- 12.Taniguchi T., Weissmann C. Inhibition of Qβ RNA 70S ribosome initiation complex formation by an oliogonucleotide complementary to the 3′-terminal region of E. coli 16S ribosomal RNA. Nature (London) 1978;275:770–772. doi: 10.1038/275770a0. [DOI] [PubMed] [Google Scholar]
- 13.Jayaraman K., McParland K., Miller P., Ts'o P. O. P. Selective inhibition of Escherichia coli protein synthesis and growth by nonionic oligonucleotides complementary to the 3′-end of 16S RNA. Proc. Natl. Acad. Sci. U.S.A. 1981;78:1537–1541. doi: 10.1073/pnas.78.3.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxena S. K., Ackerman E. J. Microinjected oligonucleotides complementary to the α-sarcin loop of 28S RNA abolish protein synthesis ion Xenopus oocytes. J. Biol. Chem. 1990;265:3263–3269. [PubMed] [Google Scholar]
- 15.Meyer H.-A., Triana-Alonso F., Spahn C. M. T., Twardowski T., Sobkiewicz A., Nierhaus K. H. Effects of antisense DNA against the α-sarcin stem-loop structure of the ribosomal 23S rRNA. Nucleic Acids Res. 1996;24:3996–4002. doi: 10.1093/nar/24.20.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demeshkina N., Repkova M., Ven'yaminova A., Graifer D., Karpova G. Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P and E sites: a cross-linking study with mRNA analogs carrying an aryl azide group at either the uracil or the guanine residue. RNA. 2000;6:1727–1736. doi: 10.1017/s1355838200000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graifer D., Karpova G. Structural and functional topography of human ribosomes deduced from crosslinking with mRNA analogs, oligoribonucleotide derivatives. Mol. Biol. (Moscow) 2001;35:496–508. [PubMed] [Google Scholar]
- 18.Vladimirov S. N., Druzina Z., Wang R., Cooperman B. S. Identification of 50S components neighbouring 23S rRNA nucleotides A2448 and U2604 within the peptidyl transferase center of Escherichia coli ribosomes. Biochemistry. 2000;39:183–193. doi: 10.1021/bi991866o. [DOI] [PubMed] [Google Scholar]
- 19.Muralikrishna P., Alexander R. W., Cooperman B. S. Placement of the α-sarcin loop within the 50S subunit: evidence derived using a photolabile oligodeoxynucleotide probe. Nucleic Acids Res. 1997;25:4562–4569. doi: 10.1093/nar/25.22.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arzumanov A., Gait M. J. Inhibition of the HIV-1 tat protein–TAR RNA interaction by 2′-O-methyl oligoribonucleotides. In: Holy A., Hocek M., editors. Collection Symposium Series, vol. 2. Academy of Sciences of the Czech Republic; 1999. pp. 168–174. [Google Scholar]
- 21.Fourmy D., Recht M. I., Blanchard S. C., Puglisi J. D. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 22.Kaul M., Barbieri C. M., Kerrigan J. E., Pilch D. S. Coupling of drug protonation to the specific binding of aminoglycosides to the A site of 16 S rRNA: elucidation of the number of drug amino groups involved and their identities. Biochemistry. 2003;326:1373–1387. doi: 10.1016/s0022-2836(02)01452-3. [DOI] [PubMed] [Google Scholar]
- 23.Spedding G. Isolation and analysis of ribosomes from prokaryotes, eukaryotes and organelles. In: Spedding G., editor. Ribosomes and Protein Synthesis. New York: Oxford University Press; 1990. pp. 1–27. [Google Scholar]
- 24.Eisen H. N., Karush F. The interaction of purified antibody with homologous hapten. J. Am. Chem. Soc. 1949;71:363–364. doi: 10.1021/ja01169a505. [DOI] [PubMed] [Google Scholar]
- 25.Wong I., Lohman T. M. A double-filter method for nitrocellulose-filter binding: application to protein–nucleic acid interactions. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5428–5432. doi: 10.1073/pnas.90.12.5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fersht A. R. New York: W. H. Freeman and Company; 1985. Enzyme Structure and Mechanism. [Google Scholar]
- 27.Zubay G. In vitro synthesis of protein in microbial systems. Annu. Rev. Genet. 1973;7:267–287. doi: 10.1146/annurev.ge.07.120173.001411. [DOI] [PubMed] [Google Scholar]
- 28.Clemons W. M., May J. L. C., Wimberley B. T., McCutcheon J. P., Capel M. S., Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature (London) 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 29.Ogle J. M., Murphy F. V., Tarry M. J., Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 30.Carter A. P., Clemons W. M., Brodersen D. E., Morgan-Warren R. J., Hartsch T., Wimberley B. T., Ramakrishnan V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 31.Schroeder R., Von Ahsen U. Interaction of aminoglycoside antibiotics with RNA. In: Eckstein F., Lilley D. M. J., editors. Nucleic Acids and Molecular Biology, vol. 10. Berlin: Springer-Verlag; 1996. pp. 53–74. [Google Scholar]
- 32.Hamy F., Asseline U., Grasby J. A., Iwai S., Pritchard C. E., Slim G., Butler P. J. G., Karn J., Gait M. J. Hydrogen-bonding contacts in the major groove are required for Human Immunodeficiency Virus type-1 tat protein recognition of TAR RNA. J. Mol. Biol. 1993;230:111–123. doi: 10.1006/jmbi.1993.1129. [DOI] [PubMed] [Google Scholar]
- 33.Iwai S., Pritchard C. E., Mann D. A., Karn J., Gait M. J. Recognition of the high affinity site in rev-response element RNA by the Human Immunodeficiency Virus type-1 rev protein. Nucleic Acids Res. 1992;20:6465–6472. doi: 10.1093/nar/20.24.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arzumanov A., Godde F., Moreau S., Toulmé J.-J., Weeds A., Gait M. J. Use of the fluorescent nucleoside analogue benzo[G]quinazoline 2′-O-methyl-β-Dribofuranoside to monitor the binding of the HIV-1 Tat protein or of antisense oligonucleotides to the TAR RNA stem-loop. Helv. Chim. Acta. 2000;83:1424–1436. [Google Scholar]
- 35.Brodersen D. E., Clemons W. M. J., Carter A. P., Morgan-Warren R. J., Wimberly B. T., Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000;103:1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- 36.Davies J., von Ahsen U., Schroeder R. Antibiotics and the RNA world: a role for low-molecular-weight effectors in biochemical evolution? In: Gesteland R. F., Atkins J. F., editors. The RNA World. New York: Cold Spring Harbor Laboratory Press; 1993. pp. 185–204. [Google Scholar]
- 37.Vicens Q., Westhof E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- 38.Vicens Q., Westhof E. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding A site. Chem. Biol. 2002;9:747–755. doi: 10.1016/s1074-5521(02)00153-9. [DOI] [PubMed] [Google Scholar]
- 39.Walker K., Elela S. A., Nazar R. N. Inhibition of protein synthesis by anti-5.8S rRNA oligodeoxyribonucleotides. J. Biol. Chem. 1990;265:2428–2430. [PubMed] [Google Scholar]
- 40.Azad A. A., Failla P., Hanna P. J. Inhibition of ribosomal subunit association and protein synthesis by oligonucleotides corresponding to defined regions of 18S rRNA and 5S rRNA. Biochem. Biophys. Res. Commun. 1998;248:51–56. doi: 10.1006/bbrc.1998.8778. [DOI] [PubMed] [Google Scholar]
- 41.Good L., Nielsen P. E. Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc. Natl. Acad. Sci. U.S.A. 1998;95:2073–2076. doi: 10.1073/pnas.95.5.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Good L., Awasthi S. K., Dryselius R., Larsson O., Nielsen P. Bactericidal antisense effects of peptide–PNA conjugates. Nat. Biotechnol. 2001;19:360–364. doi: 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- 43.Koshkin A. A., Singh S. K., Nielsen P., Rajwanshi V. K., Kumar R., Meldgaard M., Olsen C. E., Wengel J. LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 44.Arzumanov A., Walsh A. P., Rajwanshi V. K., Kumar R., Wengel J., Gait M. J. Inhibition of HIV-1 Tat-dependent trans-activation by steric block chimeric 2′-O-methyl/LNA oligoribonucleotides. Biochemistry. 2001;40:14645–14654. doi: 10.1021/bi011279e. [DOI] [PubMed] [Google Scholar]