Abstract
GRB2, or Growth Factor Receptor-Bound Protein 2, is a pivotal adaptor protein in intracellular signal transduction pathways, particularly within receptor tyrosine kinase (RTK) signaling cascades. Its crystal structure reveals a modular architecture comprising a single Src homology 2 (SH2) domain flanked by two Src homology 3 (SH3) domains, facilitating dynamic interactions critical for cellular signaling. While SH2 domains recognize phosphorylated tyrosines, SH3 domains bind proline-rich sequences, enabling GRB2 to engage with various downstream effectors. Folding and binding studies of GRB2 in its full-length form and isolated domains highlight a complex interplay between its protein-protein interaction domains on the folding energy landscape and in driving its function. Being at the crosslink of many key molecular pathways in the cell, GRB2 possesses a role in cancer pathogenesis, particularly in mediating the Ras–mitogen activated protein kinase (MAPK) pathway. Thus, pharmacological targeting of GRB2 domains is a promising field in cancer therapy, with efforts focused on disrupting protein-protein interactions. However, the dynamic interplay driving GRB2 function suggests the presence of allosteric sites at the interface between domains that could be targeted to modulate the binding properties of its constituent domains. We propose that the analysis of GRB2 proteins from other species may provide additional insights to make the allosteric pharmacological targeting of GRB2 a more feasible strategy.
Keywords: Protein-protein interactions, adaptor proteins, Intramolecular interactions, GRB2
Highlights
-
•
GRB2 is Crucial in cellular signaling pathways.
-
•
Insights into structural and functional features of GRB2.
-
•
Dynamic interplay between domains drive GRB2 function.
-
•
Possible allosteric targeting of GRB2 to modulate function.
-
•
Conservation among species and regulatory mechanisms explored.
1. Introduction
Adaptor proteins play a critical role in cellular signaling pathways by serving as central mediators, facilitating the transmission of information from extracellular signals to intracellular effectors. These proteins usually function as molecular scaffolds, enabling the assembly of multi-protein complexes. Their primary function lies in ensuring the specificity, efficiency, and fidelity of signal transduction events, thereby exerting tight control over various cellular processes. By bridging the communication between cell surface receptors and downstream signaling components, adaptor proteins regulate a myriad of fundamental biological processes, including but not limited to proliferation, differentiation, apoptosis, and metabolism. Through their dynamic interactions with key signaling molecules, adaptor proteins regulate cellular responses to diverse environmental signals, thus contributing to the maintenance of cellular homeostasis and overall organismal health. Furthermore, they possess the remarkable ability to integrate multiple signaling inputs and coordinate crosstalk between different pathways, thereby adding layers of complexity to cellular decision-making processes. Through their spatial and temporal regulation of signaling events, adaptor proteins fine-tune the cellular response to external stimuli, enabling organisms to adapt and thrive in ever-changing environments [1].
Often lacking an intrinsic catalytic activity, adaptor proteins exert their function by mediating protein-protein interactions through their modular domains, which are capable of binding to multiple partners. These partners encompass a diverse array of cellular constituents, ranging from receptors and kinases to phosphatases and an array of other signaling molecules. Through this extensive network of interactions, adaptor proteins assume the crucial task of spatially and temporally modulating signaling cascades, thereby finely tuning both the magnitude and duration of cellular responses to external stimuli [2]. Adaptor proteins play key roles in integrating signals from various extracellular stimuli by forming dynamic signaling complexes, allowing the cell to channel distinct signals into common downstream pathways or segregate overlapping signals to trigger specific cellular outcomes [1].
This review centers on GRB2, also known as Growth Factor Receptor-Bound Protein 2, a crucial adaptor protein integral to fundamental cellular pathways, with a specific emphasis on unraveling the internal dynamics that dictate its functionality. Our primary objective is to delve into the intramolecular interactions within GRB2, particularly those governing its folding and binding processes. By concentrating our efforts on this aspect, we aim to gain a comprehensive understanding of how these interactions regulate the role in fundamental cellular pathways. Additionally, we will explore the significance of GRB2 structural dynamics as a possible novel pharmacological target in cancer pathologies. Furthermore, we scrutinize variations in GRB2 sequence across different species, providing insights about how the landscape of intramolecular interactions within GRB2 may underlie its functional diversity and versatility.
1.1. GRB2 structure and folding
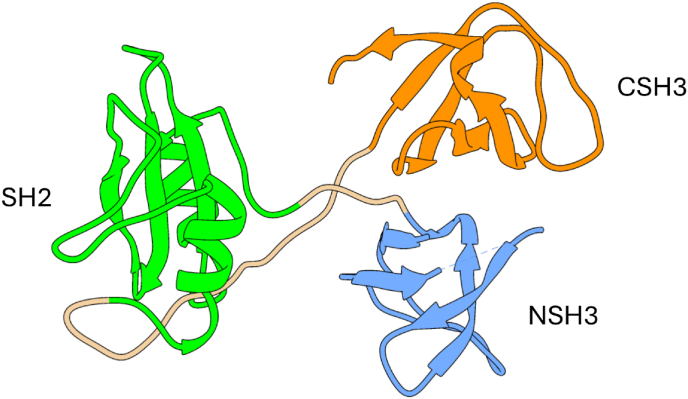
GRB2 or Growth Factor Receptor-Bound Protein 2, is a crucial signaling adaptor protein that plays a pivotal role in intracellular signal transduction pathways [3]. It represents a particularly important piece in the complicated puzzle of RTK signaling, which is integral for cellular processes such as cell growth, differentiation, and survival, as well as in the transduction of other important physiological and molecular pathways with crucial role for cell homeostasis [3]. The crystal structure of GRB2 has been solved at 3.1 Å resolution (PDB: 1GR1 – Fig. 1) and exhibits a distinctive modular architecture that confers adaptability in cellular signaling processes [4]. Its structural composition is characterized by a single Src homology 2 (SH2) domain flanked by two Src homology 3 (SH3) domains, namely the N-SH3 and C-SH3 domains. This modular arrangement not only confers specificity to GRB2-mediated signaling events but also enables the dynamic recruitment and assembly of signaling components in response to changing cellular contexts.
Fig. 1.
– Three-dimensional structure of GRB2 (PDB: 1GRI) with its modular structure comprising a N-terminal and a C-terminal SH3 domains (in blue and orange respectively) flanking a central SH2 domain (in green) is reported. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Each domain within the GRB2 structure serves a distinct yet interconnected role in orchestrating its functional repertoire. SH3 domains are small protein interaction modules composed of a β-sandwich capable of interacting with ligands presenting proline-rich sequences through a conserved binding site among the domain family [5,6]. In the case of GRB2, the binding specificities of the two SH3 domains are slightly different, with a P-X-X-P-X-R motif for the NSH3 domain and P-X-X-R-X-X-K-P for the CSH3 domain (X being any amino acid) [7] This allows GRB2 to recognize and bind to different partners in the intracellular environment. SH2 domains are protein-protein interaction domains structurally composed by two alpha-helices flanking a central beta-sheet and are typically involved in recognizing partners presenting a phosphorylated tyrosine [[8], [9], [10]]. Through these interactions and its modular architecture, GRB2 orchestrates the formation of dynamic complexes with several downstream effectors. These interactions influence the activation of signaling cascades that regulate fundamental cellular responses, including signal transduction, cell proliferation, and tumorigenesis [[11], [12], [13]].
Understanding the folding mechanism of multidomain proteins is of key importance for unraveling the intricacies of protein structure and function. The folding mechanism of multidomain proteins is a complex process that involves the sequential or concerted folding of individual domains within the protein structure. While the traditional approach to protein folding treated isolated domains as independent entities with specific equilibrium and kinetic properties, a growing body of experimental work has revealed how the presence of contiguous domains significantly influences the folding energy landscape of protein domains [[14], [15], [16], [17], [18], [19]]. This suggests a complex interplay of the domains, involving independendent and/or inter-dependent folding mechanisms. Clearly, to gain a comprehensive understanding of the folding mechanism of multidomain proteins, it is imperative to compare their folding properties with those of isolated domains that constitute the protein.
In the case of GRB2, the folding mechanism of the isolated CSH3 domain was characterized in great detail by performing extensive mutagenesis and kinetic folding experiments [20]. Data highlighted a two-state folding mechanism, without the accumulation of folding intermediates along the reaction pathway. Moreover, by exploiting thermodynamic and kinetic data as restrain for molecular dynamics simulation, it is possible to deeply understand the structural features of the transition state, compatible with a nucleation-condensation folding mechanism with a diffused transition state. By integrating computational and experimental methodologies and increasing the complexity of the system, an exploration into the folding properties of the supramodular structure in GRB2 revealed that GRB2 domains do not fold concurrently, unveiling the presence of intermediate states during the folding process, attributed to transient interdomain communication [21]. Accordingly, kinetic folding experiments identified the accumulation of transient misfolded species during the folding of GRB2 [22]. A comparison of the folding properties of full-length GRB2 with those of its three isolated domains yielded intriguing results. Notably, the analysis of folding kinetics revealed that while the isolated SH2 domain adhered to a three-state folding mechanism involving the population of a folding intermediate, the presence of its neighboring domains profoundly affected its folding process, highlighting the existence of a misfolded kinetic trap attributed to transient interdomain communication between the SH2 and CSH3 domain. Furthermore, it was observed that a decrease in the thermodynamic stability of the SH2 domain influences the refolding kinetics of the CSH3 domain within the context of a SH2-CSH3 tandem construct [22]. However, while the folding pathway of the CSH3 domain remains largely unaffected by the presence of its contiguous domain, a double mutant cycle analysis revealed the pivotal role of specific residues at the interface between the CSH3 and the SH2 domain in regulating the binding selectivity of the C-SH3 domain [23]. This observation underscores the intricate interplay between SH2 and CSH3 domains, emphasizing the potential presence of an allosteric site that could be targeted pharmacologically to modulate GRB2 function.
2. GRB2 function and implication in cancer
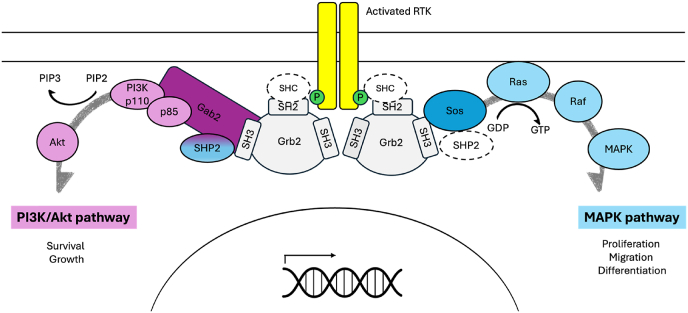
GRB2 has been identified as the linker between the epidermal growth factor receptor (EGFR) and the Ras–mitogen-activated protein kinase (MAPK) pathway, which is stimulated by RTKs [24,25] acting on the guanine nucleotide exchange factor, Son of Sevenless (SOS). Indeed, GRB2 facilitates the association between RTKs and SOS by recognizing phosphorylated tyrosines on the RTKs via its SH2 domain and a proline-rich sequence on SOS via its SH3 domains [[26], [27], [28]], promoting the GDP-GTP exchange of RAS protein and the activation of the pathway (Fig. 2). Since this pathway is upregulated in many cancer pathologies [29,30] understanding the molecular details of GRB2 binding affinity and specificity represents a field of great interest.
Fig. 2.
– Schematic representation of the involvement of GRB2 protein in different physiological pathway in the cell. Two main pathways are highlighted with different color codes and the resulting cellular events regulated are listed (see details in the text). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
In breast cancer, GRB2 functions by linking receptor tyrosine kinases (RTKs) with downstream signaling pathways, such as the Ras/MAPK pathway, which are essential for cell division and survival. The overexpression of GRB2 has been reported in breast cancer cells, strongly correlating with the progression of the disease, highlighting its key role in tumor biology [31]. Similarly, in lung cancer, a ligand of GRB2, i.e. the GRB2-associated binder 2 (Gab2) protein, is overexpressed. This overexpression hyperactivates pathways that promote cancer cell proliferation and survival [32]. The implication of GRB2 in cancer pathology has also been reported in hepatocellular carcinoma (HCC), where the coexpression of GRB2 and Gab1 is identified as an unfavorable prognostic factor, suggesting the binding between these two proteins to have a role in promoting cell growth and survival. This finding highlights the potential of targeting the binding between GRB2 and Gab1 as a possible therapeutic strategy for HCC [33]. Another example is represented by the reduction of intracellular GRB2 under non-stimulated conditions. The functional details of such events are still unclear, however it has been observed that low expression levels of GRB2 lead to the inhibition of PTEN and the consequent activation of the Akt oncoprotein. This provokes tumor formation in ovarian cancer, clearly indicating that GRB2 modulates pathways critical for cancer progression [34]. A key role of GRB2 has been also highlighted in colorectal cancer. In particular, while the overexpression of the oncoprotein Met increased tumor growth and metastasis, mutants that specifically recruited GRB2 showed slight but consistent tumor-suppressive effects, suggesting that direct GRB2 binding does not significantly drive Met-mediated tumor progression. Additionally, these mutant-expressing tumors had markedly reduced Gab1 protein levels, pointing to a post-translational regulatory process. This observation highlights the complexity of Met signaling in cancer and implies that the selective engagement of adaptor proteins like GRB2 can modulate tumor behavior depending on the context [35]. Furthermore, the implication of GRB2 in human bladder cancer is evident from the analysis of EGF receptor, GRB2, and Sos protein expression levels in four bladder cancer cell lines (T24, KU-7, UMUC-2, UMUC-6) and two cultured normal urothelial cell lines (HMKU-1, HMKU-2) [36]. While no significant difference in EGF receptor expression between cancerous and normal cells was found, GRB2 and Sos proteins were significantly overexpressed in all bladder cancer cell lines compared to normal urothelial cells. Overall, GRB2 is involved in various signaling pathways that are critical for cancer cell migration, invasion, and survival. Its widespread involvement in these processes underscores its potential as a target for developing new cancer therapies (Giubellino et al., 2008).
In the context of DNA damage response (DDR) pathways, Grb2 has been shown to form complexes with proteins like PTEN and Rad51. PTEN is a tumor suppressor protein involved in regulating prolifelation and cell survival. The interaction of Grb2 with PTEN forms a complex that is able to pass the nuclear membrane and interact with the RAD51 homolog 1 (Rad51), thereby influencing DNA repair processes and maintaining genomic stability [37]. Rad51 is an essential protein for homologous recombination repair, activating a critical pathway for the accurate repair of double-strand breaks in DNA [38]. By interacting with PTEN, Grb2 can modulate the signaling pathways that control cell cycle checkpoints and DNA repair mechanisms, ensuring that damaged DNA is effectively repaired and preventing the propagation of mutations that could lead to cancer [38].The Grb2 protein is also a pivotal component in immune responses, engaging in multiple signaling pathways essential for immune cell functionality and response. Grb2 interacts with proteins such as Gab1, BCAP, and HPK1, thereby influencing critical processes like lymphocyte differentiation, B-cell maturation, and T lymphocyte activation [[39], [40], [41]]. In B lymphocytes, the scaffolding functions of Grb2 regulate the assembly of signalosomes following antigen receptor engagement [42]. Moreover, Grb2 is crucial in actin remodeling during phagocytosis, highlighting its importance in macrophage recognition and vesicle trafficking [43].
Additional examples of important interactors of GRB2 that have been linked to cancer pathologies are GAB2 (Grb2-Associated Binding protein 2) and the fusion oncoprotein BCR-ABL (which is generated by the fusion of the Breakpoint Cluster Region on chromosome 22 with the Abelson protooncogene sequence of chromosome 9). GAB2 is a 676-residues scaffolding protein, that is composed of a pleckstrin homology (PH) domain anchoring the protein to the membrane, and a long disordered tail that presents various consensus sequences recognized for the assembly of signaling systems [[44], [45], [46]]. Since the binding between the CSH3 domain of GRB2 and GAB2 is impaired in many cancer pathologies [32,47,48], the mechanism of binding has been extensively characterized [49,50]. Importantly, a double-mutant cycle analysis highlighted a complex binding scenario in which residues far from the binding pocket regulate ligand recognition through a diffused energetic network [50], possibly representing the mechanism by how the CSH3 domain recognizes different partners in the crowded intracellular environment.
In the case of the onset of Chronic Myeloid Leukemia, the BCR-ABL oncoprotein recruits and interact with the SH2 domain of GRB2, leading to an upregulation of the RAS/MAPK pathway. The details of such interactions have been recently investigated through an in silico approach, in order to depict the molecular determinants dictating the specificity of recognition between the SH2 domain and phosphorylated BCR, a crucial information for unraveling binding selectivity and predicting potential interactions with phosphorylated receptors [51]. The results allowed the authors to propose a specific motif which optimally interacts with the SH2 domain of GRB2.
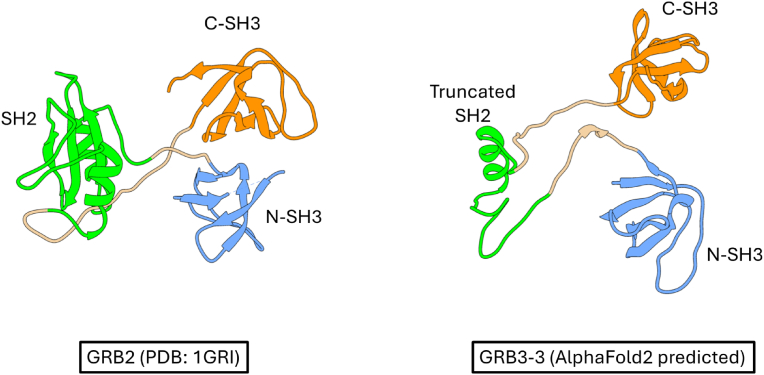
Based on these premises, it is worth noting that a natural splicing variant of GRB2, named GRB3-3, which lacks 40 residues within the SH2 domain but retains both functional and intact SH3 domains (Fig. 3), has been identified as a negative regulator of RAS activation and as a suppressor of the MAPK pathway [52,53]. Curiously, while GRB3-3 appears to compete for the binding with SOS with GRB2, it has been reported that in the absence of a functional SH2 domain, the SH3 domains of GRB3-3 acquire the ability to recognize additional sites on SOS. This phenomenon can be attributed to a mechanism orchestrated by interdomain communication within GRB2, a feature absent in GRB3-3. Specifically, in GRB2, it is proposed that the SH2 domain, upon binding to a phosphorylated target, transiently impedes the C-SH3 domain, thereby facilitating the interaction of SOS with the N-SH3 domain [27]. This intricate interplay gains further support from evidence suggesting that the expression of GRB3-3 induces apoptosis in cancer cell lines [53], highlighting its potential utility as a pharmacological tool to modulate the upregulation of the MAPK pathway. However, it is essential to acknowledge the fundamental role of the MAPK pathway, which must be maintained at a basal level to maintain key physiological events within the cell. Therefore, the lack of information about the mechanism of interaction of GRB3-3 with its ligands needs urgent attention to unravel its precise role and significance in cellular processes, potentially paving the way for novel therapeutic interventions targeting the MAPK pathway.
Fig. 3.
– The comparison of the experimentally determined three-dimensional structure of GRB2 (on the left) and the predicted three-dimensional structure (through AlphaFold2) of the splicing variant GRB3-3 (on the right) highlights the absence of a functional SH2 domain in GRB3-3 due to the truncation of 40 residues.
2.1. The interdomain communication in GRB2 hints at allosteric pharmacological strategies
In light of its crucial role in various cancer pathologies, substantial efforts have been dedicated to developing strategies that directly target the SH2 and SH3 domains of Grb2, recognizing the significance of these domains in mediating critical signaling events. Previous studies primarily focused on blocking the interaction of GRB2 with its physiological ligands by targeting the binding pockets of SH2 and SH3 with high-affinity peptides or small molecules. A NSH3 domain blocker, a peptide named HAGBP, was developed and derived from a mutated version of murine SOS protein [54]. Experiments with CML cell lines demonstrated the potency of HAGBP, although resistance was monitored in certain cases, probably due to the molecular heterogeneity of CML cells. Another example, involving a peptidomimetic molecule, is represented by the development of CGP78850, a Grb2 SH2 inhibitor designed to specifically block Ras activation [55]. The inhibitor demonstrated high specificity in vitro and inhibited the growth of human tumor cell lines with deregulated receptor tyrosine kinases. Notably, it induced cell cycle regulators and negatively regulated the cell cycle. Several phosphopeptides with high affinities for the Grb2 SH2 domain have also been developed during the years [56]. Interestingly, a hydroxy benzyl phosphinate has been developed, which exhibits higher cell permeability compared to other Pmp peptidomimetics due to its reduced negative charges and shows relatively high binding affinity (KD of 0.53 μM) to the SH2 domain [57,58]. Another notable approach has focused on peptides mimicking phospshorylated tyrosines with increased cytosolic absorption. The BC1 compound, for example, was designed as a bicyclic peptide capable of inhibiting Grb2-SH2 with an IC50 of 350 nM, although no antiproliferative effect was observed [[59], [60], [61]].
More recently, one small molecule inhibitor named AN-465-J137-985, identified through virtual screening and validated in vitro experiments showed a significant efficacy in reducing the affinity between the CSH3 domain and Gab2. Experimental validation in lung cancer cell lines A549 and H1299 confirmed the inhibitory effect [62]. Starting from these promising results, new molecules derived from AN-465-J137-985 were designed and synthesized identifying another inhibitor molecule which was tested on H1299, SKOV3 and U87 cell lines [63]. Altogether, these results corroborate the potential therapeutic relevance of disrupting the CSH3:Gab2 interaction in cancer treatment. A comprehensive list of known inhibitors of Grb2 is reported in Table 1.
Table 1.
– List of inhibitors of Grb2 protein.
| Inhibitor | Domain target | Reference |
|---|---|---|
| HAGBP | NSH3 | [54] |
| CGP78850 | SH2 | [55] |
| CGP85793 | SH2 | [64] |
| Fmoc–Glu–Tyr–Aib–Asn–NH2 | SH2 | [65] |
| Compd 2/Compd 3 | SH2 | [66] |
| SP1068 | SH2 | [67] |
| Hydroxy benzyl phosphinate | SH2 | [57] |
| BC1 | SH2 | [59] |
| Ac-N-X-V-N-I-E-amide | SH2 | [68] |
| (VPPPVPPRRR)2-K-Aha-RQIKIWFQNRRMK WKK Peptide |
CSH3 | [69] |
| GTDEVPPPVPPRR Peptide | CSH3 | [70] |
| AN-465-J137-985 | CSH3 | [62,63] |
| VPPPVPPRRR | CSH3 | [71] |
| macrocyclic peptides | CSH3 | [72] |
However, it appears evident that directly inhibiting the binding cleft of the protein-protein interaction domains of GRB2 may not be the most effective strategy for maintaining cell homeostasis, as it risks disrupting the intricate balance of signaling cascades governed by GRB2-mediated interactions. Additionally, such a strategy may lead to nonspecific inhibition, as targeting the SH2 and SH3 domains of other proteins could disrupt their physiological functions. This unintended consequence could arise from the shared structural motifs and conserved binding sites among proteins, resulting in off-target effects that disrupt critical cellular processes. On the other hand, the evidence of interdomain communication driving the function of GRB2, particularly between the SH2 and the CSH3 domains [23], suggests the potential for targeting the protein allosterically, exploiting the dynamic interplay between its domains to modulate its activity. This approach would involve targeting sites that are distinct from the typical binding pockets associated with individual protein-protein interaction domains, potentially allowing to reduce side effects by precisely influencing the function of GRB2 without directly interfering with other SH2 and SH3 domain and maintaining their functions. On the other hand, the absence of a specific binding pocket to be targeted might represent a challenging pitfall in terms of achieving optimal specificity and affinity of the inhibitory molecule. In light of this, deepening our understanding of the mechanistic properties governing transient interdomain interactions in GRB2 is imperative. By elucidating the dynamics of these interactions, researchers may gain valuable insights into potential allosteric sites that could be leveraged for targeted modulation of GRB2 activity. We propose that insights into this phenomenon could potentially be obtained from the characterization of GRB2 proteins obtained from different species. Apparently, while structure alignment suggests that the binding properties of orthologous GRB2 might be conserved, the protein function could be differently achieved through diverse dynamic interplay between domains.
2.2. Comparison of GRB2 sequence (and structure) among different species
The GRB2 protein demonstrates widespread expression across diverse animal species. Intriguingly, human GRB2 reveals structural and functional homology with the sem-5 protein from the nematode C. elegans [26]. The expression of GRB2 has been characterized in Chlonorchis sinensis, a parasitic organism belonging to the class of Platyhelminthes [73] where it plays pivotal roles in cellular functions such as meiosis, organogenesis, and energy metabolism. Moreover, the importance of GRB2 in controlling cell cycle has been characterized in Xenopus laevis oocytes [74]. Curiously, both human and X. laevis variants of GRB2 triggered the reinitiation of meiosis, indicating that they were both functional in X. laevis oocytes. The presence and function of GRB2 protein in X. laevis has been characterized in other works [75,76]. These observations underscore the conservation of GRB2 across various branches of animal evolutionary development.
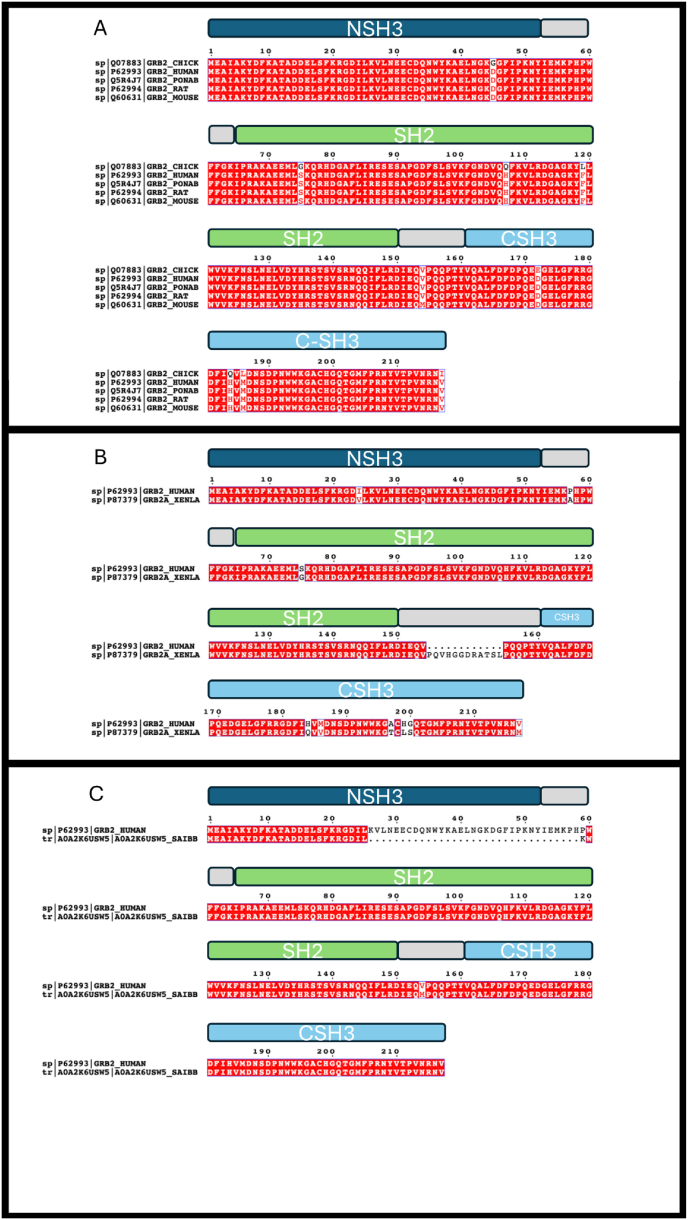
A comprehensive analysis of GRB2 protein within the Swiss-Prot reviewed entries on the Uniprot database unveils its remarkable conservation among mammals, including H. sapiens (Uniprot entry P62993), P. abelii (Uniprot entry Q5R4J7), R. norvegicus (Uniprot entry P62994), and M. musculus (Uniprot entry Q60631), as well as in the bird species G. gallus (Uniprot entry Q07883), exhibiting both high sequence identity and a consistent length of 217 amino acids (Fig. 4A). Intriguingly, when exploring the X. laevis GRB2, the conservation of SH3 and SH2 sequences is evident, yet a distinctive feature emerges—a longer linker between the central SH2 domain and the C-SH3 domain, resulting in a total of 229 amino acids (Fig. 4B), which is conserved in the X. tropicalis species (not shown). This implies that, although the binding properties of individual domains would remain conserved across species, the elongated linker may introduce a dynamic shift in intradomain communication between CSH3 and SH2 potentially influencing the regulation of binding affinity and specificity in interactions with intracellular ligands. Notably, in the primate species Saimiri boliviensis boliviensisthe GRB2 protein lacks a complete NSH3 domain (Uniprot entry A0A2K6USW5, not Swiss-prot reviewed) suggesting a distinct role for GRB2 in signal transduction within this species (Fig. 4C). The characterization of these GRB2 variants may provide a unique opportunity to delve into the functional properties of this highly dynamic protein and unravel the intricate role played by GRB2 modular structure in executing its diverse functions.
Fig. 4.
– Panel A: Sequence alignment of GRB2 proteins from H. sapiens (Uniprot entry P62993), P. abelii (Uniprot entry Q5R4J7), R. norvegicus (Uniprot entry P62994), and M. musculus (Uniprot entry Q60631), G. gallus (Uniprot entry Q07883). All the sequences are highly conserved. Panel B: Sequence alignment of GRB2 proteins from H. sapiens (Uniprot entry P62993) and X. laevis (Uniprot entry P87379), with the presence of a longer linker between the SH2 and CSH3 domain in X.laevis sequence. Panel C: Sequence alignment of GRB2 proteins from H. sapiens (Uniprot entry P62993) and S. boliviensis boliviensis (Uniprot entry A0A2K6USW5). It is possible to observe that S. boliviensis boliviensis sequence display a truncated NSH3 domain compared to the human protein.
3. Conclusions
The multifaceted roles of GRB2 in cellular signaling, particularly its involvement in RTK pathways and cancer pathogenesis, underscore its significance as a therapeutic target. The modular architecture of GRB2, characterized by its SH2 and SH3 domains, enables versatile interactions critical for signal transduction. Experimental evidence reveals the intricate interplay between domain independence and collaboration, shedding light on the structural dynamics underlying Grb2 function. Overall, the comprehensive characterization of GRB2 structure, function, and evolutionary conservation provides a foundation for further exploration of its therapeutic potential and the development of targeted therapies for cancer and other diseases. Continued research into the complex interplay between GRB2 domains and their interactions with intracellular partners is essential for unlocking its full therapeutic potential and advancing allosteric pharmacological approaches to modulate its function in pathological states. Future research should, in fact, focus on elucidating specific allosteric sites within GRB2 that can be targeted to modulate its activity without disrupting its overall function, utilizing advanced techniques such as cryo-electron microscopy (cryo-EM) and nuclear magnetic resonance (NMR) spectroscopy to map dynamic conformational changes and identify potential allosteric sites through computational modeling and mutagenesis studies. Additionally, understanding how interdomain interactions within GRB2 influence its binding affinity and specificity for various ligands can be achieved by conducting detailed kinetic and thermodynamic studies on full-length GRB2 and its isolated domains, employing Förster resonance energy transfer (FRET) and single-molecule fluorescence microscopy to observe real-time interdomain interactions. Comparative structural and functional analyses of GRB2 homologs from various species, particularly focusing on differences in linker regions and domain compositions, can reveal how structural variations affect its function and interaction networks, using site-directed mutagenesis to mimic these variations in human GRB2 and assess their impact on signaling pathways. Furthermore, characterizing the expression patterns and functional differences of GRB2 isoforms and its splicing variant GRB3-3, in different tissues and cancer types, and exploring their potential as therapeutic targets or biomarkers, using biophysical techniques and cellular assays, will enhance our understanding of the regulatory mechanism of GRB2. Overall, these efforts should pave the way for novel therapeutic strategies targeting this critical adaptor protein in cancer and other diseases.
Funding
Work supported by the Istituto Pasteur Italia – Fondazione Cenci Bolognetti (Research Program 2022–2023 Under 45 Call 2020, to A.T.) and by Sapienza University of Rome (RM12218148DA1933 to A.T.) and by the Italian MUR-PRIN 2022 grant N. 2022JY3PMB to A.T. L.P. was supported by a FIRC-AIRC fellowship for Italy.
CRediT authorship contribution statement
Francesca Malagrinò: Writing – review & editing. Elena Puglisi: Writing – review & editing. Livia Pagano: Writing – review & editing. Carlo Travaglini-Allocatelli: Writing – review & editing. Angelo Toto: Writing – review & editing, Writing – original draft, Funding acquisition, Conceptualization.
Declaration of competing interest
All the authors declare no competing interests.
References
- 1.Pawson T., Scott J.D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 2.Mayer B.J. Protein-protein interactions in signaling cascades. Methods Mol. Biol. 2006;332:79–99. doi: 10.1385/1-59745-048-0:79. [DOI] [PubMed] [Google Scholar]
- 3.Tari A.M., Lopez-Berestein G. GRB2: a pivotal protein in signal transduction. Semin. Oncol. 2001;28:142–147. doi: 10.1016/s0093-7754(01)90291-x. [DOI] [PubMed] [Google Scholar]
- 4.Maignan S., Guilloteau J.P., Fromage N., Arnoux B., Becquart J., Ducruix A. Crystal structure of the mammalian Grb2 adaptor. Science. 1995;268:291–293. doi: 10.1126/science.7716522. [DOI] [PubMed] [Google Scholar]
- 5.Saksela K., Permi P. SH3 domain ligand binding: what's the consensus and where's the specificity? FEBS Lett. 2012;586:2609–2614. doi: 10.1016/j.febslet.2012.04.042. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko T., Li L., Li S.S.-C. The SH3 domain--a family of versatile peptide- and protein-recognition module. Front. Biosci. 2008;13:4938–4952. doi: 10.2741/3053. [DOI] [PubMed] [Google Scholar]
- 7.Feller S.M., Tuchscherer G., Voss J. High affinity molecules disrupting GRB2 protein complexes as a therapeutic strategy for chronic myelogenous leukaemia. Leuk. Lymphoma. 2003;44:411–427. doi: 10.1080/1042819021000037930. [DOI] [PubMed] [Google Scholar]
- 8.Diop A., Santorelli D., Malagrinò F., Nardella C., Pennacchietti V., Pagano L., Marcocci L., Pietrangeli P., Gianni S., Toto A. SH2 domains: folding, binding and therapeutical approaches. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232415944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waksman G., Kumaran S., Lubman O. SH2 domains: role, structure and implications for molecular medicine. Expert Rev Mol Med. 2004;6:1–18. doi: 10.1017/S1462399404007331. [DOI] [PubMed] [Google Scholar]
- 10.Mayer B.J. What have We Learned from SH2 domains? Methods Mol. Biol. 2017;1555:37–43. doi: 10.1007/978-1-4939-6762-9_2. [DOI] [PubMed] [Google Scholar]
- 11.Drost J., Nonis D., Eich F., Leske O., Damrath E., Brunt E.R., Lastres-Becker I., Heumann R., Nowock J., Auburger G. Ataxin-2 modulates the levels of Grb2 and Src but not ras signaling. J. Mol. Neurosci. 2013;51:68–81. doi: 10.1007/s12031-012-9949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suen K.L., Bustelo X.R., Pawson T., Barbacid M. Molecular cloning of the mouse grb2 gene: differential interaction of the Grb2 adaptor protein with epidermal growth factor and nerve growth factor receptors. Mol. Cell Biol. 1993;13:5500–5512. doi: 10.1128/MCB.13.9.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Den Hertog J., Hunter T. Tight association of GRB2 with receptor protein-tyrosine phosphatase alpha is mediated by the SH2 and C-terminal SH3 domains. EMBO J. 1996;15:3016–3027. doi: 10.1002/j.1460-2075.1996.tb00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han J.-H., Batey S., Nickson A.A., Teichmann S.A., Clarke J. The folding and evolution of multidomain proteins. Nat. Rev. Mol. Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 15.Gautier C., Troilo F., Cordier F., Malagrinò F., Toto A., Visconti L., Zhu Y., Brunori M., Wolff N., Gianni S. Hidden kinetic traps in multidomain folding highlight the presence of a misfolded but functionally competent intermediate. Proc. Natl. Acad. Sci. U.S.A. 2020;117:19963–19969. doi: 10.1073/pnas.2004138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borgia A., Kemplen K.R., Borgia M.B., Soranno A., Shammas S., Wunderlich B., Nettels D., Best R.B., Clarke J., Schuler B. Transient misfolding dominates multidomain protein folding. Nat. Commun. 2015;6:8861. doi: 10.1038/ncomms9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borgia M.B., Borgia A., Best R.B., Steward A., Nettels D., Wunderlich B., Schuler B., Clarke J. Single-molecule fluorescence reveals sequence-specific misfolding in multidomain proteins. Nature. 2011;474:662–665. doi: 10.1038/nature10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian P., Best R.B. Structural determinants of misfolding in multidomain proteins. PLoS Comput. Biol. 2016;12 doi: 10.1371/journal.pcbi.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inanami T., Terada T.P., Sasai M. Folding pathway of a multidomain protein depends on its topology of domain connectivity. Proc Natl Acad Sci U S A. 2014;111:15969–15974. doi: 10.1073/pnas.1406244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Troilo F., Bonetti D., Camilloni C., Toto A., Longhi S., Brunori M., Gianni S. Folding mechanism of the SH3 domain from Grb2. J. Phys. Chem. B. 2018;122:11166–11173. doi: 10.1021/acs.jpcb.8b06320. [DOI] [PubMed] [Google Scholar]
- 21.Dias R.V.R., Pedro R.P., Sanches M.N., Moreira G.C., Leite V.B.P., Caruso I.P., De Melo F.A., De Oliveira L.C. Unveiling metastable ensembles of GRB2 and the relevance of interdomain communication during folding. J. Chem. Inf. Model. 2023;63:6344–6353. doi: 10.1021/acs.jcim.3c00955. [DOI] [PubMed] [Google Scholar]
- 22.Pagano L., Pennacchietti V., Diop A., Santorelli D., Pietrangeli P., Marcocci L., Nardella C., Malagrinò F., Toto A., Gianni S. Exploring the effect of tethered domains on the folding of Grb2 protein. Arch. Biochem. Biophys. 2022;731 doi: 10.1016/j.abb.2022.109444. [DOI] [PubMed] [Google Scholar]
- 23.Di Felice M., Pagano L., Pennacchietti V., Diop A., Pietrangeli P., Marcocci L., Di Matteo S., Malagrinò F., Toto A., Gianni S. The binding selectivity of the C-terminal SH3 domain of Grb2, but not its folding pathway, is dictated by its contiguous SH2 domain. J. Biol. Chem. 2024 doi: 10.1016/j.jbc.2024.107129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imperial R., Toor O.M., Hussain A., Subramanian J., Masood A. Comprehensive pancancer genomic analysis reveals (RTK)-RAS-RAF-MEK as a key dysregulated pathway in cancer: its clinical implications. Semin. Cancer Biol. 2019;54:14–28. doi: 10.1016/j.semcancer.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 25.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowenstein E.J., Daly R.J., Batzer A.G., Li W., Margolis B., Lammers R., Ullrich A., Skolnik E.Y., Bar-Sagi D., Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- 27.Kazemein Jasemi N.S., Herrmann C., Magdalena Estirado E., Gremer L., Willbold D., Brunsveld L., Dvorsky R., Ahmadian M.R. The intramolecular allostery of GRB2 governing its interaction with SOS1 is modulated by phosphotyrosine ligands. Biochem. J. 2021;478:2793–2809. doi: 10.1042/BCJ20210105. [DOI] [PubMed] [Google Scholar]
- 28.Lemmon M.A., Ladbury J.E., Mandiyan V., Zhou M., Schlessinger J. Independent binding of peptide ligands to the SH2 and SH3 domains of Grb2. J. Biol. Chem. 1994;269:31653–31658. doi: 10.1016/S0021-9258(18)31745-9. [DOI] [PubMed] [Google Scholar]
- 29.Lee S., Rauch J., Kolch W. Targeting MAPK signaling in cancer: mechanisms of drug resistance and sensitivity. Int. J. Mol. Sci. 2020;21:1102. doi: 10.3390/ijms21031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santarpia L., Lippman S.M., El-Naggar A.K. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin. Ther. Targets. 2012;16:103–119. doi: 10.1517/14728222.2011.645805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tari A.M., Hung M.C., Li K., Lopez-Berestein G. Growth inhibition of breast cancer cells by Grb2 downregulation is correlated with inactivation of mitogen-activated protein kinase in EGFR, but not in ErbB2, cells. Oncogene. 1999;18:1325–1332. doi: 10.1038/sj.onc.1202422. [DOI] [PubMed] [Google Scholar]
- 32.Xu X.-L., Wang X., Chen Z.-L., Jin M., Yang W., Zhao G.-F., Li J.-W. Overexpression of Grb2-associated binder 2 in human lung cancer. Int. J. Biol. Sci. 2011;7:496–504. doi: 10.7150/ijbs.7.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y., Li Z., Yang M., Wang D., Yu L., Guo C., Guo X., Lin N. Identification of GRB2 and GAB1 coexpression as an unfavorable prognostic factor for hepatocellular carcinoma by a combination of expression profile and network analysis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0085170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timsah Z., Ahmed Z., Ivan C., Berrout J., Gagea M., Zhou Y., Pena G.N.A., Hu X., Vallien C., Kingsley C.V., Lu Y., Hancock J.F., Liu J., Gladden A.B., Mills G.B., Lopez-Berestein G., Hung M.-C., Sood A.K., Bogdanov M., Ladbury J.E. Grb2 depletion under non-stimulated conditions inhibits PTEN, promotes Akt-induced tumor formation and contributes to poor prognosis in ovarian cancer. Oncogene. 2016;35:2186–2196. doi: 10.1038/onc.2015.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seiden-Long I., Navab R., Shih W., Li M., Chow J., Zhu C.Q., Radulovich N., Saucier C., Tsao M.-S. Gab1 but not Grb2 mediates tumor progression in Met overexpressing colorectal cancer cells. Carcinogenesis. 2008;29:647–655. doi: 10.1093/carcin/bgn009. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe T., Shinohara N., Moriya K., Sazawa A., Kobayashi Y., Ogiso Y., Takiguchi M., Yasuda J., Koyanagi T., Kuzumaki N., Hashimoto A. Significance of the Grb2 and son of sevenless (Sos) proteins in human bladder cancer cell lines. IUBMB Life. 2000;49:317–320. doi: 10.1080/15216540050033195. [DOI] [PubMed] [Google Scholar]
- 37.Demeyer A., Benhelli-Mokrani H., Chénais B., Weigel P., Fleury F. Inhibiting homologous recombination by targeting RAD51 protein. Biochim. Biophys. Acta Rev. Canc. 2021;1876 doi: 10.1016/j.bbcan.2021.188597. [DOI] [PubMed] [Google Scholar]
- 38.Hou B., Xu S., Xu Y., Gao Q., Zhang C., Liu L., Yang H., Jiang X., Che Y. Grb2 binds to PTEN and regulates its nuclear translocation to maintain the genomic stability in DNA damage response. Cell Death Dis. 2019;10:546. doi: 10.1038/s41419-019-1762-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ackermann J.A., Radtke D., Maurberger A., Winkler T.H., Nitschke L. Grb2 regulates B-cell maturation, B-cell memory responses and inhibits B-cell Ca2+ signalling: grb2-deficient mice. EMBO J. 2011;30:1621–1633. doi: 10.1038/emboj.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauenstein J.U., Udgata A., Bartram A., De Sutter D., Fisher D.I., Halabi S., Eyckerman S., Gay N.J. Phosphorylation of the multifunctional signal transducer B-cell adaptor protein (BCAP) promotes recruitment of multiple SH2/SH3 proteins including GRB2. J. Biol. Chem. 2019;294:19852–19861. doi: 10.1074/jbc.RA119.009931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huyer G., Alexander D.R. Immune signalling: SHP-2 docks at multiple ports. Curr. Biol. 1999;9:R129–R132. doi: 10.1016/s0960-9822(99)80080-3. [DOI] [PubMed] [Google Scholar]
- 42.Neumann K., Oellerich T., Urlaub H., Wienands J. The B-lymphoid Grb2 interaction code. Immunol. Rev. 2009;232:135–149. doi: 10.1111/j.1600-065X.2009.00845.x. [DOI] [PubMed] [Google Scholar]
- 43.Rajaram M.V.S., Arnett E., Azad A.K., Guirado E., Ni B., Gerberick A.D., He L.-Z., Keler T., Thomas L.J., Lafuse W.P., Schlesinger L.S., tuberculosis-Initiated M. Human mannose receptor signaling regulates macrophage recognition and vesicle trafficking by FcRγ-chain, Grb2, and SHP-1. Cell Rep. 2017;21:126–140. doi: 10.1016/j.celrep.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams S.J., Aydin I.T., Celebi J.T. GAB2—a scaffolding protein in cancer. Mol. Cancer Res. 2012;10:1265–1270. doi: 10.1158/1541-7786.MCR-12-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simister P.C., Feller S.M. Order and disorder in large multi-site docking proteins of the Gab family--implications for signalling complex formation and inhibitor design strategies. Mol. Biosyst. 2012;8:33–46. doi: 10.1039/c1mb05272a. [DOI] [PubMed] [Google Scholar]
- 46.Zhao C., Yu D.H., Shen R., Feng G.S. Gab2, a new pleckstrin homology domain-containing adapter protein, acts to uncouple signaling from ERK kinase to Elk-1. J. Biol. Chem. 1999;274:19649–19654. doi: 10.1074/jbc.274.28.19649. [DOI] [PubMed] [Google Scholar]
- 47.Bentires-Alj M., Gil S.G., Chan R., Wang Z.C., Wang Y., Imanaka N., Harris L.N., Richardson A., Neel B.G., Gu H. A role for the scaffolding adapter GAB2 in breast cancer. Nat Med. 2006;12:114–121. doi: 10.1038/nm1341. [DOI] [PubMed] [Google Scholar]
- 48.Lee S.H., Jeong E.G., Nam S.W., Lee J.Y., Yoo N.J., Lee S.H. Increased expression of Gab2, a scaffolding adaptor of the tyrosine kinase signalling, in gastric carcinomas. Pathology. 2007;39:326–329. doi: 10.1080/00313020701329773. [DOI] [PubMed] [Google Scholar]
- 49.Toto A., Bonetti D., De Simone A., Gianni S. Understanding the mechanism of binding between Gab2 and the C terminal SH3 domain from Grb2. Oncotarget. 2017;8:82344–82351. doi: 10.18632/oncotarget.19323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malagrinò F., Troilo F., Bonetti D., Toto A., Gianni S. Mapping the allosteric network within a SH3 domain. Sci. Rep. 2019;9:8279. doi: 10.1038/s41598-019-44656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., Jang H., Zhang M., Tsai C.-J., Maloney R., Nussinov R. The structural basis of BCR-ABL recruitment of GRB2 in chronic myelogenous leukemia. Biophys. J. 2022;121:2251–2265. doi: 10.1016/j.bpj.2022.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fath I., Schweighoffer F., Rey I., Multon M.C., Boiziau J., Duchesne M., Tocqué B. Cloning of a Grb2 isoform with apoptotic properties. Science. 1994;264:971–974. doi: 10.1126/science.8178156. [DOI] [PubMed] [Google Scholar]
- 53.Seiler C., Stainthorp A.K., Ketchen S., Jones C.M., Marks K., Quirke P., Ladbury J.E. The Grb2 splice variant, Grb3-3, is a negative regulator of RAS activation. Commun. Biol. 2022;5:1029. doi: 10.1038/s42003-022-03985-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kardinal C., Konkol B., Lin H., Eulitz M., Schmidt E.K., Estrov Z., Talpaz M., Arlinghaus R.B., Feller S.M. Chronic myelogenous leukemia blast cell proliferation is inhibited by peptides that disrupt Grb2-SoS complexes. Blood. 2001;98:1773–1781. doi: 10.1182/blood.v98.6.1773. [DOI] [PubMed] [Google Scholar]
- 55.Gay B., Suarez S., Caravatti G., Furet P., Meyer T., Schoepfer J. Selective GRB2 SH2 inhibitors as anti-Ras therapy. Int. J. Cancer. 1999;83:235–241. doi: 10.1002/(sici)1097-0215(19991008)83:2<235::aid-ijc15>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 56.Fretz H., Furet P., Garcia-Echeverria C., Schoepfer J., Rahuel J. Structure-based design of compounds inhibiting Grb2-SH2 mediated protein-protein interactions in signal transduction pathways. Curr Pharm Des. 2000;6:1777–1796. doi: 10.2174/1381612003398546. [DOI] [PubMed] [Google Scholar]
- 57.Furet P., Caravatti G., Denholm A.A., Faessler A., Fretz H., García-Echeverría C., Gay B., Irving E., Press N.J., Rahuel J., Schoepfer J., Walker C.V. Structure-based design and synthesis of phosphinate isosteres of phosphotyrosine for incorporation in Grb2-SH2 domain inhibitors. Part 1. Bioorg Med Chem Lett. 2000;10:2337–2341. doi: 10.1016/s0960-894x(00)00475-3. [DOI] [PubMed] [Google Scholar]
- 58.Walker C.V., Caravatti G., Denholm A.A., Egerton J., Faessler A., Furet P., García-Echeverría C., Gay B., Irving E., Jones K., Lambert A., Press N.J., Woods J. Structure-based design and synthesis of phosphinate isosteres of phosphotyrosine for incorporation in Grb2-SH2 domain inhibitors. Part 2. Bioorg Med Chem Lett. 2000;10:2343–2346. doi: 10.1016/s0960-894x(00)00476-5. [DOI] [PubMed] [Google Scholar]
- 59.Oligino L., Lung F.D., Sastry L., Bigelow J., Cao T., Curran M., Burke T.R., Wang S., Krag D., Roller P.P., King C.R. Nonphosphorylated peptide ligands for the Grb2 Src homology 2 domain. J. Biol. Chem. 1997;272:29046–29052. doi: 10.1074/jbc.272.46.29046. [DOI] [PubMed] [Google Scholar]
- 60.Quartararo J.S., Eshelman M.R., Peraro L., Yu H., Baleja J.D., Lin Y.-S., Kritzer J.A. A bicyclic peptide scaffold promotes phosphotyrosine mimicry and cellular uptake. Bioorg. Med. Chem. 2014;22:6387–6391. doi: 10.1016/j.bmc.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quartararo J.S., Wu P., Kritzer J.A. Peptide bicycles that inhibit the Grb2 SH2 domain. Chembiochem. 2012;13:1490–1496. doi: 10.1002/cbic.201200175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malagrinò F., Coluccia A., Bufano M., Regina G.L., Puxeddu M., Toto A., Visconti L., Paone A., Magnifico M.C., Troilo F., Cutruzzolà F., Silvestri R., Gianni S. Targeting the interaction between the SH3 domain of Grb2 and Gab2. Cells. 2020;9:2435. doi: 10.3390/cells9112435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bufano M., Puxeddu M., Nalli M., La Regina G., Toto A., Liberati F.R., Paone A., Cutruzzolà F., Masci D., Bigogno C., Dondio G., Silvestri R., Gianni S., Coluccia A. Targeting the Grb2 cSH3 domain: design, synthesis and biological evaluation of the first series of modulators. Bioorg. Chem. 2023;138 doi: 10.1016/j.bioorg.2023.106607. [DOI] [PubMed] [Google Scholar]
- 64.Gay B., Suarez S., Weber C., Rahuel J., Fabbro D., Furet P., Caravatti G., Schoepfer J. Effect of potent and selective inhibitors of the Grb2 SH2 domain on cell motility. J. Biol. Chem. 1999;274:23311–23315. doi: 10.1074/jbc.274.33.23311. [DOI] [PubMed] [Google Scholar]
- 65.Lung F.-D.T., Chang C.-W., Chong M.-C., Liou C.-C., Li P., Peach M.L., Nicklaus M.C., Lou B.-S., Roller P.P. Small nonphosphorylated Grb2-SH2 domain antagonists evaluated by surface plasmon resonance technology. Biopolymers. 2005;80:628–635. doi: 10.1002/bip.20209. [DOI] [PubMed] [Google Scholar]
- 66.Giubellino A., Shi Z.-D., Jenkins L.M.M., Worthy K.M., Bindu L.K., Athauda G., Peruzzi B., Fisher R.J., Appella E., Burke T.R., Bottaro D.P. Selectivity and mechanism of action of a growth factor receptor-bound protein 2 SRC homology 2 domain binding antagonist. J. Med. Chem. 2008;51:7459–7468. doi: 10.1021/jm800523u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rojas M., Yao S., Lin Y.Z. Controlling epidermal growth factor (EGF)-stimulated Ras activation in intact cells by a cell-permeable peptide mimicking phosphorylated EGF receptor. J. Biol. Chem. 1996;271:27456–27461. doi: 10.1074/jbc.271.44.27456. [DOI] [PubMed] [Google Scholar]
- 68.Ye B., Akamatsu M., Shoelson S.E., Wolf G., Giorgetti-Peraldi S., Yan X., Roller P.P., Burke T.R. L-O-(2-malonyl)tyrosine: a new phosphotyrosyl mimetic for the preparation of Src homology 2 domain inhibitory peptides. J. Med. Chem. 1995;38:4270–4275. doi: 10.1021/jm00021a016. [DOI] [PubMed] [Google Scholar]
- 69.Ye Y.-B., Lin J.-Y., Chen Q., Liu F., Chen H.-J., Li J.-Y., Liu W.-Q., Garbay C., Vidal M. The cytotoxicity of a Grb2-SH3 inhibitor in Bcr-Abl positive K562 cells. Biochem. Pharmacol. 2008;75:2080–2091. doi: 10.1016/j.bcp.2007.12.021. [DOI] [PubMed] [Google Scholar]
- 70.Terasawa H., Kohda D., Hatanaka H., Tsuchiya S., Ogura K., Nagata K., Ishii S., Mandiyan V., Ullrich A., Schlessinger J. Structure of the N-terminal SH3 domain of GRB2 complexed with a peptide from the guanine nucleotide releasing factor Sos. Nat. Struct. Biol. 1994;1:891–897. doi: 10.1038/nsb1294-891. [DOI] [PubMed] [Google Scholar]
- 71.Gril B., Liu W.Q., Lenoir C., Garbay C., Vidal M. Affinity chromatography for purification of the modular protein growth factor receptor-bound protein 2 and development of a screening test for growth factor receptor-bound protein 2 Src homology 3 domain inhibitor using peroxidase-linked ligand. Anal. Biochem. 2006;351:93–99. doi: 10.1016/j.ab.2005.12.032. [DOI] [PubMed] [Google Scholar]
- 72.Liu F., Giubellino A., Simister P.C., Qian W., Giano M.C., Feller S.M., Bottaro D.P., Burke T.R. Application of ring-closing metathesis to Grb2 SH3 domain-binding peptides. Biopolymers. 2011;96:780–788. doi: 10.1002/bip.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bai X., Lee J.-Y., Kim T.I., Dai F., Lee T.-J., Hong S.-J. Molecular cloning and characterization of growth factor receptor bound-protein in clonorchis sinensis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0085577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cailliau K., Browaeys-Poly E., Broutin-L’Hermite I., Nioche P., Garbay C., Ducruix A., Vilain J.P. Grb2 promotes reinitiation of meiosis in Xenopus oocytes. Cell. Signal. 2001;13:51–55. doi: 10.1016/s0898-6568(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 75.Browaeys-Poly E., Cailliau K., Vilain J.P. Signal transduction pathways triggered by fibroblast growth factor receptor 1 expressed in Xenopus laevis oocytes after fibroblast growth factor 1 addition. Role of Grb2, phosphatidylinositol 3-kinase, Src tyrosine kinase, and phospholipase Cgamma. Eur. J. Biochem. 2000;267:6256–6263. doi: 10.1046/j.1432-1327.2000.01710.x. [DOI] [PubMed] [Google Scholar]
- 76.Aroca P., Mahadevan D., Santos E. Functional interactions between isolated SH2 domains and insulin/Ras signaling pathways of Xenopus oocytes: opposite effects of the carboxy- and amino-terminal SH2 domains of p85 PI 3-kinase. Oncogene. 1996;13:1839–1846. [PubMed] [Google Scholar]