Abstract
The hRFC (human reduced folate carrier) is ubiquitously but differentially expressed in human tissues and its levels are regulated by up to seven non-coding regions (A1, A2, A, B, C, D and E) and at least four promoters. For the hRFC-B basal promoter, regulation involves binding of Sp (specificity protein) transcription factors to a critical GC-box. By transiently transfecting HT1080 cells with 5′- and 3′-deletion constructs spanning 1057 bp of upstream sequence, a transcriptionally important region was localized to 158 bp flanking the transcriptional start sites. By gel shift and chromatin immunoprecipitation assays, USF (upstream stimulatory factor), Sp1 and Ikaros-related proteins were bound to consensus elements (one E-box, two GC-box and three Ikaros) within this region. The functional importance of these elements was confirmed by transient tranfections of HT1080 cells with hRFC-B reporter constructs in which they were mutated, and by co-transfections of Drosophila Mel-2 cells with wild-type hRFC-B promoter and expression constructs for USF1, USF2a, Sp1 and Ikaros 2 and 8. Both USF1 and Sp1 proteins transactivated the hRFC-B promoter. Sp1 combined with USF1 resulted in a synergistic transactivation. Identical results were obtained with USF2a. Ikaros 2 was a repressor of hRFC-B promoter activity whose effects were partly reversed by the dominant-negative Ikaros 8. In HT1080 cells, transfection with Ikaros 2 decreased endogenous hRFC-B transcripts, whereas USF1 and Sp1 increased transcript levels. Ikaros 2 also decreased reporter gene activity and levels of acetylated chromatin associated with the endogenous promoter. Collectively, these results identify transcriptionally important regions in the hRFC-B promoter that include multiple GC-box, Ikaros and E-box elements. Our results also suggest that co-operative interactions between transcription factors Sp1 and USF are essential for high-level hRFC-B transactivation and imply that these effects are modulated by the family of Ikaros proteins and by histone acetylation.
Keywords: chromatin, Ikaros, methotrexate, reduced folate carrier, Sp1, upstream stimulatory factor (USF)
Abbreviations: ALL, acute lymphoblastic leukaemia; BST2, bone marrow stromal cell antigen 2; ChIP, chromatin immunoprecipitation; HDAC, histone deacetylase; Mtx, methotrexate; RFC, reduced folate carrier; hRFC, human RFC; 5′-RACE, 5′-rapid amplification of cDNA ends; Sp1, specificity protein 1; USF, upstream stimulatory factor; 5′-UTR, 5′-untranslated region; wt, wild-type
INTRODUCTION
Reduced folates are essential cofactors for one-carbon transfer reactions in anabolic pathways leading to the synthesis of purines, thymidylate, serine and methionine [1]. Since mammals lack the ability to synthesize reduced folates de novo, cellular uptake of these derivatives is essential for cell growth and survival. The primary route for the membrane transport of reduced folate cofactors is via the RFC (reduced folate carrier) [2,3].
RFC is also responsible for the cellular uptake of ‘classical’ antifolate cancer chemotherapeutic drugs, e.g. Mtx (Methotrexate), Tomudex and Pemetrexed [2,4]. Impaired transport frequently occurs accompanying exposure of human and rodent cells to antifolate drugs in vitro [5–7] and in murine tumour cells in vivo after chemotherapy with Mtx [8]. Furthermore, alterations in the levels of hRFC (human RFC) were associated with Mtx resistance in leukaemias [9] and solid tumours [10] from patients. In B-precursor ALL (acute lymphoblastic leukaemia) lymphoblasts, hRFC expression spans a wide range and is proportional to Mtx uptake [11].
Previous studies suggested a complex regulation of hRFC gene expression [12–16]. We reported recently that hRFC is ubiquitously but differentially expressed in human tissues and cell lines, and identified up to seven non-coding regions (designated A1, A2, A, B, C, D and E) for the hRFC gene in assorted human tissues spanning approx. 35 kb upstream of the major translational start [15]. After splicing, at least 18 potential hRFC transcripts would be generated with unique 5′-UTRs (5′ untranslated regions) fused to the same open reading frame. By analogy with other multi-promoter genes, the unique non-coding exons for hRFC are probably transcribed from distinct promoters. To date, promoter activity has been confirmed for the 5′ regions proximal to the non-coding A2, A, B and C regions [13–16].
To better understand the basis for varied levels of hRFC in human tissues and tumours, we have begun to identify and characterize critical transcription factors involved in regulating the hRFC promoters. We have reported previously that the hRFC-A and -B basal promoters are regulated by different families of transcription factors, including the bZip superfamily (e.g. c-Jun/c-Fos and Creb1/ATF1) and the Sp (specificity protein) family of DNA-binding proteins (e.g. Sp1 and Sp3) respectively [14]. Our results imply that cell-specific expression of these transcription factors can profoundly influence patterns of promoter and 5′-UTR usage at the basal promoter level. However, the net effect of these factors could be overshadowed or enhanced by transcription factor binding to other regulatory elements both upstream and downstream of the minimal hRFC promoter regions, as well as by the state of the surrounding chromatin structure. For instance, promoter A is transactivated by AP2 and Sp1 in co-transfection experiments, via binding to upstream elements [17]. The effects of Sp1 on promoter A activity were different, depending on the identity of the bZip protein bound to the basal region.
In the present study, we describe extensive studies of promoter B, a major hRFC promoter in immortalized cell lines and primary leukaemias from patients [15,16]. Our results document transcriptionally important regions in the hRFC-B promoter that include GC-box, E-box and Ikaros elements. Our results also suggest that co-operative interactions between the transcription factors Sp1 and USF (upstream stimulatory factor) are essential for high-level hRFC-B transactivation and imply that these effects can be modulated by the family of Ikaros proteins and by histone acetylation/deacetylation.
MATERIALS AND METHODS
Chemicals and reagents
[γ-32P]ATP (3000 Ci/mmol) was purchased from PerkinElmer (Boston, MA, U.S.A.), synthetic oligonucleotides were from Sigma-GenoSys Biotechnologies (The Woodlands, TX, U.S.A.), Lipofectin® was from Gibco Life Technologies (Gaithersburg, MD, U.S.A.), and restriction and modifying enzymes, reporter gene vectors (pGL3-Basic and pRLSV40) and other molecular biologicals were obtained from Promega (Madison, WI, U.S.A.). The pCMV-Sp1, pPacO and pPacSp1 plasmid constructs were gifts from Dr Robert Tjian (University of California, Berkeley, CA, U.S.A.). The pPacSp3 construct was provided by Dr Guntram Suske (Philipps-Universitat, Marburg, Germany).
Cell culture
The HT1080 human fibrosarcoma cell line was obtained from the A.T.C.C. (Rockville, MD, U.S.A.). HT1080 cells were grown in RPMI 1640 medium with 10% (v/v) heat-inactivated iron-supplemented calf serum (Hyclone, Logan, UT, U.S.A.), 2 mM L-glutamine, 100 units/ml penicillin and 100 μg/ml streptomycin, in an atmosphere of 5% CO2 and 95% air at 37 °C. Drosophila Mel-2 cells were purchased from Invitrogen and were maintained at ambient temperature in Schneider's insect medium, supplemented with 10% (v/v) fetal bovine serum (Gibco Life Technologies) and 2 mM glutamine plus antibiotics.
hRFC-luciferase reporter constructs
The full-length hRFC-B/−5422 construct [positions −5422 to −4364; the numbering is relative to the ATG translation start in exon 1 and is based on the nucleotide sequence of chromosome 21 contig HS21C102 (accession number AL163302)] was described by Whetstine and Matherly [14] (previously designated as hRFC-B/−2016). 5′ deleted hRFC-B constructs were prepared from hRFC-B/−5422 by successive exonuclease III and S1 nuclease digestions (hRFC-B/−4493 and hRFC-B/−4448) and restriction endonuclease digestions [hRFC-B/−4693(ApaI), hRFC-B/−4615(BstEII), hRFC-B/−4590(BsmBI) and hRFC-B/−4467(BssHII)]. A 3′-deletion was generated in hRFC-B/−5422 by restriction endonuclease digestion with BssHII. All deletion constructs were verified by automated DNA sequencing at the Wayne State University Center of Molecular Genetics Sequencing Facility.
Specific DNA-binding sites in promoter B were mutated by PCR, using the hRFC-B/−5422 construct as a template. All of the mutant constructs [hRFC-B/−4501MIK(a), hRFC-B/−4565MIK(b), hRFC-B/−4573MIK(c), hRFC-B/−4501MGC(a), hRFC-B/−4534MGC(b) and hRFC-B/−4434ME(a)] were generated by splice-overlap-extension (‘Soeing’) PCR [18] using the mutant primers shown in Table 1. Primary PCR amplifications used the mutant antisense primers with the upstream ProB (XhoI) primer [5′-CTGCCTCGAGGGTACCACGGTGGCCGTCCCCTTCTGTTCTGTGCAG-3′] and the mutant sense primer with the downstream 33–54 wt primer (5′-CTAGCTAAGCTTGGTACCTGCGACTCGGCGGGGCGGCTGCCCTGGGGCTCCCGGACCCGGCCCCGCGCACGC-3′). The primary amplicons were isolated and mixed for the secondary PCR amplifications with the ProB (XhoI) and 33–54 wt (wild-type) primers. The following PCR conditions were used: (i) for the primary PCR amplification, 95 °C for 15 s, 65 °C for 45 s and 72 °C for 60 s for 40 cycles; and (ii) for the secondary PCR amplification, 95 °C for 15 s, 68 °C for 45 s and 72 °C for 60 s for 40 cycles. For all mutants, the secondary PCR products were isolated from 2% (v/v) agarose and digested with XhoI and HindIII. The digested fragments were subcloned into pGL3-Basic in the sense orientation and each mutation was verified by automated DNA sequencing.
Table 1. Primers used to create mutant promoter B constructs.
| Construct | Position | Antisense primer* | Sense primer* |
|---|---|---|---|
| hRFC-B/−4573MIK(c) | −4573 to −4554 | 5′-CCCACGGGGCACCCCCCCAC-3′ | 5′-GTGGGGGGGTGCCCCGTGGG-3′ |
| hRFC-B/−4565MIK(b) | −4565 to −4545 | 5′-AGGAGCGCCCCCACGGGGCAC-3′ | 5′-GTGCCCCGTGGGGGCGCTCCT-3′ |
| hRFC-B/−4534MGC(b) | −4534 to −4506 | 5′-CCTGAGGGCAAAGCCTCGCGCGTGGCCGG-3′ | 5′-CCGGCCACGCGCGAGGCTTTGCCCTCAGG-3′ |
| hRFC-B/−4501MIK(a) | −4501 to −4475 | 5′-GGGGTGGGCGGGTCCGTAAAGCCGAAC-3′ | 5′-GTTCGGCTTTACGGACCCGCCCACCCC-3′ |
| hRFC-B/−4501MGC(a) | −4501 to −4475 | 5′-GGGGTAAACGGGTCCGTCCCGCCGAAC-3′ | 5′-GTTCGGCGGGACGGACCCGTTTACCCC-3′ |
| hRFC-B/−4434ME(a) | −4434 to −4414 | 5′-CCGCGCAATCGGACTCCGGGA-3′ | 5′-TCCCGGAGTCCGATTGCGCGG-3′ |
* Mutant antisense and sense primers were used for ‘Soeing’ PCR [18] to construct the mutant promoter B constructs, as described in the Materials and methods section. The mutated nucleotides are underlined.
Preparation of transcription factor expression constructs
The pCDNA3-USF1, pPacUSF1 and pPacUSF2a constructs were prepared as described by Ge et al. [20]. For preparing the Ikaros 2 and 8 expression constructs, Ikaros 2 and 8 coding sequences were PCR-amplified from cDNA prepared from CCRF-CEM cells using forward (5′-GCGGGATCCATGGATGCTGATGAGGGTCAAGACAT; BamHI site is underlined) and reverse (5′-TCCCTCGAGTTAGCTCATGTGGAAGCGGTGCTCCC; XhoI site is underlined) primers. The amplicons were digested with BamHI and XhoI and subcloned into pPacO and pCDNA3 at the BamHI and XhoI sites. All constructs were confirmed by automated DNA sequencing.
Transient transfections and reporter gene assays
HT1080 cells were transiently transfected with 3 μg of hRFC-B promoter constructs (in pGL3-Basic) and 25 ng of pRLSV40 plasmid using Lipofectin® [14]. After 48 h, lysates were prepared, and firefly luciferase activities were assayed using the dual luciferase kit (Promega) in a Turner 20/20 luminometer. Firefly luciferase activity was normalized to Renilla luciferase activity.
Fugene™ 6 reagent (Roche, Indianapolis, IN, U.S.A.) was used for transfections into Drosophila Mel-2 cells. For co-transfections, 1 μg of the hRFC-B/−5422 construct in pGL3-Basic was combined with the following expression constructs: 25–100 ng of Sp1 or Sp3 (pPacSp1 or pPacSp3 respectively); 125–500 ng of USF1 or USF2a (pPacUSF1 and pPacUSF2a respectively); and/or 100 or 200 ng of the Ikaros 2 and 8 isoforms (pPacIk2 and pPacIk8 respectively). Cells were harvested after 24 h for luciferase assays using the Single Luciferase Assay System (Promega). Luciferase activities were normalized to cellular protein contents.
For all transfections, three or more experiments were performed in duplicate.
Gel mobility-shift assays
Nuclear extracts from HT1080 cells were prepared as described previously [14], and 12–15 μg of nuclear extract were used in each binding reaction. Gel shift assays were performed exactly as described previously [14]. The [γ-32P]ATP end-labelled probes and competitive mutant oligonucleotides used in the binding reactions are summarized in Table 2. Competition experiments included 150-fold molar excess of the unlabelled wt hRFC oligonucleotides, hRFC oligonucleotides containing mutations in the DNA-binding sites for specific transcription factors or oligonucleotides containing consensus transcription elements, including IKBS1 and IKBS4 [19], GC-box and E-box (both GC- and E-box were obtained from Santa Cruz Biotechnology, Santa Cruz, CA, U.S.A.). DNA–protein complexes were supershifted with antisera to USF-1 (Santa Cruz Biotechnology; sc-229) and Sp1 (Active Motif, Carlsbad, CA, U.S.A. and Santa Cruz Biotechnology; sc-420). The gels were dried and visualized by autoradiography.
Table 2. Wt and mutant hRFC-B oligonucleotides used for gel shift assays.
| Wt | Mutant* | |||
|---|---|---|---|---|
| Position | Name | Sequence | Name | Sequence |
| −4573 to −4554 | hRFC-B/−4573 | 5′-GTGGGAGGGTGCCCCGTGGG-3′ | hRFC-B/−4573MIK(c) | 5′-GTGGGGGGGTGCCCCGTGGG-3′ |
| −4565 to −4545 | hRFC-B/−4565 | 5′-GTGCCCCGTGGGGACGCTCCT-3′ | hRFC-B/−4565MIK(b) | 5′-GTGCCCCGTGGGGGCGCTCCT-3′ |
| −4534 to −4506 | hRFC-B/−4534 | 5′-CCGGCCACGCGCGAGGCCCCGCCCTCAGG-3′ | hRFC-B/−4534MGC(b) | 5′-CCGGCCACGCGCGAGGCTTTGCCCTCAGG-3′ |
| −4501 to −4475 | hRFC-B/−4501 | 5′-GTTCGGCGGGACGGACCCGCCCACCCC-3′ | hRFC-B/−4501MIK(a) | 5′-GTTCGGCTTTACGGACCCGCCCACCCC-3′ |
| hRFC-B/−4501MGC(a) | 5′-GTTCGGCGGGACGGACCCGTTTACCCC-3′ | |||
| −4434 to −4414 | hRFC-B/−4434 | 5′-TCCCGGAGTCCGCGTGCGCGG-3′ | hRFC-B/−4434ME(a) | 5′-TCCCGGAGTCCGATTGCGCGG-3′ |
* The mutated nucleotides are underlined.
Real-time PCR quantification of hRFC-B transcripts
Total RNAs were extracted from HT1080 cells (∼80% confluent in 60 mm dishes) transfected with Ikaros 2 (3 μg), USF1 (3 μg) and Sp1 (75 ng) cDNAs in pCDNA3 or pCDNA3 mock-transfected cells using TRI Reagent (Molecular Research Center). cDNAs were prepared from 1 μg of RNA using random hexamer primers and an RT (reverse transcriptase)–PCR kit (PerkinElmer), and purified with the QIAquick PCR Purification Kit (Qiagen). hRFC-B transcripts and 18 S RNA levels were quantified using a LightCycler real-time PCR machine (Roche). The PCR mixture contained 2 μl of purified cDNA or standard plasmid, 4 mM MgCl2, 0.5 μM each of sense and antisense primers and 2 μl of FastStart DNA Master SYBR Green I enzyme-SYBR reaction mix (Roche). Primers included exon B sense (5′-CGAGTCGCAGGCACAGTGTCAG-3′) and antisense (5′-ACGAGGTGCCGCCAGGACCG-3′) primers, and 18 S RNA sense (GATGCGGGGCGTTATT-3′) and antisense (5′-TGAGGTTTCCCGTGTTGTCA-3′) primers. PCR conditions consisted of an initial denaturing step of 95 °C for 600 s (‘hot start’), amplification with 35–55 cycles of 95 °C, 59 °C for 10 s and 72 °C for 5 s, followed by melting curve analysis from 40 to 95 °C, and a final cooling step to 40 °C. External standard curves were constructed using serial dilutions of the hRFC-B and 18 S RNA amplicons, which were amplified with the above primers, cloned into pGEM T-Easy vector and linearized with SalI and ApaI respectively. Levels of hRFC-B transcripts and 18 S RNA were calculated from their respective standard curves; hRFC-B levels were normalized to 18 S RNA. Real-time PCR results were expressed as relative transcript levels±S.E.M. for three experiments.
ChIP (chromatin immunoprecipitation) assay
ChIP assays were performed in HT1080 cells as described previously [20,21] with antibodies to Sp1 (Active Motif), USF1 (Santa Cruz Biotechnology), Ikaros (sc9861; Santa Cruz Biotechnology) and acetyl histone H3 (06-599; Upstate Biotechnology, Lake Placid, NY, U.S.A.). Standard PCR for the hRFC-B promoter region was performed with forward (5′-CCGTGGGGACGCTCCTGCCGCA-3′) and reverse (5′-CGTTCCCCACCGGTACCTGCGACT-3′) primers spanning positions −4560 to −4353 using a GC-rich reagent and polymerase (Roche). An unrelated gene [BST2 (bone marrow stromal cell antigen 2)] was also amplified with forward (5′-CCTGCTCGGCTTTTCGCTTGAACAT-3′) and reverse (5′-CGGAGGGAGGCTCTGGAGGGAGAC-3′) primers to validate the specificity of the ChIP assays. PCR conditions were designed to ensure linearity and were 95 °C for 5 min, followed by 32 cycles of 95 °C for 30 s, 62 °C for 45 s and 72 °C for 45 s.
RESULTS
Deletion analysis of the hRFC-B promoter
The hRFC-B promoter is a major promoter for hRFC in immortalized cells and primary leukaemia cells [15,16]. We previously localized the minimal promoter region for hRFC-B to within 46 bp (positions −4493 to −4448) and demonstrated an important regulatory role for the Sp family of transcription factors in binding to a GC-box consensus element [designated GC-box (a)] within this region [14]. Since Sp1/Sp3 proteins are ubiquitously expressed [22], significant differences in hRFC-B transcripts between tissues [15] probably reflect contributions of other cis regulatory elements and transcription factors in regulating promoter activity.
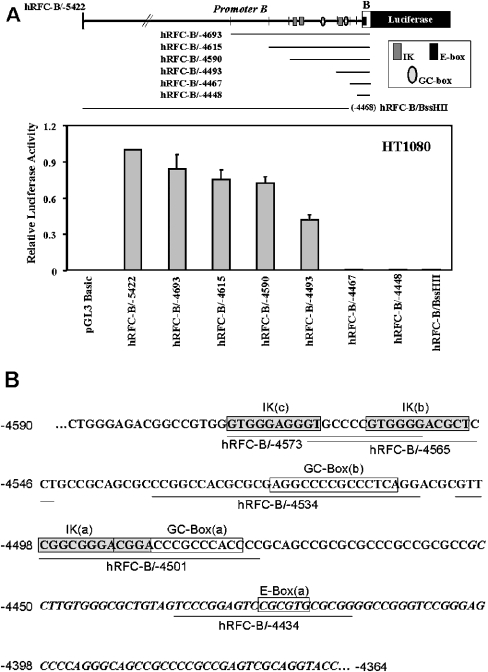
Based on the high frequency of clones with exon B sequence in HT1080 cells by 5′-RACE (5′ rapid amplification of cDNA ends) assay [15], this cell line was used to characterize further the transcriptional regulation of the hRFC-B promoter. 5′- and 3′-deletion constructs were prepared from the full-length hRFC-B/−5422 promoter construct (positions −5422 to −4364) by digesting with exonuclease III and S1 nuclease or with restriction endonucleases. The deletion constructs were subcloned into pGL3-Basic vector and transiently transfected into HT1080 human fibrosarcoma cells to compare luciferase reporter gene activities with that for the hRFC-B/−5422 construct (Figure 1A). Significant shifts in promoter activity were localized between positions −4590 and −4467 for the 5′-deletions, and between positions −4364 and −4468 (BssHII) for the 3′-deletion (Figure 1A). Putative transcription factor binding sites localized within these regions include sequentially (from the 5′ boundary), Ikaros (c), Ikaros (b), GC-box (b), Ikaros (a), GC-box (a) and E-box (a) (Figure 1B).
Figure 1. Deletion analysis of the hRFC-B promoter in HT1080 cells and promoter B sequence.
Important regulatory regions for promoter B were identified by 5′- and 3′-deletions introduced into the full-length hRFC-B/−5422 promoter construct (positions −5422 to −4364). (A) Upper panel: schematic of the 5′- and 3′-deletions, in relation to specific cis elements including Ikaros (IK), GC- and E-box elements; lower panel: relative luciferase activity for each deletion when compared with the full-length hRFC-B/−5422 construct. Promoter constructs in pGL3-Basic were transiently expressed in HT1080 cells for luciferase assays. Data are reported as relative firefly luciferase activities, normalized to Renilla luciferase activities. S.E.M. are shown by the error bars. (B) The nucleotide sequence for promoter B is shown in the sense direction and the numbering corresponds to the hRFC-B/−4590 promoter construct in (A) and is relative to the ATG translation start site in coding exon 1. The putative major cis elements are boxed. The italicized letters designate the 5′-end of exon B deduced from 5′-RACE analysis [15]. The locations of the hRFC-B oligonucleotide probes used in the gel shift analyses are also shown.
In vitro and in vivo binding of USF, Sp1 and Ikaros transcription factor families to the hRFC-B promoter
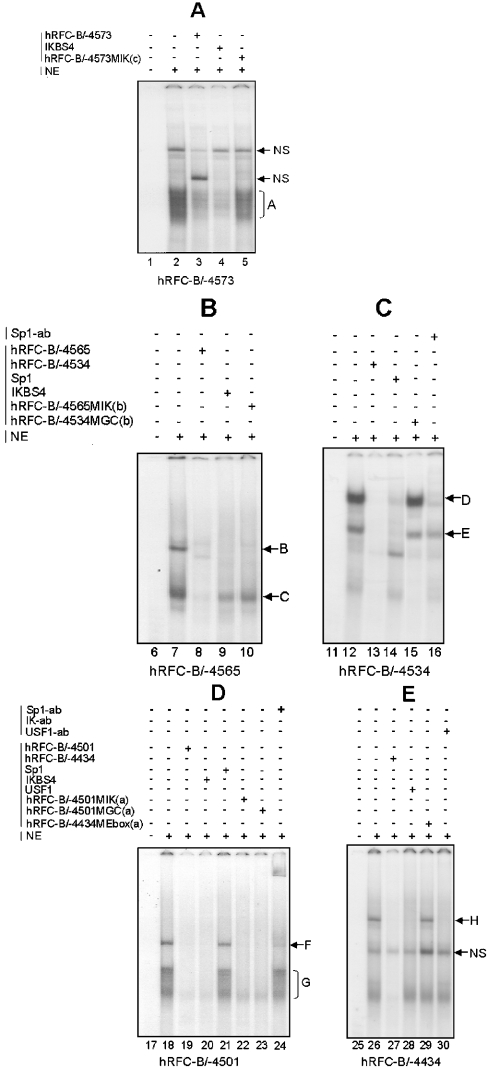
The regions that showed major fluctuations in promoter activity with progressive 5′- and 3′-deletions (Figure 1A) were obvious candidates for in vitro analyses of transcription factor binding. Sp1/Sp3 binding to GC-box (a) was previously confirmed on gel shifts using an oligonucleotide probe from positions −4493 to −4448 [14], not including the Ikaros (a) binding site. Additional gel shifts were performed with nuclear extracts prepared from HT1080 cells and double-stranded oligonucleotide probes spanning positions −4573 to −4414 (Figures 2A–2E) and including the putative transcription factor binding sites shown in Figure 1(B). A total of eight DNA–protein complexes (A–H) were identified that were effectively competed by unlabelled hRFC oligonucleotides, establishing binding specificity.
Figure 2. In vitro binding of USF, Ikaros and Sp proteins to the hRFC-B promoter.
Gel shift assays were performed with HT1080 nuclear extracts as described in the Materials and methods section. Double-stranded hRFC-B/−4573 (positions −4573 to −4554), hRFC-B/−4565 (−4565 to −4545), hRFC-B/4501 (−4501 to −4475), hRFC-B/−4534 (−4534 to −4506) and hRFC-B/−4434 (−4434 to −4414) oligonucleotides were used (Figure 1B and Table 2). Results are shown for labelled probes without HT1080 nuclear extract (lanes 1, 6, 11, 17 and 25), and with 12 μg of extract (lanes 2, 7, 12, 18 and 26). Competitions (150-fold molar excess) included unlabelled hRFC-B/−4573 (lane 3), hRFC-B/−4565 (lane 8), hRFC-B/−4534 (lane 13), hRFC-B/−4501 (lane 19) and hRFC-B/−4434 (lane 27) oligonucleotides, an Ikaros consensus oligonucleotide (IKBS4; lanes 4, 9 and 20), an Sp consensus oligonucleotide (lanes 14 and 21), an E-box consensus oligonucleotide (lane 28), hRFC-B mutant oligonucleotides (lanes 5, 10, 15, 22, 23 and 29), Sp1 supershifts (lanes 16 and 24) and USF1 supershifts (lane 30). Complexes are indicated by upper-case letters and are described in the text. NS, non-specific complex; NE, nuclear extract; ab, antibody.
With the hRFC-B/−4573 (positions −4573 to −4554), hRFC-B/−4565 (positions −4565 to −4545) and hRFC-B/−4501 (positions −4501 to −4475) oligonucleotides, a series of complexes (designated A, B, C, F and G in Figure 2, lanes 2, 7, and 18) were identified that were effectively competed by IKBS1 (not shown) and IKBS4 Ikaros consensus oligonucleotides (lanes 4, 9 and 20). For complexes A, B and C, but not F or G, competition was at least partly abolished with hRFC oligonucleotides including mutant Ikaros binding sites [hRFC-B/−4573MIK(c), hRFC-B/−4565MIK(b) and hRFC-B/−4501MIK(a); mutant sequences shown in Table 2] (lanes 5, 10 and 22). These results strongly imply that complexes A, B, C, F and G all involve binding by DNA-binding members of the Ikaros family of proteins (Ikaros 1, 2 and 3) [19] to the Ikaros (c), (b) or (a) sequence elements depicted in Figure 1(B). The failure of the mutant hRFC-B/−4501MIK(a) oligonucleotide to abolish competition at the Ikaros (a) element is considered below.
Complexes D and E (hRFC-B/−4534; lane 12) were identified as involving binding of the Sp family of proteins to GC-box (b) by competition with a Sp1 consensus oligonucleotide (lane 14), perturbation of the DNA–protein complex by Sp1 antiserum (lanes 16) and loss of competition after mutation of GC-box (b) [hRFC-B/−4534MGC(b)] (lane 15). However, for the hRFC-B/−4501 oligonucleotide, including the previously characterized GC-box (a) [14], anomalous results were obtained since (i) the IKBS4 Ikaros consensus sequence completely abolished DNA binding for both complexes F and G (lane 20), yet (ii) Sp1 consensus sequence partially competed with complex F (lane 21) and (iii) Sp1 antiserum supershifted most of complex F (lane 24). Furthermore, (iv) both Ikaros (a) and GC-box (a) mutants effectively competed for complexes F and G with wt hRFC-B/−4501 sequence (lanes 22 and 23), implying no effect on Ikaros and Sp protein binding. These anomalous results with the hRFC-B/−4501 probe probably reflect the extensive overlap between the Ikaros (a) and GC-box (a) binding sites in this region (Figure 1B). For complex F, the abolition of DNA binding in the presence of IKBS4, and the lack of complete Sp1 competition, combined with the nearly complete supershift by Sp1 antiserum, may partly reflect the presence of multiple DNA–protein complexes co-migrating as a single complex F and/or, possibly, protein–protein associations with Sp1. In spite of these ambiguities, the gel shift results with the hRFC-B/−4501 probe strongly suggest binding of Sp1 and Ikaros proteins to the overlapping Ikaros (a) and GC (a) elements in the −4501 to −4475 region.
Complex H (hRFC-B/−4434 oligonucleotide; lane 26) was identified as a USF protein bound to E-box (a) by competition with a USF1 consensus oligonucleotide (lane 28), loss of competition with mutant hRFC-B/−4434ME(a) oligonucleotide (lane 29) and perturbation of the DNA–protein complex with USF-1-specific antiserum (lane 30).
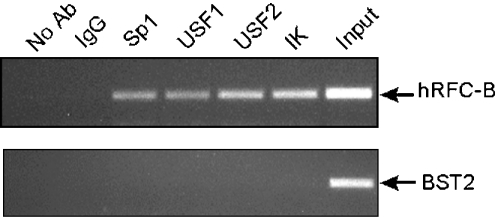
In vivo binding of USF1, USF2, Sp1 and Ikaros transcription factors to the hRFC-B promoter between positions −4560 and −4353 was confirmed by ChIP assays (Figure 3). No binding was detected in a control sequence (BST2) which did not include any transcription factor binding site.
Figure 3. In vivo binding of Ikaros, Sp1, USF-1 and USF-2 to the hRFC-B promoter.
HT1080 chromatin bound to transcription factor complexes were immunoprecipitated with Sp1, USF-1, USF-2 and Ikaros antisera. Immunoprecipitations with IgG and no antibody were used as negative controls. PCR amplifications were performed as described in the Materials and methods section. Upper panel: the PCR products for promoter B (positions: −4560 to −4353) are shown. Lower panel: a negative control for each immunoprecipitation amplified with coding exon-specific primers for the BST2 gene is shown. The amplicons were visualized on 2% agarose gels. Ab, antibody; IK, Ikaros.
Collectively, our in vitro and in vivo binding results demonstrate binding of Ikaros, Sp and USF proteins within a 158 bp region flanking the hRFC-B transcriptional start sites and including the critical GC-box (a) previously implicated in basal promoter regulation [14].
Mutagenesis of transcription factor binding elements in the hRFC-B promoter identified by gel shift assays
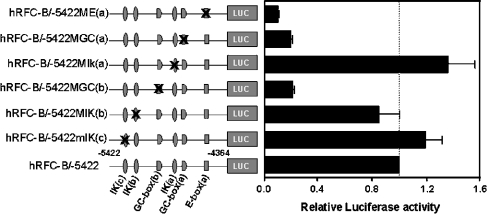
To assess further the functional importance of the major cis elements localized within the 158 bp hRFC-B core promoter (Figure 1B), full-length promoter constructs analogous to hRFC-B/−5422 were prepared containing mutations in the major cis elements identified on the gel shift assays (Table 2). With the exception of GC-box (a) and Ikaros (a), the cis element mutants (see above) significantly perturbed transcription factor binding in vitro. Promoter constructs including these same mutations in pGL3-basic were transfected into HT1080 cells, and luciferase activities were compared with that of the wt hRFC-B/−5422 promoter construct. As summarized in Figure 4, pronounced inhibitions of promoter activity were observed for mutations of both GC-boxes (a) and (b) (∼80% inhibition for both mutants) and E-box (a) (∼90% inhibition). Mutations of the Ikaros-binding sites resulted in variable and opposing effects on promoter activity, ranging from activation [Ikaros (c) and (a); ∼15–30% increase in luciferase activity] to inhibition [Ikaros (b); ∼20% decrease in luciferase activity]. Taken together, these results strongly support the notion that the cis elements depicted in Figure 1(B) and identified on gel shifts are functionally important in the transcriptional regulation of the hRFC-B core promoter.
Figure 4. Effects of mutagenesis of transcription factor binding sites on hRFC-B promoter activity.
Binding sites for the GC-boxes (a) and (b), E-box (a) and Ikaros (IK) (a), (b) and (c) elements (see Figure 1B) were mutated in the hRFC-B/−5422 construct, based on the results of our gel shift experiments. Mutant reporter constructs were transfected into HT1080 cells, and the effects on luciferase activity assayed as described in the Materials and methods section. Results are presented as changes in luciferase activities relative to that for the wt hRFC-B/−5422 promoter construct. Results are expressed as means±S.E.M. from three separate experiments.
Co-transfections of the hRFC-B promoter with Sp, USF and Ikaros expression constructs into Drosophila Mel-2 cells
The Sp and USF proteins are ubiquitously expressed transcription factors, yet are implicated in regulating tissue-specific or developmental expression of a number of genes [22–24]. Sp proteins comprise a small family of transcription factors of which Sp1 and Sp3 were previously identified as important transactivators of the hRFC-B minimal promoter via binding to the GC-box (a) element [14]. USF designates the family of basic helix–loop–helix leucine zipper (bHLH/Zip) DNA-binding proteins and is characterized by a dimerization domain composed of a helix–loop–helix motif, a leucine zipper motif, and a basic DNA-binding region [24]. USF complexes contain two proteins, USF-1 and USF-2, which can homo- and heterodimerize [25].
The Ikaros family of proteins includes eight isoforms generated by alternative splicing of exons 3–7 [19] as well as the related factors typified by Aiolos [26] and Helios [27,28]. Since at least three of the four internal zinc fingers (i.e. exons 3–5) are needed for high-affinity DNA binding, only Ikaros 1–3 significantly bind to DNA whereupon they can either activate or repress transcription [19,29]. Conversely, Ikaros 4–8 are dominant-negative isoforms that bind poorly to DNA and interfere with cellular response to the DNA-binding forms via formation of protein dimers [19,29].
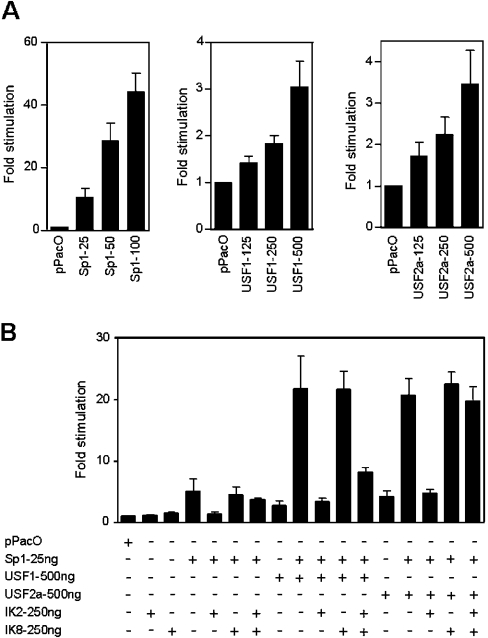
To assess directly the role of USF proteins in hRFC-B transactivation and their relationships with the Sp response, Sp and USF cDNA constructs in pPac vector were co-transfected with the hRFC-B/−4590 construct [includes GC-boxes (a) and (b) and E-box (a); Figure 1B] into Drosophila Mel-2 cells that provide a null background for both these transcription factors [30]. Sp1, USF1 and USF2a, each individually, showed a dose-dependent stimulation of luciferase activity in Drosophila Mel-2 cells over the low level seen in empty pPacO (Figure 5A). Sp3 was significantly less stimulatory (≤1.5-fold over pPacO) of hRFC-B/−4590 promoter activity than Sp1 at an equivalent dose (results not shown). Of particular interest, the transactivating effects of Sp1 and USF1 or USF2a were highly synergistic (Figure 5B shows the combined effects of 25 ng of Sp1 with 500 ng of USF1 or USF2a).
Figure 5. Effects of co-expressed Sp, USF and Ikaros proteins on hRFC-B promoter activity in Drosophila Mel-2 cells.
(A) Drosophila Mel-2 cells were co-transfected with 1 μg of the wt hRFC-B/−4590 construct in pGL3-Basic, and Sp1 (pPacSp1; 25–100 ng), USF1 (pPacUSF1; 125–500 ng) or USF2a (pPacUSF2a; 125–500 ng). (B) Drosophila Mel-2 cells were co-transfected with 1 μg of the wt hRFC-B/−4590 construct and combined with Sp1, USF1, USF2a, Ikaros 2 (IK2) and Ikaros 8 (IK8). All transfections included a total of 500 ng of total vector including expression construct and empty pPacO. Cells were harvested after 24 h for luciferase assays. Luciferase activities were normalized to cellular protein contents. Results are expressed as means±S.E.M. for three separate experiments.
Additional experiments were performed to assess the effects of Ikaros proteins on hRFC-B/−4590 promoter activity. Both Ikaros 2 and 8 (each at 250 ng) were transcriptionally inert in the absence of other additions (IK2 and IK8 in Figure 5B). However, in combination with Sp1 and USF1, Ikaros 2 was notably repressive, resulting in an approx. 90% inhibition of promoter activity. The effects of Ikaros 2 were partly reversed (∼3-fold) in the presence of the dominant-negative Ikaros 8. Whereas Ikaros 2 was only slightly less (∼85%) repressive of hRFC-B/−4590 transactivation by Sp1/USF2a, in contrast with the results of Sp1/USF1, the effects of Ikaros 2 were nearly completely reversed by Ikaros 8.
These results clearly demonstrate that Sp and USF proteins, individually, are capable of stimulating the 158 bp hRFC-B core promoter. However, both Sp1 and USF proteins are required for high levels of promoter transactivation. In addition, our results strongly imply that hRFC-B transactivation can be significantly modulated by the presence of the different Ikaros family members, including both DNA-binding and transdominant-negative forms, and that the magnitude of these effects differs slightly for USF1 and USF2a. Whereas Sp3 was less effective than Sp1 in transactivating the hRFC-B basal promoter [14], this difference was exacerbated for the longer hRFC-B/−4590 promoter construct.
Effects of Sp1, USF1 and Ikaros 2 transfections on hRFC-B transcript levels and promoter B activity in HT1080 cells
The results in Figure 5 with Drosophila Mel-2 cells co-transfected with the hRFC-B promoter and transcription factor expression constructs provide compelling evidence that both Sp1 and USF1 are each capable of stimulating the core promoter and, in combination, could elicit a highly synergistic transactivation response. Moreover, Ikaros 2 potently repressed the high-level promoter activity achieved with USF1 and Sp1, although the effects of Ikaros 2 were partially reversed by the dominant-negative Ikaros 8.
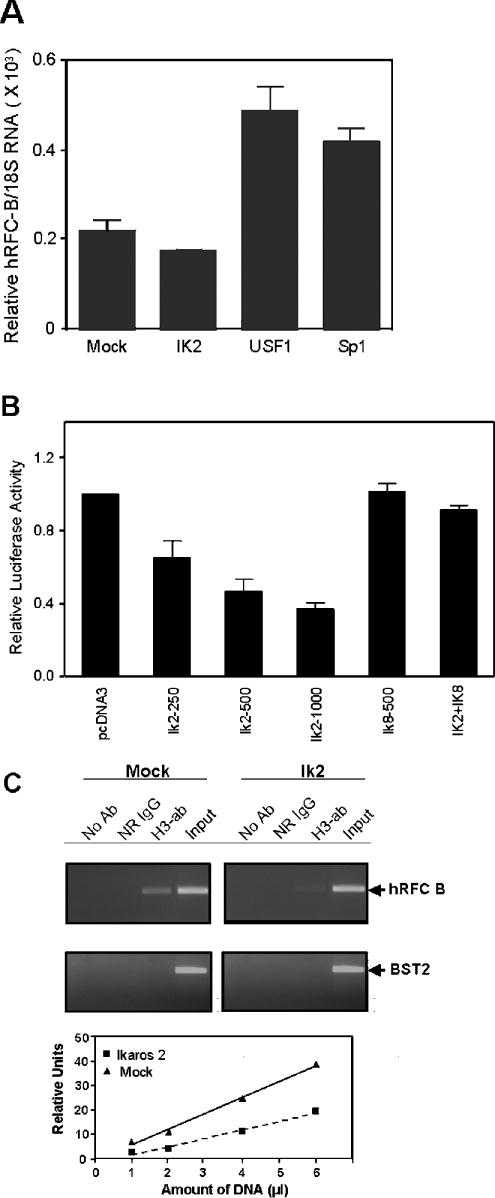
An important extension of these experiments involves the effects of these transcription factors on levels of endogenous hRFC transcripts transcribed from non-coding exon B in HT1080 cells, as a direct measure of endogenous hRFC-B promoter activity. To address this question, HT1080 cells were transiently transfected with expression constructs (in pCDNA3) for Sp1, USF1 or Ikaros 2, and after 48 h, hRFC transcripts with B 5′-UTR sequence were assayed by real-time RT–PCR with exon B-specific primers. In three experiments, both USF1 and Sp1 treatments resulted in significant and reproducible (2.2- and 1.9-fold respectively) increases in the levels of endogenous hRFC-B transcripts, whereas Ikaros 2 was repressive (∼20%) (Figure 6A). Thus the effects of these exogenous transcription factors on the endogenous hRFC-B promoter in HT1080 cells parallel the changes in hRFC-B core promoter activity in Drosophila cells transfected with the hRFC-B/−4590 reporter gene construct. Similarly, co-transfection of HT1080 cells with USF1 and Sp1 stimulated (not shown) and Ikaros 2 suppressed (Figure 6B) hRFC-B/−4590 promoter activity (Figure 6B). The repressive effects of Ikaros 2 were completely reversed in co-transfections with Ikaros 8.
Figure 6. Effects of USF1, Sp1 and Ikaros 2 on endogenous hRFC-B transcripts and effects of Ikaros 2 on hRFC-B promoter activity and histone H3 acetylation.
(A) Total RNAs, prepared from HT1080 cells (∼80% confluent, in a 60 mm dish) transfected with Ikaros (IK) 2 (3 μg), USF1 (3 μg) and Sp1 (75 ng) in pCDNA3 or empty pCDNA3 (mock transfectant) (3 μg), were reverse-transcribed, purified and amplified in real time as described in the Materials and methods section. hRFC-B transcripts were normalized to 18 S RNA levels. Results are expressed as means±S.E.M. for three experiments. (B) HT1080 cells (∼80% confluent, in a 35 mm dish) were transfected with Ikaros (IK) 2 and 8 (250, 500 or 1000 ng) or pCDNA3 (1000 ng) and the hRFC-B/−5422 promoter construct (3 μg), and assayed for reporter gene activity as described in the Materials and methods section. Total pCDNA3 plasmids equalled 1000 ng. (C) Chromatin was prepared from Ikaros 2 and mock-transfected cells and immunoprecipitated with IgG, anti-acetyl H3 antibody or no antibody. After PCR amplification, the products were visualized on 2% agarose gels with ethidium bromide. The BST2 gene was amplified as a negative control for the ChIP assays. Upper panel: ethidium bromide-stained gels; lower panel: correlation between the amounts of immunoprecipitated chromatins for the mock- and Ikaros-2-transfected cells and relative levels of hRFC-B amplicon after 32 cycles, as determined by densitometry analysis of the ethidium bromide-stained gels. Based on the slopes of the calculated lines, the relative difference in levels of acetylated histone H3 associated with the hRFC-B promoter between the mock and Ikaros 2 transfectants was 1.8-fold. Ab, antibody; H3, histone H3.
DNA-binding Ikaros proteins have been reported to achieve at least part of their transcriptional effects via interactions with chromatin remodelling complexes, by recruiting the co-repressors, Mi-2β, Sin3A and Sin3B and the class I HDACs (histone deacetylases) 1 and 2 [31,32]. If such a mechanism were operative for hRFC, loss of hRFC-B transcripts and promoter activity would be accompanied by decreased levels of acetylated chromatin associated with the B promoter. To assess the changes in acetyl histone H3 in HT1080 cells treated with Ikaros 2, we used ChIP with anti-acetyl histone H3 antibody (Lys-14). Chromatins were immunoprecipitated with IgG or acetyl H3 antibody and PCR-amplified in the linear range with primers flanking the hRFC-B core promoter region (positions −4560 to −4353). Although input signals for hRFC-B were comparable between the mock and Ikaros 2 transfectants, the level of acetyl histone H3 associated with the hRFC-B promoter decreased after treatment with Ikaros 2 (Figure 6C). As shown in the bottom panel, there was a 1.8-fold decrease in the level of acetylated histone H3, when immunoprecipitates were analysed over a 6-fold range of concentrations. No signals were detected in the absence of acetyl H3 antibody or after immunoprecipitation with IgG. For BST2 gene control amplifications, other than the input, no signals were detected. These results strongly suggest that transfections of HT1080 cells with Ikaros 2 result in significantly decreased acetyl histone H3 associated with the endogenous hRFC-B promoter accompanying decreased hRFC-B transcripts.
DISCUSSION
We previously localized the minimal promoter for hRFC-B to within 46 bp (positions −4493 to −4448) and demonstrated an important regulatory role for the Sp family of transcription factors in binding to a critical GC-box element within this region [14]. In the present study, we characterize the probable contributions of other cis elements and transcription factors, and document a potential role for chromatin remodelling in regulating promoter B activity.
Important regions involved in regulating promoter B were established by 5′- and 3′-deletions and transient transfections of HT1080 cells. From these results, oligonucleotide probes spanning a 158 bp core promoter region were designed and used in gel shift assays with HT1080 nuclear extracts to identify binding sites for key transcription factors. In vivo binding of transcription factors identified on gel shifts was confirmed by ChIP.
The important regulatory role previously described for the GC-box (a) element [14] was confirmed on gel shifts and by reporter gene assays of a GC-box (a) mutant construct. Furthermore, additional bound Sp proteins were localized to another GC-box, designated GC-box (b), and USF proteins were bound to a non-canonical E-box (a), flanking GC-box (a). When GC-box (b) and E-box (a) were individually mutated so as to impede their in vitro protein binding, promoter activity was inhibited.
Given the close proximity of the hRFC-B transcription start sites to these GC- and E-box elements, the notion of an essential transcription initiation complex between USF protein bound to E-box (a) and Sp proteins bound to GC-boxes (a) and (b) is particularly intriguing. In Sp and USF-null Drosophila Mel-2 cells, either Sp1 or USF transcription factors could transactivate the hRFC-B/−4590 reporter construct, although maximal transactivation required both Sp1 and USF1. USF2a was also capable of transactivating the hRFC-B promoter; however, Sp3 was largely inert. Both USF1 and Sp1 stimulated hRFC-B/−4590 reporter activity in HT1080 cells (not shown) and increased the levels of endogenous hRFC-B transcripts in transiently transfected HT1080 cells. Co-operative interactions between Sp1 and USF proteins were previously described for the human transcobalamin II [33], cystathionine-β-synthase 1b [34] and, most recently, deoxycytidine kinase [20] promoters. With deoxycytidine kinase, Sp1 and USF1 were co-immunoprecipitated [20], thus providing direct evidence for a physical interaction between these proteins. The synergistic stimulation of the hRFC-B promoter in Drosophila Mel-2 cells with Sp1 and USF1/2a suggests that both Sp and USF proteins are essential for high-level transactivation. Accordingly, the net level of hRFC-B promoter activity achieved would probably reflect relative levels of active Sp1, Sp3, USF1 and USF2a transcription factors within cells.
A regulatory role for the family of Ikaros proteins with hRFC-B was also suggested by our studies, via binding to three Ikaros binding sites [Ikaros (a)–(c)] upstream of GC-box (a). The functional importance of these elements was further implied by transient transfections of the Ikaros (a), (b) and (c) mutant constructs into HT1080 cells. In Drosophila Mel-2 cells, co-transfection with Ikaros 2 repressed hRFC-B transactivation by Sp1 and USF1. Whereas the dominant-negative Ikaros 8 was itself transcriptionally inert, this form reversed the repressive effects of Ikaros 2. The extent of this reversal was different in the presence of USF1 versus USF2a. Ikaros 2 also decreased the endogenous level of hRFC-B transcripts in HT1080 cells, consistent with its repressive effects on hRFC-B reporter gene activity.
T-cell malignancies in mice result from a loss of Ikaros expression or overexpression of dominant-negative Ikaros isoforms [35]. In childhood ALL (both B-precursor and T-ALL), expression of the dominant-negative Ikaros isoforms (Ikaros 4, 6–8) was also detected at high frequencies [36,37]. This provides at least a partial explanation for the extraordinarily wide range of hRFC expression previously reported in ALL lymphoblasts [11]. However, any effect of Ikaros proteins on hRFC-B promoter activity in ALL would probably be compounded by those of other transcription factors, such as the USF and the Sp families of proteins.
DNA binding by the family of Ikaros proteins has been associated both with promoter transactivation and repression [19,29]. Ikaros has been reported to associate with the SWI/SNF chromatin remodelling complex [31] and localize with inactive genes in centromeric heterochromatin [27]. Repression by DNA-binding Ikaros isoforms has been shown to occur via association with the CtBP co-repressor [38] or by association with chromatin remodelling activities such as Mi-2β and the Sin3A and Sin3B co-repressor complexes including HDACs [31,32]. It is probable that the levels and activities of the various binding partners for Ikaros determine the nature of the complexes that form and whether they repress or activate transcription. When chromatins were prepared from HT1080 cells treated with Ikaros 2 under conditions that repress hRFC-B reporter gene activity and endogenous hRFC-B transcripts, and analysed by ChIP with anti-acetyl histone H3, levels of acetylated histone H3 were decreased compared with mock-transfected cells. Thus the repressive effects of Ikaros 2 on hRFC-B transcription would seem at least in part to be reflected in decreased acetylation of the histones associated with the −4560 to −4353 core promoter region.
Interestingly, treatment of the Mtx-resistant MDA-MB-231 breast cancer subline with the HDAC inhibitor, trichostatin A, was previously reported to have no net effect on total levels of hRFC transcripts, even though loss of hRFC expression was associated with decreased acetylation of histones H3 and H4 [39]. However, these results must now be interpreted in terms of a multiplicity of hRFC promoters and 5′-UTRs [15] whereby the effects of trichostatin A may be restricted only to a limited number of 5′-UTRs/promoters with no significant effect on other hRFC 5′-UTRs/promoters. Thus the recruitment of chromatin remodelling machinery by specific transcription factors or drug treatments could represent a potentially selective mechanism for modulating hRFC 5′-UTR/promoter usage. This will be tested in future studies.
Acknowledgments
We thank Dr R. Tjian and Dr G. Suske for providing the pCMV-Sp1, pPacO, pPacSp1 and pPacSp3 expression vectors. This study was supported by grant CA53535 from the National Cancer Institute, National Institutes of Health.
References
- 1.Stokstad E. L. R. Historical perspective on key advances in the biochemistry and physiology of folates. In: Picciano M. F., Stokstad E. L. R., Gregory J. F., editors. Folic Acid Metabolism in Health and Disease. New York: Wiley-Liss; 1990. pp. 1–21. [Google Scholar]
- 2.Jansen G. Receptor- and carrier-mediated transport systems for folates and antifolates. Exploitation for folate chemotherapy and immunotherapy. In: Jackman A. L., editor. Anticancer Development Guide: Antifolate Drugs in Cancer Therapy. Totowa, NJ: Humana Press; 1999. pp. 293–321. [Google Scholar]
- 3.Matherly L. H., Goldman I. D. Membrane transport of folates. Vitam. Horm. 2003;66:403–456. doi: 10.1016/s0083-6729(03)01012-4. [DOI] [PubMed] [Google Scholar]
- 4.Goldman I. D., Zhao R. Molecular, biochemical, and cellular pharmacology of pemetrexed. Semin. Oncol. 2002;29:3–17. doi: 10.1053/sonc.2002.37461. [DOI] [PubMed] [Google Scholar]
- 5.Schuetz J. D., Matherly L. H., Westin E. H., Goldman I. D. Evidence for a functional defect in the translocation of the methotrexate transport carrier in a methotrexate resistant murine L1210 leukemia cell line. J. Biol. Chem. 1998;263:9840–9847. [PubMed] [Google Scholar]
- 6.Gong M., Yess J., Connolly T., Ivy S. P., Ohnuma T., Cowan K. H., Moscow J. A. Molecular mechanism of antifolate transport deficiency in a methotrexate resistant MOLT-3 human leukemia cell line. Blood. 1997;89:2494–2499. [PubMed] [Google Scholar]
- 7.Jansen G., Mauritz R., Drori S., Sprecher H., Kathman I., Bunni M., Priest D. G., Noordhuis P., Schornagel J. H., Pinedo H. M., et al. A structurally altered human reduced folate carrier with increased folic acid transport mediates a novel mechanism of antifolate resistance. J. Biol. Chem. 1998;273:30189–30198. doi: 10.1074/jbc.273.46.30189. [DOI] [PubMed] [Google Scholar]
- 8.Sirotnak F. M., Moccio D. M., Kelleher L. E., Goutas L. J. Relative frequency and kinetic properties of transport-defective phenotypes among methotrexate resistant L1210 clonal cell lines derived in vivo. Cancer Res. 1981;41:4442–4452. [PubMed] [Google Scholar]
- 9.Gorlick R., Goker E., Trippett T., Steinherz P., Elisseyeff Y., Mazumdar M., Flintoff W. F., Bertino J. R. Defective transport is a common mechanism of acquired methotrexate resistance in acute lymphocytic leukemia and is associated with decreased reduced folate carrier expression. Blood. 1997;89:1013–1018. [PubMed] [Google Scholar]
- 10.Guo W., Healey J. H., Meyeers P. A., Ladanyai M., Huvos A. G., Bertino J. R., Gorlick R. Mechanisms of methotrexate resistance in osteosarcoma. Clin. Cancer Res. 1999;5:621–627. [PubMed] [Google Scholar]
- 11.Zhang L., Taub J. W., Williamson M., Wong S. C., Hukku B., Pullen J., Ravindranath Y., Matherly L. H. Reduced folate carrier gene expression in childhood acute lymphoblastic leukemia: relationship to immunophenotype and ploidy. Clin. Cancer Res. 1998;4:2169–2177. [PubMed] [Google Scholar]
- 12.Zhang L., Wong S. C., Matherly L. H. Transcript heterogeneity of the human reduced folate carrier results from the use of multiple promoters and variable splicing of alternative upstream exons. Biochem. J. 1998;332:773–780. doi: 10.1042/bj3320773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tolner B., Roy K., Sirotnak F. M. Structural analysis of the human RFC-1 gene encoding a folate transporter reveals multiple promoters and alternatively spliced transcripts with 5′ end heterogeneity. Gene. 1998;211:331–341. doi: 10.1016/s0378-1119(98)00123-1. [DOI] [PubMed] [Google Scholar]
- 14.Whetstine J. R., Matherly L. H. The basal promoters for the human reduced folate carrier gene are regulated by a GC-box and a cAMP-response element/AP-1-like element. J. Biol. Chem. 2001;276:6350–6358. doi: 10.1074/jbc.M008074200. [DOI] [PubMed] [Google Scholar]
- 15.Whetstine J. R., Flatley R. M., Matherly L. H. The human reduced folate carrier gene is ubiquitously and differently expressed in normal human tissues: identification of seven non-coding exons and characterization of a novel promoter. Biochem. J. 2002;367:629–640. doi: 10.1042/BJ20020512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flatley R. M., Payton S. G., Taub J. W., Matherly L. H. Primary acute lymphoblastic leukemia cells use a novel promoter and 5′ non-coding exon for the human reduced folate carrier that encodes a modified carrier translated from an upstream translational start. Clin. Cancer Res. 2004;10:5111–5122. doi: 10.1158/1078-0432.CCR-04-0116. [DOI] [PubMed] [Google Scholar]
- 17.Whetstine J. R., Witt T. L., Matherly L. H. The human reduced folate carrier gene is regulated by the AP2 and sp1 transcription factor families and a functional 61-base pair polymorphism. J. Biol. Chem. 2002;277:43873–43880. doi: 10.1074/jbc.M208296200. [DOI] [PubMed] [Google Scholar]
- 18.Horton R. M., Cai Z. L., Ho S. N., Pease L. R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 19.Molnar A., Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell. Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge Y., Jensen T., Matherly L. H., Taub J. W. Physical and functional interactions between USF and Sp1 proteins regulated human deoxycytidine kinase activity. J. Biol. Chem. 2003;278:49901–49910. doi: 10.1074/jbc.M305085200. [DOI] [PubMed] [Google Scholar]
- 21.Ge Y., Jensen T. L., Matherly L. H., Taub J. W. Transcriptional regulation of human cystathionine beta synthase gene expression in Down syndrome and non-Down syndrome megakaryocytic leukemia cell lines. Blood. 2003;101:1551–1557. doi: 10.1182/blood-2002-07-2337. [DOI] [PubMed] [Google Scholar]
- 22.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 23.Henrion A. A., Martinez A., Mattei M. G., Kahn A., Raymondjean M. Structure, sequence, and chromosomal location of the gene for USF2 transcription factors in mouse. Genomics. 1995;25:36–43. doi: 10.1016/0888-7543(95)80107-w. [DOI] [PubMed] [Google Scholar]
- 24.Grandori C., Cowley S. M., James L. P., Eisenman R. N. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 25.Gregor P. D., Sawadogo M., Roeder R. G. The adenovirus major late transcription factor USF is a member of the helix-loop-helix group of regulatory proteins and binds to DNA as a dimer. Genes Dev. 1990;4:1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 26.Morgan B., Sun L., Avitahl N., Andrikopoulos K., Gonzales E., Nichogiannopoulou A., Wu P., Neben S., Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahm K., Cobb B. S., McCarty A. S., Brown K. E., Klug C. A., Lee R., Akashi K., Weissman I. L., Fisher A. G., Smale S. T. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley C. M., Ikeda T., Koipally J., Avitahl N., Georgopoulos K., Morgan B. A. Helios, a novel dimerization partner of Ikaros expressed in the earliest hematopoietic progenitors. Curr. Biol. 1998;8:508–515. doi: 10.1016/s0960-9822(98)70202-7. [DOI] [PubMed] [Google Scholar]
- 29.Sun L., Liu A., Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 30.Moore A. W., Barbel S., Jan L. Y., Jan Y. N. A genomewide survey of basic helix-loop-helix factors in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2000;97:10436–10441. doi: 10.1073/pnas.170301897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J., Sif S., Jones B., Jackson A., Koipally J., Heller E., Winandy S., Viel A., Sawyer A., Ikeda T., et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 32.Koipally J., Renold A., Kim J., Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li N., Seetharam B. A 69-base pair fragment derived from human transcobalamin II promoter is sufficient for high bi-directional activity in the absence of a TATA box and an initiator element in the transfected cells. J. Biol. Chem. 1998;272:28171–28177. doi: 10.1074/jbc.273.43.28170. [DOI] [PubMed] [Google Scholar]
- 34.Ge Y., Konrad M. A., Matherly L. H., Taub J. W. Transcriptional regulation of the human cystathionine-β-synthase-1b basal promoter: synergistic transactivation by transcription factors NF-Y and Sp1/Sp3. Biochem. J. 2001;357:97–105. doi: 10.1042/0264-6021:3570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winandy S., Wu P., Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell (Cambridge, Mass.) 1995;83:289–299. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 36.Sun L., Crotty M. L., Sensel M., Sather H., Navara C., Nachman J., Steinherz P. G., Gaynon P. S., Seibel N., Mao C., et al. Expression of dominant-negative Ikaros isoforms in T-cell acute lymphoblastic leukemia. Clin. Cancer Res. 1999;5:2112–2120. [PubMed] [Google Scholar]
- 37.Sun L., Goodman P. A., Wood C. M., Crotty M. L., Sensel M., Sather H., Navara C., Nachman J., Steinherz P. G., et al. Expression of aberrantly 35 spliced oncogenic ikaros isoforms in childhood acute lymphoblastic leukemia. J. Clin. Oncol. 1999;17:3753–3766. doi: 10.1200/JCO.1999.17.12.3753. [DOI] [PubMed] [Google Scholar]
- 38.Koipally J., Georgopoulos K. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem. 2000;275:19594–19602. doi: 10.1074/jbc.M000254200. [DOI] [PubMed] [Google Scholar]
- 39.Worm J., Kirkin A. F., Dzhandzhugazyan K. N., Guldberg P. Methylation-dependent silencing of the reduced folate carrier gene in inherently methotrexate-resistant human breast cancer cells. J. Biol. Chem. 2001;276:39990–40000. doi: 10.1074/jbc.M103181200. [DOI] [PubMed] [Google Scholar]